Abstract
To determine the presence of Shiga toxin-producing Escherichia coli (STEC) and other potentially diarrheagenic E. coli strains in retail meats, 7,258 E. coli isolates collected by the U.S. National Antimicrobial Resistance Monitoring System (NARMS) retail meat program from 2002 to 2007 were screened for Shiga toxin genes. In addition, 1,275 of the E. coli isolates recovered in 2006 were examined for virulence genes specific for other diarrheagenic E. coli strains. Seventeen isolates (16 from ground beef and 1 from a pork chop) were positive for stx genes, including 5 positive for both stx1 and stx2, 2 positive for stx1, and 10 positive for stx2. The 17 STEC strains belonged to 10 serotypes: O83:H8, O8:H16, O15:H16, O15:H17, O88:H38, ONT:H51, ONT:H2, ONT:H10, ONT:H7, and ONT:H46. None of the STEC isolates contained eae, whereas seven carried enterohemorrhagic E. coli (EHEC) hlyA. All except one STEC isolate exhibited toxic effects on Vero cells. DNA sequence analysis showed that the stx2 genes from five STEC isolates encoded mucus-activatable Stx2d. Subtyping of the 17 STEC isolates by pulsed-field gel electrophoresis (PFGE) yielded 14 distinct restriction patterns. Among the 1,275 isolates from 2006, 11 atypical enteropathogenic E. coli (EPEC) isolates were identified in addition to 3 STEC isolates. This study demonstrated that retail meats, mainly ground beef, were contaminated with diverse STEC strains. The presence of atypical EPEC strains in retail meat is also of concern due to their potential to cause human infections.
Escherichia coli is an important component of the intestinal microflora of humans and warm-blooded mammals. While E. coli typically harmlessly colonizes the intestinal tract, several E. coli clones have evolved the ability to cause a variety of diseases within the intestinal tract and elsewhere in the host. Those strains that cause enteric infections are generally called diarrheagenic E. coli strains, and their pathogenesis is associated with a number of virulence attributes, which vary according to pathotype (54). Currently, diarrheagenic E. coli strains are classified into six main pathotypes based on their distinct virulence determinants and pathogenic features, including enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enterohemorrhagic E. coli (EHEC)/Shiga toxin-producing E. coli (STEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusively adherent E. coli (DAEC) (37).
Among diarrheagenic E. coli strains, STEC strains are distinguished by the ability to cause severe life-threatening complications, such as hemolytic-uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (TTP) (30). Other symptoms of STEC infection include watery diarrhea, bloody diarrhea, and hemorrhagic colitis (HC). STEC strains that cause HC and HUS are also called EHEC. Although individuals of all ages are at risk of STEC infection, children younger than 5 years of age and the elderly are more likely to suffer from severe complications (51). Outbreaks and sporadic cases of STEC infections have been reported frequently worldwide.
The pathogenesis of STEC infection in humans is not fully understood. The major virulence factors implicated in STEC infection are potent Shiga toxins, which are classified into two groups: Stx1 and Stx2 (23). Additional factors that contribute to virulence have also been described, including intimin (encoded by the eae gene), an outer membrane protein involved in the attachment of E. coli to the enterocyte, and EHEC hemolysin (encoded by EHEC hlyA), which acts as a pore-forming cytolysin and causes damage to cells (41).
The first STEC O157 infections were reported in 1982, when E. coli O157:H7 was involved in outbreaks associated with two fast food chain restaurants in the United States (44). Since then, ever-increasing numbers of cases and outbreaks due to STEC O157 have been reported worldwide. Although non-O157 STEC strains have also been associated with human cases and outbreaks, few laboratories have been looking for them, and their potential in causing human infections may be underestimated (2). Recently, though, the significance of non-O157 STEC strains as human pathogens has become more recognized. In the United States alone, there were 23 reported outbreaks of non-O157 STEC infection between 1990 and 2007 (10).
Shiga toxin-producing E. coli can be transmitted through different routes, including food and water, person-to-person contact, and animal-to-person contact (9). Most human infections are caused by consumption of contaminated foods (16). Domestic and wild ruminant animals, in particular cattle, are considered the main reservoir of STEC and the main source for contamination of the food supply. Retail meats derived from animals could potentially act as transmission vehicles for STEC and other diarrheagenic E. coli strains. However, there is limited information about STEC contamination in retail meats, and fewer data exist about the presence of other diarrheagenic E. coli strains in retail meats. In the present study, we investigated 7,258 E. coli isolates from four types of meat samples (beef, chicken, pork, and turkey) collected during 2002 to 2007 to assess STEC contamination of retail meats. In addition, the presence of other potentially diarrheagenic E. coli strains was examined by detecting specific virulence determinants among E. coli isolates collected in 2006.
MATERIALS AND METHODS
Bacterial strains.
A total of 7,258 E. coli isolates (1,806 from ground beef, 2,106 from ground turkey, 2,179 from chicken breasts, and 1,167 from pork chops) from the retail meat program of the U.S. National Antimicrobial Resistance Monitoring System (NARMS) were investigated. Detailed information on sampling, isolation, and identification can be found at http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/default.htm. Briefly, retail meats were collected monthly from grocery stores in four states (Georgia, Maryland, Oregon, and Tennessee) from 2002 to 2007. For chicken and pork samples, one piece of meat was examined; for ground beef and ground turkey, 25 g of product was processed. Portions from each sample were placed in separate bags with 250 ml of buffered peptone water, and the bags were vigorously shaken. Fifty milliliters of the rinsate was mixed with 50 ml of double-strength MacConkey broth, and the contents were incubated at 35°C for 24 h. One loopful of culture was streaked onto an eosin-methylene blue (EMB) agar plate and incubated at 35°C for 24 h. The plate was examined for typical E. coli colonies, and one typical, well-isolated colony was streaked onto a blood agar plate and incubated at 35°C for 24 h. Indole-positive and oxidase-negative isolates were presumptively identified as E. coli and confirmed as E. coli by use of the Vitek 2 Compact microbial identification system (bioMérieux, Hazelwood, MO). All E. coli isolates were stored in tryptic soy broth containing 15% glycerol at −80°C until use.
DNA template preparation.
E. coli cells were recovered from frozen culture at −80°C, streaked onto blood agar, and incubated overnight at 37°C. DNA of each isolate was extracted using a previously described boiling method, with modification (46). Briefly, approximately 10 to 20 colonies were taken by use of cotton swabs and were suspended in 500 μl of distilled water. The mixture was then boiled at 100°C for 10 min. After centrifugation at 13,000 rpm for 5 min, supernatants were transferred to a new tube and stored at −20°C until used for PCR amplification.
Identification of Shiga toxin genes.
STEC was identified by the presence of stx1 and/or stx2 genes. All isolates were subjected to a multiplex PCR (assay 1) which targeted stx1 and stx2 and most of their variants (except for stx1d and stx2f, due to their considerable sequence divergence from classical stx1 and stx2, respectively) (Table 1). PCRs were performed in a 25-μl reaction mixture containing 2 μl of DNA template, 2.5 μl of 10× PCR buffer, 2 μl of a 1.25 mM mixture of deoxynucleoside triphosphates, 2.5 μl of 25 mM MgCl2, 0.25 μl (5 U) of AmpliTaq Gold DNA polymerase (Applied Biosystems, Branchburg, NJ), and 0.5 μl (25 pmol) of each oligonucleotide primer (Invitrogen, Carlsbad, CA). The thermocycling protocol included an initial denaturation step at 94°C for 10 min, followed by 30 cycles of denaturation (94°C for 30 s), annealing (58°C for 30 s), and extension (72°C for 30 s), with a final extension at 72°C for 3 min. PCR products (10 μl) were resolved by electrophoresis on a 1.5% (wt/vol) agarose gel at 100 mV for 30 min. Gels were then stained with ethidium bromide, and the DNA bands were visualized and photographed under UV illumination. E. coli EDL 933, containing stx1 and stx2, and E. coli K-12 were used as positive and negative controls, respectively. For initial screening, DNA templates of five isolates were pooled and mixed thoroughly, and the mixture was used as a template for PCR. When the mixture tested positive for a stx gene(s), DNAs of individual isolates were then tested separately to identify the stx-positive isolate(s).
TABLE 1.
Oligonucleotide primers for multiplex PCR and sequencing
| Assay | Target gene or primer name | Forward primer | Reverse primer | Product size (bp) | Reference |
|---|---|---|---|---|---|
| Assay 1 | stx1 | GTGGCATTAATACTGAATTGTCATCA | GCGTAATCCCACGGACTCTTC | 109 | 29 |
| stx2 | GGCACTGTCTGAAACTGCTCC | TCGCCAGTTATCTGACATTCTG | 255 | 47 | |
| Assay 2 | stI | TCTTTCCCCTCTTTTAGTCAGTC | CAGCACAGGCAGGATTAC | 170 | 36 |
| elt | ACGGCGTTACTATCCTCTC | TGGTCTCGGTCAGATATGTG | 274 | 48 | |
| daaE | GAACGTTGGTTAATGTGGGGTAA | TATTCACCGGTCGGTTATCAGT | 542 | 54 | |
| aafII | CACAGGCAACTGAAATAAGTCTGG | ATTCCCATGATGTCAAGCACTTC | 378 | 54 | |
| Assay 3 | eae | CTGAACCAGATCGTAACGGC | TGATAAGCTGCAGTCGAATCC | 229 | 36 |
| bfp | AACCGTTACTGCCGGTGTGA | GTTGCCGCCTCAGCAGGAGT | 450 | 36 | |
| ipaH | CTCGGCACGTTTTAATAGTCTGG | GTGGAGAGCTGAAGTTTCTCTGC | 933 | 54 | |
| Assay 4 | eae | CTGAACCAGATCGTAACGGC | TGATAAGCTGCAGTCGAATCC | 229 | 36 |
| EHEC hlyA | AGCCGGAACAGTTCTCTCAG | CCAGCATAACAGCCGATGT | 526 | 35 | |
| stx1 sequencing | Paton 1 | TCGCATGAGATCTGACC | AACTGACTGAATTGAGATG | 1,470 | 5 |
| Paton 2 | ATAAATCGCCATTCGTTGACTAC | AGAACGCCCACTGAGATCATC | 180 | 5 | |
| Gannon | ACACTGGATGATCTCAGTGG | CTGAATCCCCCTCCATTATG | 603 | 5 | |
| Vidiya | TCGCATGAGATCTGACC | AATAAGCCGTAGATTATT | 448 | 5 | |
| stx2 sequencing | Stx2-1 | TTCTGAGCAATCGGTCACTG | CGGCGTCATCGTATACACAG | 779 | 55 |
| Stx2-2 | GTCACAGCAGAAGCCTTACG | ACCCACATACCACGAATCAG | 714 | 55 |
DNA sequence and phylogenetic analysis of stx genes.
Shiga toxin genes were amplified and sequenced using the primers listed in Table 1. The stx1 genes were amplified with primer Paton 1 to generate a 1,470-bp product which covered both the A and B subunits of stx1. Two overlapping DNA fragments (779 and 714 bp), which together cover the entire stx2 gene, were amplified with primers Stx2-1 and Stx2-2, respectively. The PCR products were purified using 96-well multiscreen filter plates (Millipore Corp., Billerica, MA), and purified amplicons were sequenced on both strands. Sequencing reactions were performed using a Big Dye Terminator cycle sequencing ready reaction DNA sequencing kit and electrophoresed on an AB3730 DNA sequencer (Applied Biosystems, Foster City, CA). DNA sequence data were compiled and analyzed using DNA Sequencher 4.0 software (Gene Codes Corp., Ann Arbor, MI). Nucleotide phylogenetic trees were generated by multiple sequence alignment and neighbor-joining analysis of the alignment, using the MAFFT program (http://align.bmr.kyushu-u.ac.jp/mafft/software/). Reference DNA and amino acid sequences for Shiga toxins were obtained from GenBank and are indicated in the phylogenetic tree by their accession numbers.
Vero cell cytotoxicity assay.
The STEC isolates were examined for cytotoxicity on Vero cells according to previously published protocols (20, 55). First, 96-well microtiter plates were seeded with approximately 104 Vero cells/well and incubated at 37°C for 24 h in the presence of 5% CO2. The tissue culture medium (Eagle's minimum essential medium [EMEM] containing 10% fetal bovine serum; ATCC, Manassas, VA) was aspirated and replaced with 100 μl of fresh medium prior to the addition of bacterial supernatant dilutions. The bacterial isolates were inoculated into 5 ml LB broth and incubated overnight at 37°C with shaking. The cell concentration of the overnight bacterial culture was adjusted with LB broth to approximately 109 CFU/ml (optical density at 600 nm [OD600] = 1). The culture was centrifuged at 10,000 × g for 10 min, and the supernatant was filtered through a 0.45-μm-pore-size membrane filter. The filtrate was serially diluted (1:5) in tissue culture medium. One hundred microliters of each dilution was inoculated into triplicate wells of the 96-well microtiter plate with Vero cells. Control wells, which refer to wells containing cells not inoculated with toxin, were included on each plate for a nonintoxicated cell background. After incubation at 37°C in a 5% CO2 atmosphere for 48 h, detached cells, medium, and toxin were removed by vigorous shaking. Remaining Vero cells were fixed with 2% formalin in 0.067 M phosphate-buffered saline (pH 7.2) for 1 min, the fixative was removed, and the plate was stained with 0.13% crystal violet in 5% ethanol for 30 min. Excess stain was removed by rinsing, and the plates were air dried. For quantification, stain was eluted with 200 μl of 50% ethanol, and the color intensity of each well was measured with an Elx800 microplate reader (Bio-Tek Instruments, Winooski, VT) at a 600-nm wavelength. The color intensity was proportional to the number of viable, attached cells in the well. The absorbance values were then plotted against the log of the toxin dilution. To determine the toxin dilution resulting in 50% cell detachment (CD50), the zero-detachment dye absorbance value was obtained from control wells with nonintoxicated cells on each plate, and the CD50 value was determined by extrapolating one-half of this value to the log scale of toxin dilution. E. coli EDL933 and E. coli K-12 were used as positive and negative controls, respectively. All assays were conducted in triplicate and independently repeated three times.
eae and EHEC hlyA genes.
The STEC isolates were further tested for the presence of eae and EHEC hlyA genes, using a multiplex PCR (assay 4) (Table 1). The PCR conditions used were the same as those described above.
Genomic DNA fingerprinting using PFGE.
Genomic DNA fingerprints of the STEC isolates were further determined using pulsed-field gel electrophoresis (PFGE) according to a standard protocol developed by PulseNet for E. coli O157:H7 (19). Briefly, agarose-embedded DNA was digested with 50 U of XbaI for 3 h in a water bath at 37°C. DNA fragments were separated by electrophoresis in 0.5× Tris-borate-EDTA buffer at 14°C for 18 h on a CHEF-III Mapper electrophoresis system, with a pulse time of 2.2 s to 54.2 s. The gels were stained with ethidium bromide, and images were taken under UV transillumination. The images were analyzed with Bionumerics software by using Dice coefficients and the unweighted-pair group method to achieve dendrograms with a 1.5% band position tolerance.
Identification of virulence determinants of other diarrheagenic E. coli strains.
To assess the presence of other potentially diarrheagenic E. coli strains in retail meats, 1,275 E. coli isolates recovered from retail meats in 2006 were chosen and examined using two multiplex PCRs (assays 2 and 3) (Table 1) to detect the following virulence gene markers: eae for intimin of EPEC, bfp for the bundle-forming pilus of EPEC, elt and stI for the heat-labile and heat-stable enterotoxins of ETEC, respectively, ial for the invasion-associated locus of EIEC, aafII for aggregative adherence fimbriae II in EAEC, and daaE for F1845 fimbriae in DAEC. Each of the two PCR assays was performed in a 25-μl reaction mixture containing 2 μl of template DNA, 2.5 μl of 10× PCR buffer, 2 μl of a 1.25 mM mixture of deoxynucleoside triphosphates, 4 μl of 25 mM MgCl2, 0.25 μl (5 U) of AmpliTaq Gold DNA polymerase (Applied Biosystems, Branchburg, NJ), and 0.5 μl of each oligonucleotide primer (Invitrogen, Carlsbad, CA). The thermocycling conditions were as follows: 95°C for 12 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 3 min, with a final 10-min extension at 72°C. PCR products (10 μl) were examined in a 2% (wt/vol) agarose gel at 120 mV for 30 min. E. coli strains ATCC 35401 (containing elt and stI), ATCC 43893 (ipaH), ATCC 43887 (eae bfp), 042 (aafII), and F1845 (daaE) were used as positive controls, while E. coli K-12 served as a negative control.
Serotyping.
All STEC and other potentially diarrheagenic E. coli isolates identified in the study were sent to the E. coli Reference Center at Pennsylvania State University to determine their O antigen and H antigen.
Statistical analysis.
Differences in percentage data were analyzed by the chi-square test, using SPSS software (version 12.0; SPSS Inc., Chicago, IL). A P value of <0.05 was considered significant for all comparisons.
Nucleotide sequence accession numbers.
The nucleotide sequences of the complete stx1 operons from isolates 20177, 22813, N2688, N11354, N11355, N13844, and N15018 were submitted to GenBank and given accession numbers GQ429154 to GQ429160. The nucleotide sequences of the complete stx2 operons from isolates N22813, 23765, N2688, N2743, N2746, N4854, N5545, N5578, N11354, N11355, N11682, and N15432 and a partial stx2 operon from isolate N5789 were also deposited in the GenBank database under accession numbers GQ429161 to GQ429173.
RESULTS
Presence of STEC in retail meats.
Among the 7,258 E. coli isolates recovered from retail meats collected in four states from 2002 to 2007, 17 (0.23%) tested positive for the presence of a stx gene(s) and were considered STEC (Table 2). Almost all STEC isolates (n = 16) were from ground beef, accounting for 0.89% of 1,806 E. coli isolates from this product. One STEC isolate (0.09%) was identified among 1,167 E. coli isolates from pork chops. No STEC strains were found in ground turkey or chicken breast. Five STEC isolates were detected among 1,306 E. coli isolates collected in 2005, followed by three STEC isolates each in 2004, 2006, and 2007, two STEC isolates in 2002, and one in 2003. The percentages of STEC isolates among the E. coli isolates ranged from 0.08% to 0.38% annually during the 6-year period, but there was no significant difference in percentage by year. Nearly 60% of the STEC (n = 10) isolates were identified among E. coli isolates from Maryland, where a higher percentage (0.57%) of STEC strains was observed than those for other states (0.05 to 0.22%) (P < 0.05).
TABLE 2.
Characteristics of STEC and atypical EPEC isolates from retail meatsa
| Isolate | Serotype | Presence of gene |
Source | State | Year | ||||
|---|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | EHEC hlyA | bfp | |||||
| 22813 | O83:H8 | + | + | − | + | Ground beef | MD | 2002 | |
| N2688 | O88:H38 | + | + | − | + | Ground beef | MD | 2004 | |
| N11354 | O83:H8 | + | + | − | + | Ground beef | GA | 2006 | |
| N11355 | O83:H8 | + | + | − | + | Ground beef | GA | 2006 | |
| N15018 | O15:H27 | + | + | − | − | Ground beef | GA | 2007 | |
| 20177 | O8:H16 | + | − | − | − | Ground beef | TN | 2002 | |
| N13844 | ONT:H51 | + | − | − | − | Pork chop | OR | 2007 | |
| N5577 | ONT:H46 | − | + | − | + | Ground beef | MD | 2005 | |
| N5578 | ONT:H46 | − | + | − | + | Ground beef | MD | 2005 | |
| N5789b | O15:H16 | − | + | − | + | Ground beef | MD | 2005 | |
| 23765 | ONT:H2 | − | + | − | − | Ground beef | OR | 2003 | |
| N2743 | O83:H8 | − | + | − | − | Ground beef | MD | 2004 | |
| N2746 | O83:H8 | − | + | − | − | Ground beef | MD | 2004 | |
| N4854 | ONT:H10 | − | + | − | − | Ground beef | MD | 2005 | |
| N5545 | ONT:H7 | − | + | − | − | Ground beef | MD | 2005 | |
| N11682 | O83:H8 | − | + | − | − | Ground beef | MD | 2006 | |
| N15432 | O83:H8 | − | + | − | − | Ground beef | OR | 2007 | |
| N11475 | O154:H9 | + | − | Chicken breast | GA | 2006 | |||
| N11537 | O2:H27 | + | − | Chicken breast | GA | 2006 | |||
| N11573 | O26:H9 | + | − | Chicken breast | GA | 2006 | |||
| N11575 | O123:H51 | + | − | Chicken breast | GA | 2006 | |||
| N12563 | ONT:H7 | + | − | Chicken breast | TN | 2006 | |||
| N11452 | ONT:H8 | + | − | Ground beef | GA | 2006 | |||
| N12148 | ONT:H8 | + | − | Ground beef | OR | 2006 | |||
| N12174 | O15:H2 | + | − | Ground beef | OR | 2006 | |||
| N12475 | O10:H2 | + | − | Ground beef | TN | 2006 | |||
| N11710 | O18:H16 | + | − | Pork chop | MD | 2006 | |||
| N12051 | O81:H7 | + | − | Pork chop | OR | 2006 | |||
Common virulence genes include stx1, stx2, eae, and EHEC hlyA for STEC and eae and bfp for EPEC.
This strain was negative for Vero cell cytotoxicity, but whether it could produce Stx was not tested by other assays. It was tentatively called STEC here based on the presence of stx.
Characterization of STEC.
Serotyping results showed that H antigens were successfully typed for all 17 STEC isolates, whereas the O antigens of 6 STEC isolates could not be determined (Table 2). Only five serotypes were found for 11 typeable isolates. Many STEC isolates (7 of 11 isolates) belonged to serotype O83:H8, whereas the other four typeable isolates exhibited different serotypes: O8:H16, O15:H16, O15:H27, and O88:H38. Different H types were observed in six O-nontypeable isolates, among which isolates N5577 and N5578 reacted with the same H antiserum (H46).
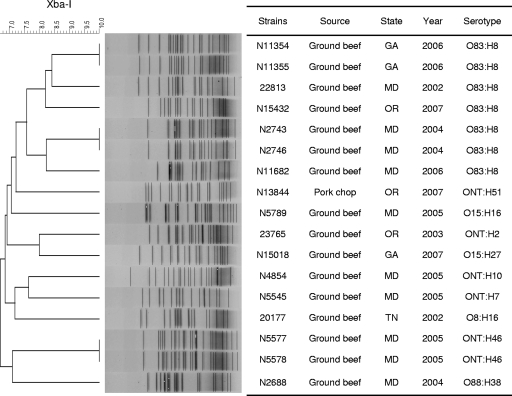
Digestion of genomic DNAs from 17 STEC isolates by use of the XbaI restriction enzyme and analysis using PFGE revealed 14 distinct profiles (Fig. 1). Although isolates of the same serotypes tended to cluster together, polymorphism of the genome sequence was also observed between some isolates of the same serotype, according to different PFGE patterns. Three pairs of STEC isolates (N11354 and N11355, N2743 and N2746, and N11354 and N11355) showed identical PFGE profiles and serotypes. Isolates within each pair were recovered from the same food source (i.e., ground beef) and the same geographic locale. Presumably, they were the same clones. All other isolates had their own specific PFGE profiles, with similarity indexes ranging from 67% to 84%.
FIG. 1.
Dendrogram of PFGE profiles obtained by XbaI digestion for 17 STEC isolates from retail meat. Similarities of PFGE profiles were calculated using the Dice algorithm, with a 1.5% tolerance level.
STEC virulence genes and Vero cell cytotoxicity.
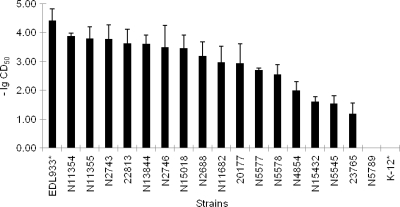
Among the 17 STEC isolates, 5 contained both stx1 and stx2 genes, whereas 2 contained stx1 only and 10 contained stx2 only (Table 2). None of the STEC isolates in this study carried the eae gene, while seven (41%) STEC isolates were EHEC hlyA positive. Cytotoxicities of the STEC isolates were examined on Vero cells. Sixteen isolates were considered toxic to Vero cells compared to E. coli K-12. Only one isolate (N5789) showed no cytotoxicity to Vero cells. For the cytotoxic STEC isolates, the CD50s ranged from 10−1.2 to 10−3.9 (Fig. 2). Overall, STEC isolates with both stx1 and stx2 displayed greater toxicity (CD50 < 10−3.2) than did those with only one of the stx genes, with the exception of N2743/N2746 and N13844.
FIG. 2.
Vero cell cytotoxicity of Shiga toxin-producing E. coli isolates from retail meats. The CD50 is expressed as the toxin dilution that causes 50% Vero cell detachment compared with untreated cells (control). The value on the y axis indicates the log of the reciprocal of the CD50. The data shown are averages for three independent assays. *, strains EDL933 and K-12 served as positive and negative controls, respectively.
stx gene sequence and phylogenetic analysis.
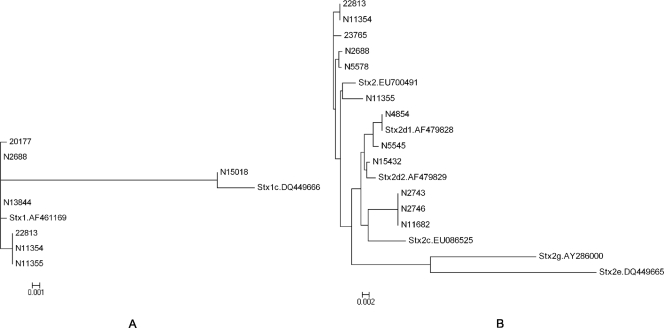
All stx1 genes from seven STEC isolates were successfully amplified and sequenced. A portion of the stx2 gene from N5789 could not be amplified despite repeated efforts. In addition, chromatograms of stx2 sequences amplified from isolates N5577 and N15018 exhibited two peaks at several positions, suggesting the presence of more than one allele of stx2. Shiga toxin 2 gene sequences from these two isolates were not determined further in this study and were excluded from phylogenetic analysis. Two previously described stx1 sequences (one classical stx1 and one stx1c sequence) and seven stx1 sequences determined in this study were aligned and used to construct a phylogenetic tree (Fig. 3A). Sequences were aligned from the start codon of stx1a to the stop codon of stx1b. Most (6 of 7 sequences) of the stx1 sequences in this study were closely related to classical stx1, whereas stx1 from N15018 was very similar to stx1c. Isolates N2688 and N13844 shared an identical stx1 gene sequence. Isolates 22813, N11354, and N11355 also shared an identical stx1 gene sequence, but it was different from the stx1 gene sequence found in N2688 and N13844.
FIG. 3.
Phylogenetic trees of stx1 sequences (A) and stx2 sequences (B) determined in this study and sequences of previously described stx genes and their variants. stx1d and stx2f were not included due to their considerable sequence divergence from classical stx1 and stx2, respectively. The horizontal bar indicates 0.001 (A) and 0.002 (B) nucleotide substitution per site. Reference DNA sequences for stx genes were obtained from GenBank and are identified in the phylogenetic trees by their accession numbers.
Six previously described sequences of stx2 and its variants and the 12 stx2 sequences determined in this study were aligned (Fig. 3B). None of the stx2 sequences determined in this study were close to stx2e or stx2g (< 93% similarity) (data not shown). Isolates 22813 and N11354 shared an identical stx2 gene sequence, as did N2743, N2746, and N11682. Putative amino acid sequence analysis revealed that Stx toxins from five isolates (N2743, N2746, N4854, N11682, and N15432) possessed two amino acid substitutions compared to classical Stx2, namely, Ser291 and Glu297 in the Stx2A2 subunit (data not shown), which are characteristic of mucus-activatable Stx2d.
Presence of virulence genes specific for other diarrheagenic E. coli strains.
In addition to 3 STEC isolates identified among the 1,275 E. coli isolates collected in 2006, 11 E. coli isolates were eat+ bfp− and were classified as atypical EPEC (Table 2). The positive rates of atypical EPEC strains among E. coli isolates from different types of meat were 1.2% for chicken breast (5/415 isolates), 1.4% for ground beef (4/293 isolates), 1.1% for pork chop (2/180 isolates), and 0% (0/387 isolates) for ground turkey. With the exception of three atypical EPEC isolates whose O serogroups could not be determined, none of the eight typeable EPEC isolates belonged to the same serotype (Table 2). No virulence genes specific for ETEC, EIEC, EAEC, or DAEC were detected among the E. coli isolates.
DISCUSSION
In this study, we analyzed 7,258 E. coli isolates recovered from retail meats collected by the NARMS program for the presence of stx genes, and 1,275 E. coli isolates recovered in 2006 were further examined for virulence determinants of other diarrheagenic E. coli strains. To our knowledge, this was the largest survey of virulence factors in E. coli isolates nonselectively recovered from retail meats. Only a small number (17) of the E. coli isolates were identified as STEC. No DAEC, EIEC, or ETEC strains were detected in the 2006 collection of E. coli isolates, although 11 atypical EPEC isolates were identified.
Shiga toxin-producing E. coli strains are mostly commensal bacteria in animals, with a high potential for food-borne transmission to humans (9). Ruminants, primarily cattle, are the predominant reservoir of STEC, and beef products serve as one of the most important sources of food-borne STEC infections (9). This consensus was supported by the result in the present study that almost all STEC isolates were recovered from ground beef. Contamination of beef by STEC has been examined by other researchers around the world. In the United States, a recent study by Samadpour et al. reported STEC in 3.5% of 1,750 retail ground beef samples collected from stores in Seattle, WA (45). In other countries, STEC was detected in 4% of beef samples in France (43) and 3% of raw beef samples in Australia (11). Fantelli and Stephan reported STEC in 1.75% of minced beef samples in Switzerland (17), and Lee et al. found STEC in 1.5% of beef samples in Korea (32). Many factors, such as geographical location and sampling, isolation, and testing methods, make comparisons of different studies difficult. In a study using a protocol similar to ours, Lee et al. detected a higher rate of STEC in beef in Korea, possibly indicating the influence of regional differences or different processing technologies in different countries (32). Culture confirmation is also an important factor affecting the results from different studies. In some research, samples were considered to contain STEC based only on positive PCR results for enrichment broth (43, 45), while in other studies, culture confirmation was performed to assess the real occurrence of STEC in meat samples (6, 17). Such variations could lead to a big difference in results, since isolation of STEC from stx-positive samples was found to be relatively difficult due to the small number of bacterial cells or the occurrence of free stx-carrying phages in meat samples (43, 45). In terms of isolation method, instead of picking multiple colonies randomly from one plate and testing each of them (17, 26), colony hybridization with a stx probe would more conveniently detect any STEC in all colonies on each plate and yield more accurate results (18, 45). The level of STEC contamination (<1%) in the NARMS ground beef appeared low compared to reports from other investigators, but caution should be exercised in interpreting this low prevalence. The NARMS program was designed to determine the prevalence of generic E. coli, not STEC. Moreover, only one E. coli isolate was picked from each E. coli-positive meat sample in the NARMS program. Consequently, studies specifically designed to determine the prevalence of specific pathotypes, such as STEC, would likely detect a much greater prevalence. Nevertheless, the screening of over 7,000 randomly selected E. coli isolates from four states over a 6-year period provides useful data on the presence of STEC in retail meats against the background of generic E. coli populations. Moreover, the analysis of a large number of samples enables us to estimate the extent of STEC contamination in different meats.
There is a paucity of data on contamination by STEC in retail meats other than beef. In this study, only one STEC isolate was found among 1,168 E. coli isolates from pork chops, whereas none of the E. coli isolates from chicken (n = 2,181) or turkey (n = 2,106) was identified as STEC. Swine has been suggested as a potential reservoir of STEC strains, and the presence of STEC in pork has been reported before (18). In a study by Samadpour et al., 9 of 51 pork samples collected in local grocery stores in the Seattle area were positive for STEC (45). In New Zealand, Brooks et al. detected one STEC isolate in 35 pork samples (6). In contrast to swine, poultry is generally not considered a source of STEC. The lack of STEC in poultry meat from the NARMS program was consistent with reports from Schroeder et al., who failed to detect STEC in retail chicken (51 ground chicken samples and 212 whole chickens) and turkey (50 ground turkey samples and 194 turkey breasts) samples from the Washington, DC, area (46). Similarly, Brooks et al. failed to isolate STEC from 36 chicken samples in New Zealand (6, 46), and Heuvelink et al. did not find STEC O157 in raw chicken (n = 744) and poultry products (n = 75) in The Netherlands (26). However, Samadpour et al. recovered 4 STEC isolates from 33 chicken breasts and 1 STEC isolate from 15 turkey samples (12), and Doyle and Schoeni found STEC O157 in 4 of 263 poultry products (15). These isolations were previously thought to possibly be due to exposure to infected ruminants, but recent isolation of STEC from laying hens indicated that poultry may be a source of contamination (14). As discussed above, contamination rates in different studies could be affected by many factors, and attention needs to be paid in comparing data from different studies.
Although O157:H7 is the most common STEC serotype that causes human illness in the United States, there is growing concern over the emergence of highly virulent non-O157 STEC serotypes that are globally distributed, several of which are associated with outbreaks and/or severe human illness, such as HUS and HC (2, 7). Although E. coli O157 was reported to be present in 0.7% of ground beef samples in a USDA study and 1.1% of beef samples in another study in Washington (38, 45), none of the STEC isolates in the present study belonged to serotype O157, which was in agreement with another study conducted in the United States (50). Among the typeable STEC isolates, it is interesting that there was relatively common recovery of serotype O83:H8, which was not reported previously for food or animals. Although the seven isolates belonged to the same serotype, they did not have identical PFGE profiles and stx genotypes, except for N11354 and N11355, indicating a nonclonal spread. The fact that STEC O83 has been associated with human illness (2) raises the possibility that O83:H8 (with diverse genotypes and cytotoxicities) might be transmitted to humans via meat products. Additionally, isolate N15018 belonged to O15:H27, a serotype which has also been implicated in human illness (42). The failure to determine the O type for several STEC isolates suggests the complexity of STEC isolates present in retail meats. Since many STEC isolates were nontypeable by serotyping, it is important to improve the current typing scheme and to develop new typing technologies to encompass serogroups that are untypeable at present.
Since not all STEC strains are equally pathogenic to humans, evaluation of virulence-associated factors is necessary to assess an individual isolate's potential to cause human illness. Our results showed that none of the STEC isolates carried the eae gene, which is consistent with findings of other studies, in which no eae carriage was observed among non-O157 STEC isolates from beef (24, 43). Studies have shown that most STEC isolates from healthy cattle do not carry the eae gene (22, 27), and since beef products are generally derived from healthy cattle, this may explain the absence of eae in STEC isolates from NARMS beef samples. Although eae is considered important for causing attaching and effacing lesions in human intestinal epithelial cells, it may not be essential for STEC pathogenicity, since eae-negative STEC strains have also been reported to cause severe human infections (39). It was postulated that eae-negative STEC may utilize additional adherence factors, such as Saa (an autoagglutinating adhesin), Iha (an adherence-conferring protein), Efa1 (an EHEC factor for adherence), and LP (the long polar fimbria protein), in the disease process (52). Interestingly, the EHEC hlyA gene was observed frequently (41% [7 of 17 isolates]) in the STEC isolates in the present study. Similar frequencies (40 to 51%) of this gene were observed by Slanec et al., in STEC strains isolated from food samples (40%), and by Aidar-Ugrinovich et al., in STEC isolates from calves (51%) (1, 49). EHEC hlyA is located in a large plasmid which many human-pathogenic STEC strains harbor. The gene product may contribute to pathogenesis by acting as a pore-forming cytolysin on eukaryotic cells. The presence of this gene may enhance the virulence potential of STEC isolates from retail meats.
Shiga toxins differ in toxicity, toxin receptor, and amino acid sequence (33). Nucleotide sequence analysis of stx1 and stx2 genes present in our STEC isolates confirmed this finding. stx2 gene sequences exhibited much more sequence diversity than did stx1 sequences (Fig. 3). Three stx2 sequences were identical to a previously published stx2 gene sequence (GenBank accession no. AY443058.1) from a human isolate, and two of them were identical to an stx2 allele in a bovine isolate (AY443054.1). No other stx2 sequences were identical at the nucleotide level. All mucus-activatable stx2d sequences clustered together with two previously described activatable stx2d sequences (stx2d1 and stx2d2), but with some sequence difference (Fig. 3B). stx2c and stx2 from N5545 were also included in the cluster, but they did not have the characteristic amino acid substitutions common in activatable stx2d. There were two isolates (N5577 and N15018) whose stx2 chromatograms showed two peaks at multiple sites, which indicates that these two strains may carry more than one allele, an interesting phenomenon that was also observed by other researchers (3). All STEC isolates were shown to be toxic to Vero cells, except for one isolate (N5789). The stx2 sequence could not be obtained for this isolate, since it could not be amplified by the first pair of sequencing primers. Several primers were tried to amplify a fragment which would cover the Stx2A subunit of the gene, but part of the gene could never be amplified. It is possible that a large intervening insertion may be present that prevents successful PCR amplification. It is also possible that this insertion abolishes the activity of Stx2A and consequently renders the isolate less toxic than other STEC strains.
Shiga toxin types were suggested to correlate with the clinical symptoms of STEC infection (28). Mucus-activatable Stx2d is associated with high virulence and the ability to cause HUS (4, 21, 28). The particular attribute of this variant is that it can be cleaved by elastase in the intestinal mucosa, causing an increase in cytotoxicity of up to 1,000-fold (31). This character is attributed to two amino acid substitutions relative to the sequence of classical Stx2, namely, Ser291 and Glu297. Based on predicted amino acid sequence analysis, 5 of 15 stx2-containing isolates in this study harbored mucus-activatable stx2d. The relatively large number of mucus-activatable stx2d genes found in STEC strains isolated from retail meat deserves attention. Studies found that eae-negative, mucus-activatable Stx2d-producing STEC strains were involved in sporadic and outbreak cases of HC and HUS (28, 40). It was also shown that although eae-negative STEC strains are normally isolated from persons with no or mild disease, most eae-negative STEC strains associated with severe symptoms harbor mucus-activatable stx2d as the sole stx gene (4). The pathogenic potential of the five isolates with mucus-activatable stx2d should not be underestimated. Currently, very limited data are available concerning the presence of STEC strains harboring mucus-activatable stx2d in food or livestock sources. Zheng et al. identified seven STEC isolates carrying activatable stx2d among 153 STEC strains isolated from food, cattle, and humans, and none of them contained the eae gene (55). Gobius et al. investigated 311 STEC strains possessing stx2 from food and livestock and found 12 STEC isolates carrying activatable stx2d, all of which did not have eae either (21). It has been suggested that the expression of an activatable toxin may compensate for the lack of intimin (34). Due to their strong association with severe clinical outcomes, more surveillance of STEC strains expressing activatable stx2d in food and human illness is warranted.
Another interesting finding of this study was the identification of several atypical EPEC isolates. EPEC is a leading cause of infant diarrhea in developing countries and is also an important cause of diarrhea in developed countries (53). Typical EPEC strains contain both eae and bfp, while atypical EPEC strains contain only eae. In industrialized countries, typical EPEC infections have decreased and atypical E. coli infections seem to have increased in recent years (25, 53). Unlike typical EPEC strains, which are found only in humans, atypical EPEC strains have been isolated from a variety of animal species, such as cattle, goats, sheep, chickens, pigeons, and gulls (13). In addition to chicken and beef, we also identified atypical EPEC isolates among E. coli isolates from pork, indicating that pigs may also be potential reservoirs for the pathogen. Atypical EPEC strains found in this study belonged to a variety of serogroups, most of which were not found in atypical EPEC strains involved in human infections. However, isolate N11573 belonged to O26, a serogroup that is frequently found in classic human EPEC strains (8). Further studies are needed to determine whether atypical EPEC strains of animal origin can actually cause human infections when ingested.
In conclusion, retail meats, especially ground beef, were contaminated with STEC, although at a very low frequency, and some of the strains contained Shiga toxins associated with high potential to cause severe human disease. Moreover, the identification of atypical EPEC strains in retail meats is noteworthy, and the potential role of animal-derived atypical EPEC strains in causing human infections requires further investigation.
Acknowledgments
This study was made possible by grants from the Joint Institute for Food Safety and Applied Nutrition (JIFSAN) of the University of Maryland and the U.S. Food & Drug Administration.
We greatly thank James P. Nataro of the University of Maryland School of Medicine for providing E. coli strains 042 and F1845.
Footnotes
Published ahead of print on 15 January 2010.
REFERENCES
- 1.Aidar-Ugrinovich, L., J. Blanco, M. Blanco, J. E. Blanco, L. Leomil, G. Dahbi, A. Mora, D. L. Onuma, W. D. Silveira, and A. F. Pestana de Castro. 2007. Serotypes, virulence genes, and intimin types of Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) isolated from calves in Sao Paulo, Brazil. Int. J. Food Microbiol. 115:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim, K. A. 2007. The non-O157 Shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33:67-87. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., A. Miko, G. Krause, K. Pries, S. Haby, K. Steege, and N. Albrecht. 2007. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. [DOI] [PubMed] [Google Scholar]
- 5.Brett, K. N., V. Ramachandran, M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2003. stx1c is the most common Shiga toxin 1 subtype among Shiga toxin-producing Escherichia coli isolates from sheep but not among isolates from cattle. J. Clin. Microbiol. 41:926-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, H. J., B. D. Mollison, K. A. Bettelheim, K. Matejka, K. A. Paterson, and V. K. Ward. 2001. Occurrence and virulence factors of non-O157 Shiga toxin-producing Escherichia coli in retail meat in Dunedin, New Zealand. Lett. Appl. Microbiol. 32:118-122. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, J. T., E. G. Sowers, J. G. Wells, K. D. Greene, P. M. Griffin, R. M. Hoekstra, and N. A. Strockbine. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192:1422-1429. [DOI] [PubMed] [Google Scholar]
- 8.Campos, L. C., M. R. Franzolin, and L. R. Trabulsi. 2004. Diarrheagenic Escherichia coli categories among the traditional enteropathogenic E. coli O serogroups—a review. Mem. Inst. Oswaldo Cruz 99:545-552. [DOI] [PubMed] [Google Scholar]
- 9.Caprioli, A., S. Morabito, H. Brugere, and E. Oswald. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289-311. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 27 July 2009, accession date. Outbreak surveillance data. CDC, Atlanta, GA. http://www.cdc.gov/foodborneoutbreaks/outbreak_data.htm.
- 11.Chattopadhyay, U. K., S. Dutta, A. Deb, and D. Pal. 2001. Verotoxin-producing Escherichia coli—an environment-induced emerging zoonosis in and around Calcutta. Int. J. Environ. Health Res. 11:107-112. [DOI] [PubMed] [Google Scholar]
- 12.Cobeljic, M., B. Dimic, D. Opacic, Z. Lepsanovic, V. Stojanovic, and S. Lazic. 2005. The prevalence of Shiga toxin-producing Escherichia coli in domestic animals and food in Serbia. Epidemiol. Infect. 133:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes, C., R. De la Fuente, J. Blanco, M. Blanco, J. E. Blanco, G. Dhabi, A. Mora, P. Justel, A. Contreras, A. Sanchez, J. C. Corrales, and J. A. Orden. 2005. Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 110:67-76. [DOI] [PubMed] [Google Scholar]
- 14.Dipineto, L., A. Santaniello, M. Fontanella, K. Lagos, A. Fioretti, and L. F. Menna. 2006. Presence of Shiga toxin-producing Escherichia coli O157:H7 in living layer hens. Lett. Appl. Microbiol. 43:293-295. [DOI] [PubMed] [Google Scholar]
- 15.Doyle, M. P., and J. L. Schoeni. 1987. Isolation of Escherichia coli O157:H7 from retail fresh meats and poultry. Appl. Environ. Microbiol. 53:2394-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson, M. C., and M. P. Doyle. 2007. Food as a vehicle for transmission of Shiga toxin-producing Escherichia coli. J. Food Prot. 70:2426-2449. [DOI] [PubMed] [Google Scholar]
- 17.Fantelli, K., and R. Stephan. 2001. Prevalence and characteristics of Shiga toxin-producing Escherichia coli and Listeria monocytogenes strains isolated from minced meat in Switzerland. Int. J. Food Microbiol. 70:63-69. [DOI] [PubMed] [Google Scholar]
- 18.Fratamico, P. M., L. K. Bagi, E. J. Bush, and B. T. Solow. 2004. Prevalence and characterization of Shiga toxin-producing Escherichia coli in swine feces recovered in the National Animal Health Monitoring System's Swine 2000 study. Appl. Environ. Microbiol. 70:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentry, M. K., and J. M. Dalrymple. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J. Clin. Microbiol. 12:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobius, K. S., G. M. Higgs, and P. M. Desmarchelier. 2003. Presence of activatable Shiga toxin genotype (stx(2d)) in Shiga toxigenic Escherichia coli from livestock sources. J. Clin. Microbiol. 41:3777-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guth, B. E., I. Chinen, E. Miliwebsky, A. M. Cerqueira, G. Chillemi, J. R. Andrade, A. Baschkier, and M. Rivas. 2003. Serotypes and Shiga toxin genotypes among Escherichia coli isolated from animals and food in Argentina and Brazil. Vet. Microbiol. 92:335-349. [DOI] [PubMed] [Google Scholar]
- 23.Gyles, C. L. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45-E62. [DOI] [PubMed] [Google Scholar]
- 24.Hazarika, R. A., D. K. Singh, K. N. Kapoor, R. K. Agarwal, A. B. Pandey, and Purusottam. 2007. Verotoxic Escherichia coli (STEC) from beef and its products. Indian J. Exp. Biol. 45:207-211. [PubMed] [Google Scholar]
- 25.Hernandes, R. T., W. P. Elias, M. A. Vieira, and T. A. Gomes. 2009. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 297:137-149. [DOI] [PubMed] [Google Scholar]
- 26.Heuvelink, A. E., J. T. Zwartkruis-Nahuis, R. R. Beumer, and E. de Boer. 1999. Occurrence and survival of verocytotoxin-producing Escherichia coli O157 in meats obtained from retail outlets in The Netherlands. J. Food Prot. 62:1115-1122. [DOI] [PubMed] [Google Scholar]
- 27.Irino, K., M. A. Kato, T. M. Vaz, I. I. Ramos, M. A. Souza, A. S. Cruz, T. A. Gomes, M. A. Vieira, and B. E. Guth. 2005. Serotypes and virulence markers of Shiga toxin-producing Escherichia coli (STEC) isolated from dairy cattle in Sao Paulo State, Brazil. Vet. Microbiol. 105:29-36. [DOI] [PubMed] [Google Scholar]
- 28.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 29.Jinneman, K. C., K. J. Yoshitomi, and S. D. Weagant. 2003. Multiplex real-time PCR method to identify Shiga toxin genes stx1 and stx2 and Escherichia coli O157:H7/H− serotype. Appl. Environ. Microbiol. 69:6327-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405-418. [DOI] [PubMed] [Google Scholar]
- 31.Kokai-Kun, J. F., A. R. Melton-Celsa, and A. D. O'Brien. 2000. Elastase in intestinal mucus enhances the cytotoxicity of Shiga toxin type 2d. J. Biol. Chem. 275:3713-3721. [DOI] [PubMed] [Google Scholar]
- 32.Lee, G. Y., H. I. Jang, I. G. Hwang, and M. S. Rhee. 2009. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry, and pork in Korea. Int. J. Food Microbiol. 134:196-200. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. E., J. Reed, M. S. Shields, K. M. Spiegel, L. D. Farrell, and P. P. Sheridan. 2007. Phylogenetic analysis of Shiga toxin 1 and Shiga toxin 2 genes associated with disease outbreaks. BMC Microbiol. 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melton-Celsa, A. R., S. C. Darnell, and A. D. O'Brien. 1996. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64:1569-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng, J., S. Zhao, and M. P. Doyle. 1998. Virulence genes of Shiga toxin-producing Escherichia coli isolated from food, animals and humans. Int. J. Food Microbiol. 45:229-235. [DOI] [PubMed] [Google Scholar]
- 36.Moyo, S. J., S. Y. Maselle, M. I. Matee, N. Langeland, and H. Mylvaganam. 2007. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect. Dis. 7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naugle, A. L., K. G. Holt, P. Levine, and R. Eckel. 2005. Food safety and inspection service regulatory testing program for Escherichia coli O157:H7 in raw ground beef. J. Food Prot. 68:462-468. [DOI] [PubMed] [Google Scholar]
- 39.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierard, D., D. Stevens, L. Moriau, H. Lior, and S. Lauwers. 1997. Isolation and virulence factors of verocytotoxin-producing Escherichia coli in human stool samples. Clin. Microbiol. Infect. 3:531-540. [DOI] [PubMed] [Google Scholar]
- 43.Pradel, N., V. Livrelli, C. De Champs, J. B. Palcoux, A. Reynaud, F. Scheutz, J. Sirot, B. Joly, and C. Forestier. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J. Clin. Microbiol. 38:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 45.Samadpour, M., M. W. Barbour, T. Nguyen, T. M. Cao, F. Buck, G. A. Depavia, E. Mazengia, P. Yang, D. Alfi, M. Lopes, and J. D. Stopforth. 2006. Incidence of enterohemorrhagic Escherichia coli, Escherichia coli O157, Salmonella, and Listeria monocytogenes in retail fresh ground beef, sprouts, and mushrooms. J. Food Prot. 69:441-443. [DOI] [PubMed] [Google Scholar]
- 46.Schroeder, C. M., D. G. White, B. Ge, Y. Zhang, P. F. McDermott, S. Ayers, S. Zhao, and J. Meng. 2003. Isolation of antimicrobial-resistant Escherichia coli from retail meats purchased in Greater Washington, DC, U.S.A. Int. J. Food Microbiol. 85:197-202. [DOI] [PubMed] [Google Scholar]
- 47.Schroeder, C. M., C. Zhao, C. DebRoy, J. Torcolini, S. Zhao, D. G. White, D. D. Wagner, P. F. McDermott, R. D. Walker, and J. Meng. 2002. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl. Environ. Microbiol. 68:576-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sjoling, A., G. Wiklund, S. J. Savarino, D. I. Cohen, and A. M. Svennerholm. 2007. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J. Clin. Microbiol. 45:3295-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slanec, T., A. Fruth, K. Creuzburg, and H. Schmidt. 2009. Molecular analysis of virulence profiles and Shiga toxin genes in food-borne Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 75:6187-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarr, P. I., N. T. Tran, and R. A. Wilson. 1999. Escherichia coli O157:H7 in retail ground beef in Seattle: results of a one-year prospective study. J. Food Prot. 62:133-139. [DOI] [PubMed] [Google Scholar]
- 51.Thorpe, C. M. 2004. Shiga toxin-producing Escherichia coli infection. Clin. Infect. Dis. 38:1298-1303. [DOI] [PubMed] [Google Scholar]
- 52.Toma, C., E. Martinez Espinosa, T. Song, E. Miliwebsky, I. Chinen, S. Iyoda, M. Iwanaga, and M. Rivas. 2004. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 42:4937-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trabulsi, L. R., R. Keller, and T. A. Tardelli Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidal, M., E. Kruger, C. Duran, R. Lagos, M. Levine, V. Prado, C. Toro, and R. Vidal. 2005. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J. Clin. Microbiol. 43:5362-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, J., S. Cui, L. D. Teel, S. Zhao, R. Singh, A. D. O'Brien, and J. Meng. 2008. Identification and characterization of Shiga toxin type 2 variants in Escherichia coli isolates from animals, food, and humans. Appl. Environ. Microbiol. 74:5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]