Abstract
By placing the anode of a sediment microbial fuel cell (SMFC) in the rhizosphere of a rice plant, root-excreted rhizodeposits can be microbially oxidized with concomitant current generation. Here, various molecular techniques were used to characterize the composition of bacterial and archaeal communities on such anodes, as influenced by electrical circuitry, sediment matrix, and the presence of plants. Closed-circuit anodes in potting soil were enriched with Desulfobulbus-like species, members of the family Geobacteraceae, and as yet uncultured representatives of the domain Archaea.
Living plants release substantial amounts of carbon in the soil as rhizodeposits, which are to a large extent transformed into the greenhouse gas methane in wetlands (21). It was recently demonstrated (8, 33) that the rhizodeposits can be harvested by plant microbial fuel cells (plant MFCs) and transformed into electricity. In its most straightforward form, a plant MFC is an adaptation of a sediment MFC (SMFC), which has an anode buried in (planted) sediment, allowing (microbial) oxidation of reduced compounds, and a cathode in the overlying water.
The roots and surrounding rhizosphere in a plant SMFC add an extra parameter to the as yet multifaceted SMFC system. In the present study, two molecular profiling techniques (denaturing gradient gel electrophoresis [DGGE] and terminal restriction fragment length polymorphism [T-RFLP]) will be applied to evaluate the effect of plant presence, support material, operation of the electrical circuit, and anode depth on the bacterial and archaeal communities associated with rice SMFC anodes. Phylogenetic analysis will give further insight in their composition.
Experimental setup and operation.
Several groups of SMFCs planted with rice were set up, operated, and electrochemically evaluated as previously described (8). Two series (A and B) of SMFCs were installed during subsequent summers as replication in time. Both series consisted of one group of reactors filled with vermiculite (exfoliated vermiculite; Sibli SA, Andenne, Belgium) and one group of reactors filled with potting soil (structural professional type 1; M. Snebbout N.V., Kaprijke, Belgium) as support for plant growth. The potting soil was based on NPK-enriched peat (peat enriched with nitrogen, phosphorus, and potassium) with a mean of 150 mg SO42− liter−1 and 20% organic substances. In the reactors of series A, two anodic carbon felts were placed at 6 and 14 cm (depth indicated as H for high and L for low, respectively) below the support surface (one anode at 6 cm in open-circuit reactors). For the more-extensive series B, three anodic carbon felts were placed at 5-, 11-, and 17-cm depths (H, M for medium, and L, respectively). The reactors were inoculated with effluent from an acetate-fed MFC (series A and B) and with a methanogenic culture (presettler of constructed wetland, Wontergem, Belgium) (only series A). At the end of the reactor runs, the pH was 6.2 ± 0.6 for reactors with soil and 7.0 ± 0.5 for those with vermiculite.
Apart from the support type used (in reactor names indicated with V for vermiculite and S for [potting] soil) and the experimental period (a and b for series A and B), there were three types of reactors: (i) P-CC reactors, with plants and closed electrical circuit, allowing the harvest of electrical current; (ii) NP-CC control reactors, without plants, but with closed circuit; (iii) P-OC control reactors, with plants, but with open circuit (no electron harvest). Table S1 in the supplemental material shows the overall reactor setup, nomenclature, and biological replicates.
The electrochemical performance of series A and part of series B was previously reported (8) and is summarized in Table S2 in the supplemental material. Vermiculite reactors produced electricity only in the presence of plants. The current output of soil reactors was three times higher with plants. Series B with vermiculite was not reported before. Plant growth as well as current output remained limited for this group (lower by a factor of 60 compared to the current output of series A [see Table S2 in the supplemental material]), but it was higher by a factor of 3 in the presence of plants.
Molecular profiling techniques.
The anodes were removed and stored at −20°C once all plants per series had started to senesce, i.e., about 195 and 140 days after SMFC startup for series A and B, respectively. For DGGE analysis, DNA from 2 g (wet weight) anode was extracted by standard methods (6). 16S rRNA gene fragments were amplified with primers P338f-GC and P518r for Bacteria (26). 16S rRNA gene fragments were amplified with primers Ar3f and Ar9r, followed by Saf-GC and Parch 519r (nested PCR) (27) for Archaea from series A and primers Arc915f and 1352ar-GC (nonnested PCR) (30) for Archaea from series B. PCR products were analyzed by DGGE with a denaturing gradient from 45 to 60% for Bacteria (8% acrylamide, 16 h at 38V) (5) and 55 to 70% for Archaea (7% acrylamide, 30 min at 40 V, 16 h at 70V) (30). Gel patterns were normalized with Bionumerics 5.1 (Applied Maths). For T-RFLP analysis, three parallel DNA extracts were made from ∼0.5 g anode using bead-beating (25), DNA was mixed, and T-RFLP analysis was performed by the method of Egert et al. (10). Briefly, 16S rRNA genes were amplified using 5′ 6-carboxyfluorescein-labeled primers Ba27f and Ba907r for Bacteria and primers Ar109f and Ar912r for Archaea. Amplicons (∼100 ng) were digested with restriction enzymes MspI for Bacteria and TaqI for Archaea (Promega). Electrophoresis was performed on an ABI PRISM 3130 genetic analyzer (Applera Deutschland GmbH, Darmstadt, Germany). Electropherograms were analyzed with GeneScan 4.0 (Applied Biosystems). Only peaks from 40 to 900 bp were considered; peak heights were standardized to the minimum (9). Cluster analysis of DGGE and T-RFLP profiles was performed with Bionumerics 5.1 and based on the Pearson correlation matrix and unweighted-pair group method using average linkages (UPGMA) algorithm, and cluster cutoff was conducted by using point-bisectional correlation.
Clone libraries and band excision.
For the clone libraries, 16S rRNA genes were amplified using primers Ba27f and Ba907r for Bacteria and primers Ar109f and Ar912r for Archaea. PCR fragments were cloned using the pGEM-T vector system II (Promega). Sequence analysis (ADIS, Max Planck Institute for Plant Breeding Research, Cologne, Germany) of randomly selected clones resulted in a total of 133 sequences for Bacteria and 52 for Archaea. The clone libraries were screened for chimeras using the Bellerophon server (16) and Mallard software (3); 44 putative chimeras for Bacteria and 2 for Archaea were verified by fractional treeing (23) and excluded from further analysis. The values for diversity coverage (35) of the libraries were 84% and 8% for Bacteria and Archaea, respectively. Alternatively, bands were excised from DGGEs (1). The final product was amplified without GC clamp, purified (Qiagen PCR purification kit), sequenced (IIT Biotech, Bielefeld, Germany), and checked using Chromas 2.33. The sequences obtained through cloning and band excision were compared to GenBank sequences using BLAST (2) (NCBI, October 2009). Phylogenetic analysis of the clone sequences was conducted with ARB software (http://www.arb-home.de); the sequences were added to the database and aligned with the Fast Aligner tool (corrected January 2004, released January 2005), and phylogenetic trees were constructed by fast parsimony and neighbor joining with Jukes Cantor correction.
Molecular fingerprints and factors affecting the microbial communities.
Clustering of the fingerprint analyses of series A reactors, consisting of SMFCs with vermiculite and with soil, revealed that the type of support had a key influence on the composition of the bacterial community (Fig. 1a and b) as well as the archaeal community (Fig. 1c and d). Both supports received the same inoculum mixture. However, whereas the anodes in exfoliated vermiculite were primarily influenced by the added inoculum (direct or indirect through rice transplantation), the anodes in soil were also influenced by endogenous microorganisms. The difference in chemical (e.g., mineral status) and physical parameters further affected the residing microbial communities.
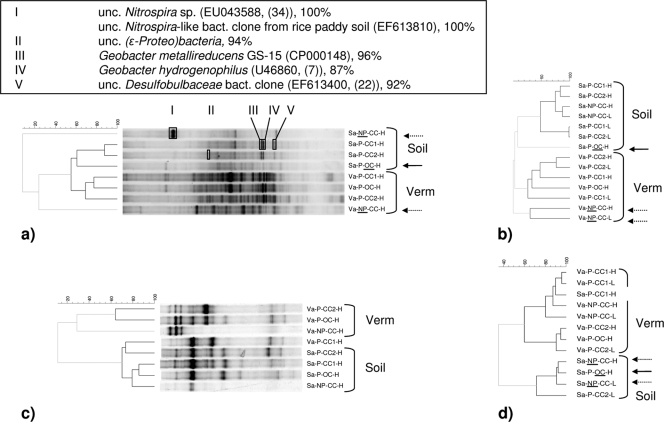
FIG. 1.
Clustering with Pearson correlation of bacterial and archaeal 16S rRNA gene profiles of anodes of reactor series A with potting soil and vermiculite (Verm) as support material. (a) Bacterial DGGE profiles; (b) bacterial T-RFLP profiles; (c) archaeal DGGE profiles; (d) archaeal T-RFLP profiles. Dashed branches refer to cluster cutoff. Significant effects caused by the absence of plants are marked by dashed arrows, while open circuits are marked by solid black arrows. Excised DGGE bands I to V are marked, and information regarding the closest affiliation and similarity is given in the box above the gel in panel a. The closest affiliation is shown (unc., uncultured; bact., bacterial), and the GenBank accession number and relevant reference (references 7, 22, and 34) are shown in parentheses after the closest species or clone. The similarity is shown as a percentage after the closest species or clone. The reactor samples are named as follows: potting soil (Sa) and vermiculite (Va); P means with plants, and NP means without plants (no plants). CC means closed circuit, and OC means open circuit; and H means high and L means low anode position.
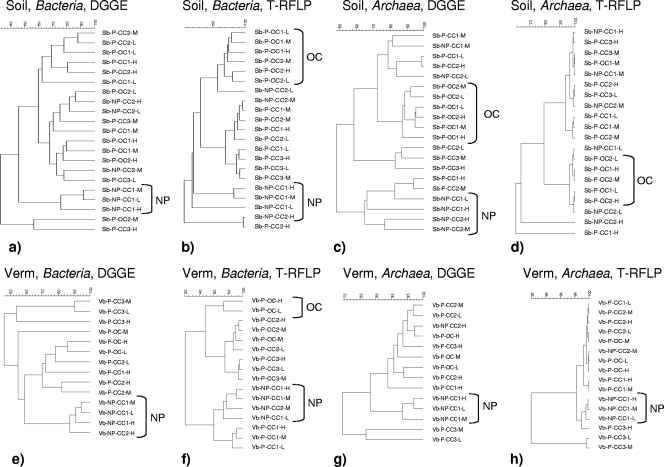
The presence of plants was of major importance. This was especially apparent for the bacterial communities found in reactors with the inert vermiculite, where the plants were the only source of organic compounds (Fig. 1a and b, dashed arrows in the absence of plants [series A], and Fig. 2e and f [larger series B]). The plant effect was to a large extent also applicable for reactors with soil (Fig. 1a, b, 2a, and b), but this was only clear from DGGE (Fig. 1a) and not from T-RFLP (Fig. 1b) for series A. Plants, which release a range of organic compounds, stimulate the growth of soil microorganisms considerably (13). Moreover, several studies suggest that plants select for taxonomic and functional groups in the rhizosphere (32). For the archaeal communities (e.g., Fig. 2g and h), an effect of plants could also be observed, but it was less pronounced. For vermiculite series B reactors, there was a high similarity between the archaeal DGGE (Fig. 2g) and T-RFLP (Fig. 2h) clusters, while the effect of plants in soil series B could be noticed only in the DGGE profiles (Fig. 2c).
FIG. 2.
Clustering with Pearson correlation of bacterial and archaeal 16S rRNA gene profiles of anodes of reactor series B with potting soil (a to d) and vermiculite (Verm) (e to h) as support material. The Pearson correlation values are given as a percentage. DGGE and T-RFLP profiles are given for Bacteria and Archaea. The reactor samples are named as follows: potting soil (Sb) or vermiculite (Vb); P means with plants, and NP means without plants (no plants); CC means closed circuit, and OC means open circuit; and H means high, M means middle, and L means low anode position. The actual DGGE profiles are shown in Fig. S1 in the supplemental material.
Closing the electrical circuit, allowing a capture of electrons by the anode, resulted in a clear shift in the bacterial community of soil reactors (Fig. 1a and b, solid black arrows for open circuit, and Fig. 2b), which was also observed in conventional nonplanted SMFCs (15). The microbial community on anodes is considered responsible for the generation of electrical current (28) and hence fulfils a pivotal role in MFCs. For vermiculite, the shift was not as clear. It can be noted that samples from reactor Vb-P-CC1, producing a negative current near sampling time, formed a distinct cluster (T-RFLP; Fig. 2f) or clustered with samples from an open circuit reactor (DGGE; Fig. 2e). Furthermore, the clustering demonstrated an effect of the electrical circuit on the Archaea. The Archaea were less influenced than the Bacteria were. The effect was apparent only when the reactor contained soil (versus vermiculite) (Fig. 1d, 2c, and d).
Reimers et al. (29) found that the diversity of bacterial communities increased with anode depth. In the present research, the effect of anode depth was minor and could be noticed only in soil series A (T-RFLP, Bacteria; Fig. 1b). This lack of trend could be related to the interruption of the typical redox gradient due to the dense root systems, unequally releasing oxygen and organic substrates into the support matrix (13).
Overall, the dendrograms obtained through DGGE and T-RFLP were comparable. Some effects were evident from both analyses (e.g., support, plants in vermiculite), while other were only evident from one analysis (e.g., plants in soil). These results show that the techniques were complementary and allows us to discern the weight of the factors influencing microbial communities.
Phylogenetic community analysis of Bacteria.
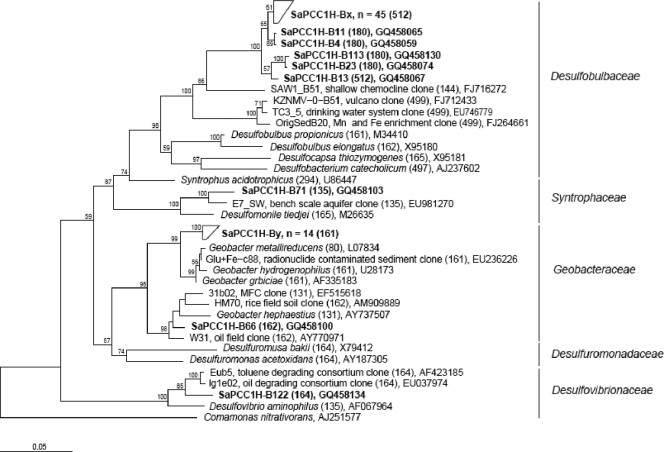
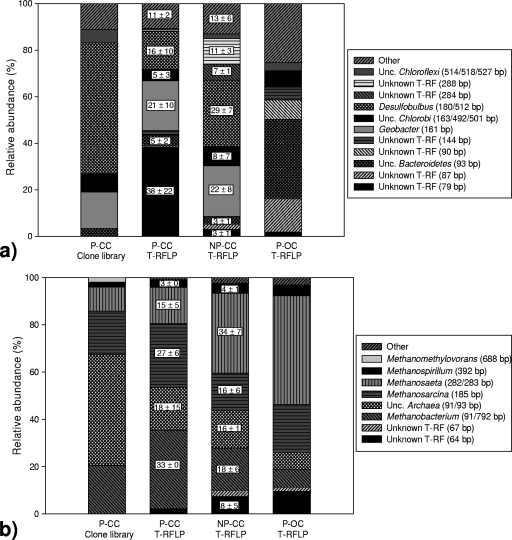
Clone libraries were made for the bacterial and archaeal communities residing on the anode of a current-producing rice SMFC with soil and are represented by phylogenetic trees in Fig. 3 and Fig. S2 in the supplemental material. The relative abundance of the most important phylogenetic groups found in different operational conditions, based on the clone libraries and T-RFLP profiles, is shown in Fig. 4. The most common bacterial groups on the closed-circuit anode with plants (Fig. 3 and 4a) were Desulfobulbus species (56% of all clones) and members of the family Geobacteraceae (16%). Deltaproteobacteria made up a total of 75% of all bacterial clones. Furthermore, members of the Desulfobulbaceae and Geobacteraceae families were not detected on non-current-producing anodes (T-RFLP; Fig. 4a). The enrichment of these groups upon current generation was also shown through excision of DGGE bands; bands III, IV, and V (Fig. 1) were stronger on current-producing anodes and showed 92, 96.5, and 90% sequence identity with the clones related to Geobacter (bands III and IV) and Desulfobulbus (band V), respectively. Deltaproteobacteria and more specifically Geobacteraceae have often been found enriched on closed-circuit anodes (15, 19). The latter are known for anaerobic respiration of organic compounds, such as acetate with concomitant reduction of insoluble Fe(III), often replaceable by a solid electrode (4). Desulfobulbus (and/or Desulfocapsa) species have also been found enriched on anodes (15, 29). The sulfate-reducing Desulfobulbus propionicus was found to be able to oxidize organic compounds (but not acetate) with electrode reduction (14), but its role was also suggested to be linked to the ability to oxidize S0 to sulfate with the electrode as electron acceptor and/or the ability to disproportionately oxidize S0 to sulfate and sulfide (31). The Desulfobulbus-related sequences found here might represent a new species as they were only 89% similar with Desulfobulbus propionicus.
FIG. 3.
Phylogenetic tree of Deltaproteobacteria 16S rRNA gene sequences from clones retrieved from the upper anode of a sediment MFC planted with rice, with potting soil as the anodic support layer and operated with a closed electrical circuit (series A; sample Sa-P-CC1-H). The bar indicates 5% sequence divergence. Bootstrap values higher than 50% (for 1,000 iterations) are shown at the nodes of the trees. Clones are shown in boldface type. The numbers in parentheses represent the in silico T-RF in base pairs. The GenBank accession numbers for grouped clones can be found in the supplemental material. The phylogenetic tree for the archaeal clone library can be found in Fig. S2 in the supplemental material.
FIG. 4.
Comparison of relative abundance of phylogenetic groups on anodes (closed circuit and open circuit) with and without rice plants. The relative abundance of Bacteria (a) and Archaea (b) on closed-circuit (CC) and open-circuit (OC) anodes with rice plants (P) and without rice plants (no plants [NP]) is shown. The analyzed samples originated from potting soil series A, i.e., Sa-P-CC1-H (n = 1) for the first (leftmost) column, Sa-P-CC1-H and Sa-P-CC2-L (n = 2) for the second Archaea column (supplemented with Sa-P-CC1-L and Sa-P-CC2-H [n = 4] for the second Bacteria column), Sa-NP-CC-H and Sa-NP-CC-L (n = 2) for the third column, and Sa-P-OC-H (n = 1) for the fourth (rightmost) column, with n the number of anode replicates. For columns with n > 1, the averages ± standard deviations are shown in the figure. The phylogenetic affiliations given are the closest relatives. For the T-RFLP profiles, the closest relatives were obtained through comparison with the in silico T-RFs from the corresponding clone library. For Bacteria, “Other” contains all groups with an abundance of <6%, comprising uncultured (unc.) OD1, Sphingobacteria, Desulfomonile, (unc.) Spirochaeta, unc. Deltaproteobacteria, Desulfovibrio, unc. Planctomycetes, unc. OP11, and unknown T-RFs. For Archaea, “Other” contains all groups with an abundance of <2%, comprising unknown T-RFs. Unknown T-RF (fragment length in base pairs) means that the affiliation of the fragment could not be deduced.
Other affiliations of importance on a current-producing anode were Chlorobi (8% of all clones), Chloroflexi (6%), and Bacteroidetes (3%). Chloroflexi have been found enriched on the anode of a cellulose-fed MFC (18), but their current relevance for a closed-circuit anode was not clear when T-RFLP profiles were compared. The bacterial species found here do not correspond with those found important in earlier research regarding rice SMFCs (20) (Natronocella, Beijerinckiaceae, Rhizobiales). That study, however, employed a rice paddy field and no inocula.
On the basis of the T-RFLP profiles (Fig. 4a), the non-current-producing anode was dominated by uncultured Bacteroidetes. More phylogenetic groups could be detected in the absence of plants, involving a Nitrospira-related species (DGGE band I).
Phylogenetic community analysis of Archaea.
Almost half (47%) of the archaeal clone sequences derived from the closed-circuit anode (Fig. 4b; see Fig. S2 in the supplemental material) were most closely related to uncultured Archaea, and not to any of the known methanogenic lineages (11) or novel rice cluster lineages (24). These sequences clustered in two groups, accounting for 35 and 12% of all clones. The archaeal clones that could be assigned (Fig. 4b; see Fig. S2 in the supplemental material) belonged to a few methanogenic groups, with the most dominant groups being Methanobacteriaceae (20% of all clones), Methanosarcinaceae (18%), and Methanosaetaceae (10%). These are also found important in genuine rice paddy soil (12).
When T-RFLP fingerprints for closed and open circuits were compared, a shift in the archaeal community could be observed (Fig. 4b). The production of a current led to a (variable, but up to 4-fold) increase in the relative abundance of the uncultured Archaea. Within the methanogens, there was a 4-fold increase for Methanobacterium (CH4 production from H2 and CO2 and/or formate) and a 3-fold decrease for the strictly acetotrophic Methanosaetaceae. There was a small increase (from 20% to 27%) for the generalist Methanosarcina (CH4 production from H2 and CO2, acetate, and/or C1 compounds). These changes might reflect an increased importance of hydrogenotrophic methanogenesis compared to acetotrophic methanogenesis combined with possible growth promotion of a group of (uncultured) Archaea upon current generation. Archaeal anodic communities are largely unexplored so far. Ishii et al. (17) did find less (methanogenic) Euryarchaeota and suppressed methanogenesis in closed-circuit anodes compared to open-circuit anodes. To verify the specific effects of plant SMFCs on the methanogenic rate and pathways and on the overall metabolism in rhizospheres, follow-up experiments, involving, for example 13CO2, are appropriate.
This research showed that despite the strong effect of support type and plants, an effect of the electrical circuit could also be observed, both on bacterial and archaeal communities. These findings can accordingly guide future practical and fundamental work regarding plant MFCs.
Nucleotide sequence accession numbers.
All 16S rRNA gene sequences obtained were deposited in GenBank under accession numbers GQ458057 to GQ458194 (clone libraries) and GQ422145 to GQ422149 (DGGE bands I to V).
Supplementary Material
Acknowledgments
L.D.S. was supported by a Ph.D. grant from the Bijzonder Onderzoeks Fonds of Ghent University (grant 01D24405), A.C. was supported by a Ph.D. grant from the Deutscher Akademischer Austausch Dienst (DAAD), and M.W.F. was supported by the Max Planck Society and Fonds der Chemischen Industrie.
We thank Petra Van Damme, Leen Van Den Bossche, and Bianca Pommerenke for their excellent technical assistance. The useful comments of David van der Ha and Jan Arends are kindly acknowledged.
Footnotes
Published ahead of print on 22 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aelterman, P., K. Rabaey, H. T. Pham, N. Boon, and W. Verstraete. 2006. Continuous electricity generation at high voltages and currents using stacked microbial fuel cells. Environ. Sci. Technol. 40:3388-3394. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond, D. R., D. E. Holmes, L. M. Tender, and D. R. Lovley. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483-485. [DOI] [PubMed] [Google Scholar]
- 5.Boon, N., W. De Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. [DOI] [PubMed] [Google Scholar]
- 6.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl. Environ. Microbiol. 66:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coates, J. D., E. J. P. Phillips, D. J. Lonergan, H. Jenter, and D. R. Lovley. 1996. Isolation of Geobacter species from diverse sedimentary environments. Appl. Environ. Microbiol. 62:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Schamphelaire, L., L. van den Bossche, H. S. Dang, M. Höfte, N. Boon, K. Rabaey, and W. Verstraete. 2008. Microbial fuel cells generating electricity from rhizodeposits of rice plants. Environ. Sci. Technol. 42:3053-3058. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egert, M., B. Wagner, T. Lemke, A. Brune, and M. W. Friedrich. 2003. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6659-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erkel, C., D. Kemnitz, M. Kube, P. Ricke, K.-J. Chin, S. Dedysh, R. Reinhardt, R. Conrad, and W. Liesack. 2005. Retrieval of first genome data for rice cluster I methanogens by a combination of cultivation and molecular techniques. FEMS Microbiol. Ecol. 53:187-204. [DOI] [PubMed] [Google Scholar]
- 12.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinsinger, P., C. Plassard, and B. Jaillard. 2006. Rhizosphere: a new frontier for soil biogeochemistry. J. Geochem. Explor. 88:210-213. [Google Scholar]
- 14.Holmes, D. E., D. R. Bond, and D. R. Lovley. 2004. Electron transfer by Desulfobulbus propionicus to Fe(III) and graphite electrodes. Appl. Environ. Microbiol. 70:1234-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, D. E., D. R. Bond, R. A. O'Neill, C. E. Reimers, L. R. Tender, and D. R. Lovley. 2004. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb. Ecol. 48:178-190. [DOI] [PubMed] [Google Scholar]
- 16.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 17.Ishii, S., Y. Hotta, and K. Watanabe. 2008. Methanogenesis versus electrogenesis: morphological and phylogenetic comparisons of microbial communities. Biosci. Biotechnol. Biochem. 72:286-294. [DOI] [PubMed] [Google Scholar]
- 18.Ishii, S., T. Shimoyama, Y. Hotta, and K. Watanabe. 2008. Characterization of a filamentous biofilm community established in a cellulose-fed microbial fuel cell. BMC Microbiol. 8:6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung, S., and J. M. Regan. 2007. Comparison of anode bacterial communities and performance in microbial fuel cells with different electron donors. Appl. Microbiol. Biotechnol. 77:393-402. [DOI] [PubMed] [Google Scholar]
- 20.Kaku, N., N. Yonezawa, Y. Kodama, and K. Watanabe. 2008. Plant/microbe cooperation for electricity generation in a rice paddy field. Appl. Microbiol. Biotechnol. 79:43-49. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, M., J. Murase, and Y. H. Lu. 2004. Carbon cycling in rice field ecosystems in the context of input, decomposition and translocation of organic materials and the fates of their end products (CO2 and CH4). Soil Biol. Biochem. 36:1399-1416. [Google Scholar]
- 22.Kleinsteuber, S., K. M. Schleinitz, J. Breitfeld, H. Harms, H. H. Richnow, and C. Vogt. 2008. Molecular characterization of bacterial communities mineralizing benzene under sulfate-reducing conditions. FEMS Microbiol. Ecol. 66:143-157. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 24.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 26.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicol, G. W., L. A. Glover, and J. I. Prosser. 2003. The impact of grassland management on archaeal community structure in upland pasture rhizosphere soil. Environ. Microbiol. 5:152-162. [DOI] [PubMed] [Google Scholar]
- 28.Rabaey, K., and W. Verstraete. 2005. Microbial fuel cells: novel biotechnology for energy generation. Trends Biotechnol. 23:291-298. [DOI] [PubMed] [Google Scholar]
- 29.Reimers, C. E., P. Girguis, H. A. Stecher, L. M. Tender, N. Ryckelynck, and P. Whaling. 2006. Microbial fuel cell energy from an ocean cold seep. Geobiology 4:123-126. [Google Scholar]
- 30.Rooney-Varga, J. N., M. W. Giewat, K. N. Duddleston, J. P. Chanton, and M. E. Hines. 2007. Links between archaeal community structure, vegetation type and methanogenic pathway in Alaskan peatlands. FEMS Microbiol. Ecol. 60:240-251. [DOI] [PubMed] [Google Scholar]
- 31.Ryckelynck, N., H. A. Stecher, and C. E. Reimers. 2005. Understanding the anodic mechanism of a seafloor fuel cell: interactions between geochemistry and microbial activity. Biogeochemistry 76:113-139. [Google Scholar]
- 32.Singh, B. K., P. Millard, A. S. Whiteley, and J. C. Murrell. 2004. Unravelling rhizosphere-microbial interactions: opportunities and limitations. Trends Microbiol. 12:386-393. [DOI] [PubMed] [Google Scholar]
- 33.Strik, D., H. V. M. Hamelers, J. F. H. Snel, and C. J. N. Buisman. 2008. Green electricity production with living plants and bacteria in a fuel cell. Int. J. Energy Res. 32:870-876. [Google Scholar]
- 34.Tarlera, S., K. Jangid, A. H. Ivester, W. B. Whitman, and M. A. Williams. 2008. Microbial community succession and bacterial diversity in soils during 77,000 years of ecosystem development. FEMS Microbiol. Ecol. 64:129-140. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, M., A. Loy, R. Nogueira, U. Purkhold, N. Lee, and H. Daims. 2002. Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek 81:665-680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.