Abstract
The structure and composition of the oocyst wall are primary factors determining the survival and hydrologic transport of Cryptosporidium parvum oocysts outside the host. Microscopic and biochemical analyses of whole oocysts and purified oocyst walls were undertaken to better understand the inactivation kinetics and hydrologic transport of oocysts in terrestrial and aquatic environments. Results of microscopy showed an outer electron-dense layer, a translucent middle layer, two inner electron-dense layers, and a suture structure embedded in the inner electron-dense layers. Freeze-substitution showed an expanded glycocalyx layer external to the outer bilayer, and Alcian Blue staining confirmed its presence on some but not all oocysts. Biochemical analyses of purified oocyst walls revealed carbohydrate components, medium- and long-chain fatty acids, and aliphatic hydrocarbons. Purified walls contained 7.5% total protein (by the Lowry assay), with five major bands in SDS-PAGE gels. Staining of purified oocyst walls with magnesium anilinonaphthalene-8-sulfonic acid indicated the presence of hydrophobic proteins. These structural and biochemical analyses support a model of the oocyst wall that is variably impermeable and resistant to many environmental pressures. The strength and flexibility of oocyst walls appear to depend on an inner layer of glycoprotein. The temperature-dependent permeability of oocyst walls may be associated with waxy hydrocarbons in the electron-translucent layer. The complex chemistry of these layers may explain the known acid-fast staining properties of oocysts, as well as some of the survival characteristics of oocysts in terrestrial and aquatic environments. The outer glycocalyx surface layer provides immunogenicity and attachment possibilities, and its ephemeral nature may explain the variable surface properties noted in oocyst hydrologic transport studies.
Previous studies of the survival of Cryptosporidium parvum under natural and laboratory conditions have shown that the oocyst phase is a durable stage in the life cycle of this apicomplexan parasite and is crucial for parasite transmission. A major public health problem is the resistance of oocysts to chlorine at normal concentrations used in water treatment systems. Oocysts have the reputation of being tough, durable structures; however, they can be inactivated by many physical and chemical disinfectants, including UV radiation, ozone, ammonia, high temperature, desiccation, freezing, and exposure to extreme alkaline or acidic conditions (10, 11, 12, 17, 18, 22, 35). Low temperatures above freezing extend oocyst viability and infectivity for very long times (12, 18, 19, 20, 35). Environmental temperature is a major factor controlling oocyst survival (23, 24, 32). While there have been many studies documenting the significance of temperature for oocyst survival and the influence of temperature on stored energy reserve utilization has been recognized (see references 10 and 32 for reviews), the effects of temperature on the key oocyst wall structures and macromolecules have not been well investigated.
While oocyst wall structure and macromolecular chemistry have been investigated in some detail (10, 14, 31, 33, 34, 41) and survival and transport in natural environments have been studied (5, 6, 8, 10, 23, 24), neither the underlying mechanisms by which oocysts resist environmental pressures nor the surface properties that control environmental transport have been well characterized (23, 24).
In this study, we investigated details of the ultrastructure and chemical composition of the C. parvum oocyst wall with the aim of understanding the key physical and chemical properties of the oocyst wall that may confer environmental resistance and affect environmental transport.
MATERIALS AND METHODS
Oocyst purification.
Oocysts were collected from the feces of Cryptosporidium-infected 6- to 20-day-old Holstein calves and purified by a continuous-flow differential density sucrose flotation method (17). After purification, oocysts were stored in distilled water containing 100 μg of streptomycin sulfate ml−1, 0.25 μg of amphotericin B ml−1 of suspension, and 100 U of penicillin G sodium ml−1 at 4°C.
In vitro excystation.
Approximately 5.0 × 109 oocysts were divided into aliquots containing 5.0 × 108 oocysts each and placed into 1.5-ml Eppendorf microcentrifuge tubes. Oocysts were washed once with 1 ml of Hanks' balanced salt solution (HBSS; Sigma Chemical Co., St. Louis, MO) and resuspended in 1 ml of acidified HBSS (100 μl of 1 M HCl in 10 ml of HBSS, pH 2.5) as described by Robertson et al. (36). The oocysts were incubated for 3 h at 37°C. After incubation, oocysts were pelleted by centrifugation, washed three times with 1 ml of 0.1 M phosphate-buffered saline (PBS; 0.028 M NaH2PO4·H2O, 0.072 M Na2HPO4, 0.145 M NaCl, pH 7.2), and washed once with 1 ml of HBSS. Oocysts were then resuspended in 100 μl of HBSS. Ten microliters of 2.2% sodium bicarbonate in HBSS and 10 μl of 1% sodium deoxycholate in Hanks' minimal essential medium (Sigma) were added to each sample. The samples were resuspended on a Vortex mixer and incubated at 37°C for 3 h (17). After incubation, the excysted suspensions were washed three times in distilled water and brought to a final volume of 1 ml.
Wall purification.
The 1-ml suspensions of excysted oocysts were added to 2.0-ml screw-cap microcentrifuge tubes (LPS, Rochester, NY) containing 1.5 g of 0.5-mm glass beads (BioSpec Products, Inc., Bartlesville, OK), to which 0.1 M PBS was added to obtain a final volume in each tube of 2.0 ml. The microcentrifuge tubes were then shaken at 1,600 rpm for 1.5 min on a Mini-Beadbeater cell disrupter (BioSpec Products, Inc.). After bead beating, the samples were centrifuged at 11,300 × g for 3 min to pellet the suspensions. Approximately 500 μl of solution was withdrawn, and 500 μl of 0.1% PBS-Tween (100 μl Tween 80 in 100 ml 0.1 M PBS) was added to each sample. The samples were resuspended and emptied into a 50-ml conical screw-top centrifuge tube (Sarstedt, Newton, NC). The suspension was underlain with 20 ml of sucrose (4°C) with a specific gravity of 1.22 and centrifuged on a model TJ-6 centrifuge with a TH-4 swinging-bucket rotor (Beckman) at 1,500 × g for 20 min. After centrifugation, the turbid sucrose-water interface was removed by a syringe and deposited into an empty 50-ml centrifuge tube. Thirty milliliters of 0.1 M PBS was added, and the sample was centrifuged at 1,500 × g for 15 min. The supernatant was removed by aspiration, leaving the pellet in approximately 2 ml of solution. The pellet was resuspended in the remaining 2 ml of solution and underlain with 20 ml of sucrose (4°C) with a specific gravity of 1.18. The sample was centrifuged at 1,500 × g for 20 min. After centrifugation, a visible band (of oocyst walls) was present approximately 5 mm below the sucrose-water interface. This band was removed by a syringe and placed into a 50-ml centrifuge tube. The sample was diluted in a volume of 50 ml with 0.1 M PBS and subjected to a vortex. The sample was then centrifuged at 1,500 × g for 15 min, and the supernatant was aspirated, leaving the pellet to be resuspended in 2 ml of solution. The wall suspension was transferred into a 2.0-ml microcentrifuge tube and washed three times with distilled water. The purity of the wall suspensions was verified by differential interference contrast (DIC) microscopy (3). The suspensions of purified walls were stored at −20°C until further analysis.
TEM.
Whole-oocyst suspensions and subsamples of purified oocyst walls were rinsed in 0.1 M Na-cacodylate buffer with 1.0 mM CaCl2 (pH 6.8) and centrifuged at 11,300 × g for 30 s. Samples were fixed in a pure formaldehyde-glutaraldehyde mixture containing each aldehyde at 2.5% in the Na-cacodylate buffer for 30 min at room temperature plus 3 h at 4°C. Samples were washed twice in cold buffer and fixed in 2% osmium tetroxide in buffer overnight at 4°C. After two washes in buffer, each sample was enrobed in 1.5% NuSieve agarose in buffer. Samples were then dehydrated with 10-min exposures to successive 10, 30, 50, 70, 90, and 100% concentrations of pure ethanol. The samples were treated with 100% ethanol for 25 min and fresh 100% ethanol for 15 min. The samples were infiltrated with Araldite-Embed 812 embedding resin for 3.5 days and formed into blocks overnight at 68 to 70°C. Thin sections were cut using a diamond knife mounted on an LKB ultramicrotome. Sections were picked up onto Formvar-carbon-coated or bare copper transmission electron microscopy (TEM) grids, contrasted with lead citrate and uranyl acetate, and examined with a Phillips 301 TEM operated at 80 kV with instrument magnifications of ×7 to 45,000.
Freeze fracture TEM.
Oocyst walls were prepared for freeze fracture according to the method described by Yoshikawa and Iseki (46). Oocyst walls were fixed for 30 min at room temperature with 0.05 M cacodylate buffer containing 2.5% glutaraldehyde, 2.5% formaldehyde, and 0.5 mM calcium, pH 6.8. After being washed in 10-, 5-, and 0.7-ml volumes of buffer, the sample was treated with 0.005 ml of glycerol, mixed, and allowed to stand for 10 min. A further 0.005 ml of glycerol was added, and the sample was mixed and allowed to stand for 5 min. Two hundred microliters of glycerol was then added, and the sample was allowed to stand for 1 h. To achieve a total percent glycerol of 31.7%, 0.1 ml of glycerol was added to the sample. The sample was centrifuged, and the supernatant was removed. After resuspension, the pellets were loaded into several gold sample cups and plunged into propane in a liquid nitrogen bath. Samples were then transferred into liquid nitrogen and processed in a Baltec 400K freeze-fracturing apparatus. Samples were viewed with a Philips 300 or 201 TEM at 100 kV.
Freeze-substitution TEM.
Purified whole oocysts were suspended in an 18% glycerol solution in distilled water for 2 h, centrifuged to form a pellet, and taken up into a drop of 2% warm, molten Noble agar. Samples were mixed on a Vortex mixer and then spread in a thin layer on cellulose ester filter paper. After the agar solidified, a small triangle was cut from the filter paper and plunge-frozen in liquid propane cooled by liquid nitrogen as described above for freeze fracture TEM. Frozen samples were transferred into a substitution mixture containing 2% osmium tetroxide and 1% uranyl acetate dissolved in acetone for substitution for 2 days at −90°C. The samples were then gradually warmed, washed twice over a period of 2 h in fresh acetone at 4°C, and infiltrated with and embedded in the Araldite 502-Embed 812 epoxy mixture. Thin sectioning and TEM were done as described above for TEM thin sectioning.
Light microscopy.
A Nikon E600 light microscope equipped with oil immersion 60× and 100× objective lenses and appropriate optics and filters for DIC, epifluorescence, and bright-field viewing was used for all light microscopy. Images were captured electronically using a Hamamatsu C5810 digital camera.
Ethanolic Alcian Blue staining.
A drop of the oocyst suspension was placed on a glass slide and mixed with a drop of ethanolic Alcian Blue staining solution (0.5% Alcian Blue 8 GX [Sigma], 40% ethanol, 5% acetic acid). The mixture was mounted under a glass cover and examined under bright-field or DIC optics. Clumps of oocysts were observed for several minutes, until the Alcian Blue reactions had reached their peak.
FITC-labeled concanavalin A staining.
A drop of the whole-oocyst or purified oocyst wall suspension was mixed with a drop of a 1-mg/ml solution of fluorescein isothiocyanate (FITC)-labeled concanavalin A (Sigma) dissolved in PBS. Samples were mounted on agar-coated slides and observed under DIC and epifluorescence optics using a filter combination appropriate for FITC.
Mg ANS staining.
A 1-mg/ml solution of magnesium anilinonaphthalene-8-sulfonic acid (Mg ANS; Sigma) was mixed with a drop of purified oocyst envelopes, and the sample was observed under epifluorescence optics using the FITC filter combination.
Protein analysis.
The total protein content of purified oocyst walls was determined by the Lowry protein assay using subsamples of the oocyst wall suspensions (13, 15).
Proteins in purified oocyst walls and their residues after lipid extraction were identified by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12). Samples were mixed 1:3 (vol/vol) with sample buffer (1.0 ml 0.5 M Tris-HCl, pH 6.8, 1.6 ml 10% [wt/vol] SDS, 0.8 ml glycerol, 0.4 ml 2-mercaptoethanol, 0.4 ml 5% bromophenol blue) and boiled for 10 min. Samples were then centrifuged at 11,300 × g for 30 s to remove any insoluble particulates. Broad-range protein standards (Kaleidoscope) were loaded at the concentration recommended by the manufacturer (Bio-Rad). Sample aliquots of 15 μl were loaded onto Ready Gel 4 to 20% polyacrylamide gels (Bio-Rad) and run at 200 V for 45 min in a Mini-Protean II gel apparatus (Bio-Rad). The gels were rinsed three times with distilled water and stained with Gel Code reagent (Pierce, Rockford, IL) for 1 h. After staining, the gels were washed in distilled water for 3 h to remove background staining and documented with a charge-coupled device camera (43).
Carbohydrate analysis.
A subsample of the purified oocyst wall suspension was lyophilized, and the dry weight was determined. The pellet was placed in a glass ampoule to which 1 ml of 8 M trifluoroacetic acid (Sigma) was added. The ampoule was sealed by heating in a methane-oxygen flame and incubated for 1 h at 110°C. After incubation, the ampoule was opened and the trifluoroacetic acid was evaporated under a stream of nitrogen gas. The remaining residue was redissolved in 1 ml of pyridine (Mallinckrodt, Phillipsburg, NJ).
A 50-μl subsample was removed, mixed with 50 μl of N-methyl-bis(trifluoroacetamide) (BSTFA; Sigma), and incubated at 70°C for 15 min to form trimethylsilyl (TMS) derivatives of the monosaccharides. After incubation, the sample was analyzed by gas chromatography-mass spectrometry (GC-MS).
A subsample of purified walls in pyridine was subjected to evaporation under a stream of nitrogen gas, and the residue was resuspended in 0.5 ml of distilled water. Twenty microliters of the sample was injected into a Rabbit high-performance liquid chromatograph (HPLC; Rainin Instrument Co., Emeryville, CA) equipped with a 100- by 7.8-mm HPLC fast acid analysis column. The carrier liquid was 6.5 mM H2SO4 at a flow rate of 1 ml/min. Compounds were detected with a Knauer differential refractive index detector, and chromatograms were recorded on a PerkinElmer LCI-100 laboratory computing integrator.
The total hexose content was measured by the anthrone reaction (13).
Lipid extraction.
To prevent contamination and false identifications, all glassware was acid washed to remove traces of organic matter and all solvents were checked for impurities by GC-MS. Total lipids were extracted from suspensions of purified walls by a modification of the Bligh and Dyer procedure (4, 21). Standards of palmitic acid and 12-hydroxyoctadecanoic acid were used as controls. Oocyst wall suspensions were pelleted and resuspended in 100 μl of distilled water, to which 375 μl of 2:1 (vol/vol) methanol-chloroform was added. Samples were left at room temperature for 5 h with intermittent shaking. Samples were then centrifuged at 11,300 × g for 1 min to pellet wall residues, and the supernatants were decanted into a 2-ml sterile microcentrifuge tube. The pelleted residue was resuspended in 475 μl of 2:1:0.8 (vol/vol/vol) methanol-chloroform-water, subjected to a vortex, and centrifuged in a microcentrifuge at 11,300 × g for 1 min. The two supernatants were combined, and 250-μl volumes of chloroform and water were added to the combined supernatants. The samples were resuspended and centrifuged at 11,300 × g for 2 min. After centrifugation, the aqueous phase was removed and the bottom organic phase was diluted with four drops of benzene to remove residual water. The samples were heated in a temperature block at 30 to 35°C to evaporate the organic solvents. The dry residues were immediately resuspended in 300 μl of chloroform and stored in a desiccator at −20°C until further analysis.
Formation of FAMEs.
Total lipids were converted into their respective fatty acid methyl esters (FAMEs) by acid methanolysis (13). The chloroform solvent was evaporated under a stream of nitrogen gas at room temperature. Once the samples were dry, 1 ml of 5% (wt/vol) H2SO4 in methanol was added to each sample. Samples were incubated in a temperature block at 70°C for 2 h. After samples were cooled to room temperature, 1 ml of n-hexane (Fisher) was added to each sample and the samples were vigorously shaken for 10 min. The hexane phase was removed with a glass Pasteur pipette and placed in an acid-washed screw-cap glass vial. The remaining methanol phase was reextracted with 1 ml of n-hexane, and the hexane phases were pooled for GC-MS analysis.
TMS derivatization of FAMEs.
To ensure that all methyl esters were suitable for GC-MS analysis and that hydroxyl groups were protected, subsamples of the pooled methyl esters were subjected to trimethylsilylation (2) before being injected into the GC-MS system.
GC-MS analysis.
FAMEs were analyzed using a 5890 series II gas chromatograph (Hewlett-Packard, Avondale, PA) equipped with a Hewlett Packard 5971 mass selective detector and a 30-m by 0.25-mm-diameter Rtx-5 fused-silica capillary column (Restek, Bellefonte, PA). Splitless injections of 0.9 μl were made with a 7001 series syringe (Hamilton, Reno, NV). The column temperature was programmed to ramp from 100 to 270°C at 5°C/min; the injector temperature was kept at 250°C, and the detector temperature was maintained at 280°C. Component peaks were identified with HP Chemstation software and the HP National Institute of Standards and Technology/Environmental Protection Agency/National Institutes of Health mass spectral database. FAME standards (GLC-30 FAME mix; Supelco, Bellefonte, PA) and hydrocarbon standards (diesel range organics [DRO] mix; Florida total recoverable petroleum hydrocarbon [TRPH] standard [Restek]) were utilized to confirm retention times for identified wall components.
Alternative fatty acid identification.
As an alternative method of identifying fatty acids present in oocyst walls, total lipid extract was analyzed with the Sherlock microbial identification system according to the saponification and methylation protocol suggested by the manufacturer (MIDI, Newark, DE).
Thin-layer chromatography (TLC).
To further characterize the lipids present in oocyst walls, subsamples of total lipid extract were applied to silica gel plates (20 by 20 cm, with a silica layer thickness of 250 μm; Whatman Inc., Clifton, NJ). Plates were developed in a 30:8:1 (vol/vol/vol) chloroform-methanol-water mixture as outlined by Schrum et al. (38). Glycolipids were identified by using alpha-naphthol and heating at 105°C. Phospholipids were identified with Phospray reagent (Supelco), while amino compounds were detected with ninhydrin reagent (Supelco). Choline-containing compounds were detected with Dragendorff stain (Supelco).
RESULTS
Ultrastructure and surface polymers (glycocalyx) of whole oocyst walls.
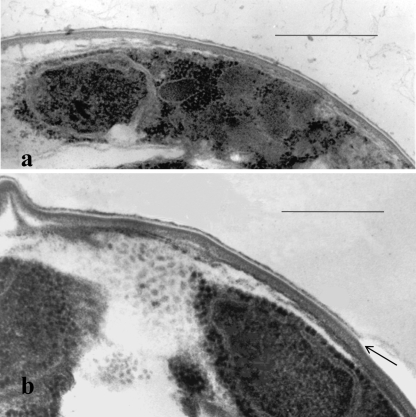
TEM analysis of thin-sectioned glutaraldehyde-formaldehyde-fixed whole oocysts confirmed previous results from others (see reference 10 for a review) showing three electron-dense layers and an intermediate electron-translucent layer. In conventional glutaraldehyde-formaldehyde-fixed samples (Fig. 1 and 2a), an irregular outer electron-dense layer (8.5 ± 0.6 nm) was separated from two underlying dense layers (13.0 ± 0.5 nm and 28.6 ± 1.6 nm, respectively) by a thin electron-translucent middle layer (4.0 ± 0.2 nm). The average thickness of the oocyst wall measured in perpendicular cross sections was 54.1 ± 4.1 nm. The irregular outer dense layer and translucent middle layer appeared to overlay the complex suture structure (the oocyst wall structure that opens upon excystation and from which four infective sporozoites contained within can exit), with two prominent electron-dense vertical bands separated by an approximately 50-nm zone of medium electron density within the two inner dense wall layers (Fig. 2a).
FIG. 1.
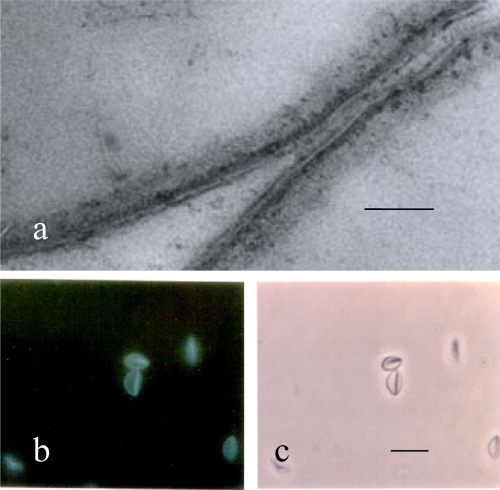
Transmission electron micrographs of thin sections of peripheral portions of glutaraldehyde-formalin-fixed C. parvum whole oocysts showing a peripheral sporozoite with a nucleus (a) and perpendicular cross sections through the oocyst wall (a and b). Note the four-layer structure of the wall and, in the outer portion of the wall, a membrane-like electron-translucent layer, which appears to be split along the axis of the translucent zone (as indicated by the arrow in panel b). Bars, 0.5 μm.
FIG. 2.
(a and b) Transmission electron micrographs of thin sections of peripheral portions of a glutaraldehyde-formalin-fixed C. parvum whole oocyst showing the wall layers with a suture complex embedded in the inner layers beneath the middle translucent layer and outer dense layer (a) and a freeze-substituted whole oocyst showing a split in the weak middle translucent layer of the wall (compare with Fig. 1b) and an extensive polymer matrix (glycocalyx) extending outward from the translucent layer of the wall (b). (c) Freeze fracture replica of a whole oocyst showing a smooth convex fracture face containing the zipper-like suture complex seen in cross section in panel a. Bars, 0.5 μm.
Freeze fracture preparations of whole oocysts revealed fracture sites that appeared to be uniformly smooth surfaces (Fig. 2c). The absence of particles in this smooth fracture face indicates that there were no translayer proteins present in the fractured layer and no active transoocyst transport. The fracture apparently occurred in the membrane-like electron-translucent layer seen in thin sections of both conventionally fixed and freeze-substituted oocysts (Fig. 1 and 2a and b). The smooth fracture faces seen in freeze-fractured oocysts (e.g., in Fig. 2c) likely represented fractures along the electron-translucent middle layer, which was often observed to split along its weak axis in thin sections of conventionally fixed and freeze-substituted oocysts (Fig. 1b and 2b, arrows). Fractures along smooth, convex oocyst surfaces (Fig. 2c) revealed new details of the suture structure, which appeared in these preparations as a broad zipper-like structure containing a central 50-nm groove bordered by parallel zones that formed a band approximately 165 nm wide. The central 50-nm groove in the freeze-fractured walls corresponded in dimensions to the two vertical dense bands of the complex suture structure seen in thin sections (Fig. 2a).
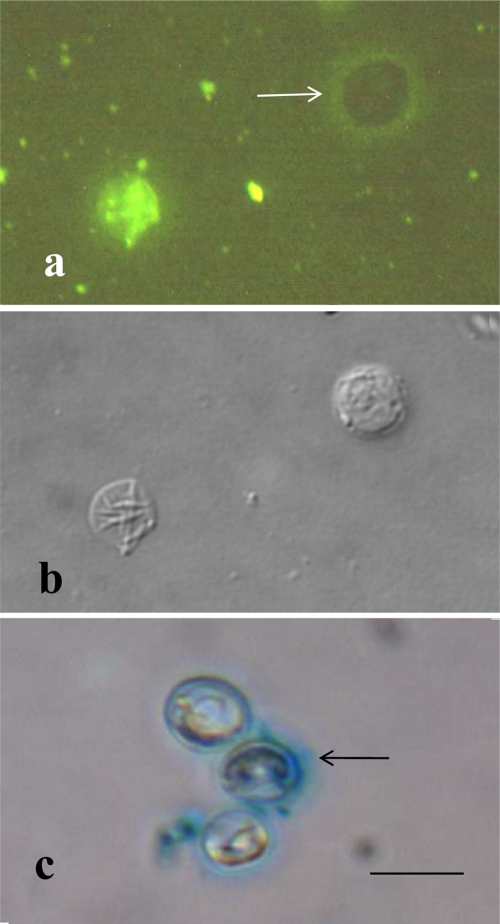
Freeze-substitution did not adequately preserve wall structures, and internal structures of whole oocysts were not preserved in our samples. However, freeze-substitution did preserve an extensive layer of polymers (a glycocalyx) external to the electron-dense and translucent middle layers (Fig. 2b). Comparison of an epifluorescence image (Fig. 3a) with a DIC image of the same field (Fig. 3b) showed that FITC-labeled concanavalin A stained excysted oocyst walls brightly. The flattened sac in the DIC image (Fig. 3b) is brightly stained in the epifluorescence image (Fig. 3a), but the unexcysted oocyst (Fig. 3a, arrow, and b) is not stained. Instead, a halo of FITC-stained material surrounds the unstained oocyst, suggesting that the concanavalin A was binding to glucose or mannose residues in a polysaccharide matrix bound to the oocyst surface. Ethanolic Alcian Blue staining confirmed the presence of an acidic polysaccharide matrix (Fig. 3c, arrow) associated with some but not all whole oocysts. The results of freeze-substitution and staining are consistent with the interpretation that an ephemeral glycocalyx matrix of acidic polysaccharides originally covered the surfaces of whole unpurified oocysts and that this glycocalyx would be removed to some degree during the oocyst purification process.
FIG. 3.
(a and b) Paired epifluorescence (a) and DIC (b) images of the same microscopic field treated with FITC-labeled concanavalin A. The flattened empty oocyst sac in panel b is brightly stained in panel a, indicating that concanavalin A was bound to polysaccharide material in the wall. The intact oocyst in panel b is not stained, showing that the concanavalin A did not react strongly with the outside of the oocyst. Instead, a halo of lightly stained glycocalyx material (arrow) surrounds the intact oocyst in panel a. (c) Bright-field image confirming that the oocysts stained with ethanolic Alcian Blue, which revealed a matrix of acidic polymers (arrow) surrounding some but not all oocysts. Bar, 0.5 μm.
Ultrastructure and Mg ANS staining of purified oocyst walls.
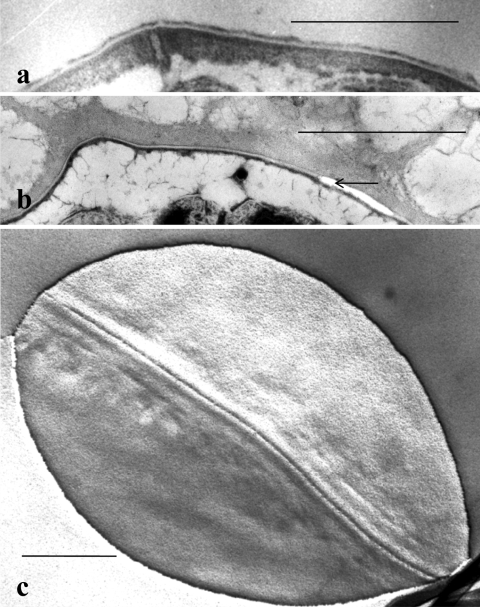
The fixed walls in perpendicular cross sections (Fig. 4a) showed the same four layers, with dimensions similar to those seen in the whole oocysts described above. The purified walls stained uniformly with Mg ANS (Fig. 4b), a fluorescent hydrophobic dye known for its affinity for hydrophobic proteins (40), and they were seen to be free of residual bodies (membrane-bounded structures containing metabolites for the infective sporozoites within the oocyst [10]) when examined by phase-contrast microscopy (Fig. 4c).
FIG. 4.
(a) Thin-section electron micrograph of purified oocyst walls showing two parallel wall sections joined by their outer layers. Note the similarity of the purified wall profiles to those of whole oocysts in Fig. 1 and 2a. Bar, 100 nm. (b and c) Paired epifluorescence (b) and phase-contrast (c) light micrographs showing uniform staining of purified oocyst walls with Mg ANS (b) and the absence of residual bodies in the purified walls (c). Bar, 0.5 μm.
SDS-PAGE analysis of proteins.
Residual-body-free, purified oocyst walls (Fig. 4) contained 7.5% total protein. Five major protein bands from the purified oocyst walls at approximately 87.5, 65.3, 42.5, 17.0, and 11.0 kDa were identified by SDS-PAGE. After lipid extraction, 12 major bands were found at approximately 173.2, 87.5, 77.8, 65.3, 55.8, 50.6, 46.0, 38.5, 17.0, 12.4, 11.0, and 10.8 kDa (data not shown).
Identification of oocyst wall carbohydrates.
C. parvum oocyst walls were determined to contain approximately 2% hexose. GC-MS analysis identified glucose, galactose, mannose, talose, glucofuranose, d-glucopyranose, and D-mannopyranose in the purified oocyst walls. The presence of both glucose and galactose was confirmed by HPLC.
Identification of fatty acids, hydrocarbons, and alcohols.
GC-MS analysis of the total lipid extract from purified oocyst walls yielded saturated fatty acids, including hexadecanoic acid, octadecanoic acid, and 11,14-eicosadienoic acid (Table 1). Straight-chain aliphatic hydrocarbons ranging from 11 to 34 carbons in length were also identified (Table 2). Exact identifications for C21 and larger hydrocarbons, however, could not be obtained because their retention times did not match those of analytical standards, although the mass spectrometer identified these hydrocarbons with a high-quality index (>80). Several long-chain alcohols, including 2-decanol and 1-tetracosanol, were also identified in oocyst walls (Table 3).
TABLE 1.
Fatty acid compositions of C. parvum whole oocysts and oocyst walls
| Fatty acid | Melting point (°C)a | Occurrenceb in: |
|
|---|---|---|---|
| Oocyst walls | Whole oocysts | ||
| Dodecanoic acid | 44.2 | + | |
| Tetradecanoic acid | 53.9 | + | |
| 6-Hexadecenoic acid | ND | + | |
| cis-9-Hexadecenoic acid | ND | + | |
| Hexadecanoic acid | 63.1 | + | + |
| 6-Octadecenoic acid | 33.0 | + | |
| cis-9-Octadecenoic acid | 16.3 | + | |
| 9-(Z)-Octadecenoic acid | 44.5 | + | |
| Octadecanoic acid | 69.6 | + | + |
| 3-OH-octadecanoic acid | ND | + | |
| 9,15-Octadecadienoic acid | ND | + | |
| cis-Eicosadienoic acid | ND | + | |
| 11,14-Eicosadienoic acid | ND | + | |
| 3-OH-tetracosanoic acid | ND | + | |
Data are from CRC Handbook of Microbiology (26). ND, not determined.
+ indicates the presence of the fatty acid.
TABLE 2.
Hydrocarbon compositions of C. parvum whole oocysts and oocyst walls
| Hydrocarbona | Melting point (°C)b | Occurrencec in: |
|
|---|---|---|---|
| Oocyst walls | Whole oocysts | ||
| C11H24 (undecane) | −25.6 | + | |
| C12H26 (dodecane) | −9.7 | + | |
| C15H32 (pentadecane) | 10.0 | + | + |
| C16H34 (hexadecane) | 18.1 | + | + |
| C17H36 (heptadecane) | 22.0 | + | + |
| C19H38 (nonadecane) | 32.0 | + | |
| C20H42 (eicosane) | 36.4 | + | |
| C21H44 b | 40.4 | + | + |
| C22H46 b | 44.4 | + | |
| C24H50 b | 51.1 | + | |
| C31H64 b | 66.0-81.0 | + | |
| C32H66 b | 66.0-81.0 | + | |
| C34H70 b | 66.0-81.0 | + | |
Identifications are approximate for C21H44 through C34H70. Although GC-MS identified these hydrocarbons with a high-quality index (>80), retention times did not match those of analytical standards.
Data are from Rodd's Chemistry of Carbon Compounds (37).
+ indicates the presence of the hydrocarbon.
TABLE 3.
Fatty alcohol compositions of C. parvum whole oocysts and oocyst walls
| Fatty alcohol | Melting point (°C)a | Occurrenceb in: |
|
|---|---|---|---|
| Oocyst walls | Whole oocysts | ||
| 2-Decanol | ND | + | |
| Dodecanol | 24 | + | |
| Tridecanol | ND | + | + |
| 1-Octadecanol | 56-60 | + | |
| 1-Tetracosanol | 77-79 | + | |
| Cholesterol | ND | + | |
Data are from The Merck Index (44). ND, not determined.
+ indicates the presence of the fatty alcohol.
Identification of lipids.
Alpha-naphthol staining of TLC plates revealed the presence of glycolipids. Ninhydrin, Phospray reagent, and Dragendorff stain failed to reveal amino-containing compounds, phospholipids, and choline-containing compounds, respectively; however, these compounds may be absent or present at levels below our limits of detection.
DISCUSSION
C. parvum oocysts have been shown to be resistant to several environmental pressures commonly encountered in natural waters and sediments (11, 12, 17, 18, 35). An analysis of the chemical composition and structure of the oocyst wall is fundamental to understanding the mechanism by which oocysts remain robust in natural environments.
To ensure an accurate analysis of oocyst wall chemistry, our purification protocols did not include harsh detergents or solvents that may alter or degrade proteins and lipids in the oocyst wall. Oocyst purification techniques commonly utilize potassium dichromate, chloroform, or ethyl ether (9). The effects of these compounds on oocyst wall chemistry are not known, but evidence from electrophoretic mobility studies suggests that oocysts purified with formalin, ethyl acetate, and Percoll have their surface chemistry altered (5). Brush et al. (5) demonstrated that the electrophoretic mobility of C. parvum oocysts purified from feces by methods that maintained the integrity of the outer glycocalyx layer of the oocyst wall did not resolve an isoelectric point between pH 2 and 10. Oocysts purified from feces by methods using chemicals such as ethyl acetate and Percoll-sucrose had different electrophoretic mobilities from those purified by methods maintaining glycocalyx integrity and had an isoelectric point of 2.37, indicating that the outer glycocalyx layer was altered. Because the adhesive properties of oocysts are governed by their electrophoretic mobility (5, 6), alterations of the outer glycocalyx layer of the oocyst wall by artificial or natural environmental processes will affect their interactions with soil and sediment particles and, thus, affect their hydrologic transport in soil and aquifer materials. The demonstrated immunogenicity of the oocyst glycocalyx (31) may also be altered by exposure to environmental pressures, which subsequently affects oocyst pathogenicity.
Conventional thin-section electron microscopy analysis of whole oocysts revealed a four-layered structure in the oocyst wall (Fig. 5) with structural dimensions similar to those reported previously for both whole oocysts and purified oocyst walls (10, 14, 33, 34). The oocysts analyzed were the thick-walled oocysts that sporulate within a host cell and are passed in feces (7). Thin-walled oocysts, which develop concurrently with thick-walled oocysts, are characterized by a single-unit membrane, and are a source of autoinfection (7), were not observed. We did, however, note slight differences between the cross-sectional dimensions of layers in purified walls and those in whole-oocyst walls (compare Fig. 1 and 2 with 4a, and see data for and figures showing purified walls in references 33 and 34). On average, purified walls appeared to be somewhat thinner in cross section than walls of whole oocysts prepared for electron microscopy by the same protocol. This finding may have been due to enhanced extraction effects of detergents employed during the wall purification protocol or to effects of dehydration solvents used in preparing samples for electron microscopy.
FIG. 5.
Proposed model for the C. parvum oocyst wall based on data presented in this paper.
We also noted that the prominent electron-translucent wall layer was frequently the location of separations in thin sections of both conventionally fixed and freeze-substituted walls (Fig. 1b and 2b, arrows), suggesting that this layer was a weak point in the wall structure. A fracture along the plane of this layer probably accounts for the smooth appearance of the oocyst wall in freeze-fractured oocysts (Fig. 2c), which revealed clear views of the suture structure in the fracture plane. The arrangement of wall layers relative to the suture structure, including the suture complex, was clearly seen in the cross-sections of the wall (Fig. 2a). Such sections clearly showed that the electron-translucent layer covers the entire suture structure, which appears to be embedded primarily in the structural polysaccharide layer of the wall. Directly beneath the translucent layer is a thin, electron-dense layer (Fig. 1, 2a, and 4) which appears to be the locus of the main structural proteins of the wall (see below) and is thought to provide much of the strength and flexibility characteristic of the oocyst wall (14, 33, 39, 41).
The existence of a glycocalyx on the outer surface of the wall has been noted previously by numerous investigators (10, 14, 31, 33, 34); however, the actual extent of the glycocalyx matrix and its structural and chemical properties have not been determined precisely. This is due likely to the difficulty of preserving and analyzing the delicate glycocalyx structure composed of heteropolymers. Nanduri et al. (31) reported that the oocyst glycocalyx is composed mainly of glucose, with galactose, mannose, xylose, and ribose being the next most abundant components. The freeze-substitution fixation technique we used clearly did preserve a substantial glycocalyx matrix extending outward from the outer surfaces of the walls of many oocysts (Fig. 2b). However, this extended glycocalyx matrix was found to be ephemeral. It surrounded many but not all freeze-fractured oocysts. It appeared as a thin, electron-dense layer of variable thickness in conventionally fixed oocysts (Fig. 1 and 2a). We did find evidence of a halo of glycocalyx material around FITC-concanavalin A-stained unexcysted oocysts (Fig. 3a, arrow). Furthermore, a negatively charged glycocalyx was observed in clumps of some purified whole oocysts stained with ethanolic Alcian Blue (Fig. 3c). These results suggest that the glycocalyx surrounding oocysts can be extensive but that it is easily stripped off during preparations for microscopy. It is likely that the surface chemistry of oocysts, as well as their immunogenicity, may change with the loss of the majority of the glycocalyx matrix. Such changes may affect the net surface charge and, thus, the properties of oocyst binding to particles as well as host tissue. Changes in the amount of surface glycocalyx would have profound effects on the transport behavior of oocysts in natural and engineered systems. Stripping off the majority of the glycocalyx matrix could alter the oocyst's interaction with an environmental matrix, especially if the underlying layer is hydrophobic, as would be expected since the underlying electron-translucent layer appeared to be composed of complex lipids as discussed below. Such changes may account for the variable surface properties of oocysts prepared by different purification protocols (5, 6, 8, 24).
Proteins present in Cryptosporidium isolates have been characterized previously (28); however, amylopectin granules and residual bodies were not removed from oocyst walls prior to SDS-PAGE analysis. Because the residual bodies have not been fully characterized, SDS-PAGE analysis of excysted oocysts containing residual bodies may produce bands that are not truly associated with the oocyst wall. Our analysis of oocyst wall proteins yielded 5 major bands, while wall residues obtained after lipid extraction yielded 12 major bands. Our results can be compared to those of Harris and Petry (14), who reported finding seven protein bands under 66 kDa at 54, 48, 46, 43, 38, 21, and 14.5 kDa in samples of similarly purified oocyst walls. While it is difficult to reconcile the differences between our results and theirs because of the different methods we employed, it is significant that our results for post-lipid extraction wall residues showed 12 major proteins, with nine bands under 66 kDa, similar to the results of Harris and Petry (14), who for starting material used oocysts purified after treatment with ethyl ether to extract lipids. Our results for purified walls not subjected to extraction showed only four bands under 66 kDa. It is likely that the increased number of bands found in post-lipid extraction oocyst walls may have resulted from the release of lipoproteins during the lipid extraction process.
A gene encoding a Cryptosporidium oocyst wall protein (COWP) has been successfully cloned into Escherichia coli, and the protein has been localized to the inner wall layers by immunogold electron microscopy (39). The COWP was characterized by tandem arrays of cysteine-rich domains, and Spano et al. (39) proposed that these intermolecular disulfide bonds are a source of oocyst wall rigidity. Templeton et al. (41) identified nine additional COWP genes, all encoding products characterized by tandem arrays of cysteine-rich domains, and demonstrated the expression of these genes in the in vitro development of the oocyst wall. They further demonstrated that the oocyst wall protein COWP8, like the COWP that Spano et al. characterized (39), is located inside the oocyst wall of C. parvum. Based on the amino acid sequence, the mature COWP polypeptide was predicted to have a molecular mass of 174 kDa (39). Our analysis of wall residues yielded a protein band meeting this molecular mass criterion. Future N-terminal sequencing of this protein monomer should confirm the identity of this oocyst wall protein.
To our knowledge, the carbohydrate composition of purified, intact oocyst walls has not been determined. Oocyst walls were found to contain several polysaccharides. While these compounds may be present as a polysaccharide matrix, it is likely that these sugars are associated with the glycolipids identified by TLC, noted to be components of the glycocalyx (31). Although Nanduri et al. (31) detected the presence of N-acetyl-d-galactosamine in the oocyst glycocalyx, we detected no amino sugars.
The phospholipid composition of whole oocysts has been well documented. Whole oocysts have been reported to contain phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine, sphingomyelin, and cardiolipin (30). We were unsuccessful in locating these lipids in samples of oocyst walls. The most likely explanation is that the concentrations of these compounds were below our limits of detection.
This study is the first report of the fatty acid composition of purified oocyst walls. Fatty acids in whole oocysts have been characterized previously (30). Our results indicate that whole oocysts and oocyst walls have many fatty acids in common (Table 1). Hexadecanoic, octadecanoic, 9-octadecenoic, and 11,14-eicosandienoic acids are the most abundant fatty acids in the oocyst wall. The abundances of these fatty acids corresponded with the neutral lipid fatty acid profile of whole oocysts as described by Mitschler et al. (30).
Fatty alcohols in another coccidian, Eimeria tenella, may confer resistance to chemical treatments and dehydration (42). Fatty alcohols were not found in the neutral lipid fraction from whole oocysts (30); however, we have found at least two fatty alcohols in whole oocysts and four fatty alcohols in the oocyst wall (Table 3). Separation of the total lipid extract on a silica column and analysis of each fraction would prove valuable; however, the substantial amount of wall material that would be required for this procedure would be difficult to obtain. Cholesterol was also found in whole oocysts (Table 3) (30) but not in oocyst walls. This finding indicates that cholesterol is localized to either sporozoites or residual bodies.
Acid-fast staining has been utilized to detect C. parvum oocysts in fecal specimens (9); therefore, the possibility exists that oocyst walls and mycobacterium cell walls may have similar compositions and structures. Mycolic acid, a complex branched-chain hydroxy lipid, is the primary component required for acid-fast staining (29). The cell wall of Mycobacterium chelonae has been found to contain 16:0, 16:1, and 18:1 fatty acids and glycopeptidolipids that may contain C28 to C34 fatty acids (27). Our analyses demonstrate that the C. parvum oocyst wall lipid profile shares many of these fatty acids.
Based on the data from freeze fracture and transmission electron micrographs, we propose a structural model for the C. parvum oocyst wall (Fig. 5). The smooth fracture face (Fig. 3) suggests that the fracture occurred within the transparent lipid-like layer depicted in the model. The dark band separating the proposed lipid layer from the inner carbohydrate layer strongly resembles protein layers characterized in other transmission electron micrographs.
The influence of oocyst wall chemistry on impermeability in natural environments may be directly related to temperature. In previous studies, oocyst infectivity has been shown to decrease upon exposure to temperatures ranging from 50 to 80°C (9). In addition, oocyst viability has been found to decrease with increasing temperature from 30 to 50°C (20). The melting points of the fatty acids and hydrocarbons present in oocyst walls are within the range of temperatures that produce decreases in viability. The melting of the lipid components in the oocyst wall may be responsible for increased wall permeability and subsequent decreases in viability and infectivity. Mycobacterial cell walls are generally impermeable to lipophilic antibiotics and chemotherapeutic agents (16). A relationship between temperature and loss of impermeability in M. chelonae, in which cell wall lipids experience significant thermal changes between 30 and 60°C, has been established previously (27). The similarities in lipid composition and acid-fast staining between C. parvum and Mycobacterium suggest that the two organisms may share chemical and structural mechanisms that confer resistance to natural and artificial agents. The outer glycocalyx and lipid layer of the C. parvum oocyst wall appear to be analogous to the asymmetrical bilayer of M. chelonae, which Liu et al. (27) described as having a moderately fluid outer layer and an inner layer of extremely low fluidity. Both structures appear to govern wall permeability and provide the organisms' robust ability to survive.
Genomic analyses (1, 25, 45, 47) show that the small genomes of C. parvum and its human counterpart, C. hominis, like those of other Apicomplexa, are ripe with evidence of horizontal gene transfer. However, C. parvum and C. hominis have highly streamlined metabolic-function genes (47), which makes the parasites highly dependent on nutrient salvage from the animal host for survival in their native environment, the intestinal epithelium. The C. parvum genome also indicates the expansion of functions of surface antigens (mucins) and transporters, emphasizing the value of these factors for sporozoite survival. In addition, there is evidence of dependence on amylopectin utilization by glycolysis as a sole source of ATP for the oocyst. Furthermore, the presence of long-chain fatty acid synthesis genes suggests that long-chain fatty acids are important for survival. But the absence of genes for beta-oxidation of fatty acids indicates that fatty acids are not an energy source for oocysts (48).
Acknowledgments
This work is dedicated to Mercedes R. Edwards and Terry Beveridge, two giants of electron microscopy who provided inspiration and advice.
This study was supported in part by the USDA and the Cornell University College of Agriculture and Life Sciences.
The assistance of Rhea Garen with electron microscopy and contributions from undergraduate research students Seth Axelrad, Jacqueline Bassett, David Graham, Jeremy Levanthal, Merton Lee, David Mista, and Matthew Sullivan are gratefully acknowledged.
Footnotes
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu. C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. [DOI] [PubMed] [Google Scholar]
- 2.Alugupalli, S., F. Portaels, and L. Larsson. 1994. Systematic study of the 3-hydroxy fatty acid composition of mycobacteria. J. Bacteriol. 176:2962-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anguish, L. J., and W. C. Ghiorse. 1997. Computer-assisted laser scanning and video microscopy for analysis of Cryptosporidium parvum oocysts in soil, sediment, and feces. Appl. Environ. Microbiol. 63:724-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 5.Brush, C. F., M. F. Walter, L. J. Anguish, and W. C. Ghiorse. 1998. Influence of pretreatment and experimental conditions on electrophoretic mobility and hydrophobicity of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 64:4439-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brush, C. F., W. C. Ghiorse, L. J. Anuish, J.-Y. Parlange, and H. G. Grimes. 1998. Transport of Cryptosporidium parvum oocysts through saturated columns. J. Environ. Qual. 28:809-815. [Google Scholar]
- 7.Current, W. L., and N. C. Reese. 1986. A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J. Protozool. 33:98-108. [DOI] [PubMed] [Google Scholar]
- 8.Darnault, C. J. G., T. S. Steenhuis, P. Garnier, Y.-J. Kim, M. Jenkins, W. C. Ghiorse, P. C. Baveye, and J.-Y. Parlange. 2004. Preferential flow and transport of Cryptosporidium parvum oocysts through the vadose zone: experiments and modeling. Vadose Zone J. 3:262-270. [Google Scholar]
- 9.Dubey, J. P., C. A. Speer, and R. Fayer. 1990. Cryptosporidiosis of man and animals. CRC Press, Boca Raton, FL.
- 10.Fayer, R. 2008. General biology, p. 1-42. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press, Boca Raton, FL.
- 11.Fayer, R. 1994. Effect of high temperature on infectivity of Cryptosporidium parvum oocysts in water. Appl. Environ. Microbiol. 60:2732-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fayer, R., and T. Nerad. 1996. Effects of low temperatures on viability of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 62:1431-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.). 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 14.Harris, J. R., and F. Petry. 1999. Cryptosporidium parvum structural components of the oocyst wall. J. Parasitol. 85:839-849. [PubMed] [Google Scholar]
- 15.Holme, D. J., and H. Peck. 1993. Analytical biochemistry. Longman Scientific and Technical, Harlow, England.
- 16.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins, M. B., L. J. Anguish, D. D. Bowman, M. J. Walker, and W. C. Ghiorse. 1997. Assessment of a dye permeability assay for determination of inactivation rates of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 63:3844-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins, M. B., D. D. Bowman, and W. C. Ghiorse. 1998. Inactivation of Cryptosporidium parvum oocysts by ammonia. Appl. Environ. Microbiol. 64:784-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins, M. B., M. J. Walker, D. D. Bowman, L. C. Anthony, and W. C. Ghiorse. 1999. Use of a sentinel system for field measurements of Cryptosporidium parvum oocyst inactivation in soil and animal waste. Appl. Environ. Microbiol. 65:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins, M. B., D. D. Bowman, E. A. Fogarty, and W. C. Ghiorse. 2002. Cryptosporidium parvum oocyst inactivation in three soil types at various temperatures and water potentials. Soil Biol. Biochem. 34:1101-1109. [Google Scholar]
- 21.Kates, M. 1972. Laboratory techniques in biochemistry and molecular biology, vol. 3, part 2. Techniques in lipidology: isolation, analysis and identification. American Elsevier Publishing Co., Inc., New York, NY.
- 22.Keller, S. L., M. B. Jenkins, and W. C. Ghiorse. 2004. Simulating the effect of liquid CO2 on Cryptosporidium parvum oocysts in aquifer material. J. Environ. Eng. 130:1547-1551. [Google Scholar]
- 23.King, B. J., A. R. Keegan, P. T. Monis, and C. P. Saint. 2005. Environmental temperature controls Cryptosporidium oocyst metabolic rate and associated retention of infectivity. Appl. Environ. Microbiol. 71:3848-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, B. J., and P. T. Monis. 2007. Critical processes affecting Cryptosporidium oocyst survival in the environment. Parasitology 134:309-323. [DOI] [PubMed] [Google Scholar]
- 25.Kissinger, J. C. 2008. Genomics, p. 43-53. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press, Boca Raton, FL.
- 26.Laskin, A. I., and H. A. Lechevalier (ed.). 1977. CRC handbook of microbiology. CRC Press, Cleveland, OH.
- 27.Liu, J., E. Y. Rosenberg, and H. Nikaido. 1995. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc. Natl. Acad. Sci. U. S. A. 92:11254-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lumb, R., J. A. Lanser, and P. J. O'Donoghue. 1988. Electrophoretic and immunoblot analysis of Cryptosporidium oocysts. Immunol. Cell Biol. 66:369-376. [DOI] [PubMed] [Google Scholar]
- 29.Madigan, M. T., J. M. Martinko, P. V. Dunlap, and D. P. Clark. 2009. Brock biology of microorganisms. Pearson Benjamin Cummings, San Francisco, CA.
- 30.Mitschler, R. R., R. Welti, and S. J. Upton. 1994. A comparative study of lipid compositions of Cryptosporidium parvum (Apicomplexa) and Madin-Darby bovine kidney cells. J. Eukaryot. Microbiol. 41:8-12. [DOI] [PubMed] [Google Scholar]
- 31.Nanduri, J., S. Williams, T. Aji, and T. P. Flannigan. 1999. Characterization of an immunogenic glycocalyx on the surfaces of Cryptosporidium parvum oocysts and sporozoites. Infect. Immun. 67:2022-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng, X., T. Murphy, and N. M. Holden. 2008. Evaluation of the effect of temperature on the die-off rate for Cryptosporidium parvum oocysts in water, soil, and feces. Appl. Environ. Microbiol. 74:7101-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petry, F. 2004. Structural analysis of Cryptosporidium parvum. Microsc. Microanal. 10:586-601. [DOI] [PubMed] [Google Scholar]
- 34.Reduker, D. W., C. A. Speer, and J. A. Blixt. 1985. Ultrasructural changes in the oocyst wall during excystation of Cryptosporidium parvum (Apicomplexa; Eucoccidiorida). Can. J. Zool. 63:1982-1986. [Google Scholar]
- 35.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1992. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 58:3494-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1993. In vitro excystation of Cryptosporidium parvum. Parasitology 106:13-19. [DOI] [PubMed] [Google Scholar]
- 37.Rodd, E. H., and S. Coffey (ed.). 1964. Rodd's chemistry of carbon compounds: a modern comprehensive treatise, vol. 1. Elsevier, New York, NY.
- 38.Schrum, D. P., S. Alugupalli, S. T. Kelly, D. C. White, and R. Fayer. 1997. Structural characterization of a “signature” phosphatidylethanolamine as the major 10-hydroxy stearic acid-containing lipid of Cryptosporidium parvum oocysts. Lipids 32:789-793. [DOI] [PubMed] [Google Scholar]
- 39.Spano, F., C. Purp, L. Ranucci, L. Putignani, and A. Crisanti. 1996. Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual parasite development. Parasitology 114:427-437. [DOI] [PubMed] [Google Scholar]
- 40.Stryer, L. 1965. The interaction of a naphthalene dye with apomyoglobin and apoheoglobin. A fluorescent probe of non-polar binding sites. J. Mol. Biol. 13:482-495. [DOI] [PubMed] [Google Scholar]
- 41.Templeton, T. T., C. A. Lancto, V. Vigdorovich, C. Liu, N. R. London, K. Z. Hadsall, and M. S. Abrahamsen. 2004. The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in Toxoplasma. Infect. Immun. 72:980-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weppelman, R. M., W. J. A. Vanden Heuvel, and C. C. Wang. 1976. Mass spectrometric analysis of the fatty acids and nonsaponifiable lipids of Eimeria tenella oocysts. Lipids 11:209-215. [DOI] [PubMed] [Google Scholar]
- 43.Winans, S. C., and M. J. Rooks. 1993. Sensitive, economical laboratory photodocumentation using a standard video camera and thermal printer. Biotechniques 14:902. [PubMed] [Google Scholar]
- 44.Windholz, M., S. Budavari, R. F. Blumetti, and E. S. Otterbein (ed.). 1983. The Merck index, 10th ed. Merck & Co., Inc., Rahway, NJ.
- 45.Xu, P., G. Widmer, Y. Wang, L. S. Ozaki, J. M. Alves, M. G. Serrano, D. Puiu, P. Manque, D. Akiyoshi, A. J. Mackey, W. R. Pearson, P. H. Dear, A. T. Bankier, D. L. Peterson, M. S. Abrahamsen, V. Kapur, S. Tzipori, and G. A. Buck. 2004. The genome of Cryptosporidium hominis. Nature 431:1107-1112. [DOI] [PubMed] [Google Scholar]
- 46.Yoshikawa, H., and M. Iseki. 1991. Freeze-fracture studies of Cryptosporidium muris. J. Protozool. 38:171S-172S. [PubMed] [Google Scholar]
- 47.Zhu, G. 2008. Biochemistry, p. 57-77. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press, Boca Raton, FL.
- 48.Zhu, G. 2004. Current progress in the fatty acid metabolism in Cryptosporidium parvum. J. Eukaryot. Microbiol. 51:381-388. [DOI] [PubMed] [Google Scholar]