Abstract
Listeria monocytogenes was previously shown to form biofilms composed of a network of knitted chains under continuous-flow conditions. Here we show that the SOS response is activated under these conditions and that deletion of its regulon member yneA results in diminished biofilm formation under continuous-flow conditions.
The food-borne pathogen Listeria monocytogenes is widely distributed in the environment and is able to grow in soil and on plant materials, thereby facilitating environmental transmission of this pathogen. L. monocytogenes is therefore frequently encountered in food processing facilities, on food contact surfaces, in pipelines, on floors, and in drains, which in turn may result in contamination of food products. It is expected that the formation of biofilms and subsequent dispersal plays an important role in recontamination processes. Biofilms are structured communities of microorganisms adhering to a surface that may be encapsulated within a self-produced protective and adhesive matrix of extracellular polymeric substances (EPS) (9). Most studies of L. monocytogenes biofilm formation focus on biofilm formation under static conditions on polystyrene, glass, or stainless steel surfaces. L. monocytogenes biofilms on polystyrene and glass consist of a homogeneous layer, while on stainless steel L. monocytogenes biofilms consist of single attached cells or microcolonies (2, 6, 11). The small, rod-shaped morphology of these static biofilm cells is very similar to the morphology of planktonic cells. However, L. monocytogenes biofilms formed under continuous-flow conditions, conceivably encountered in industrial pipelines, consist of a dense network of knitted chains composed of elongated cells and surrounding ball-shaped microcolonies (10). Recently, it has been shown that activation of the L. monocytogenes SOS response factor YneA resulted in cell elongation (14). The SOS response is involved in DNA repair, restart of stalled replication forks (3, 8), and mutagenesis (12). It is regulated by RecA (activator) and LexA (repressor) and furthermore contains DNA repair systems and translesion DNA polymerases such as DinB (15). To prevent transaction of the genome during replication fork stalling, septum formation at the midcell is inhibited by YneA, which results in cell elongation (7, 14). Recently, RecA-dependent genetic recombination was described for Pseudomonas aeruginosa biofilm cells harvested from a drip flow reactor, pointing to activation of the SOS response under these conditions (1). In this study, we investigated whether the SOS response is activated during L. monocytogenes EGD-e biofilm formation and whether there is a role for YneA in knitted chain biofilm formation.
L. monocytogenes EGD-e (5), its isogenic in-frame ΔrecA and ΔyneA deletion mutants, its yneA complementation mutant, and its recA and yneA promoter reporter mutants (14) were grown in brain heart infusion (BHI; Difco) broth. No significant difference in planktonic growth between wild-type and mutant cultures was observed (results not shown). Continuous-flow biofilm formation experiments were performed as described previously (10) with small modifications. Biofilms were grown in a flow cell (BST FC 281; Biosurface Technologies Corporation) at 20°C, using BHI with a flow rate of 10 ml/h. Static biofilm experiments were performed as described previously (4) with small modifications. Biofilms were grown in BHI in 12-well polystyrene microtiter plates (Greiner) using a 1% inoculum of an overnight-grown culture. For quantification, the biofilm cells were harvested in phosphate-buffered saline (PBS), serially diluted in PBS, and plated on BHI agar. Colonies were enumerated after 2 days of incubation at 30°C. Quantitative real-time PCR analysis was performed as described previously (13) using primers shown in appendix S1 in the supplemental material. Shortly, biofilms were quenched in RNAprotect (Qiagen) following the manufacturer's protocol and harvested. Expression levels were normalized using the housekeeping genes tpi, rpoB, and 16S rRNA. Biofilm formation experiments and quantitative PCR (Q-PCR) analysis were performed in two independent biological experiments using two replicates each. Statistically significant differences were identified using a two-tailed Student t test (P < 0.05).
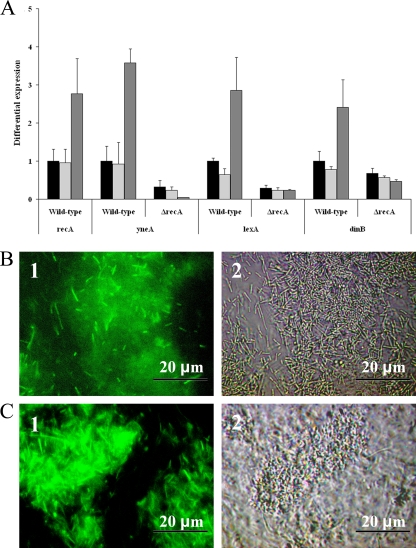
To investigate activation of L. monocytogenes EGD-e SOS response during continuous-flow biofilm formation, Q-PCR analysis of the SOS response genes recA, lexA, yneA, and dinB and promoter reporter studies using the promoters for recA and yneA were performed (Fig. 1). Compared with the reference (planktonic cells from a 48-h liquid culture), all four tested SOS response genes were upregulated in wild-type strain cells isolated from a 48-h continuous-flow biofilm (P < 0.05, t test), but not in cells isolated from a 48-h static biofilm (Fig. 1A). Furthermore, the ΔrecA mutant strain did not show upregulation of yneA and the other SOS response genes during continuous-flow biofilm formation, indicating that RecA is required for activation of the SOS response during continuous-flow biofilm formation. Furthermore, continuous-flow biofilm formation also resulted in visible expression of enhanced green fluorescent protein (EGFP) for both yneA and recA promoter reporters (Fig. 1B and C). Expression of EGFP was not observed for these promoter reporters in planktonic cells grown in liquid culture or during static biofilm formation (results not shown). These results indicate that the SOS response is specifically activated during continuous-flow biofilm formation.
FIG. 1.
Activation of the SOS response during biofilm formation. (A) The graph shows differential expression of four SOS response genes in the wild-type and ΔrecA mutant strain between 48-h stationary-phase cultures (black), 48-h static biofilms (light gray), and 48-h continuous-flow biofilms (dark gray). Expression for each SOS response gene in the wild-type 48-h stationary-phase cultures is set at 1. (B and C) Micrographs show fluorescence (1) and phase-contrast (2) pictures of cells expressing EGFP from the recA (B) and yneA (C) promoters after 48 h of biofilm formation in BHI at 20°C under continuous-flow conditions.
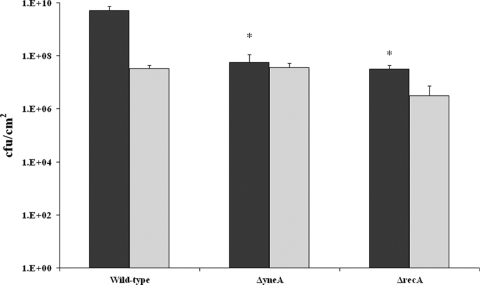
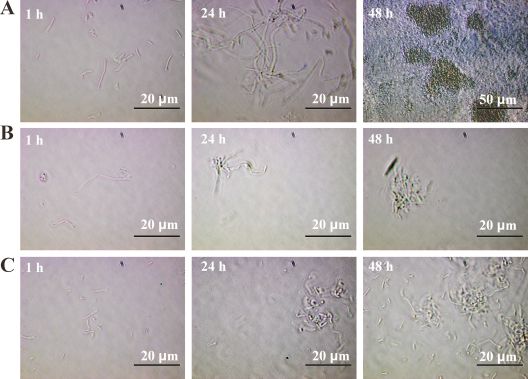
The impact of RecA and YneA on biofilm formation was assessed using the wild-type strain and in-frame ΔyneA and ΔrecA strains (Fig. 2). Both ΔrecA and ΔyneA mutants showed a significant deficiency in total biofilm produced under continuous-flow conditions (P < 0.05, t test), which was approximately 100-fold lower than that of the wild-type strain. No significant difference in static biofilm formation between wild-type and mutant strains was observed. Apparently, YneA and RecA are not required for static biofilm formation, which is in line with the lack of activation of the SOS response under these conditions. The wild-type, ΔyneA, and ΔrecA strains were microscopically examined during continuous-flow biofilm formation (Fig. 3). Analysis of the number of adherent cells 1 h after the start of the experiment did not reveal differences between the wild-type strain and the two mutants, which indicates that initial attachment is similar. After 24 h, the wild-type strain biofilm appeared to be composed of a complex structure of elongated cells forming a network of knitted chains, which after 48 h had developed into a denser network containing ball-shaped microcolonies. These results are in concordance with the study by Rieu et al. (10). However, both ΔyneA and ΔrecA mutant strains showed only some patches of adherent cells after 24 h, which developed into very small microcolonies after 48 h. Thus, formation of elongated cells in a network of knitted chains was not observed for these mutants. These results indicate that RecA and YneA are required to form this type of biofilm. To verify the specific role of YneA in continuous-flow biofilm formation, a yneA complementation mutant was constructed, which indeed showed biofilm formation capacity similar to that of the wild type, under both continuous-flow and static conditions (results not shown).
FIG. 2.
Comparative analysis of biofilm formation between wild-type strain and ΔrecA and ΔyneA mutants under continuous-flow and static conditions. The graph shows the amount of biofilm produced by wild-type and mutant strains after 48 h of biofilm formation at 20°C under continuous-flow (dark gray) and static (light gray) conditions. *, significantly different from wild-type strain (P < 0.05, t test).
FIG. 3.
Knitted chain biofilm formation under continuous-flow conditions is dependent on RecA and YneA. The micrographs show biofilms formed after 1, 24, and 48 h in BHI at 20°C for the wild-type strain (A), the ΔrecA mutant strain (B), and the ΔyneA mutant strain (C).
This study established a clear link between the SOS response and knitted chain biofilm formation under continuous-flow conditions. RecA-dependent activation of the SOS response and in particular of yneA under continuous-flow conditions resulted in cell elongation and the formation of knitted chain biofilms. The signals that activate the L. monocytogenes SOS response are currently being studied and may provide tools for control of biofilm formation under continuous-flow conditions.
Supplementary Material
Footnotes
Published ahead of print on 22 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chae, M. S., H. Schraft, L. Truelstrup Hansen, and R. Mackereth. 2006. Effects of physicochemical surface characteristics of Listeria monocytogenes strains on attachment to glass. Food Microbiol. 23:250-259. [DOI] [PubMed] [Google Scholar]
- 3.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 4.Djordjevic, D., M. Wiedmann, and L. A. McLandsborough. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 6.Kalmokoff, M. L., J. W. Austin, X. D. Wan, G. Sanders, S. Banerjee, and J. M. Farber. 2001. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 91:725-734. [DOI] [PubMed] [Google Scholar]
- 7.Kawai, Y., S. Moriya, and N. Ogasawara. 2003. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol. Microbiol. 47:1113-1122. [DOI] [PubMed] [Google Scholar]
- 8.Maul, R. W., and M. D. Sutton. 2005. Roles of the Escherichia coli RecA protein and the global SOS response in effecting DNA polymerase selection in vivo. J. Bacteriol. 187:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 10.Rieu, A., R. Briandet, O. Habimana, D. Garmyn, J. Guzzo, and P. Piveteau. 2008. Listeria monocytogenes EGD-e biofilms: no mushrooms but a network of knitted chains. Appl. Environ. Microbiol. 74:4491-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez, A., W. R. Autio, and L. A. McLandsborough. 2008. Effect of surface roughness and stainless steel finish on Listeria monocytogenes attachment and biofilm formation. J. Food Prot. 71:170-175. [DOI] [PubMed] [Google Scholar]
- 12.Schlacher, K., and M. F. Goodman. 2007. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat. Rev. Mol. Cell Biol. 8:587-594. [DOI] [PubMed] [Google Scholar]
- 13.van der Veen, S., T. Hain, J. A. Wouters, H. Hossain, W. M. de Vos, T. Abee, T. Chakraborty, and M. H. Wells-Bennik. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 153:3593-3607. [DOI] [PubMed] [Google Scholar]
- 14.van der Veen, S., S. van Schalkwijk, D. Molenaar, W. M. de Vos, T. Abee, and M. H. J. Wells-Bennik. 2010. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology 156:374-384. [DOI] [PubMed] [Google Scholar]
- 15.Wagner, J., P. Gruz, S. R. Kim, M. Yamada, K. Matsui, R. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.