Abstract
Research into archaea will not achieve its full potential until systems are in place to carry out genetics and biochemistry in the same species. Haloferax volcanii is widely regarded as the best-equipped organism for archaeal genetics, but the development of tools for the expression and purification of H. volcanii proteins has been neglected. We have developed a series of plasmid vectors and host strains for conditional overexpression of halophilic proteins in H. volcanii. The plasmids feature the tryptophan-inducible p.tnaA promoter and a 6×His tag for protein purification by metal affinity chromatography. Purification is facilitated by host strains, where pitA is replaced by the ortholog from Natronomonas pharaonis. The latter lacks the histidine-rich linker region found in H. volcanii PitA and does not copurify with His-tagged recombinant proteins. We also deleted the mrr restriction endonuclease gene, thereby allowing direct transformation without the need to passage DNA through an Escherichia coli dam mutant.
Over the past century, our understanding of fundamental biological processes has grown exponentially, and this would have been impossible without the use of organisms that are amenable to experimental manipulation. Model species, such as Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Arabidopsis thaliana, have become a byword for scientific progress (15). The rational choice of a model organism is critically important, and certain features are taken for granted, such as ease of cultivation, a short generation time, and systems for genetic manipulation. This list has now grown to include a genome sequence and methods for biochemical analysis of purified proteins in vitro.
Research into archaea has lagged behind work on bacteria and eukaryotes but has nonetheless yielded profound insights (2). One hurdle has been the paucity of archaeal organisms suitable for both biochemistry and genetics. For example, Methanothermobacter thermautotrophicus is a stalwart of archaeal biochemistry but has proved resistant to even the most rudimentary genetic manipulation (2). Progress has recently been made with another biochemical workhorse, Sulfolobus spp., and a few genetic tools are now available (6, 13, 37). Methanosarcina spp. and Thermococcus kodakaraensis offer alternative systems with an increasing array of techniques (16, 35, 36), but sophisticated genetics has traditionally been the preserve of haloarchaea, of which Haloferax volcanii is the organism of choice (39). It is easy to culture, the genome has been sequenced (19), and there are several selectable markers and plasmids for transformation and gene knockout (3, 7, 31), including a Gateway system (14), as well as reporter genes (20, 33) and a tightly controlled inducible promoter (26).
The genetic prowess of H. volcanii is not yet fully matched by corresponding systems for protein overexpression and purification. Like other haloarchaea, H. volcanii grows in high salt concentrations (2 to 5 M NaCl), and to cope with the osmotic potential of such environments, it accumulates high intracellular concentrations of potassium ions (12). Consequently, halophilic proteins are adapted to function at high salt concentrations and commonly feature a large excess of acidic amino acids; the negative surface charge is thought to be critical to solubility (28). This can pose problems for expression in heterologous hosts, such as E. coli, since halophilic proteins can misfold and aggregate under conditions of low ionic strength. The purification of misfolded halophilic enzymes from E. coli has relied on the recovery of insoluble protein from inclusion bodies, followed by denaturation and refolding in hypersaline solutions (8, 11). This approach is feasible only where the protein is well characterized and reconstitution of the active form can be monitored (for example, by an enzymatic assay). Furthermore, archaeal proteins expressed in heterologous bacterial hosts lack posttranslational modifications, such as acetylation or ubiquitination (4, 22), which are critical to understanding their biological function.
Systems for expression of halophilic proteins in a native haloarchaeal host are therefore required. A number of studies have successfully purified recombinant proteins with a variety of affinity tags after overexpression in H. volcanii. For example, Humbard et al. employed tandem affinity tagging to purify 20S proteasomal core particles from the native host (23). However, the protein expression constructs used in these studies were custom made and somewhat tailored to the application in question. We report here the development of “generic” plasmid vectors and host strains for conditional overexpression of halophilic proteins in H. volcanii. The plasmids feature a tryptophan-inducible promoter derived from the tnaA gene of H. volcanii (26). We demonstrate the utility of these vectors by overexpressing a hexahistidine-tagged recombinant version of the H. volcanii RadA protein. Purification was greatly facilitated by a host strain in which the endogenous pitA gene was replaced by an ortholog from Natronomonas pharaonis. The latter protein lacks the histidine-rich linker region found in H. volcanii PitA (5) and therefore does not copurify with His-tagged recombinant proteins. Finally, we deleted the mrr gene of H. volcanii, which encodes a restriction enzyme that cleaves foreign DNA methylated at GATC residues. The mrr deletion strain allows direct transformation of H. volcanii without the need to passage plasmid DNA through an E. coli dam mutant (21).
MATERIALS AND METHODS
Unless stated otherwise, chemicals were from Sigma and restriction enzymes were from New England Biolabs. Standard molecular techniques were used (34).
Strains and plasmids.
H. volcanii strains (Table 1) were grown at 45°C on complete (Hv-YPC) or Casamino Acids (Hv-Ca) agar or in Hv-YPC or Hv-Ca broth, as described previously (3, 17). Gene deletion/replacement mutants were constructed using a knockout system described previously (3, 7). The plasmids for gene deletion/replacement and protein overexpression are shown in Table 2 and were generated by PCR using the primers in Table 3. E. coli strains XL1-Blue MRF′ (ΔmcrA183 ΔmcrCB-hsdSMR-mrr173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10]) and GM121 (F− dam-3 dcm-6 ara-14 fhuA31 galK2 galT22 hdsR3 lacY1 leu-6 thi-1 thr-1 tsx-78) were grown in Luria-Bertani medium with 100 μg/ml ampicillin where appropriate. The latter strain was used to prepare unmethylated DNA for transformation of H. volcanii.
TABLE 1.
H. volcanii strains used
| Strain | Genotypea | Source or referenceb |
|---|---|---|
| H26 | ΔpyrE2 | 3 |
| H98 | ΔpyrE2 ΔhdrB | 3 |
| H133 | ΔpyrE2 ΔtrpA ΔleuB ΔhdrB | 3 |
| H989 | ΔpyrE2 ΔhdrB {p.tnaA::6×His tag pyrE2+hdrB+} | H98 pTA963 |
| H1045 | ΔpyrE2 ΔhdrB {p.tnaA::6×His tag::radA+pyrE2+hdrB+} | H98 pTA1041 |
| H1154 | ΔpyrE2-pitANph | H26 (pTA1106) |
| H1155 | ΔpyrE2 ΔhdrB pitANph | H98 (pTA1106) |
| H1172 | ΔpyrE2 ΔhdrB pitANph {p.tnaA::6×His tag pyrE2+hdrB+} | H1155 pTA963 |
| H1173 | ΔpyrE2 ΔhdrB pitANph {p.tnaA::6×His tag::radA+pyrE2+hdrB+} | H1155 pTA1041 |
| H1174 | ΔpyrE2 ΔhdrB pitANph {p.tnaA::6×His tag::radB+pyrE2+hdrB+} | H1155 pTA1043 |
| H1206 | ΔpyrE2 Δmrr | H26 (pTA1150) |
| H1207 | ΔpyrE2 pitANph Δmrr | H1154 (pTA1150) |
| H1208 | ΔpyrE2 ΔhdrB Δmrr | H98 (pTA1150) |
| H1209 | ΔpyrE2 ΔhdrB pitANph Δmrr | H1155 (pTA1150) |
| H1227 | ΔpyrE2 ΔhdrB pitANph Δmrr {p.tnaA::6×His tag::radA(A196V) pyrE2+hdrB+} | H1209 pTA1182 |
| H1228 | ΔpyrE2 ΔhdrB pitANph Δmrr cdc48d+::[Δcdc48d pyrE2+] | H1209 pTA1180c |
| WR755 | ΔpyrE2 ΔtrpA ΔleuB ΔhdrB pitA+::[ΔpitA pyrE2+] | H133 pMM1231c |
| WR756 | ΔpyrE2 ΔtrpA ΔleuB ΔhdrB pitA+::[ΔpitA::hdrB+pyrE2+] | H133 pMM1232c |
Episomal plasmids are indicated by braces ({}); plasmids integrated on the chromosome are indicated by brackets ([]).
Deletion constructs used successfully are indicated by parentheses.
Pop-in strain from which a pop-out strain could not be obtained.
TABLE 2.
Plasmids used
| Plasmid | Relevant properties | Source or referencea |
|---|---|---|
| pTA131 | Integrative vector based on pBluescript, with pyrE2 marker | 3 |
| pTA187 | Integrative vector based on pUC19, with hdrB marker | 3 |
| pTA230 | Shuttle vector derived from pTA131, with pyrE2 marker and pHV2 replication origin | 3 |
| pTA354 | Shuttle vector based on pBluescript, with pyrE2 marker and pHV1/4 replication origin | 31 |
| pTA927 | Overexpression vector with pyrE2 marker and pHV2 origin, derived from pTA230 by insertion of 131-bp t.L11e terminator at KpnI site, 224-bp p.tnaA promoter at ApaI and ClaI sites, and 35-bp t.Syn terminator at NotI and BstXI sites | This study; FN645893 |
| pTA929 | Overexpression vector with 6×His tag, pyrE2 marker, and pHV2 origin, derived from pTA927 by insertion of 26-bp fragment containinga His tag (CAC)6 tract at the NdeI and ClaI sites | This study; FN645892 |
| pTA949 | Shuttle vector derived from pTA230, with pyrE2 and hdrB markers and pHV2 replication origin | This study; FN645894 |
| pTA962 | Overexpression vector with pyrE2 and hdrB markers and pHV2 origin, derived from pTA949 by insertion of t.L11e terminator, p.tnaA promoter and t.Syn terminator (see pTA927 for details) | This study; FN645891 |
| pTA963 | Overexpression vector with 6×His tag, pyrE2 and hdrB markers, and pHV2 origin, derived from pTA962 by insertion of a His tag (CAC)6 tract (see pTA929 for details) | This study; FN645890 |
| pTA1041 | pTA963 with insertion of 1.0-kb NcoI-BamHI radA+ fragment at the PciI and BamHI sites; for overexpression of 6×His-tagged RadA | This study |
| pTA1043 | pTA963 with insertion of 0.7-kb BspHI-BamHI radB+ fragment at the PciI and BamHI sites; for overexpression of 6×His-tagged RadB | This study |
| pTA1106 | pTA131 with 3.8-kb pitANph gene replacement construct, consisting of HindIII-EcoRI upstream ΔpitAHvo fragment, EcoRI-XbaI pitANph+ fragment, and XbaI-NotI downstream ΔpitAHvo fragment inserted at HindIII and NotI sites | This study |
| pTA1150 | pTA131 with 1.3-kb Δmrr construct consisting of a KpnI-BamHI upstream Δmrr fragment and a BamHI-XbaI downstream Δmrr fragment inserted at KpnI and XbaI sites | This study |
| pTA1180 | pTA131 with 878-bp Δcdc48d construct consisting of KpnI-BamHI upstream Δcdc48d fragment and BamHI-EcoRI downstream Δcdc48d fragment inserted at KpnI and EcoRI sites | This study |
| pTA1182 | pTA963 with insertion of 1.0-kb NcoI-BamHI radA(A196V) fragment at PciI and BamHI sites; for overexpression of 6×His-tagged RadA(A196V) | This study |
| pMM1231 | pTA131 with 2.0-kb ΔpitAHvo construct consisting of a HindIII-EcoRI upstream ΔpitAHvo fragment and an EcoRI-NotI downstream ΔpitAHvo fragment; inserted at HindIII and NotI sites | This study |
| pMM1232 | pTA131 with 2.7-kb ΔpitAHvo::hdrB+ construct consisting of a HindIII-EcoRI upstream ΔpitAHvo fragment, an EcoRI-XbaI fragment of pTA187 with an hdrB marker, and an XbaI-NotI downstream ΔpitAHvo fragment; inserted at HindIII and NotI sites | This study |
The accession number is given where appropriate.
TABLE 3.
Oligonucleotides used
| Primer | Sequence (5′-3′)a | Relevant properties | Plasmid(s) |
|---|---|---|---|
| TERF | GACGGTACCGACTTCGACGACTACTTCGACG | t.L11e forward primer; KpnI site | pTA927 pTA962 |
| TERR | GGCGGTACCGGGTCGAATCGGGTCGGTG | t.L11e reverse primer; KpnI site | pTA927 pTA962 |
| PtnaApa2 | CGAGTTCTGGGCCCGTTCTCGTCG | p.tnaA forward primer; ApaI site | pTA927 pTA962 |
| PtnaNde3 | CCTTGATCGATTTCATATGCGCAATAGGTCC | p.tnaA reverse primer; ClaI and NdeI sites | pTA927 pTA962 |
| t.syn F | GGCCGCACCTCTGGACCATCGCATTTTTCGGCGCG | Top strand of t.Syn fragment; NotI and BstXI ends | pTA927 pTA962 |
| t.syn R | CCGAAAAATGCGATGGTCCAGAGGTGC | Bottom strand of t.Syn fragment | pTA927 pTA962 |
| HisndeI | TATGCACCACCACCACCACCACATGT | Top strand of 6×His tag; NdeI end; PciI site | pTA929 pTA963 |
| HisclaI | CGACATGTGGTGGTGGTGGTGGTGCA | Bottom strand of 6×His tag; ClaI end; PciI site | pTA929 pTA963 |
| radANcoF | GAACGACTGGCCATGGCAGAAGACG | radA forward primer; NcoI site | pTA1041 pTA1182 |
| radABamR | CCGACGGATCCACGGCTTACTCGG | radA reverse primer; BamHI site | pTA1041 pTA1182 |
| radBBsF | CCTCCTGTCATGACAGAGTCAGTCTCC | radB forward primer; BspHI site | pTA1043 |
| radBBamR2 | CCGCGTGGATCCCTTTTCTACACG | radB reverse primer; BamHI site | pTA1043 |
| mrrUSF | GGATGGTACCGCCGTAGAACAGCG | Δmrr upstream external primer; KpnI site | pTA1150 |
| mrrUSR | CCGGGGATCCCGTCATCGCTCGAC | Δmrr upstream internal primer; BamHI site | pTA1150 |
| mrrDSF | GGCGGATCCGTCGGCATTTGGCTC | Δmrr downstream internal primer; BamHI site | pTA1150 |
| mrrDSR | CCGCTCTAGAAGGCCGAGGAGGCC | Δmrr downstream external primer; XbaI site | pTA1150 |
| cdc48dUF | ACGGGTACCCACGTTGCTGG | Δcdc48d upstream external primer; KpnI site | pTA1180 |
| cdc48dUR | GGACGGATCCGTCGAACCGAG | Δcdc48d upstream internal primer; BamHI site | pTA1180 |
| cdc48dDF | CACGGATCCCCCAGAAATTGC | Δcdc48d downstream internal primer; BamHI site | pTA1180 |
| cdc48dDR | GCCGAATTCGAGCCGAGGTGG | Δcdc48d downstream external primer; EcoRI site | pTA1180 |
| 5′Up HindIII | AAGCTTTCCGACCCGATTCGCGTGAC | ΔpitAHvo upstream external primer; HindIII site | pMM1231 pMM1232 pTA1106 |
| 3′Up EcoRI | GAATTCACGAGGGCCTAGGGAGTCATCC | ΔpitAHvo upstream internal primer; EcoRI site | pMM1231 pMM1232 pTA1106 |
| 5′Down EcoRI | GAATTCCTTCGCGACCATCGGGAGC | ΔpitAHvo downstream internal primer; EcoRI site | pMM1231 |
| 3′Down NotI | GCGGCCGCCGGCATCGACCGCTTCGAC | ΔpitAHvo downstream external primer; NotI site | pMM1231 pMM1232 pTA1106 |
| 5′Down XbaI | TCTAGACTTCGCGACCATCGGGAGC | ΔpitAHvo downstream internal primer; XbaI site | pMM1232 pTA1106 |
| 5′NatroEcoRI | GAATTCATGCCACAACGCCAACCAC | pitANph forward primer;, EcoRI site | pTA1106 |
| 3′NatroXbaI | TCTAGATCAGGCGAGGAAGACGTGG | pitANph reverse primer; XbaI site | pTA1106 |
| pitAF | GGAAAATCAAGCAGGTCATCGC | Forward primer specific to pitAHvo | NA |
| pitAR | GTAGAACATCCCCATCGTGCC | Reverse primer specific to pitAHvo | NA |
| NphPitAF | GCAGTATGCCGACAAGGTCTCC | Forward primer specific to pitANph | NA |
| NphPitAR | CCCGCTCGTTTTTCCACAG | Reverse primer specific to pitANph | NA |
| mrrF | TGGGCGTTCAGGCGAAGC | Forward primer for mrr probe | NA |
| mrrR | CGGGTGAGCGACCAGCGG | Reverse primer for mrr probe | NA |
Restriction endonuclease sites used in cloning are underlined; internal sites used subsequently are in boldface. NA, not applicable.
Protein overexpression and purification.
A starter culture was grown overnight in 40 ml Hv-Ca broth to an optical density at 650 nm (OD650) of ∼0.6 and used to inoculate 360 ml Hv-YPC broth containing 1 mM tryptophan (Trp) to induce protein expression. The culture was incubated at 42°C with shaking (175 rpm) for 5 to 6 h to an OD650 of ∼0.5, when protein expression was further induced by adding 36 ml prewarmed 25 mM Trp dissolved in 18% salt water (SW) (3), and the culture was incubated at 42°C with shaking for a further 1 h. The culture was then centrifuged at 3,300 × g for 10 min at 4°C, and the cells were resuspended in 7 ml ice-cold binding buffer (2 M NaCl, 20 mM HEPES, pH 7.5, 20 mM imidazole, 1 mM phenylmethanesulfonyl fluoride) and lysed by sonication on ice until the suspension was no longer turbid. The cell lysate was clarified by centrifugation at 16,000 × g for 15 min at 4°C and incubated overnight at 4°C with 0.5 ml of IMAC Sepharose 6 FastFlow beads (GE Healthcare) that had been charged with Ni2+ and equilibrated in binding buffer. The slurry was applied to a Poly-Prep column (Bio-Rad), and the flowthrough was collected and reloaded onto the column, followed by three washes with 4 ml of ice-cold binding buffer and one wash with 1 ml ice-cold binding buffer containing 50 mM imidazole. Bound protein was eluted with 1 ml binding buffer containing 500 mM imidazole. Samples were analyzed on 12.5% SDS-PAGE gels with PageRuler size marker (Fermentas), and quantification of protein bands stained with Coomassie blue was carried out using ImageGauge v4.22 (Fuji).
Mass spectrometry.
Proteins in gel bands were reduced, carboxyamidomethylated, and digested with Trypsin Gold (Promega) on a robotic platform for protein digestion (MassPREP station; Waters). The resulting peptides were analyzed by electrospray ionization-tandem mass spectrometry (MS/MS) after on-line separation on a PepMap C18 reversed-phase, 75-μm-inner-diameter, 15-cm column (LC Packings) on a CapLC system attached to a Q-TOF2 mass spectrometer equipped with a nanolockspray source (Waters) and operated with MassLynx version 4.0 acquisition software. ProteinLynxGlobalSERVER software version 2.1 (Waters) was used to generate a peak list file of uninterpreted fragment mass data, which was used to search the Swissprot 57.1 and NCBInr databases using the MASCOT search engine (32). Only protein identifications with probability-based MOWSE scores above a threshold of P < 0.05 were accepted. The H. volcanii genome sequence (19) is not currently available for interrogation with uninterpreted mass spectral data; therefore, protein identifications relied on matches to publicly available sequences from related haloarchaea (see Table 4), which were used to find corresponding sequences in the H. volcanii genome at HaloLex (https://www.halolex.mpg.de/public/).
TABLE 4.
Mass spectrometry identification of H. volcanii proteins obtained by immobilized metal affinity chromatography
| Protein name | NCBI or Swiss-Prot entry | MASCOT score | Coverage (%) | No. of unique peptides | Peptide sequences | Predicted mass (kDa) | Observed mass (kDa) | HaloLex entry |
|---|---|---|---|---|---|---|---|---|
| PitA | gi‖224820302 (Natrialba magadii chlorite dismutase) | 63 | 3 | 2 | SIDWDAWR | 56.1 | 60.9 | HVO_1871 |
| DVLADRPR | ||||||||
| Cdc48d | gi‖110668990 (Haloquadratum walsbyi AAA-type ATPase) | 369 | 12 | 5 | ILFVGPPGTGK | 53.4 | 55.7 | HVO_1907 |
| EAVLEALTEER | ||||||||
| ILFIDEFDSVAKa | ||||||||
| LSMITSQYLGETAK | ||||||||
| RFDEIVNFPKPDR | ||||||||
| RadA | Q48328 (Haloferax volcanii DNA repair and recombination protein RadA) | 546 | 32 | 9 | ADIGSSTASDIINAA | 38.3b | 41.9 | HVO_0104 |
| DAADVGGFETGSMVLER | ||||||||
| GLEDEALEATLDDR | ||||||||
| EMEGSIDDEETIK | ||||||||
| ALVDDFLDK | ||||||||
| VDDFLDKIHVAKa | ||||||||
| AGEHEDTEWPVRa | ||||||||
| LVDAPNLADGEAIMR | ||||||||
| VQDAGLKPE |
Semitryptic peptide.
Including the hexahistidine tag.
Protein sequence alignment.
Alignment of the Mrr core domain was carried out in MacVector using ClustalW (BLOSUM; penalty for open gap = 10; extend gap = 2). A neighbor-joining tree was built using Schizosaccharomyces pombe SPAC824.03c as an outgroup.
Nucleotide sequence accession numbers.
The sequences of the major plasmid vectors constructed in this study have been deposited in the EMBL nucleotide sequence database under accession numbers FN645893 (pTA927), FN645892 (pTA929), FN645894 (pTA949), FN645891 (pTA962), and FN645890 (pTA963).
RESULTS
Plasmid vectors for conditional overexpression of hexahistidine-tagged proteins.
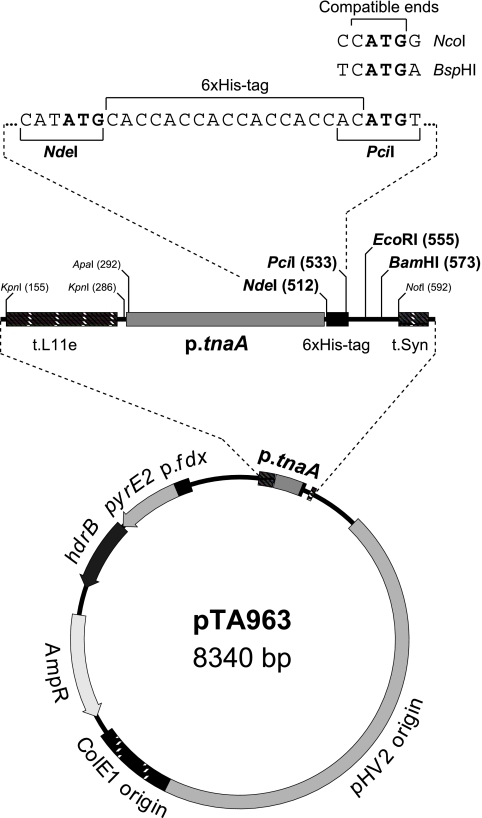
A series of plasmid shuttle vectors for conditional protein overexpression that utilized the p.tnaA tryptophanase promoter of H. volcanii were constructed (Fig. 1). Genes under the control of the p.tnaA promoter show rapid and strong induction of expression upon addition of ≥1 mM tryptophan (26). For expression of proteins with an N-terminal hexahistidine (6×His) tag, a (CAC)6 tract was incorporated downstream of the p.tnaA promoter; plasmid variants without the 6×His tag were also generated (Fig. 1). To ensure that the gene was insulated from read-through transcription initiated elsewhere on the plasmid, the expression cassette was flanked by two transcriptional terminators, the L11e rRNA terminator (t.L11e) (38) and a synthetic terminator (t.Syn) comprising a T tract flanked by G/C-rich sequences. The vectors were based on pTA230 (3) and used the pHV2 replication origin, which maintained the plasmid at a copy number of ∼6 per genome equivalent (10). Selection was based on the pyrE2 and hdrB markers, which allowed growth on media lacking uracil and thymidine, respectively (3, 7). The former marker complemented the pyrE2 deletion found in almost all laboratory strains of H. volcanii (3), while the latter marker allowed the plasmid to be maintained in rich medium (Hv-YPC); plasmid variants are available with the pyrE2 marker only (Fig. 1).
FIG. 1.
Conditional overexpression vector pTA963. pTA963 features a tryptophan-inducible promoter from the tnaA gene (p.tnaA) (26) flanked by the L11e rRNA terminator (t.L11e) (38) and a synthetic terminator (t.Syn). For expression of native proteins, the coding sequence is inserted between the NdeI site downstream of the promoter and either the EcoRI or BamHI site. For N-terminal 6×His-tagged proteins, the 5′ end of the gene is ligated instead with the PciI site located downstream of a (CAC)6 tract. PciI-compatible ends are generated by NcoI and BspHI and are used where the second codon starts with G and A, respectively. pTA963 uses a pHV2 replication origin and has hdrB and pyrE2 selectable markers; variants are available with pyrE2 only (pTA929) and without a (CAC)6 tract (pTA962, or pTA927 for pyrE2 only).
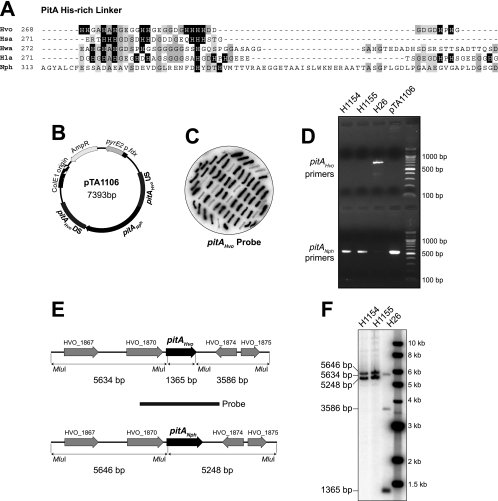
Replacement of the H. volcanii pitA gene with an ortholog from Natronomonas pharaonis.
The H. volcanii PitA protein is a fusion of chlorite dismutase-like and antibiotic synthesis monooxygenase-like domains within a single open reading frame (5). PitA is unique to haloarchaea and in almost all species features a histidine-rich linker between the conserved N- and C-terminal domains (Fig. 2A). Owing to the numerous histidines in this region, PitA is a major contaminant of His-tagged recombinant proteins purified from H. volcanii by immobilized metal affinity chromatography (5). We attempted to delete pitA by the pop-in/pop-out technique based on the pyrE2 counterselectable marker (7), using the deletion constructs pMM1231 and pMM1232. In the latter construct, the pitA coding sequence is replaced with an hdrB marker, allowing direct selection for gene deletion events (3). With either construct (in strains WR755 and WR756, respectively), we were unable to recover cells with a pitA deletion (data not shown), indicating that this gene is most likely essential.
FIG. 2.
Replacement of the pitA gene. (A) Protein sequence alignment of the central region of PitA (5), linking the N-terminal chlorite dismutase-like and C-terminal antibiotic biosynthesis monooxygenase-like domains, from selected species of haloarchaea. Histidine residues are indicated by a black background; conserved residues are indicated by gray shading. Hvo, H. volcanii; Hsa, Halobacterium salinarum; Hwa, Haloquadratum walsbyi; Hla, Halorubrum lacusprofundi; Nph, N. pharaonis. (B) Gene replacement construct pTA1106, containing pitA from N. pharaonis (pitANph) flanked by upstream (US) and downstream (DS) regions of H. volcanii pitA (pitAHvo). (C) Colony hybridization of 5-fluoroorotic acid (5-FOA)-resistant H. volcanii clones, after pop-in/pop-out gene replacement with pTA1106. H. volcanii pitA sequences (amplified with pitAF/pitAR primers) were used as a probe, and clones failing to hybridize therefore carried the N. pharaonis pitA gene. (D) Verification of pitA replacement in H1154 and H1155 using PCR with primers specific for either H. volcanii (pitAF/pitAR) or N. pharaonis (NphPitAF/NphPitAR) genes. H26 genomic DNA was used as a negative control and pTA1106 plasmid DNA as a positive control. (E) Map of the H. volcanii pitA region, indicating MluI sites and restriction fragment sizes of the native pitA gene (pitAHvo) and the replacement with N. pharaonis pitA (pitANph). (F) Verification of pitA replacement in H1154 and H1155 by MluI digestion and Southern blotting. The probe used is indicated in panel E.
The PitA ortholog from the haloalkaliphile Natronomonas pharaonis is unique in that it does not feature a high number of histidines in the central linker region. We reasoned that replacing H. volcanii pitA (pitAHvo) with the N. pharaonis gene (pitANph) would prevent copurification with His-tagged recombinant proteins. Gene replacement with the construct in pTA1106 (Fig. 2B) was carried out using the pop-in/pop-out technique (3, 7). Successful replacement of pitAHvo with pitANph was established by colony lift and verified by both PCR and Southern blotting of a restriction digest (Fig. 2C to F). The resulting H. volcanii strains, H1154 and H1155, with the pitANph gene replacement showed no obvious growth defects in Hv-YPC and Hv-Ca broth and agar.
Overexpression of 6×His-tagged RadA in pitAHvo and pitANph strains.
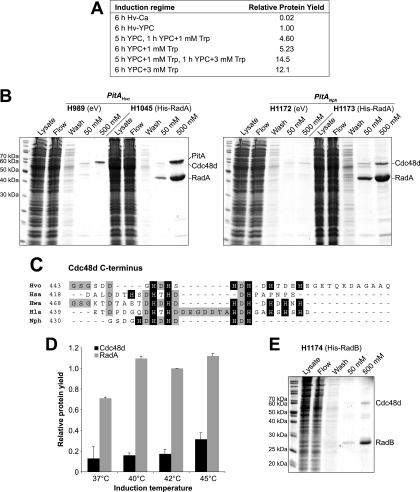
6xHis-tagged RadA was used to test the conditional protein overexpression system. RadA is the archaeal RecA family recombinase; it forms a nucleoprotein filament with single-stranded DNA and catalyzes strand exchange during homologous recombination (18, 40). Purification of recombinant 6×His-tagged RadA from E. coli has proved problematic. The protein copurifies with DNA, and removal of the latter requires harsh conditions (denaturation and benzonase treatment) that interfere with RadA activity (K. Bunting, personal communication).
The radA coding sequence was cloned in pTA963 and used to transform H. volcanii strains H98 and H1155. To determine the optimal induction regime for protein overexpression, the conditions shown in Fig. 3A were used. Since concentrations of >1 mM tryptophan affect the growth of H. volcanii (even without the RadA expression construct), it proved best to delay full induction (with 3 mM tryptophan) until 1 h before the cells were harvested. Metal affinity chromatography was used to purify 6×His-tagged RadA in the presence of 2 M NaCl, using an IMAC Sepharose column charged with Ni2+. We had previously established that the yield of 6×His-tagged protein from columns charged with Co2+ was very low when the buffers contained >1 M NaCl (data not shown). The 6×His-tagged RadA purified from the pitAHvo strain H1045 showed significant contamination with PitA, while this was not seen when 6×His-tagged RadA was purified from the pitANph strain, H1173 (Fig. 3B); the identities of both proteins were confirmed by mass spectrometry (Table 4).
FIG. 3.
Overexpression of RadA in a pitANph replacement strain. (A) Induction regime for protein overexpression using the tryptophan-inducible p.tnaA promoter. H1173 was grown for the times indicated in either Hv-Ca or Hv-YPC broth supplemented with tryptophan at the concentrations shown. 6×His-tagged RadA was purified, and the relative protein yield was determined by quantification of Coomassie blue-stained bands displayed on polyacrylamide gels as in panel B. (B) 6×His-tagged RadA was overexpressed in strains with either the native pitA gene (PitA Hvo; H1045) or replacement with N. pharaonis pitA (PitA Nph; H1173). The induction regime used was 5 h in Hv-YPC plus 1 mM tryptophan and 1 h in Hv-YPC plus 3 mM tryptophan at 42°C. The control strains H989 and H1172 contained the empty vector (eV) pTA963. 6×His-tagged RadA was purified from the soluble fraction (Lysate) by affinity chromatography on an Ni2+ chelating column, and samples were taken from the flowthrough (Flow) and after washing with 20 mM imidazole (Wash). Bound protein was eluted using 50 mM and 500 mM imidazole. Two additional bands were identified by mass spectrometry, PitA and Cdc48d. (C) Protein sequence alignment of the C termini of Cdc48d from selected species of haloarchaea. Histidine residues are indicated by a black background; conserved residues are indicated by gray shading. (D) Contamination by Cdc48d is reduced by growth at <45°C. 6×His-tagged RadA was overexpressed in H1173 and purified by affinity chromatography on a Ni2+ chelating column as in panel B. The relative yields of RadA and Cdc48d (standardized to the RadA yield at 42°C) were determined by quantification of Coomassie blue-stained bands displayed on polyacrylamide gels. The averages and standard errors of two experiments are shown. (E) Overexpression of 6×His-tagged RadB results in less contamination by Cdc48d. 6×His-tagged RadB was overexpressed in strain H1174 and purified by affinity chromatography on a Ni2+ chelating column as in panel B.
The absence of PitAHvo in cell lysates of H1173 revealed an additional contaminant, which was identified by mass spectrometry as Cdc48d (HVO_1907); Cdc48d is also present in H1045 but is difficult to distinguish in size from PitA (Fig. 3B). HVO_1907 is one of four H. volcanii isoforms of a putative AAA+ ATPase that show homology to the yeast Ccd48 and E. coli FtsH proteins. In eukaryotes, Cdc48 is a ubiquitin-dependent chaperone that is involved in protein degradation (29), while bacterial FtsH is a Zn2+ metalloprotease that degrades a set of short-lived proteins (24); both are thought to regulate cell division at the level of protein stability. Notably, H. volcanii Cdc48d features a histidine-rich C terminus (Fig. 3C) that is almost certainly responsible for its copurification on metal affinity chromatography columns.
We attempted to delete cdc48d by the pop-in/pop-out technique (3, 7), using the deletion construct pTA1180 (in strain H1228). We were unable to recover cells with a cdc48d deletion (data not shown), indicating that the gene is essential. Unfortunately, all known haloarchaeal homologs of Cdc48d feature the conserved histidine-rich C terminus (Fig. 3C), ruling out the gene replacement strategy that had proved successful in eliminating PitA contamination. Increased concentrations of imidazole in the binding and wash buffers (>20 mM) were not helpful, and the yield of 6×His-tagged RadA was reduced significantly without eliminating Cdc48d contamination. However, growth at lower temperatures was more successful; copurification of Cdc48d was reduced by 50% when cultures were grown at 40°C or 42°C, rather than the standard cultivation temperature of 45°C (Fig. 3D). Contamination by Cdc48d was also less pronounced during the purification of other recombinant proteins, such as 6×His-tagged RadB (Fig. 3E).
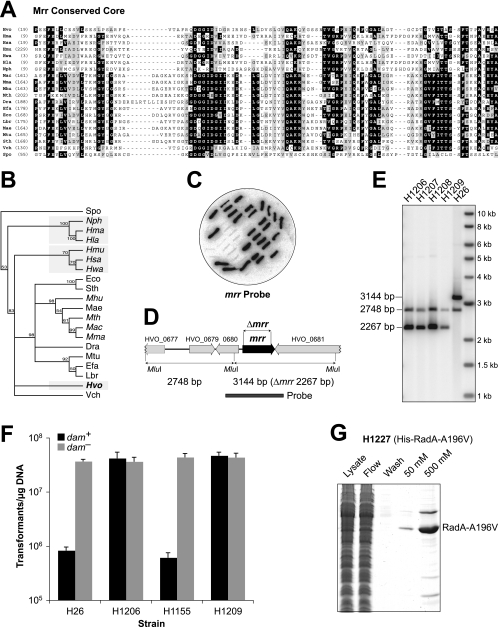
Deletion of the Haloferax volcanii mrr restriction endonuclease gene.
It has long been suspected that H. volcanii encodes a methylation-sensitive restriction enzyme that targets 5′-GATC-3′ sequences. Transformation of H. volcanii is much more efficient when the DNA is purified from E. coli dam mutants, which are unable to methylate 5′-GATC-3′, than from dam+ strains (21). Furthermore, transformation of H. volcanii with DNA methylated at 5′-GATC-3′ sequences leads to frequent plasmid loss, presumably due to cutting by restriction enzymes followed by recombination with chromosomal sequences (21). We hypothesized that the H. volcanii restriction endonuclease might be HVO_0682, which belongs to the Mrr family of enzymes that recognize and cleave N6-methyladenine- and 5-methylcytosine-containing DNA (9). The conserved core of HVO_0682 shows homology to other Mrr family members from archaea, bacteria, and yeast (Fig. 4A). However, in a phylogenetic tree, the H. volcanii Mrr protein does not group with homologs from other haloarchaea (Fig. 4B).
FIG. 4.
Deletion of the mrr gene improves transformation with dam-methylated DNA. (A) Protein sequence alignment of the conserved cores of Mrr homologs from selected archaea and bacteria and Schizosaccharomyces pombe SPAC824.03c (9). Conserved residues are indicated by gray shading, and identical residues are indicated by a black background. Hvo, H. volcanii; Hma, Haloarcula marismortui; Hsa, H. salinarum; Hmu, Halomicrobium mukohataei; Hwa, H. walsbyi; Hla, H. lacusprofundi; Nph, N. pharaonis; Mac, Methanosarcina acetivorans; Mhu, Methanospirillum hungatei; Mth, Methanothermobacter thermautotrophicus; Dra, Deinococcus radiodurans; Efa, Enterococcus faecalis; Eco, E. coli; Lbr, Lactobacillus brevis; Mae, Microcystis aeruginosa; Mtu, Mycobacterium tuberculosis; Sth, Salmonella thyphimurium; Vch, Vibrio cholerae; Spo, S. pombe. (B) Phylogenetic tree representing evolutionary relationships between Mrr homologs, constructed using neighbor joining and rooted using S. pombe SPAC824.03c. Support for individual branches is indicated by bootstrap values (1,000-fold resampling); values of <50% are not recorded. Pairwise distances between sequences are uncorrected. (C) Colony hybridization of 5-FOA-resistant H. volcanii clones after pop-in/pop-out gene deletion with pTA1150. H. volcanii mrr sequences (amplified with mrrF/mrrR primers) were used as a probe, and clones failing to hybridize therefore had the mrr gene deleted. (D) Map of the H. volcanii mrr region indicating MluI sites and restriction fragment sizes. (E) Verification of mrr deletion in H1206 to H1209 by MluI digestion and Southern blotting; the probe used is indicated in panel D. (F) Transformation efficiencies of Δmrr strains H1206 and H1209 compared to the mrr+ strains H26 and H1155 using a methylated pTA354 plasmid purified from an E. coli dam+ strain or unmethylated pTA354 DNA from an E. coli dam mutant (31). The averages and standard errors of three experiments are shown. (G) 6×His-tagged RadA(A196V) was overexpressed in the Δmrr strain H1209 and purified by affinity chromatography on a Ni2+ chelating column as in Fig. 3B.
We successfully deleted the mrr gene (from both pitAHvo and pitANph strains) by the pop-in/pop-out technique (3, 7), using the construct pTA1150. Successful deletion of mrr was established by colony lift and verified by Southern blotting of a restriction digest (Fig. 4C to E). The H. volcanii Δmrr mutants H1206 to H1209 showed no obvious growth defects in Hv-YPC and Hv-Ca broth and agar. To test the methylation-dependent restriction barrier, mrr+ and Δmrr strains (and the pitANph equivalents) were transformed with the shuttle vector pTA354 (31), which had been purified from either an E. coli dam mutant or a dam+ strain. The number of transformants obtained in an mrr+ strain was ∼50-fold higher with DNA from the E. coli dam mutant than with DNA from the dam+ strain, while the Δmrr mutants were transformed efficiently regardless of the source of the DNA (Fig. 4F). Therefore, deletion of the mrr gene removed the need to passage DNA through an E. coli dam mutant prior to transformation of H. volcanii. Furthermore, it allowed direct transformation of H. volcanii with a DNA ligation, which might be useful for cloning of protein overexpression constructs that are not tolerated by E. coli.
To test this possibility, we PCR amplified a mutant allele of radA [radA(A196V)], ligated the cut product with pTA963, and transformed the H. volcanii Δmrr strain H1209. Although only ∼200 transformants were obtained (from ∼1 μg vector DNA), 3 out of the 6 tested contained the correct plasmid construct (pTA1182). In contrast, we had failed on six previous occasions to construct this plasmid in E. coli, even though the PCR primers and restriction digests used were identical to those that had been used successfully to construct the radA+ overexpression plasmid pTA1041. This suggests that in E. coli, the p.tnaA promoter is leaky and RadA(A196V) is toxic. To confirm that 6×His-tagged RadA(A196V) could be purified from H. volcanii, we induced overexpression with tryptophan and carried out metal affinity chromatography as usual using strain H1227 (Fig. 4G). There was no significant difference in the level of expression of mutant RadA(A196V) compared to the wild-type protein.
DISCUSSION
Existing methods for expression of halophilic enzymes in E. coli are far from ideal; the protein is often insoluble and must be reactivated by denaturation and refolding, which is not always successful (11). Even when the protein is soluble, problems can arise. For example, H. volcanii RadA expressed in E. coli is bound to host DNA that cannot be removed easily. Purification from the native host is the obvious solution, but until now, systems for overexpression in H. volcanii have failed to capitalize on recent advances in haloarchaeal genetics (2, 39). For example, existing vectors feature constitutive gene promoters and rely on mevinolin resistance or novobiocin resistance markers for selection (30), which are far from ideal, since the use of these antibiotics impairs cell growth.
We have harnessed the conditional p.tnaA promoter to develop a series of plasmid vectors (Fig. 1) for rapid and strong induction of protein expression upon addition of tryptophan (26). Our constructs use pyrE2 and hdrB markers that maintain plasmids in rich medium (Hv-YPC) without the use of antibiotics (3); they are available with and without an in-frame 6×His tag for protein purification by metal affinity chromatography. This technique is ideal for purification of halophilic proteins, since it is compatible with the high salt concentrations used. However, it has until now been problematic in H. volcanii due to contamination by PitA, a protein with a histidine-rich linker region. We have replaced pitA with the gene from N. pharaonis, which encodes an ortholog lacking the histidine-rich region (Fig. 2). Proteins purified from the pitANph gene replacement strain are free of PitA contamination (Fig. 3). Interestingly, contamination by H. volcanii PitA is more pronounced in strains overexpressing 6×His-tagged RadA than in strains containing the empty vector (Fig. 3B). This is also true for Cdc48d, another histidine-rich contaminant that we identified. PitA and Cdc48d might be upregulated in response to cell stress, in this case due to overexpression of a recombinant protein, which is consistent with our observation that contamination by Cdc48d is reduced by growth at lower temperatures (Fig. 3D). Furthermore, proteins that are expressed at lower levels than 6×His-tagged RadA, such as 6×His-tagged RadB, exhibit less contamination by Cdc48d (Fig. 3E).
Finally, we have deleted the mrr gene, which encodes a homolog of the methylation-sensitive restriction enzyme Mrr (9). In contrast to the wild type, H. volcanii Δmrr strains exhibit high transformation efficiencies regardless of whether the plasmid DNA is methylated at 5′-GATC-3′ sites by E. coli Dam methylase (Fig. 4F). Therefore, Δmrr strains remove the need to passage plasmid DNA through an E. coli dam mutant before transformation (21). Not only does this save time, it also reduces the risk of incurring mutations during growth in E. coli dam mutants, which are deficient in DNA mismatch repair (27). Furthermore, H. volcanii Δmrr strains may be transformed directly with a DNA ligation, which permits the cloning of protein overexpression constructs that are toxic to E. coli.
We urge caution regarding the use of Δmrr strains in routine genetic experiments, since they have not been phenotyped extensively and deletion of the mrr gene might have pleiotropic effects. For example, Salmonella enterica serovar Typhimurium mutants with a constitutively active mrr gene display increased basal SOS induction (1). Some restriction modification systems act as selfish mobile genetic elements that resist their loss from the host, leading to cell death (25). However, mrr is unlikely to have been acquired by lateral gene transfer, since the synonymous codon usage of this gene is identical to the genome average (data not shown). We consider it more probable that Mrr acts in cellular defense against foreign DNA. Furthermore, we have demonstrated that Δmrr strains are fit for purpose when used in protein overexpression (Fig. 4G), thereby bringing closer the goal of an archaeal model organism that is suitable for both biochemistry and genetics.
Acknowledgments
We are grateful to the BBSRC, Wellcome Trust, Leverhulme Trust, and MRC for funding and to the Royal Society for a University Research Fellowship awarded to T.A.
We thank Stéphane Delmas, Bob Lloyd, Ed Bolt, Geoff Briggs, and Karen Bunting for helpful advice and Charles Daniels for the t.Syn sequence.
S.L., K.W., M.M., and T.A. wrote the paper; M.M. and T.A. designed the experiments; S.B., K.W., and T.A. performed the microbiological and biochemical experiments; S.L. carried out the mass spectrometry; and S.L. and T.A. analyzed the data.
Footnotes
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Aertsen, A., M. Tesfazgi Mebrhatu, and C. W. Michiels. 2008. Activation of the Salmonella typhimurium Mrr protein. Biochem. Biophys. Res. Commun. 367:435-439. [DOI] [PubMed] [Google Scholar]
- 2.Allers, T., and M. Mevarech. 2005. Archaeal genetics—the third way. Nat. Rev. Genet. 6:58-73. [DOI] [PubMed] [Google Scholar]
- 3.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70:943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altman-Price, N., and M. Mevarech. 2009. Genetic evidence for the importance of protein acetylation and protein deacetylation in the halophilic archaeon Haloferax volcanii. J. Bacteriol. 191:1610-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bab-Dinitz, E., H. Shmuely, J. Maupin-Furlow, J. Eichler, and B. Shaanan. 2006. Haloferax volcanii PitA: an example of functional interaction between the Pfam chlorite dismutase and antibiotic biosynthesis monooxygenase families? Bioinformatics 22:671-675. [DOI] [PubMed] [Google Scholar]
- 6.Berkner, S., and G. Lipps. 2008. Genetic tools for Sulfolobus spp.: vectors and first applications. Arch. Microbiol. 190:217-230. [DOI] [PubMed] [Google Scholar]
- 7.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blecher, O., S. Goldman, and M. Mevarech. 1993. High expression in Escherichia coli of the gene coding for dihydrofolate reductase of the extremely halophilic archaebacterium Haloferax volcanii. Reconstitution of the active enzyme and mutation studies. Eur. J. Biochem. 216:199-203. [DOI] [PubMed] [Google Scholar]
- 9.Bujnicki, J. M., and L. Rychlewski. 2001. Identification of a PD-(D/E)XK-like domain with a novel configuration of the endonuclease active site in the methyl-directed restriction enzyme Mrr and its homologs. Gene 267:183-191. [DOI] [PubMed] [Google Scholar]
- 10.Charlebois, R. L., W. L. Lam, S. W. Cline, and W. F. Doolittle. 1987. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. U. S. A. 84:8530-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connaris, H., J. B. Chaudhuri, M. J. Danson, and D. W. Hough. 1999. Expression, reactivation, and purification of enzymes from Haloferax volcanii in Escherichia coli. Biotechnol. Bioeng. 64:38-45. [PubMed] [Google Scholar]
- 12.Danson, M. J., and D. W. Hough. 1997. The structural basis of protein halophilicity. Comp. Biochem. Physiol. A Physiol. 117:307-312. [Google Scholar]
- 13.Deng, L., H. Zhu, Z. Chen, Y. X. Liang, and Q. She. 2009. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13:735-746. [DOI] [PubMed] [Google Scholar]
- 14.El Yacoubi, B., G. Phillips, I. K. Blaby, C. E. Haas, Y. Cruz, J. Greenberg, and V. de Crecy-Lagard. 2009. A Gateway platform for functional genomics in Haloferax volcanii: deletion of three tRNA modification genes. Archaea 2:211-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields, S., and M. Johnston. 2005. Cell biology. Whither model organism research? Science 307:1885-1886. [DOI] [PubMed] [Google Scholar]
- 16.Guss, A. M., M. Rother, J. K. Zhang, G. Kulkarni, and W. W. Metcalf. 2008. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2:193-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy, C. P., S. Haldenby, A. Brindley, D. A. Walsh, G. S. Briggs, M. J. Warren, T. Allers, and E. L. Bolt. 2006. Interactions of RadB, a DNA repair protein in archaea, with DNA and ATP. J. Mol. Biol. 358:46-56. [DOI] [PubMed] [Google Scholar]
- 18.Haldenby, S., M. F. White, and T. Allers. 2009. RecA family proteins in archaea: RadA and its cousins. Biochem. Soc. Trans. 37:102-107. [DOI] [PubMed] [Google Scholar]
- 19.Hartman, A. L., C. Norais, J. Badger, S. Delmas, S. Haldenby, R. Madupu, J. Robinson, H. Khouri, Q. Ren, T. Lowe, J. Maupin-Furlow, M. Pohlschroder, C. Daniels, T. Allers, F. Pfeifer, and J. A. Eisen. The complete genome sequence of Haloferax volcanii DS2 a model archaeon. PLoS One, in press. [DOI] [PMC free article] [PubMed]
- 20.Holmes, M. L., and M. L. Dyall-Smith. 2000. Sequence and expression of a halobacterial beta-galactosidase gene. Mol. Microbiol. 36:114-122. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, M. L., S. D. Nuttall, and M. L. Dyall-Smith. 1991. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J. Bacteriol. 173:3807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humbard, M. A., H. V. Miranda, J. Lim, D. J. Krause, J. R. Pritz, G. Zhou, S. Chen, L. Wells, and J. Maupin-Furlow. 2010. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humbard, M. A., G. Zhou, and J. A. Maupin-Furlow. 2009. The N-terminal penultimate residue of 20S proteasome alpha1 influences its N(alpha) acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J. Bacteriol. 191:3794-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, K., and Y. Akiyama. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59:211-231. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Large, A., C. Stamme, C. Lange, Z. Duan, T. Allers, J. Soppa, and P. A. Lund. 2007. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol. Microbiol. 66:1092-1106. [DOI] [PubMed] [Google Scholar]
- 27.Lobner-Olesen, A., O. Skovgaard, and M. G. Marinus. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8:154-160. [DOI] [PubMed] [Google Scholar]
- 28.Mevarech, M., F. Frolow, and L. M. Gloss. 2000. Halophilic enzymes: proteins with a grain of salt. Biophys. Chem. 86:155-164. [DOI] [PubMed] [Google Scholar]
- 29.Meyer, H., and O. Popp. 2008. Role(s) of Cdc48/p97 in mitosis. Biochem. Soc. Trans. 36:126-130. [DOI] [PubMed] [Google Scholar]
- 30.Nieuwlandt, D. T., and C. J. Daniels. 1990. An expression vector for the archaebacterium Haloferax volcanii. J. Bacteriol. 172:7104-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norais, C., M. Hawkins, A. L. Hartman, J. A. Eisen, H. Myllykallio, and T. Allers. 2007. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 33.Reuter, C. J., and J. Maupin-Furlow. 2004. Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl. Environ. Microbiol. 70:7530-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Santangelo, T. J., L. Cubonova, and J. N. Reeve. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.She, Q., C. Zhang, L. Deng, N. Peng, Z. Chen, and Y. X. Liang. 2009. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 37:92-96. [DOI] [PubMed] [Google Scholar]
- 38.Shimmin, L. C., and P. P. Dennis. 1996. Conserved sequence elements involved in regulation of ribosomal protein gene expression in halophilic archaea. J. Bacteriol. 178:4737-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soppa, J. 2006. From genomes to function: haloarchaea as model organisms. Microbiology 152:585-590. [DOI] [PubMed] [Google Scholar]
- 40.Woods, W. G., and M. L. Dyall-Smith. 1997. Construction and analysis of a recombination-deficient (radA) mutant of Haloferax volcanii. Mol. Microbiol. 23:791-797. [DOI] [PubMed] [Google Scholar]