Abstract
For Hypocrea jecorina (anamorph Trichoderma reesei), a filamentous fungus used for hydrolase production in different industries, it has been a long-term practice to use d-xylose as an inducing substance. We demonstrate in this study that the degree of xylanase-encoding gene induction strictly depends on the concentration of d-xylose, which was found to be optimal from 0.5 to 1 mM for 3 h of cultivation. At higher concentrations of d-xylose, a reduced level of xylanase gene expression was observed. In the present study, we also provide evidence that the d-xylose concentration-dependent induction is antagonized by carbon catabolite repressor 1. This repressor mediates its influence on d-xylose indirectly, by reducing the expression of xylanase regulator 1, the main activator of most hydrolase-encoding genes. Additionally, a direct influence of the repressor on xylanase 1 expression in the presence of d-xylose was found. Furthermore, we show that d-xylose reductase 1 is needed to metabolize d-xylose to achieve full induction of xylanase expression. Finally, a strain which expresses xylanase regulator 1 at a constant level was used to partially overcome the negative influence exerted by carbon catabolite repressor 1 on d-xylose.
The filamentous ascomycete Hypocrea jecorina (anamorph Trichoderma reesei) is of industrial importance mainly because it secretes a broad range of hydrolytic enzymes. These comprise, among others, the two major specific endo-β-1,4-xylanases, XYNI and XYNII (EC 3.2.1.8) (11), and one β-xylosidase, BXLI (EC 3.2.1.37) (12, 18). The entire set of enzymes works together to completely degrade xylan, which is a predominant biopolymeric substrate encountered by the fungus. The xylanases catalyze hydrolysis to smaller, soluble oligo- and monosaccharides, which finally act as the low-molecular-weight inducers xylobiose and d-xylose (18, 30). Therefore, d-xylose is a cheap and renewable carbon source for microorganism cultivation and an attractive starting material for synthesis of value-added chemicals (1, 21).
In H. jecorina, the expression of xyn1 is induced by d-xylose and repressed by glucose; this is mediated via carbon catabolite repressor 1 (Cre1) (15, 23, 26). Furthermore, it was demonstrated that low constitutive xyn2 expression levels can be induced by xylobiose and sophorose (29). In Aspergillus, the xylanolytic and cellulolytic system is strictly coregulated via the inducer d-xylose (e.g., see references 9 and 10). Notably, d-xylose concentrations higher than 1 mM have been reported to reduce expression of a number of xylanase-encoding genes in Aspergillus niger (6). Carbon catabolite repression (CCR) was suggested as the responsible mechanism, since repression was associated with the carbon catabolite repressor protein CreA (7) in studies using a CreA mutant strain. Accordingly, carbon catabolite-repressing effects of d-xylose have been reported for Aspergillus nidulans (22), even though it simultaneously acts as an inducer. So far, nothing is known about the influence of d-xylose concentration on xylanase induction and the relevant mechanisms in H. jecorina.
In 2006, we identified the main activator of hydrolases in H. jecorina, namely, Xyr1 (xylanase regulator 1) (27). Xyr1 is a central regulatory protein responsible for activation of all important hydrolytic enzyme-encoding genes, including xyn1, xyn2, and bxl1. It also contributes to the regulation of d-xylose metabolism because it has a strong impact on the formation of d-xylose reductase 1 (Xyl1 [25]) activity (27). In addition to Xyr1-mediated induction, Cre1 has been described as a widely acting repressor of particular hydrolase-encoding genes. In H. jecorina, only one endoxylanase-encoding gene, namely, xyn1, is under direct Cre1 control (14, 15), whereas the other xylanolytic enzyme-encoding gene, xyn2, is not. In addition, the activator Xyr1 is itself under Cre1-mediated carbon catabolite repression on glucose, which means a Cre1-dependent double lock on xyn1 expression (16).
Moreover, isolation of the transcription factors AceI (activator of cellulases 1) and Ace2 (activator of cellulases 2), involved in the regulation of hydrolase formation in H. jecorina, has been reported (3, 24). While the previously described repressor AceI (2) has been proven to antagonize Xyr1 function directly, by competing for one of its binding sites in the xyn1 promoter (23), Ace2 was found to contact the Xyr1-binding sites in the xyn2 promoter, and it is needed for continuing xyn2 gene expression. It also antagonizes early xyn2 induction (28).
In this study, we report the contrasting effects of d-xylose on the induction of the xylanase transcriptome, which are dependent on its concentration. We report for the first time that xyn2 can also be induced via d-xylose, using an optimal concentration of 0.5 to 1 mM. The transcription of xylanase-encoding genes depends on the d-xylose concentration and is reduced in a Cre1-mediated manner, although this influence is less pronounced than that with glucose. The influence of Cre1 occurs via regulation of xyr1 transcription and by antagonizing xyn1 transcription. Transcription of xylanase-encoding genes was always found to be strongly reduced or absent if Xyl1 was missing, indicating a need for d-xylose metabolism for subsequent induction. Finally, we partly overcame the negative influence of Cre1 at high d-xylose concentrations by using a strain constitutively expressing xyr1.
MATERIALS AND METHODS
Strains and growth conditions.
The following H. jecorina strains were used throughout this study: QM9414 (parental strain; ATCC 26921), nx7 strain (a recombinant strain constitutively expressing xyr1) (16), Δxyl1 strain (a xyl1 deletion strain) (25), and Δcre1 strain (a cre1 deletion strain; kindly provided by C. P. Kubicek, TU Vienna). All strains were maintained on malt agar, with the exception of the Δcre1 strain, which was maintained on potato dextrose agar.
For replacement experiments, mycelia were precultured in 1-liter Erlenmeyer flasks on a rotary shaker (250 rpm) at 30°C for 18 h in 250 ml of Mandels-Andreotti (MA) medium (17), applying 1% (wt/vol) glycerol as the sole carbon source. An inoculum of 109 conidia per liter (final concentration) was used. Pregrown mycelia were washed, and thereafter, equal amounts were resuspended in MA media containing 1% (wt/vol) glucose, d-xylose at specified concentrations, or a mixture of both as the carbon source. Mycelia were also replaced on MA medium containing 1% (wt/vol) glycerol and on medium without a carbon source as reference conditions. Incubation was continued for up to 8 h, and 15-ml samples were taken after 3 and 8 h.
RNA extraction and reverse transcription (RT).
Harvested mycelia were homogenized in 1 ml of peqGOLD TriFast DNA/RNA/protein purification system reagent (Peqlab Biotechnologie, Erlangen, Germany), using a FastPrep FP120 Bio101 ThermoSavant cell disrupter (Qbiogene, Carlsbad, CA). RNA was isolated according to the manufacturer's instructions.
Synthesis of cDNA from mRNA was carried out using a RevertAid H Minus first-strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany) according to the manufacturer's instructions.
qPCR analysis.
All quantitative PCRs (qPCRs) were performed in a Mastercycler ep realplex2 machine (Eppendorf, Hamburg, Germany). The software realplex2.2 was used to compile PCR protocols and to define plate setups. All PCRs were carried out in triplicate in 25-μl reaction mixtures. For xyn1, xyn2, and bxl1 transcription analysis, a SYBR green assay with reference to the actin gene (act1) was performed, and reaction mixtures included iQ SYBR green Supermix (Bio-Rad), 0.1 μM forward primer, 0.1 μM reverse primer, and the cDNA as template. Primer pairs are given in Table 1. The following PCR program was used: 3 min of initial denaturation at 95°C, followed by 45 cycles of 15 s at 95°C, 15 s at 59°C, and 15 s at 72°C. Results of transcription analyses are given as relative transcript ratios with reference to the actin gene. The values given in the figures are means for three independent experiments. Error bars indicate standard deviations. The amounts always refer to one reference sample within an experiment (e.g., QM9414, no carbon source, 3 h), as indicated by an asterisk in each figure.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′-3′) | Use |
|---|---|---|
| act1f | TGAGAGCGGTGGTATCCACG | act1 qPCR |
| act1r | GGTACCACCAGACATGACAATGTTG | act1 qPCR |
| bxl1f | GCCAACTTCGCCACCAAGG | bxl1 qPCR |
| bxl1r | CGGCAATCTGGTGGATCAATGTG | bxl1 qPCR |
| Taqxyn1f | CAGCTATTCGCCTTCCAACAC | xyn1 qPCR |
| Taqxyn1r | CCAAAGTTGATGGGAGCAGAA | xyn1 qPCR |
| Taqxyn2r | CCGAGAAGTTGATGACCTTGTTC | xyn2 qPCR |
| Taqxyn2f | GGTCCAACTCGGGCAACTTT | xyn2 qPCR |
HPLC analysis.
Analyses were performed using a Thermo Finnigan Surveyor high-performance liquid chromatography (HPLC) instrument (Thermo Fisher Scientific, MA). All 10-μl samples were injected onto a Rezex RHM-monosaccharide column (H+, 8%, 150 × 7.8 mm; Phenomenex, CA). Water was used as the mobile phase, and elution was performed at 85°C, applying a flow rate of 0.6 ml/min for 20 min. The concentration was determined using d-xylose as a standard.
RESULTS
Induction of xylanase-encoding genes is dependent on d-xylose concentration.
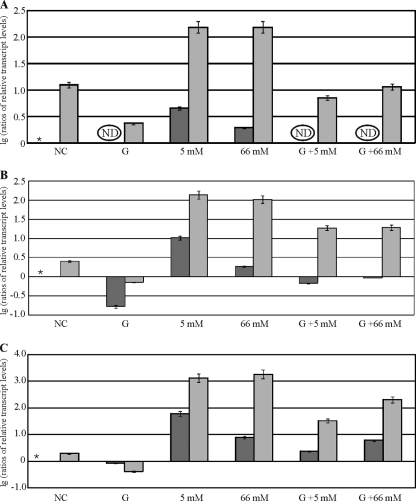
In H. jecorina, the induction of xylanolytic enzyme-encoding gene expression, in particular that of xyn1, has been studied to date using d-xylose at a concentration of 1% (wt/vol), which corresponds to 66 mM. Because d-xylose concentrations above 1 mM have been reported to repress xylanase-encoding genes in Aspergillus niger (6), this was the first point of investigation in the present study. Therefore, H. jecorina QM9414 was precultured in glycerol and transferred to MA medium containing 0.5, 5, 10, or 66 mM d-xylose as the sole carbon source. The controls were MA medium without a carbon source and MA medium with 1% (wt/vol) glycerol, which was previously reported to be a neutral carbon source (13). Dual controls were chosen to account for potential starvation/derepression (low d-xylose concentrations) and growth (high d-xylose concentrations) conditions. Cultivation was followed for 3 h. It is noteworthy that differences in xyn1, xyn2, and bxl1 expression could be observed between cultures derived from the two control conditions mentioned above, which in all cases were about 1 order of magnitude higher for xylanase gene expression on medium without a carbon source (due to derepression). Transcriptional analysis of xyn1 revealed decreasing transcript levels with increasing d-xylose concentrations. The best induction was achieved using 0.5 mM d-xylose (Fig. 1). Higher d-xylose concentrations led to reduced induction but never reverted to the levels under derepressed conditions (no carbon source) (Fig. 1). Continuing the incubation for up to 8 h led to a second induction peak for all three genes at a 5 mM initial concentration of d-xylose (data not shown). An HPLC analysis of the remaining d-xylose level confirmed a concentration below 0.5 mM in this sample after 8 h of cultivation. Obviously, this again would correspond to an optimal induction concentration. In summary, it can be deduced that in H. jecorina the optimal induction of xylanase-encoding genes by d-xylose can be achieved at a concentration of 0.5 mM for 3 h. However, xylanase-encoding gene expression is elevated even at higher d-xylose concentrations compared to that under derepressing conditions (no carbon source).
FIG. 1.
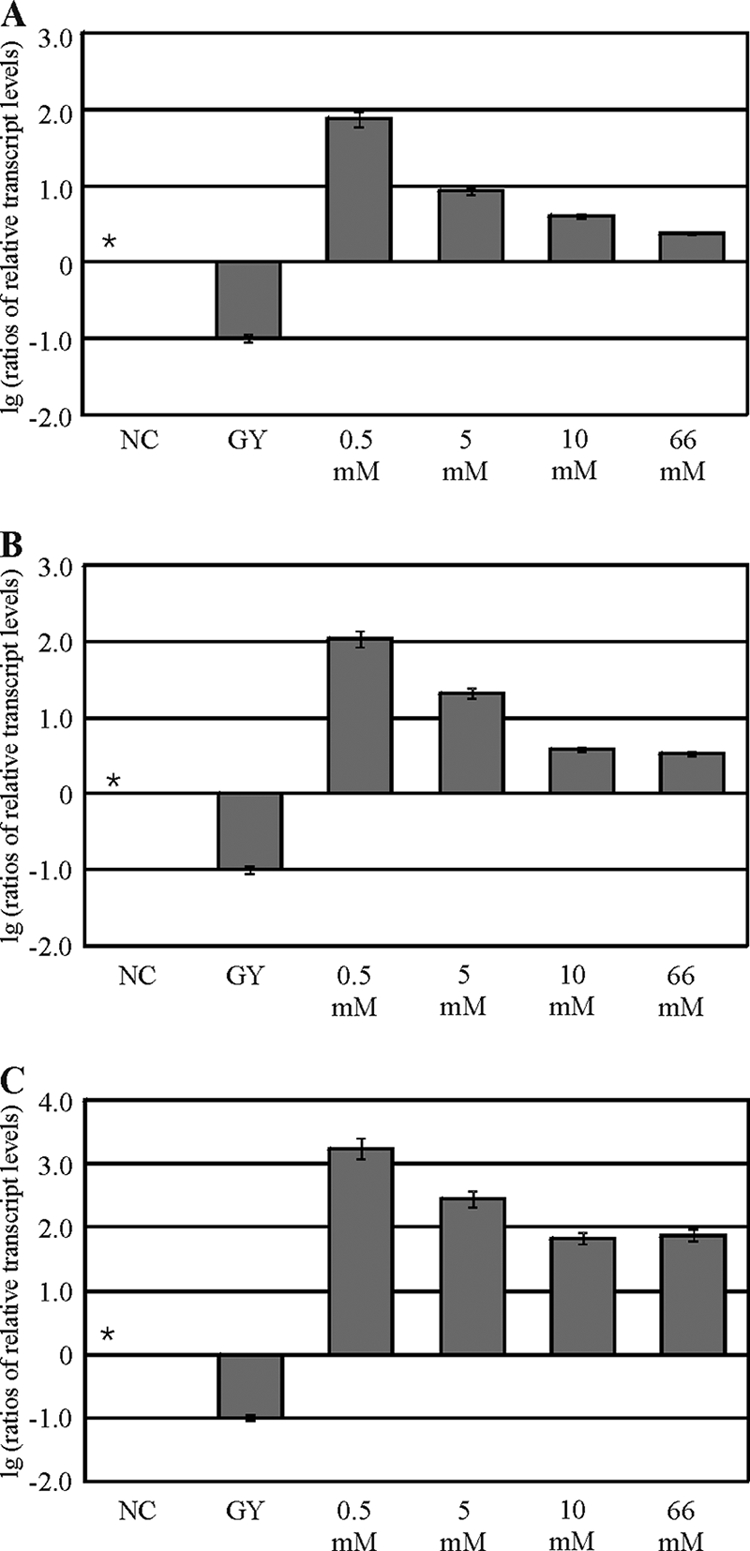
Analysis of d-xylose concentration-dependent transcription of xylanase-encoding genes in H. jecorina. The parental strain QM9414 was precultured on glycerol and was thereafter transferred to MA medium without a carbon source (NC) or containing 1% (wt/vol) glycerol (GY) or the indicated d-xylose concentration as the sole carbon source and was incubated for 3 h. Transcription analyses of xyn1 (A), xyn2 (B), and bxl1 (C) were performed via qPCR, using actin transcription for normalization. Results are given as relative transcript ratios on a logarithmic scale (lg). The values provided in the figures are means from three independent experiments. Error bars indicate standard deviations. Transcript levels always refer to one reference sample within an experiment, which is indicated by an asterisk.
High transcript levels of xylanase-encoding genes at low d-xylose concentrations are not caused by starvation.
To verify that the transcription peaks of xyn1, xyn2, and bxl1 obtained with 0.5 mM d-xylose for 3 h were not due to starvation, another replacement experiment was performed. H. jecorina QM9414 was pregrown on glycerol and subsequently incubated on MA medium without a carbon source. Transcription was investigated after 0.5, 1, 1.5, 2, 2.5, and 3 h of incubation. For all three genes (xyn1, xyn2, and bxl1), only marginal transcription was observed, with the earliest starting after 1.5 h; however, transcript levels at all time points were lower than those after 3 h of incubation (data not shown). These data suggest that the observed transcription peak obtained using d-xylose is due to induction, which can be achieved using low concentrations. Because at this stage it was not completely clear if the low transcription level obtained using higher d-xylose concentrations was caused by CCR and if Cre1 played a role in the corresponding regulatory mechanism, this was the next point of investigation.
Cre1 provokes less-pronounced CCR of xylanase-encoding genes at high d-xylose concentrations than that observed with glucose.
To examine whether the lower levels of transcription of xylanase-encoding genes at high d-xylose concentrations were due to CCR, we precultured the parental strain QM9414 and a corresponding cre1 deletion strain. Both strains were transferred to MA medium supplemented with 1% (wt/vol) glucose or high d-xylose concentrations (5 and 66 mM) or to medium without a carbon source (derepressed conditions). In addition, mixed cultures were grown, in which 1% (wt/vol) glucose was combined with either 5 or 66 mM d-xylose. A transcript analysis of xyn1 in QM9414 yielded no transcript under all conditions when glucose was used. Furthermore, strongly reduced xyn1 levels for all conditions tested were observed compared to those in the Δcre1 strain (Fig. 2A). Interestingly, xyn1 transcription in the Δcre1 strain reached a constant, induced level independent of the d-xylose concentration. A similar effect was observed for the mixed cultures. Nevertheless, induction of xyn1 expression at high d-xylose concentrations in the presence of glucose in a cre1-negative background remained 1 order of magnitude lower than that in culture without glucose. xyn1 is already known to be subject to direct CCR by Cre1 in glucose (15), but obviously the mechanism with high concentrations of d-xylose is not as pronounced, although it is Cre1 dependent. Whereas glucose fully repressed xyn1 transcription in QM9414, high d-xylose concentrations led to higher xyn1 transcript levels than those obtained under derepressing conditions (Fig. 2A). Transcript levels of xyn2 (Fig. 2B) and bxl1 (Fig. 2C) in QM9414 indicated a very low level of basal transcription in glucose, which is in accordance with previous results (29). Furthermore, similar to the case for xyn1, constant induction levels of xyn2 and bxl1 were reached in a Cre1-negative strain, both at a high d-xylose concentration and on a mixed carbon source. The latter again exhibited a reduction of transcript levels, by 1 order of magnitude. In summary, we concluded that the expression of xyn1, xyn2, and bxl1 is regulated by a dual mechanism at high d-xylose concentrations: on the one hand, there is d-xylose concentration-dependent CCR mediated by Cre1, and on the other hand, there is a concentration-independent inducer function of d-xylose. Clearly, Cre1-mediated repression overrules the induction mechanism of d-xylose, probably by simultaneously regulating xyr1 gene expression. Both mechanisms together cause the initially observed d-xylose concentration-dependent induction.
FIG. 2.
Transcription analysis of d-xylose-dependent Cre1 influence on xylanase-encoding genes in H. jecorina. The parental strain QM9414 (dark gray bars) and a Δcre1 strain (light gray bars) were precultured on glycerol and were thereafter transferred to MA medium containing 1% (wt/vol) glucose (G) or the indicated d-xylose concentration as the sole carbon source, mixtures of 1% (wt/vol) glucose with the indicated d-xylose concentrations, or no carbon source (NC) and incubated for 3 h. Transcription analyses of xyn1 (A), xyn2 (B), and bxl1 (C) were performed via qPCR, using actin transcription for normalization. Results are given as relative transcript ratios on a logarithmic scale (lg). The values given in the figures are means for three independent experiments. Error bars indicate standard deviations. Transcript levels always refer to one reference sample within an experiment, which is indicated by an asterisk. ND, no transcript detected.
d-Xylose-dependent influence of Cre1 on xylanase-encoding genes is indirectly exerted via Xyr1.
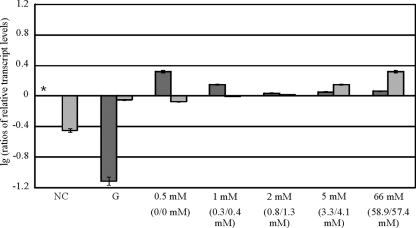
Because the influence of Cre1 on xyn1, xyn2, and bxl1 transcription using d-xylose was demonstrated in this study and the repressing influence of Cre1 on xyr1 transcription on glucose was recently reported (16), we consequently investigated if Cre1 indirectly affects xylanase-encoding genes by withdrawing their activator, Xyr1. Therefore, QM9414 and a Δcre1 strain were pregrown and then transferred to MA medium containing glucose (1% [wt/vol]) or 0.5, 1, 2, 5, or 66 mM d-xylose as the carbon source or containing no carbon source for derepressing conditions. As described previously (16), repression of xyr1 transcription was noted in glucose, which is released in a Δcre1 strain (Fig. 3). Remarkably, a slight d-xylose concentration-dependent response of xyr1 transcription was observed (Fig. 3). Similar to the case for xylanase-encoding genes, even at high d-xylose concentrations there was no repression of xyr1 transcription as occurred in glucose. On the contrary, xyr1 transcript levels at all d-xylose concentrations were higher than those without a carbon source. Nevertheless, Cre1 also influenced the transcription of xyr1 in a d-xylose concentration-dependent manner (Fig. 3), leading to the assumption that Cre1 exerts its influence on xylanase-encoding genes by causing a reduced function of their activator, Xyr1.
FIG. 3.
Analysis of Cre1-dependent d-xylose influence on xyr1 transcription in H. jecorina. The parental strain QM9414 (dark gray bars) and a Δcre1 strain (light gray bars) were precultured on glycerol and were thereafter transferred to MA medium containing 1% (wt/vol) glucose (G) or the indicated d-xylose concentration as the sole carbon source or no carbon source (NC) and then incubated for 3 h. Remaining concentrations of d-xylose after 3 h of incubation are given in parentheses (QM9414/Δcre1 strain) below the starting concentration. Transcription analysis of xyr1 was performed via qPCR, using actin transcription for normalization. Results are given as relative transcript ratios on a logarithmic scale (lg). The values given in the figures are means for three independent experiments. Error bars indicate standard deviations. Transcript levels always refer to one reference sample within an experiment, which is indicated by an asterisk.
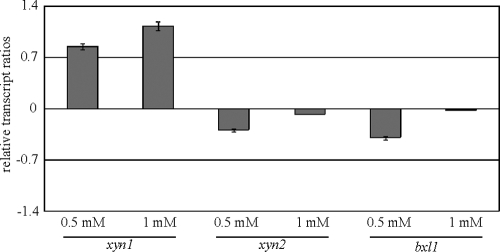
Optimal d-xylose-dependent induction of xyn1 transcription is further increased in the absence of Cre1.
On the one hand, xyn1 (but not xyn2 or bxl1) transcription is subject to CCR directly mediated by Cre1 (15, 29); however, on the other hand, we demonstrate in this study a Cre1-related reduction of transcription for all three xylanase-encoding genes in d-xylose. This prompted us to examine the transcription of those genes in a Cre1-dependent manner, using an optimal d-xylose concentration for induction. We performed a transfer of QM9414 and Δcre1 strains to MA media containing optimal d-xylose concentrations (0.5 and 1 mM) for 3 h of cultivation. The xyn1, xyn2, and bxl1 transcript ratios of the Δcre1 strain to QM9414 are given in Fig. 4. An increase of transcript formation in the Δcre1 strain compared to that in QM9414 was seen only for the xyn1 gene. This correlates with the fact that functional (double) Cre1 sites are present only in the xyn1 promoter. Thus, the observed, strongly positive effect on xyn1 transcription when Cre1 was absent may have been due to a double release from Cre1 influence. As already mentioned, the Δcre1 strain had higher xyn1 expression levels in 0.5 and 1.0 mM d-xylose than did strain QM9414. However, it is clear in Fig. 3 that at these d-xylose concentrations, the xyr1 transcript level was lower in the Δcre1 strain than in QM9414. In other words, in a cre1-negative background, xyn1 was expressed more strongly under conditions that caused xyr1 to be expressed at lower levels. This suggests, in our opinion, that even at 0.5 mM d-xylose, the release from CCR has such a pronounced effect that it compensates for lower Xyr1-mediated induction. It is noteworthy that previous investigations demonstrating that no de novo synthesis of Xyr1 is necessary for an initial induction of xyn1 expression (16) perfectly fit this assumption.
FIG. 4.
Transcription analysis of optimal d-xylose induction of xylanase-encoding genes in a Cre1-negative background of H. jecorina. The parental strain QM9414 and a Δcre1 strain were precultured on glycerol and were thereafter transferred to MA medium containing the indicated d-xylose concentration as the sole carbon source and then incubated for 3 h. Transcription analyses of xyn1, xyn2, and bxl1 were performed via qPCR, using actin transcription for normalization. Results are given as the logarithmic transcript ratio for the Δcre1 strain to QM9414. The values given in the figures are means for three independent experiments. Error bars indicate standard deviations. Transcript levels always refer to one reference sample within an experiment (QM9414, no carbon source, 3 h).
Influence of Xyl1 on xylanase expression in the presence of d-xylose.
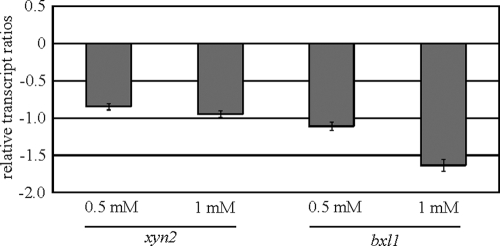
We next asked whether d-xylose must be metabolized via Xyl1 (25) to be able to exert its inducing influence on xylanase-encoding gene expression. To answer this question, the parental strain QM9414 and a Δxyl1 strain were precultured and transferred to MA media containing optimal inducing concentrations of d-xylose (0.5 and 1 mM). No xyn1 transcript could be detected in the Δxyl1 strain (data not shown), in contrast to the parental strain, suggesting that functional Xyl1 is required for d-xylose metabolism. As can be inferred from Fig. 5, the xyn2 and bxl1 ratios for the Δxyl1 strain to QM9414 were negative, also implying lower transcript levels in the xyl1 deletion strain (Fig. 5). In transferring both strains to medium without a carbon source, no such differences in xyn2 and bxl1 transcript levels were observed (relative ratios were between 0.4 and 1.8). This strongly indicates that neither a starvation effect in the Δxyl1 strain nor the simple presence of d-xylose, but rather its metabolism, is necessary to induce the expression of xylanase-encoding genes.
FIG. 5.
Transcription analysis of optimal d-xylose induction of xylanase-encoding genes in a Xyl1-negative background of H. jecorina. The parental strain QM9414 and a Δxyl1 strain were precultured on glycerol and were thereafter transferred to MA medium containing the indicated d-xylose concentration as the sole carbon source and then incubated for 3 h. Transcription analyses of xyn2 and bxl1 were performed via qPCR, using actin transcription for normalization. Results are given as the logarithmic transcript ratio for the Δxyl1 strain to QM9414. The values given in the figures are means for three independent experiments. Error bars indicate standard deviations. Transcript levels always refer to one reference sample within an experiment (QM9414, no carbon source, 3 h).
Constitutive expression of xyr1 can partly overcome Cre1-mediated antagonism.
We demonstrated in this study that Cre1 causes a reduced transcription of xylanase-encoding genes that is dependent on the d-xylose concentration. Furthermore, evidence was given that this influence is exerted indirectly, by antagonizing the activator Xyr1. Consequently, it can be questioned if the effect of Cre1 on xylanase expression can be overcome in a strain bearing a constitutively expressed and deregulated (Cre1-independent) xyr1 gene. Therefore, we precultured the parental strain QM9414 and the nx7 recombinant strain (a strain constitutively expressing xyr1 [16]) on glycerol, transferred them to medium containing 66 mM d-xylose, and followed the cultures for 3 h. Quantitative RT-PCR analysis of xyn1 revealed nearly the same transcript levels in the nx7 strain as in QM9414 (Fig. 6). Obviously, in this case, it was not possible to overcome the Cre1 influence simply by providing a constant basal level of Xyr1, perhaps due to an additional direct influence of Cre1. Unexpectedly, the same effect was found for transcription of xyn2 (Fig. 6). In contrast to that of xyn1 and xyn2, bxl1 transcription was found to be clearly higher if xyr1 was constitutively expressed than that in the parental strain (Fig. 6). It can therefore be stated that the indirect influence of Cre1 on bxl1 expression can be overcome by constitutive expression of xyr1.
FIG. 6.
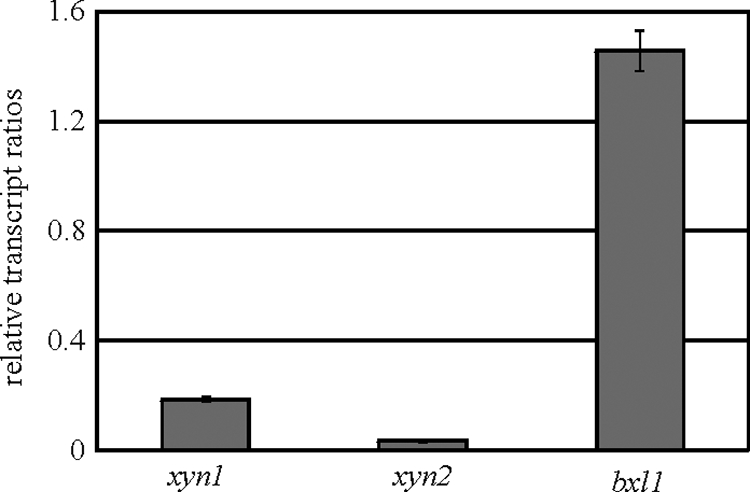
Transcription analysis of xylanase-encoding genes in H. jecorina at high d-xylose concentrations when xyr1 was constitutively expressed. The parental strain QM9414 and the nx7 strain were precultured on glycerol and were thereafter transferred to MA medium containing 66 mM d-xylose as the sole carbon source and then incubated for 3 h. Transcription analyses of xyn1, xyn2, and bxl1 were performed via qPCR, using actin transcription for normalization. Results are given as the logarithmic transcript ratio for the nx7 strain to QM9414. The values given in the figures are means for three independent experiments. Error bars indicate standard deviations. Transcript levels always refer to one reference sample within an experiment (QM9414, no carbon source, 3 h).
DISCUSSION
In Hypocrea and Aspergillus, d-xylose at a concentration of 66 mM (corresponding to 1% [wt/vol]) has been used as a xylanase-inducing substance for a long time (5, 8, 12, 15, 19, 23, 27). Because it was reported that d-xylose concentrations above 1 mM lead to carbon catabolite repression of xylanase expression in A. niger (6) and because evidence for a dual role of d-xylose as a repressor and inducer was given for A. nidulans (22), it was questioned whether the same may be true for H. jecorina. In H. jecorina, xylanase gene expression was induced by culture in d-xylose even at high concentrations such as 66 mM, because under all conditions tested, the addition of d-xylose increased transcript levels compared to those under derepressed conditions. However, the induction of xyn1, xyn2, and bxl1 gene expression was optimal at low d-xylose concentrations (i.e., 0.5 to 1 mM) for 3 h of cultivation and was decreased at higher concentrations of d-xylose. In particular, it is a new finding that xyn2 transcription can be induced by d-xylose, because the opposite was previously reported (30). The former observations may have been due to the high d-xylose concentration that was used and/or to a less sensitive experimental setup (Northern slot blot analysis instead of qPCR analysis).
In this study, the observed d-xylose concentration-dependent degree of induction of xylanase-encoding genes was found to be influenced by Cre1. Interestingly, for H. jecorina in d-xylose, Cre1 does not mediate as pronounced CCR as that on glucose (14, 15) or as the ortholog CreA does in Aspergillus on glucose and d-xylose (6, 20, 22). However, high d-xylose concentrations led to reduced xylanase expression mediated by Cre1, even if transcript levels of xylanase-encoding genes were still higher than those on medium without a carbon source. The influence of Cre1 on xylanase-encoding genes was primarily exerted indirectly, by decreasing the expression of xyr1. Reduction of Xyr1, which is an indispensable activator of the xylanase-encoding genes (27), in all probability diminishes expression of xylanases. The regulation of xyn1 expression poses an exception, because for this xylanase-encoding gene, a stronger impact of release from Cre1 influence was noted. This observation, taken together with the fact that xyn1 expression is under direct Cre1 control on glucose (15, 23), points to a direct involvement of Cre1 in xyn1 transcriptional regulation on d-xylose as well. A summary of these findings is provided in Fig. 7.
FIG. 7.
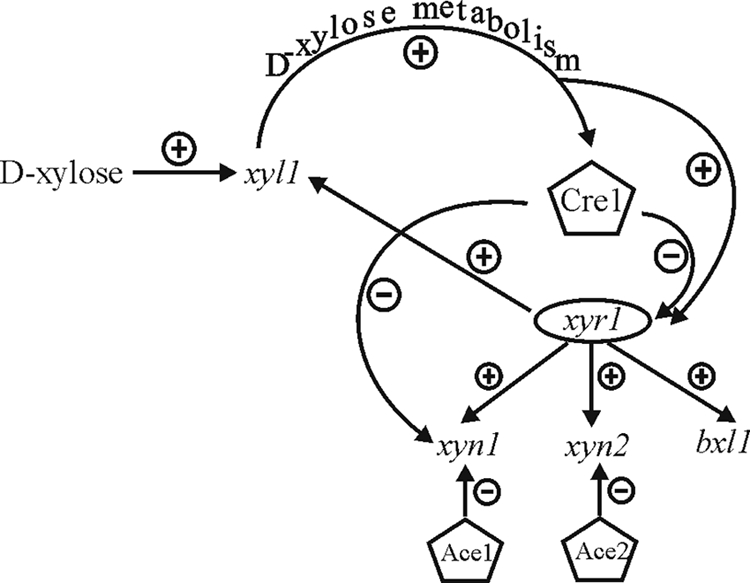
Schematic drawing of interrelated mechanisms underlying the effects of d-xylose on xylanase expression in H. jecorina. +, positive regulation; −, negative regulation.
In a cre1-negative background, d-xylose exerts its inducer function independent of the respective concentration applied. Constant transcript levels in mixtures of d-xylose and glucose were still clearly higher than those observed in glucose alone, although induction was 1 order of magnitude lower than that in d-xylose. This effect of reduced induction due to the addition of glucose must be seen as independent. Possible explanations might include the impact of glucose on d-xylose uptake, as postulated for A. niger (6), or the impact of glucose and/or cre1 deletions on d-xylose metabolism, as demonstrated for A. nidulans (4).
The insight that Cre1 exerts its antagonistic function on xylanase expression indirectly, via Xyr1, led to the hypothesis that this can be overcome by providing a constant level of Xyr1. Such a Cre1-independently xyr1-expressing strain was already demonstrated to influence expression of xyn1, xyn2, and bxl1 (16) and of a number of other enzymes involved in xylan degradation (A. R. Mach-Aigner, M. G. Steiger, and R. L. Mach, unpublished data), in a positive way. Surprisingly, only bxl1 expression was increased if this constitutively xyr1-expressing strain was cultured at high d-xylose concentrations. It can be assumed that this observation again results from a direct influence that Cre1 exerts on the xyn1 promoter. In addition, there is another repressor involved in the regulation of xyn1 expression, namely, AceI (23). A similar argument may explain the observed effects on xyn2 expression, which is governed by the transcription factor Ace2 and by Xyr1. Ace2 has antagonizing effects on xyn2 expression at early induction stages (28). In contrast, it should be noted that there are no potentially functional Cre1 sites in the bxl1 promoter (tandem or inverted repeats [14, 15]), neither is another antagonist of bxl1 expression known.
Until now, it was not clear if d-xylose itself can function as the actual inducer or if it needs to be metabolized. The enzyme responsible for the first step of d-xylose metabolism in H. jecorina is d-xylose reductase (25), which converts d-xylose to d-xylitol. The expression of xyl1 was proven to be strongly dependent on activation by Xyr1 (27). It is not likely that the observed, clearly reduced transcription of all xylanase-encoding genes in the xyl1-negative background was due to starvation effects, because this strain is able to grow on d-xylose (25). Rather, this points to the need for d-xylose metabolism to generate the real inducer that triggers xylanase expression. These known facts, together with the results we have obtained here, are depicted in Fig. 7.
Acknowledgments
The cre1 and xyl1 deletion strains were kindly provided by C. P. Kubicek, Vienna University of Technology, which is gratefully acknowledged.
M. E. Pucher is the recipient of a DOC-fFORTE fellowship from the Austrian Academy of Sciences at the Institute of Chemical Engineering of the Vienna University of Technology. This study was supported by grant P20192-B03 from the Austrian Fonds zur Förderung Wissenschaftlicher Forschung.
Footnotes
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Aden, A., J. Bozell, J. Holladay, J. White, and A. Manheim. 2004. Top value added chemicals from biomass, vol. I. U.S. Department of Energy, Washington, DC.
- 2.Aro, N., M. Ilmén, A. Saloheimo, and M. Penttilä. 2003. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69:56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aro, N., A. Saloheimo, M. Ilmen, and M. Penttilä. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 4.David, H., A. M. Krogh, C. Roca, M. Akesson, and J. Nielsen. 2005. CreA influences the metabolic fluxes of Aspergillus nidulans during growth on glucose and xylose. Microbiology 151:2209-2221. [DOI] [PubMed] [Google Scholar]
- 5.de Graaff, L. H., H. C. van den Broeck, A. J. van Ooijen, and J. Visser. 1994. Regulation of the xylanase-encoding xlnA gene of Aspergillus tubigensis. Mol. Microbiol. 12:479-490. [DOI] [PubMed] [Google Scholar]
- 6.de Vries, R. P., J. Visser, and L. H. de Graaff. 1999. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 150:281-285. [DOI] [PubMed] [Google Scholar]
- 7.Drysdale, M. R., S. E. Kolze, and J. M. Kelly. 1993. The Aspergillus niger carbon catabolite repressor encoding gene, creA. Gene 130:241-245. [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Espinar, M., F. Piñaga, L. de Graaff, J. Visser, D. Ramón, and S. Vallés. 1994. Purification, characterization and regulation of the synthesis of an Aspergillus nidulans acidic xylanase. Appl. Microbiol. Biotechnol. 42:555-562. [Google Scholar]
- 9.Gielkens, M. M., E. Dekkers, J. Visser, and L. H. de Graaff. 1999. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl. Environ. Microbiol. 65:4340-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36:193-200. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann, M. C., M. Vrsanska, M. Jurickova, J. Hirsch, P. Biely, and C. P. Kubicek. 1997. The beta-d-xylosidase of Trichoderma reesei is a multifunctional beta-d-xylan xylohydrolase. Biochem. J. 321:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrmova, M., P. Biely, and M. Vrsanska. 1986. Specificity of cellulase and β-xylanase induction in Trichoderma reesei QM 9414. Arch. Microbiol. 144:307-311. [Google Scholar]
- 13.Ilmén, M., A. Saloheimo, M. L. Onnela, and M. E. Penttilä. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilmén, M., C. Thrane, and M. Penttilä. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 15.Mach, R. L., J. Strauss, S. Zeilinger, M. Schindler, and C. P. Kubicek. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21:1273-1281. [DOI] [PubMed] [Google Scholar]
- 16.Mach-Aigner, A. R., M. E. Pucher, M. G. Steiger, G. E. Bauer, S. J. Preis, and R. L. Mach. 2008. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl. Environ. Microbiol. 74:6554-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandels, M. 1985. Applications of cellulases. Biochem. Soc. Trans. 13:414-416. [DOI] [PubMed] [Google Scholar]
- 18.Margolles-Clark, E., M. Ilmén, and M. Penttilä. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 19.Orejas, M., A. P. MacCabe, J. A. Perez-Gonzalez, S. Kumar, and D. Ramon. 2001. The wide-domain carbon catabolite repressor CreA indirectly controls expression of the Aspergillus nidulans xlnB gene, encoding the acidic endo-beta-(1,4)-xylanase X(24). J. Bacteriol. 183:1517-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orejas, M., A. P. MacCabe, J. A. Perez Gonzalez, S. Kumar, and D. Ramon. 1999. Carbon catabolite repression of the Aspergillus nidulans xlnA gene. Mol. Microbiol. 31:177-184. [DOI] [PubMed] [Google Scholar]
- 21.Parajó, J., H. Dominguez, and J. Dominguez. 1998. Biotechnological production of xylitol. 3. Operation in culture media made from lignocellulose hydrolysates. Bioresour. Technol. 66:25-40. [Google Scholar]
- 22.Prathumpai, W., M. McIntyre, and J. Nielsen. 2004. The effect of CreA in glucose and xylose catabolism in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 63:748-753. [DOI] [PubMed] [Google Scholar]
- 23.Rauscher, R., E. Würleitner, C. Wacenovsky, N. Aro, A. R. Stricker, S. Zeilinger, C. P. Kubicek, M. Penttilä, and R. L. Mach. 2006. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot. Cell 5:447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saloheimo, A., N. Aro, M. Ilmén, and M. Penttilä. 2000. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275:5817-5825. [DOI] [PubMed] [Google Scholar]
- 25.Seiboth, B., C. Gamauf, M. Pail, L. Hartl, and C. P. Kubicek. 2007. The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for beta-galactosidase and cellulase induction by lactose. Mol. Microbiol. 66:890-900. [DOI] [PubMed] [Google Scholar]
- 26.Strauss, J., R. L. Mach, S. Zeilinger, G. Hartler, G. Stoffler, M. Wolschek, and C. P. Kubicek. 1995. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 376:103-107. [DOI] [PubMed] [Google Scholar]
- 27.Stricker, A. R., K. Grosstessner-Hain, E. Würleitner, and R. L. Mach. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stricker, A. R., P. Trefflinger, N. Aro, M. Penttilä, and R. L. Mach. 2008. Role of Ace2 (activator of cellulases 2) within the xyn2 transcriptosome of Hypocrea jecorina. Fungal Genet. Biol. 45:436-445. [DOI] [PubMed] [Google Scholar]
- 29.Würleitner, E., L. Pera, C. Wacenovsky, A. Cziferszky, S. Zeilinger, C. P. Kubicek, and R. L. Mach. 2003. Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryot. Cell 2:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeilinger, S., R. L. Mach, M. Schindler, P. Herzog, and C. P. Kubicek. 1996. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271:25624-25629. [DOI] [PubMed] [Google Scholar]