Abstract
The ubiquitous opportunistic human pathogen Pseudomonas aeruginosa secretes a viscous extracellular polysaccharide, called alginate, as a virulence factor during chronic infection of patients with cystic fibrosis. In the present study, it was demonstrated that the outer membrane protein AlgE is required for the production of alginate in P. aeruginosa. An isogenic marker-free algE deletion mutant was constructed. This strain was incapable of producing alginate but did secrete alginate degradation products, indicating that polymerization occurs but that the alginate chain is subsequently degraded during transit through the periplasm. Alginate production was restored by introducing the algE gene. The membrane topology of the outer membrane protein AlgE was assessed by site-specific insertions of FLAG epitopes into predicted extracellular loop regions.
Pseudomonas aeruginosa is an ubiquitous opportunistic human pathogen responsible for chronic infections of the lungs of patients with cystic fibrosis (CF), in whom it is the leading cause of mortality and morbidity (9). The establishment of a chronic infection in the lungs of patients with CF coincides with the switch of P. aeruginosa to a stable mucoid variant, producing copious amounts of the exopolysaccharide alginate; this is typically a poor prognostic indicator for these patients (24, 31). Alginate is a linear unbranched exopolysaccharide consisting of 1,4-linked monomers of β-d-mannuronic acid and its C-5 epimer, α-l-guluronic acid, which is known to be produced by only two bacterial genera, Pseudomonas and Azotobacter (34). The switch to a mucoid phenotype coincides with the appearance of a 54-kDa protein in the outer membrane; this protein has been identified and has been designated AlgE (13, 31).
The genes encoding the alginate biosynthesis machinery are located within a 12-gene operon (algD-alg8-alg44-algK-algE-algG-algX-algL-algI-algJ-algF-algA). AlgA and AlgD, along with AlgC (not encoded in the operon), are involved in precursor synthesis (34). Alg8 is the catalytic subunit of the alginate polymerase located at the inner membrane (35). AlgG is a C-5 mannuronan epimerase (19). AlgK contains four putative Sel1-like repeats, similar to the tetratricopeptide repeat motif often found in adaptor proteins involved in the assembly of multiprotein complexes (3, 10). AlgX shows little homology to any known protein, and its role is unclear (14). Knockout mutants of AlgK, AlgG, and AlgX have nonmucoid phenotypes, although they produce short alginate fragments, due to the activity of the alginate lyase (AlgL), which degrades the nascent alginate (1, 14, 19-21, 36). AlgF, AlgI, and AlgJ are involved in acetylation of alginate, but they are not ultimately required for its production (12). The membrane-anchored protein, Alg44, is required for polymerization and has a PilZ domain for the binding of c-di-GMP, a secondary messenger essential for alginate production (16, 25, 33). The periplasmic C terminus of Alg44 shares homology with the membrane fusion proteins involved in the bridging of the periplasm in multidrug efflux pumps (11, 43). The periplasmic alginate lyase, AlgL, appears to be required for the translocation of intact alginate across the periplasm (1, 26). AlgE is an outer membrane, anion-selective channel protein through which alginate is presumably secreted (30). A protein complex or scaffold through which the alginate chain can pass and be modified and which spans the periplasm bridging the polymerase located (Alg8) at the outer membrane pore (AlgE) has been proposed (21). Indeed, it has been demonstrated that both the inner and the outer membranes are required for the in vitro polymerization of alginate (35).
The requirement of AlgE for the biosynthesis of alginate in P. aeruginosa was first observed by complementation of an alginate-negative mutant derived by chemical mutagenesis with a DNA fragment containing algE (8) Secondary structure predictions suggested that AlgE forms an 18-stranded β barrel with extended extracellular loops. Several of these loops show high densities of charged amino acids, suggesting a functional role in the translocation of the anionic alginate polymer (29, 30). Preliminary analysis of AlgE crystals has been reported (48).
In this study, the role of AlgE in alginate biosynthesis was investigated and the membrane topology of AlgE was assessed by site-directed insertion mutagenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains, plasmids, and oligonucleotides used in the present study are listed in Table 1. The Escherichia coli strains were grown in LB medium at 37°C. E. coli S17-1 was used for conjugative transfer of suicide plasmid pEX100T:ΔalgEGm and flipase-encoding plasmid pFLP2. When they were required, the following antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; and streptomycin, 30 μg/ml. The P. aeruginosa strains were grown in LB medium or Pseudomonas isolation agar (PIA) medium (20 g of peptone, 10 g of K2SO4, 1.4 g MgCl2, 0.025 g of Triclosan, 20 ml of glycerol per liter) at 37°C, and when they were required, antibiotics were added at the appropriate concentrations. The antibiotic concentrations used for the P. aeruginosa strains were as follows: gentamicin, 300 μg/ml, and carbenicillin, 300 μg/ml. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
TABLE 1.
Bacterial strains, plasmids and oligonucleotide used in this study
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PDO300 | mucA22 isogenic mutant derived from PAO1 | 18 |
| PDO300ΔalgE | Isogenic algE deletion mutant derived from PDO300 | This study |
| E. coli | ||
| TOP10 | E. coli cloning strain | Invitrogen |
| S17-1 | thi-1 proA hsdR17 (rK− mK+) recA1; tra gene of plasmid RP4 integrated in chromosome | 39 |
| Plasmids | ||
| pGEM-T Easy | Apr, Plac | Invitrogen |
| pBBR1MCS-5 | Gmr; broad-host-range vector; Plac | 23 |
| pBBR1MCS-5:algE | HindIII-BamHI fragment comprising algE inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:algEDelL7 | algE fragment with 57-bp deletion in region encoding putative surface exposed loop 7 inserted into pBBR1MCS-5 | This study |
| pBBRMCS-5:algEL1FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 189th bp of the ORF | This study |
| pBBRMCS-5:algEL2FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 336th bp of the ORF | This study |
| pBBRMCS-5:algEL3FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 468th bp of the ORF | This study |
| pBBRMCS-5:algEL4FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 582nd bp of the ORF | This study |
| pBBRMCS-5:algEL5FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 711th bp of the ORF | This study |
| pBBRMCS-5:algEL6FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 882nd bp of the ORF | This study |
| pBBRMCS-5:algEL7FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 1035th bp of the ORF | This study |
| pBBRMCS-5:algEL8FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 1269th bp of the ORF | This study |
| pBBRMCS-5:algEL9FLAG | algE fragment with a 24-bp insertion (encoding the FLAG epitope) inserted after the 1428th bp of the ORF | This study |
| pEX100T | Apr Cbr; gene replacement vector containing sacB gene for counterselection | 17 |
| pEX100T:ΔalgEGm | Apr Cbr Gmr; vector pEX100T with SmaI-inserted algE deletion construct | This study |
| pPS865 | Apr Gmr; source of 1,100-bp BamHI fragment comprising aacC1 gene flanked by Flp recombinase target site signal sequences | 17 |
| pPFLP2 | Apr Cbr; broad-host-range vector encoding Flp recombinase | 17 |
Isolation, analysis, and manipulation of DNA.
General cloning procedures were performed as described previously (37). All pBBR1MCS-5-derived plasmids were transferred to P. aeruginosa strains via electroporation, as described previously (7). DNA primers, deoxynucleoside triphosphates, and Taq and platinum Pfx polymerases were purchased from Invitrogen. The DNA sequences of the plasmid constructs were confirmed by DNA sequencing.
Construction and confirmation of algE deletion mutants.
Two regions of the algE gene were amplified by using Taq polymerase and primers algE1N-Ec5, algE1C-Ba, algE2N-Ba, and algE2C-Ec5. Region algEN (469 bp) comprised bases 136 to 585 and region algEC (445 bp) comprised bases 1047 to 1472 relative to the designated algE-coding region (41). Both PCR products were hydrolyzed by using BamHI and were inserted into the pGEM-T Easy vector (Promega). Vector pPS856 (17) was hydrolyzed with BamHI. The fragment containing the aacC1 gene (which encodes gentamicin acetyltransferase) flanked by two Flp recombinase target sites was inserted into the BamHI site of plasmid pGEM-TEasy:algENC, resulting in plasmid pGEM-TEasy:ΔalgEGm. The DNA of the 1,989-bp algEGm fragment was amplified by using Pfx polymerase and primers algE1N-Ec5 and algE2C-Ec5, and the corresponding PCR product was inserted into SmaI site of vector pEX100T (17), resulting in plasmid pEX100T:ΔalgEΩGm.
E. coli S17-1 was used as the donor for the transfer of plasmid pEX100T:ΔalgEΩGm into the P. aeruginosa strains, and transconjugants were selected on mineral salt medium (38) containing gentamicin and 5% (wt/vol) sucrose. Cells growing on this selective medium should have emerged from double-crossover events. Gene replacement was confirmed after subculture of the cells on PIA medium containing gentamicin and by PCR with primers algEup and algEdown.
E. coli S17-1 was used to transfer the Flp recombinase-encoding vector pFLP2 (17) into P. aeruginosa PDO300Δalg8ΩGm strains, and after 24 h of cultivation on PIA medium containing 5% (wt/vol) sucrose, the gentamicin- and carbenicillin-sensitive cells were analyzed by PCR for the loss of the gentamicin resistance-conferring cassette.
Complementation of isogenic algE deletion mutants.
For complementation of the algE deletion mutants, the algE open reading frame (ORF) of P. aeruginosa PAO1 was amplified by using Pfx polymerase and primers algEN(HiSDNd) and algEC(Ba). This product was then inserted into the pGEM-T Easy vector, resulting in plasmid pGEMTEasy:algE. The algE fragment was released by hydrolysis with HindIII and BamHI and was inserted into the HindIII and BamHI sites of broad-host-range vector pBBR1MCS-5 (23), resulting in plasmid pBBR1MCS-5:algE. In addition, a plasmid encoding AlgE with a 9-amino-acid deletion in the highly charged putative surface-exposed loop 7 region was constructed. This was constructed by amplifying the region upstream of the deletion point by using Pfx polymerase and primers algEN(HiSDNd) and algEDel7N(Ba) and the region downstream of the deletion point by using primers algEDel7C(Ba) and algEC(SaI). The two products were hydrolyzed with HindIII and BamHI or BamHI and SacI, respectively, and inserted into the HindIII and SacI sites of pBBR1MCS-5, resulting in plasmid pBBR1MCS-5:algEDelL7.
In order to probe the membrane topology of AlgE, insertions of the FLAG (DYKDDDDK) epitope were introduced into all of the nine putative surface-exposed loops. The insertions were constructed by site-directed, ligase-independent mutagenesis (SLIM) (6). Briefly, two PCRs were completed for each construct by using plasmid pGEMTEasy:algE as the template: one with the “FFLAG” and the corresponding “RS” primer (algEL1FFLAG and algEL1RS, algEL2FFLAG and algEL2RS, algEL3FFLAG and algEL3RS, algEL4FFLAG and algEL4RS, algEL5FFLAG and algEL5RS, algEL6FFLAG and algEL6RS, algEL7FFLAG and algEL7RS, algEL8FFLAG and algEL8RS, and algEL9FFLAG and algEL9RS) and one with the FS and the corresponding RFLAG primer (algEL1FS and algEL1RFLAG, algEL2FS and algEL2RFLAG, algEL3FS and algEL3RFLAG, algEL4FS and algEL4RFLAG, algEL5FS and algEL5RFLAG, algEL6FS and algEL6RFLAG, algEL7FS and algEL7RFLAG, algEL8FS and algEL8RFLAG, and algEL9FS and algEL9RFLAG). The plasmid template was removed by hydrolysis with DpnI. The two PCR products were mixed in equimolar amounts, and hybridization was achieved by incubation in H buffer (150 mM NaCl, 25 mM Tris, 20 mM EDTA, pH 8.0) at 99°C for 3 min, followed by three cycles of 65°C for 5 min and 30°C for 40 min. The resulting mixture was used to transform competent E. coli TOP 10 cells. Selection for cells coating the new plasmid was performed on ampicillin-containing medium, and the plasmids were extracted. Insertion of the 24-bp FLAG-encoding region was confirmed by DNA sequencing of the ORF, resulting in plasmids pGEMTEasy:algEL1FLAG to pGEMTEasy:algEL9FLAG. The algE (FLAG)-containing fragments were hydrolyzed and inserted into pBBR1MCS-5, as described above, resulting in plasmids pBBRMCS-5:algEL1FLAG, pBBRMCS-5:algEL2FLAG, pBBRMCS-5:algEL3FLAG, pBBRMCS-5:algEL4FLAG, pBBRMCS-5:algEL5FLAG, pBBRMCS-5:algEL6FLAG, pBBRMCS-5:algEL7FLAG, pBBRMCS-5:algEL8FLAG, and pBBRMCS-5:algEL9FLAG.
Alginate production assays.
Two milliliters of an overnight culture was harvested at 4°C and washed twice with saline. Then, 200 μl of the cell suspension was plated onto PIA medium and incubated 72 h at 37°C. Cells were scraped off of two agar plates by using a sterile spatula and were washed twice with 100 ml of saline (retaining the alginate-containing supernatant for subsequent precipitation). The cellular sediments were freeze-dried, and the final weight was determined. The alginate-containing supernatants were precipitated with 1 volume of ice-cold isopropanol, and the alginate was harvested and freeze-dried. For further purification, the precipitated alginate was redissolved in 0.05 M Tris-HCl-10 mM MgCl2 (pH 7.4) to a final concentration of 0.5% (wt/vol), followed by incubation with 15 μg of DNase I/ml and 15 μg of RNase A/ml at 37°C for 6 h. Pronase E was added to a final concentration of 20 μg/ml, and this solution was incubated for a further 18 h at 37°C. The solutions were dialyzed against 5 liters of ultrapure H2O for 48 h. Alginate was precipitated with 1 volume of ice-cold isopropanol and freeze-dried for quantification and uronic acid analysis.
The levels of free uronic acids (alginate degradation products) in the supernatants of 2 ml of overnight cultures were measured. The total uronic acid content of the supernatant was determined as described below; the supernatants were filtered with Amicon Ultra-0.5 (Millipore) centrifugal filter devices (nominal molecular mass cutoff, 10 kDa), and the flowthrough was collected. The uronic acid content of the flowthrough (which contained free uronic acids and short-length alginate degradation products) was determined as described below.
Uronic acid assay.
The alginate concentrations were assayed by the uronic acid assay described previously (4) by using alginic acid from brown seaweed (Sigma-Aldrich) as the standard. Briefly, the alginate samples were dissolved in 200 μl ultrapure H2O at concentrations of between 0.25 and 0.05 mg/ml. The sample was mixed with 1.2 ml tetraborate solution (0.0125 M disodium tetraborate in concentrated sulfuric acid), and the mixture was incubated on ice for 10 min. The mixtures were incubated at 100°C for 5 min and then cooled down on ice for a further 5 min. Twenty microliters of m-hydroxybiphenyl reagent (0.15% m-hydroxybiphenyl in 0.125 M NaOH) was added and the reaction mixtures were mixed for 1 min. For each sample or dilution, a negative control was assayed by using 0.0125 M NaOH instead of the hydroxybiphenyl reagent. The uronic acid concentrations were determined spectrophotometrically at a wavelength of 520 nm.
Purification of outer membranes.
Strains of P. aeruginosa were grown overnight in LB medium containing the appropriate antibiotics. The cells were harvested by centrifugation (at 5,000 × g for 1 h) and washed twice with 1 volume of 10 mM HEPES (pH 7.4). The cells were placed in 15 ml 10 mM HEPES with Roche complete mini-EDTA-free protease inhibitor and sonicated on ice for 12 cycles of 15 s of sonication, followed by a 15-s cooldown. Cellular debris and the remaining intact cells were sedimented by centrifugation (at 5,000 × g for 1 h). The total membrane fraction was then isolated by centrifugation at 100,000 × g for 2 h. The supernatant (soluble fraction) was removed, the sediment was resuspended in 1 volume of 10 mM HEPES containing 0.7% (wt/vol) N-lauroylsarcosine, and the suspension was incubated at room temperature with shaking for 2 h to selectively solubilize the cytoplasmic membrane. This was then centrifuged at 100,000 × g for 2 h; the resulting sediment represents the outer membrane fraction. The total protein concentration of the respective fractions was determined by using a Quant-iT protein assay kit (Invitrogen).
Analysis of outer membrane proteins.
Fifteen micrograms of total protein was then separated by SDS-PAGE on 8% polyacrylamide gels. The resulting gels were then either stained with Coomassie brilliant blue or transferred to a nitrocellulose membrane by using an iBlot dry blotting system (Invitrogen). The nitrocellulose membrane was blocked with 5% (wt/vol) skim milk powder in Tris-buffered saline with 0.1% Tween 20 for 1 h and was subsequently probed with 1 μg/ml anti-FLAG (M2) tag antibody conjugated to horseradish peroxidase (HRP; Abcam, Cambridge, United Kingdom). The membrane was washed, and the bound antibodies were resolved with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL) and developed on X-ray film. Bands suspected of being AlgE or the AlgE mutants were identified by tryptic peptide fingerprinting by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS).
RESULTS
Construction of an isogenic knockout mutant of algE.
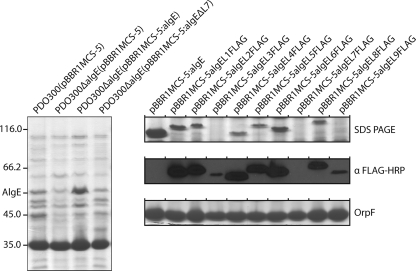
To investigate the requirement for AlgE in alginate biosynthesis, a marker-free algE deletion mutant of alginate-overproducing strain P. aeruginosa PDO300 was generated. This mutant showed a nonmucoid phenotype when it was grown on solid medium. The outer membrane protein profiles showed that AlgE was absent from PDO300ΔalgE (Fig. 1, left panel). It has been shown that the lack of mucoidity associated with some deletion mutants (algK, algG, and algX) is distinct from the biosynthesis/polymerization of alginate. These mutants secrete alginate degradation products (free uronic acids), which have been shown to be the products of alginate lyase, AlgL (19-21, 36). To address this for the ΔalgE mutant, the culture supernatants of the respective mutants were filtered and the uronic acid contents of the filtrates (containing alginate degradation products) were determined. Alginate degradation products could be detected in PDO300ΔalgE(pBBR1MCS-5) at levels 9.7 times that of PDO300(pBBR1MCS-5) and 3.2 times that of the positive control, PDO300ΔalgX(pBBR1MCS-5) (Table 2). Only small amounts of uronic acid were detected from the negative control, PDO300Δalg8(pBBR1MCS-5) (Table 2). These results suggest that the ΔalgE mutant is capable of the synthesis/polymerization of alginate but that it is subsequently degraded in the periplasm.
FIG. 1.
(Left panel) Outer membrane profiles of the ΔalgE mutant and the ΔalgE mutant harboring various plasmids showing the absence or presence of AlgE. Faint bands with the same apparent molecular mass (indicated on the left, in thousands) as AlgE present in the PDO300ΔalgE(pBBR1MCS-5) and PDO300ΔalgE(pBBR1MCS-5:algEΔL7) outer membrane profiles were identified as flagellin type B protein by MALDI-TOF MS. (Right panels) Presence of tagged AlgE proteins in the outer membrane, as indicated by Western immunoblotting (middle). Constitutively produced OprF was used to standardize the samples (bottom).
TABLE 2.
Production of alginate and free uronic acids by ΔalgE mutants
| Strain | Mean alginate concn (g/g CDWa) ± SD | Free uronic acids |
|
|---|---|---|---|
| Sourceb | Mean concn (g/g CDW) ± SD | ||
| PDO300(pBBR1MCS-5) | 0.216 ± 0.027 | Total | 0.524 ± 0.076 |
| Filtrate | 0.044 ± 0.009 | ||
| PDO300ΔalgE(pBBR1MCS-5) | NDc | Total | 0.486 ± 0.050 |
| Filtrate | 0.425 ± 0.039 | ||
| PDO300ΔalgE(pBBR1MCS-5:algE) | 1.096 ± 0.251 | Total | 0.500 ± 0.014 |
| Filtrate | 0.268 ± 0.010 | ||
| PDO300ΔalgE(pBBR1MCS-5:algEΔL7) | ND | Total | 0.249 ± 0.007 |
| Filtrate | 0.261 ± 0.031 | ||
| PDO300ΔalgX(pBBR1MCS-5) | ND | Total | 0.138 ± 0.002 |
| Filtrate | 0.131 ± 0.001 | ||
| PDO300Δalg8(pBBR1MCS-5) | ND | Total | 0.012 ± 0.002 |
| Filtrate | 0.013 ± 0.001 | ||
CDW, cellular dry weight.
Total, concentration of uronic acids in the unfiltered culture media (including alginate and free uronic acids); Filtrate, concentration of uronic acids in the filtered culture media (free uronic acids).
ND, none detected.
Complementation of the PDO300ΔalgE isogenic knockout mutant.
To verify that the observed loss of mucoidity was not due to polar effects on other genes in the alginate biosynthesis operon, a plasmid containing the algE ORF (pBBR1MCS-5:algE) was used to complement the mutant in trans. This plasmid was able to restore the mucoid phenotype as well as the production of alginate to levels beyond (five times greater) that of the parent strain containing the vector control PDO300(pBBR1MCS-5) (Table 2). This indicates that there were no polar effects in the deletion mutant and demonstrates the requirement of AlgE for the mucoid phenotype and the formation of intact and extracellular alginate. Interestingly, complemented strain PDO300ΔalgE(pBBR1MCS-5:algE) produced more alginate degradation products than PDO300(pBBR1MCS-5) (53.6% and 8.4% total uronic acids, respectively) (Table 2). The presence of AlgE in the outer membrane was also restored (Fig. 1, left panel).
Predicted topology of AlgE.
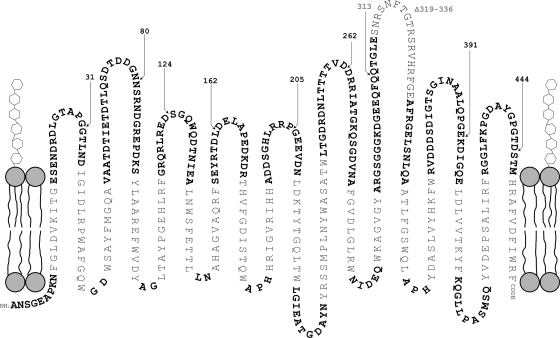
The PRED-TMBB transmembrane β-strands prediction program for Gram-negative bacteria outer membrane proteins (2) predicts that the mature AlgE protein has 18 transmembrane β strands with nine extended surface-exposed loops and eight short periplasmic turns (Fig. 2). This is similar to what was predicted previously (29, 30). A search through the HHpred program (40) did not reveal homology to any annotated protein families or domains with known structures with which to refine this model.
FIG. 2.
Predicted topology of the outer membrane β-barrel AlgE. Predicted transmembrane β sheets are indicated in regular text (nonboldface), extracellular loops at the top are indicated in boldface, and periplasmic turns at the bottom are indicated in boldface. Locations where the FLAG epitopes were inserted are indicated by triangles, and the positions (relative to the position of the cleavage of the signal peptide) are indicated; black numbers, the insertion was permissive, i.e., alginate production was restored; gray numbers, the insertion was nonpermissive. The deleted region of loop 7 is indicated by regular gray text (nonboldface).
The extracellular loop 7 region of AlgE is proposed to be required for the translocation of the alginate polymer.
The proposed surface-exposed loop 7 is significantly longer than the other loops and contains a high density of positively charged amino acid residues which are conserved among all Pseudomonas species in the Pseudomonas Genome Database sequenced (49) (data not shown), as well as AlgJ from Azotobacter vinelandii (29). A conserved cluster of these positively charged residues is present at the apex of the loop region, suggesting that this region may be folded into the pore, facilitating the translocation of the anionic alginate chain through the pore. A vector encoding AlgE with an 18-amino-acid deletion in this conserved region at the apex of loop 7 (pBBR1MCS-5:algEΔL7) was constructed in an attempt to analyze any role that this region may have in alginate translocation and/or protein folding/structure. This plasmid could not restore alginate production in the ΔalgE mutant, nor could the AlgE variant be localized to the outer membrane (Table 2; Fig. 1). This strain produced free uronic acids, but at levels 1.6 times lower than those for the algE deletion mutant, PDO300ΔalgE(pBBR1MCS-5).
AlgE membrane topology analysis.
To test the predicted AlgE model presented in Fig. 2, site-directed epitope insertion mutagenesis targeting the proposed extracellular loops was done. This method utilizes the observation that insertions in the hydrophilic loop regions distant from the β-barrel core are usually tolerated, permitting the correct formation of the β barrel required for insertion of the protein into the outer membrane (27, 32). The FLAG epitope was inserted into the predicted loop regions for loops 1 to 9 by SLIM (6) at the positions indicated in Fig. 2. Plasmids encoding these tagged proteins were introduced into the ΔalgE mutant strain, the outer membrane protein profiles were obtained by SDS-PAGE, and the respective proteins were subjected to Western immunoblotting with anti-FLAG HRP-conjugated antibody. Insertion of the FLAG epitope into loops 1 to 6, 8, and 9 were tolerated, and the protein could be detected in the respective outer membranes. Interestingly, although the tagged proteins have identical deduced molecular masses of 52.2 kDa, they migrated significantly differently by SDS-PAGE: insertions in loops 1, 6, and 9 at 53.5 kDa; loop 2 at 54.7 kDa; loop 3 at 52.3 kDa; loop 4 at 51.4 kDa; loop 5 at 55 kDa; and loop 8 at 56.5 kDa. Additionally, AlgEL3FLAG and AlgEL9FLAG appeared to be present at significantly smaller quantities than the other AlgE proteins (Fig. 1, right panels). AlgE variants harboring insertions in loops 1 to 6, 8, and 9 restored alginate production in the ΔalgE mutant (Table 3), indicating that the core protein structure was not disrupted and that no functional regions essential for alginate formation were affected. Surprisingly, insertion of the epitope into the proposed loop 7 region was not tolerated. This protein could not be detected in the outer membrane and did not restore alginate formation (Table 3).
TABLE 3.
Alginate production of ΔalgE complemented with various epitope tagged AlgE variants
| Strain | Mean alginate concn (g/g CDWa) ± SD |
|---|---|
| PDO300ΔalgE(pBBR1MCS-5:algEL1FLAG) | 0.728 ± 0.080 |
| PDO300ΔalgE(pBBR1MCS-5:algEL2FLAG) | 1.261 ± 0.236 |
| PDO300ΔalgE(pBBR1MCS-5:algEL3FLAG) | 0.874 ± 0.081 |
| PDO300ΔalgE(pBBR1MCS-5:algEL4FLAG) | 0.835 ± 0.067 |
| PDO300ΔalgE(pBBR1MCS-5:algEL5FLAG) | 1.091 ± 0.053 |
| PDO300ΔalgE(pBBR1MCS-5:algEL6FLAG) | 0.796 ± 0.146 |
| PDO300ΔalgE(pBBR1MCS-5:algEL7FLAG) | NDb |
| PDO300ΔalgE(pBBR1MCS-5:algEL8FLAG) | 0.654 ± 0.105 |
| PDO300ΔalgE(pBBR1MCS-5:algEL9FLAG) | 0.892 ± 0.158 |
CDW, cellular dry weight.
ND, none detected.
DISCUSSION
The role of AlgE in alginate production and its membrane topology were investigated in the present study. The isogenic marker-free algE deletion mutant was nonmucoid and did not produce alginate (Table 2). Alginate secretion could be restored by introduction of only the algE gene in trans. This indicates that AlgE is essential for the mucoid phenotype. The algE deletion mutant secreted free uronic acids, indicating that alginate is still being polymerized but is presumably degraded by AlgL in the periplasm (Table 2). Thus, like AlgK, AlgX, AlgG, and AlgL, AlgE is not required for the polymerization or translocation of alginate across the inner membrane but is required for the successful translocation of alginate through the periplasm and across the outer membrane (1, 14, 19-21, 36). This supports the hypothesis that alginate is guided through the periplasm by a multiprotein complex or scaffold containing periplasmic proteins AlgK, AlgX, AlgG, and AlgL and outer membrane protein AlgE. The loss of any of these proteins results in the lack of the integrity of this complex and degradation of the alginate chain. It is possible that the predicted protein scaffold is attached to the outer membrane via AlgE, perhaps via the tetratricopeptide repeat motif of AlgK often found in adaptor proteins involved in the assembly of multiprotein complexes and/or the membrane fusion domain of Alg44 (3, 33). Indeed, mutants lacking Alg44 show reduced levels of AlgE in the outer membrane (28).
The in trans complementation of the ΔalgE mutant still resulted in the secretion of free uronic acids (albeit less than the level of secretion by the ΔalgE mutant) (Table 2). Since AlgE is overproduced in the outer membrane (Fig. 1) of the complemented ΔalgE mutant, it could interfere with the stoichiometry of proteins in the proposed scaffold complex. Hence, the presence of an increased AlgE copy number might, to a certain extent, increase the number of functional multiprotein complexes contributing to alginate overproduction, while the additional copy number might also lead to dysfunctional multiprotein complex formation, causing the degradation of some of the alginate as it traverses the periplasm.
Deletion of the conserved extracellular loop 7 region likely interfered with the folding pathway of AlgE, as evidenced by its absence in the outer membrane and its inability to restore alginate production (Fig. 1; Table 3). This suggests that the deleted region is essential for the functional folding of AlgE. The deleted region contains a relatively high concentration of conserved (among all sequences of Pseudomonas spp. in the Pseudomonas Genome Database [49] as well as AlgJ of Azotobacter vinelandii) positively charged amino acid residues, and therefore, it was proposed that this region is involved in the active translocation of the anionic alginate chain through the outer membrane (Fig. 2). Alternatively, it cannot be ruled out that the topology prediction could be incorrect and that the deletion site may be in an unpredicted transmembrane region.
Site-directed epitope insertion mutagenesis of all predicted extracellular loop regions was completed to probe the predicted membrane topology of AlgE. Similar methods have been utilized to probe the topologies of various outer membrane proteins (27, 32, 46, 47). As expected, insertion of the FLAG epitope into eight of nine loop regions was tolerated and did not disrupt the β-barrel structure, allowing insertion into the outer membrane (Fig. 1). These proteins were also capable of complementing the ΔalgE mutant by restoring alginate production (Table 3). Hence, supporting evidence that these tagged regions are loop regions not involved in the formation of the β barrel through which alginate is exported was provided. When the FLAG epitope was inserted into the predicted loop 7 region, it was not tolerated, even though this insertion site is the most distant from any predicted β sheets among the loop insertions. However, it was consistent with the finding that the deletion of an 18-amino-acid region 5 amino acid residues upstream of the insertion site in this loop was not tolerated (Fig. 1; Table 3). Loop 7, which was proposed to be actively involved in the transport of alginate, might be folded and might bend into the hydrophobic β-barrel core, similar to the “eyelet” region described in other porins. Indeed, this loop 7 contains a concentration of charged residues similar to that found in other eyelet loops (5, 22, 42, 44, 45). Consequently, insertion of the highly hydrophilic FLAG tag into this loop could disrupt the hydrophobic core of the barrel and prevent folding.
Under the conditions used in this study, AlgE did not migrate aberrantly as a 54-kDa protein, as described elsewhere (8, 30), but migrated at its deduced molecular mass of 51.2 kDa. Additionally, even though they had identical deduced molecular masses, the epitope-tagged AlgE variants migrated differently, an observation similar to that reported previously for OprH (32) (Fig. 1). This can be explained by the observation that outer membrane proteins are known for their extraordinary stability and have been reported to exhibit heat-modifiable characteristics, resulting in differences in their migration patterns when they are subjected to SDS-PAGE (15).
In conclusion, experimental evidence was provided for the requirement of AlgE for the secretion of alginate, whereas it is not required for alginate polymerization and translocation into the periplasm. Furthermore, it was demonstrated that loop 7 is required for the functional folding of AlgE, while the other insertion mutants support the proposed membrane topology as a β barrel with 18 β strands and 9 loops. Overall, the data suggest that AlgE not only might be involved in the secretion of alginate but also contributes to the proposed protein scaffold, guiding and protecting the nascent alginate chain through the periplasm and outer membrane.
Acknowledgments
This study was supported by research grants to B.H.A.R. from the Institute of Molecular BioSciences at Massey University and the Deutsche Forschungsgemeinschaft (Re 1097/6-1). I.D.H. is funded by a Massey University doctoral scholarship. Z.U.R. is supported by a doctoral scholarship and research grant from the Higher Education Commission of Pakistan.
We are grateful to Uwe Remminghorst for the construction of plasmid pEX100T:ΔalgEGm.
Footnotes
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Albrecht, M. T., and N. L. Schiller. 2005. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:3869-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagos, P. G., T. D. Liakopoulos, I. C. Spyropoulos, and S. J. Hamodrakas. 2004. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 32:W400-W404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 4.Blumenkrantz, N., and G. Asboe-Hansen. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484-489. [DOI] [PubMed] [Google Scholar]
- 5.Bornet, C., N. Saint, L. Fetnaci, M. Dupont, A. Davin-Regli, C. Bollet, and J. M. Pages. 2004. Omp35, a new Enterobacter aerogenes porin involved in selective susceptibility to cephalosporins. Antimicrob. Agents Chemother. 48:2153-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu, J., D. Tillett, I. W. Dawes, and P. E. March. 2008. Site-directed, ligase-independent mutagenesis (SLIM) for highly efficient mutagenesis of plasmids greater than 8kb. J. Microbiol. Methods 73:195-198. [DOI] [PubMed] [Google Scholar]
- 7.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 8.Chu, L., T. B. May, A. M. Chakrabarty, and T. K. Misra. 1991. Nucleotide sequence and expression of the algE gene involved in alginate biosynthesis by Pseudomonas aeruginosa. Gene 107:1-10. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 9:50-52. [DOI] [PubMed] [Google Scholar]
- 10.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 11.Elkins, C. A., and H. Nikaido. 2003. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J. Bacteriol. 185:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabert, E., J. Wingender, and U. K. Winkler. 1990. An outer membrane protein characteristic of mucoid strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 56:83-87. [DOI] [PubMed] [Google Scholar]
- 14.Gutsche, J., U. Remminghorst, and B. H. A. Rehm. 2005. Biochemical analysis of alginate biosynthesis protein AlgX from Pseudomonas aeruginosa: purification of an AlgX-MucD (AlgY) protein complex. Biochimie 88:245-251. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E. W., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay, I. D., U. Remminghorst, and B. H. A. Rehm. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75:1110-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 18.Holloway, B. W., H. Matsumoto, and P. V. Phibbs, Jr. 1986. The chromosome map of Pseudomonas aeruginosa PAO. Acta Microbiol. Pol. 35:161-164. [PubMed] [Google Scholar]
- 19.Jain, S., M. J. Franklin, H. Ertesvag, S. Valla, and D. E. Ohman. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47:1123-1133. [DOI] [PubMed] [Google Scholar]
- 20.Jain, S., and D. E. Ohman. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain, S., and D. E. Ohman. 2005. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect. Immun. 73:6429-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jap, B. K., P. J. Walian, and K. Gehring. 1991. Structural architecture of an outer membrane channel as determined by electron crystallography. Nature 350:167-170. [DOI] [PubMed] [Google Scholar]
- 23.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 24.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876-895. [DOI] [PubMed] [Google Scholar]
- 26.Monday, S. R., and N. L. Schiller. 1996. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J. Bacteriol. 178:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen, K. A., J. Zylicz, P. Szczesny, A. Sroka, N. Hunter, and J. Potempa. 2009. Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oglesby, L. L., S. Jain, and D. E. Ohman. 2008. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154:1605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rehm, B. H. A. 1996. The Azotobacter vinelandii gene algJ encodes an outer-membrane protein presumably involved in export of alginate. Microbiology 142(Pt 4):873-880. [DOI] [PubMed] [Google Scholar]
- 30.Rehm, B. H. A., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehm, B. H. A., E. Grabert, J. Hein, and U. K. Winkler. 1994. Antibody response of rabbits and cystic fibrosis patients to an alginate-specific outer membrane protein of a mucoid strain of Pseudomonas aeruginosa. Microb. Pathog. 16:43-51. [DOI] [PubMed] [Google Scholar]
- 32.Rehm, B. H. A., and R. E. W. Hancock. 1996. Membrane topology of the outer membrane protein OprH from Pseudomonas aeruginosa: PCR-mediated site-directed insertion and deletion mutagenesis. J. Bacteriol. 178:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remminghorst, U., and B. H. A. Rehm. 2006. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 580:3883-3888. [DOI] [PubMed] [Google Scholar]
- 34.Remminghorst, U., and B. H. A. Rehm. 2006. Bacterial alginates: from biosynthesis to applications. Biotechnol. Lett. 28:1701-1712. [DOI] [PubMed] [Google Scholar]
- 35.Remminghorst, U., and B. H. A. Rehm. 2006. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:298-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robles-Price, A., T. Y. Wong, H. Sletta, S. Valla, and N. L. Schiller. 2004. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7369-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 38.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch. Mikrobiol. 38:209-222. (In German.) [PubMed] [Google Scholar]
- 39.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 40.Soding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 42.Struyve, M., J. Visser, H. Adriaanse, R. Benz, and J. Tommassen. 1993. Topology of PhoE porin: the ‘eyelet’ region. Mol. Microbiol. 7:131-140. [DOI] [PubMed] [Google Scholar]
- 43.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Gelder, P., N. Saint, P. Phale, E. F. Eppens, A. Prilipov, R. van Boxtel, J. P. Rosenbusch, and J. Tommassen. 1997. Voltage sensing in the PhoE and OmpF outer membrane porins of Escherichia coli: role of charged residues. J. Mol. Biol. 269:468-472. [DOI] [PubMed] [Google Scholar]
- 45.Van Gelder, P., N. Saint, R. van Boxtel, J. P. Rosenbusch, and J. Tommassen. 1997. Pore functioning of outer membrane protein PhoE of Escherichia coli: mutagenesis of the constriction loop L3. Protein Eng. 10:699-706. [DOI] [PubMed] [Google Scholar]
- 46.Visudtiphole, V., D. A. Chalton, Q. Hong, and J. H. Lakey. 2006. Determining OMP topology by computation, surface plasmon resonance and cysteine labelling: the test case of OMPG. Biochem. Biophys. Res. Commun. 351:113-117. [DOI] [PubMed] [Google Scholar]
- 47.Wang, S. K., I. Sa'-Correia, A. Darzins, and A. M. Chakrabarty. 1987. Characterization of the Pseudomonas aeruginosa alginate (alg) gene region II. J. Gen. Microbiol. 133:2303-2314. [DOI] [PubMed] [Google Scholar]
- 48.Whitney, J. C., A. M. Neculai, D. E. Ohman, and P. L. Howell. 2009. Expression, refolding, crystallization and preliminary X-ray analysis of Pseudomonas aeruginosa AlgE. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winsor, G. L., T. Van Rossum, R. Lo, B. Khaira, M. D. Whiteside, R. E. Hancock, and F. S. Brinkman. 2009. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37:D483-D488. [DOI] [PMC free article] [PubMed] [Google Scholar]