Abstract
The purpose of this work was to evaluate the evolutionary history of Campylobacter coli isolates derived from multiple host sources and to use microarray comparative genomic hybridization to assess whether there are particular genes comprising the dispensable portion of the genome that are more commonly associated with certain host species. Genotyping and ClonalFrame analyses of an expanded 16-gene multilocus sequence typing (MLST) data set involving 85 isolates from 4 different hosts species tentatively supported the development of C. coli host-preferred groups and suggested that recombination has played various roles in their diversification; however, geography could not be excluded as a contributing factor underlying the history of some of the groups. Population genetic analyses of the C. coli pubMLST database by use of STRUCTURE suggested that isolates from swine form a relatively homogeneous genetic group, that chicken and human isolates show considerable genetic overlap, that isolates from ducks and wild birds have similarity with environmental water samples and that turkey isolates have a connection with human infection similar to that observed for chickens. Analysis of molecular variance (AMOVA) was performed on these same data and suggested that host species was a significant factor in explaining genetic variation and that macrogeography (North America, Europe, and the United Kingdom) was not. The microarray comparative genomic hybridization data suggested that there were combinations of genes more commonly associated with isolates derived from particular hosts and, combined with the results on evolutionary history, suggest that this is due to a combination of common ancestry in some cases and lateral gene transfer in others.
Campylobacter species are a leading bacterial cause of gastroenteritis within the United States and throughout much of the rest of the developed world. According to the CDC, there are an estimated 2 million to 4 million cases of Campylobacter illness each year in the United States (37). Campylobacter jejuni is generally recognized as the predominant cause of campylobacteriosis, responsible for approximately 90% of reported cases, while the majority of the remainder are caused by the closely related sister species Campylobacter coli (27). Not surprisingly, therefore, the majority of research on Campylobacter has centered on C. jejuni, and C. coli is a less studied organism.
A multilocus sequence typing (MLST) scheme of C. jejuni was first developed by Dingle et al. (13) on the basis of the genome sequence of C. jejuni NCTC 11168. There have also been a number of studies using the genome sequence data to develop microarrays for gene presence/absence determination across strains of C. jejuni and to identify the core genome components for the species (6, 15, 32, 33, 42, 43, 53, 57). Although C. coli is responsible for fewer food-borne illnesses than C. jejuni, the impact of C. coli is still substantial, and there is also evidence that C. coli may carry higher levels of resistance to some antibiotics (1). C. coli and C. jejuni also tend to differ in their relative prevalences in animal host species and various environmental sources (4, 48, 58), and there is some evidence that both taxa may include groups of host-specific putative ecotype strains (7, 36, 38, 39, 52, 56). At present, there is only a single draft genome sequence available for C. coli, and there are no microarray comparative genomic hybridization data for C. coli strains. Thus, there is no information on intraspecies variability in gene presence/absence in C. coli and how such variability might correlate with host species.
The purpose of this work was to develop and apply an expanded 16-locus MLST genotyping scheme to evaluate the evolutionary history of Campylobacter coli isolates derived from multiple host sources and to use microarray comparative genomic hybridization to assess whether there are particular genes comprising the dispensable portion of the genome that are more commonly associated with isolates derived from different host species.
MATERIALS AND METHODS
Bacterial strains.
A collection of 84 Campylobacter coli isolates from diverse geographic origins (the United States, Canada, the United Kingdom, Poland, Switzerland, Sweden, Israel, Belgium, Slovenia, and Bosnia Herzegovina), including the sequenced strain RM2228 and 11 isolates from swine, 19 from bovines, 12 from chickens, 16 from turkeys (chickens and turkeys are hereinafter occasionally grouped in the host category “poultry”), and 26 from humans, were selected for MLST genotyping and comparative genomic comparisons.
Multilocus sequence typing and analysis.
MLST was performed by sequencing the seven housekeeping genes (protein products are shown in parentheses) aspA (aspartase A), glnA (glutamine synthetase), gltA (citrate synthase), glyA (serine hydroxymethyltransferase), pgm (phosphoglucomutase), tkt (transketolase), and uncA (ATP synthase α subunit) according to the method of Dingle et al. (12, 13). To increase the genotyping resolution, nine additional housekeeping loci, scattered through the C. coli chromosome, were selected from the complete and draft sequences of C. jejuni strain NCTC 11168 and C. coli strain RM2228. The chromosomal locations of these housekeeping loci were chosen such that it was unlikely for any of these loci to be coinherited in the same recombination event. Details regarding these nine loci can be found in Table 1 .
TABLE 1.
Information relevant to the nine-gene MLST loci new to this study
| Locus | Size (bp) | No. of alleles | % Variable sites | dN/dS ratioa | Coordinatesb | Primer sequence (5′-3′) |
|
|---|---|---|---|---|---|---|---|
| Forward | Reverse | ||||||
| adk | 471 | 7 | 1.49 | 1.0086 | 599978-600448 | TAATTATAGGTGCACCAGGAAGCG | GGTTCGATAGTGCGTTCTCCATC |
| aroE | 675 | 7 | 3.26 | 0.0809 | 369933-370607 | ACAATGCCATTCAAGCTCTCAAGC | CAGAGCTTCTTGCATGGCATTT |
| mdh | 630 | 20 | 14.29 | 0.2716 | 1096816-1097445 | GGGTTTTGTGGGTGCAGCAA | CAAATTCGGTTCTTCCTTTGCG |
| nadP | 606 | 28 | 7.43 | 0.1061 | 1478490-1479096 | AATGGTCGCATTCAAAGCAAGG | ACGCCCATAGCCAAAGCATC |
| pheS | 594 | 11 | 2.53 | 0.096 | 839250-838653 | AGATTTAGCGGGCGAAGAAAAG | CACAACCCTTGCAAAATACACACG |
| pgi | 594 | 18 | 2.19 | 0.7806 | 1466697-1467290 | TGGTTCAAGTTGCGGTGTTAAAGC | TGCTGATCCCTTGCTCCTATAAGAG |
| recA | 615 | 11 | 13.33 | 0.0234 | 1594041-1594655 | CAGGTTCTGTAGGACTTGATCTTGC | TCAGCTTGTCTAAATGGAGGTGC |
| carB | 603 | 9 | 9.29 | 0.0283 | 642616-643218 | TTGGCACTTGATTTAACCGCAG | GCAGCGTCAAGATCAGGAAAGATA |
| trmU | 561 | 25 | 10.87 | 0.0276 | 68933-69497 | TTCTAGTCGCTATGAGCGGTGG | CGTGTTTTCCTACAACCTTACCGCT |
Ratio of nonsynonymous to synonymous evolutionary changes.
The genomic coordinates are relative to those of the Campylobacter jejuni NCTC 11168 genome.
The evolutionary history of the C. coli isolates was evaluated using eBURST (http://eburst.mlst.net/) (21) and ClonalFrame version 1.1 (11). Sequence types (STs) were grouped into clonal complexes (CCs) by using eBURST version 3, and phylogenetic analysis was performed using ClonalFrame version 1.1, including all 16 loci. ClonalFrame has received wide use in the assessment of evolutionary relationships of strains of the same species of bacteria, including C. jejuni and C. coli (e.g., references 2, 10, 14, 23, and 52). Two of the many benefits of reconstructing the evolutionary history of bacterial clonal lines via ClonalFrame analysis are that bacterial recombination can be taken into consideration when the history is reconstructed and that the time, in coalescent units, to the most recent common ancestor of different groups can be estimated (11). To assess the influence of recombination, two 50% consensus trees were created for all 84 isolates with 16 loci, one estimating parameters of recombination and the other with recombination parameters fixed at zero. Five independent runs were performed for each model, with each run consisting of 100,000 burn-in iterations plus 200,000 sampling iterations. The first half of the chains was discarded, and the second half was sampled at intervals of 100 iterations. Convergence was estimated based on the Gelman-Rubin statistic (25).
To examine the effects of host/environment and geography on C. coli population structure, a large data set of 969 isolates, including the 84 isolates in this study and the 885 isolates available on pubMLST, from different host species and geographic regions (see Table S1 in the supplemental material) were assigned to bacterial populations by using the linkage model of the program STRUCTURE 2.2 (18, 19, 44). STRUCTURE has been used in similar analyses involving a range of species of bacteria, including C. jejuni (e.g., references 20, 36, and 54). The data set was assembled by treating each of the 1,683 polymorphic sites from the seven MLST genes as a single locus. Map distances, used by the linkage model, were assumed to be proportional to the number of base pairs between sites, except for sites in different gene fragments, which were treated as being unlinked. The number of bacterial populations, K, was determined by comparing the posterior probability from multiple runs, assuming that 2 was ≤K and that K was ≤14. Three individual runs (100,000 burn-in iterations and 200,000 sampling iterations) were performed for each value of K. An additional examination of these pubMLST data, focusing on assessing the importance of host species and geography in structuring the genetic variation, was conducted using the analysis-of-molecular-variance (AMOVA) approach in Arlequin (17, 49).
Microarray description.
Combimatrix CustomArray 4X2K was used in this study (26). This array is divided into 4 sectors, each of which contains 2,240 in situ-synthesized oligonucleotide probes (spots) with the same probe design and layout. On the basis of the sequence of Campylobacter coli strain RM2228, oligonucleotide probes were designed to have a similar annealing temperature of 56°C and a length of 35 to 40 bp. Two separate designs were used in this study; both included 100 control probes (20 negative controls with sequences from plants and Escherichia coli phage, each with 5 replicate spots) as well as loci from the RM2228 genome. Because of the strict criteria for probe design, not all open reading frames (ORFs) could be covered in this analysis. The first design included 1,942 of the 1,967 protein-coding genes described to occur in the unfinished sequence of C. coli strain RM2228. The second-generation design was based on genes that were not clearly present (loci with low intensity or no hybridization for at least one strain) in the hybridization results involving the first design and included a total of 615 loci. Two to five additional probes, separated from one another in order to span the entire gene, for each of these 615 ambiguous loci were synthesized in situ to occupy the 2,240 independent microarray spots.
Microarray DNA isolation, labeling, and hybridization.
Genomic DNA was digested by sonication to sizes of 200 to 400 bp, as visualized by agarose gel electrophoresis, and then purified by using a Qiagen Qiaquick PCR purification kit. Purified fragments (1 to 2 μg) were labeled with biotin, using a Mirus Label IT μArray biotin labeling kit (Mirus Corp., Madison, WI) according to the manufacturer's instructions, followed by removal of unincorporated dyes by use of QIAquick (Qiagen) columns.
The standard hybridization conditions for the biotinylated target included preblocking for 30 min at 50°C with 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) containing 0.05% Tween 20, 5× Denhardt's solution, and 100 ng/μl salmon sperm DNA, followed by hybridization of the biotinylated target in hybridization solution (6× SSPE, 20 to 50 ng/μl labeled DNA, and 0.05% SDS) overnight at 50°C. The arrays were then washed once for at least 15 min with SSPE wash 1 (6× SSPE, 0.05% Tween 20) and then for 1 min each with SSPE wash 2 (3× SSPE, 0.05% Tween 20), SSPE wash 3 (0.5× SSPE, 0.05% Tween 20), and PBST wash (2× phosphate-buffered saline [PBS], 0.1% Tween 20), followed by a final 2× PBS wash at room temperature. The hybridized array was then blocked with 5× casein-PBS buffer (BioFX Laboratories, Owings Mills, MD) for 15 min at room temperature and labeled for 30 min with Cy5-streptavidin (GE Healthcare, Amersham Biosciences, Piscataway, NJ) diluted 1:1,000 in 5× casein-PBS buffer. The arrays were scanned after they were washed twice each with the PBST and PBS solutions.
Array scanning and data analysis.
Hybridized microarrays were scanned with a GenePix 4000B laser scanner (Axon Instruments, Union City, CA) at a wavelength of 635 nm to obtain raw Cy5 fluorescence intensity. Replicate or triplicate arrays were hybridized for 65 strains tested in this study. Background intensity was estimated as the 75th percentile of all negative-control probes and subtracted on a log2 scale from the foreground Cy5 intensity of all spots on the array. Such normalization made the magnitudes of signal values of different arrays more comparable, with absent genes on each array centering around 0 (see Fig. S1 in the supplemental material). An expectation maximization (EM) algorithm was then applied on the adjusted probe signals of each array to estimate the means and standard deviations of present genes and absent genes, with the following parameters at initiation: the means ± standard deviations for the genes present and absent were 5.0 ± 1.0 and 0 ± 0.5, respectively, with the percentage of absent genes at 10%. The EM algorithm was run for 100 iterations or stopped when the magnitude of change in mean estimate was less than 0.001 between iterations. The EM algorithm fitted a well-established Gaussian mixture model to the normalized signals of each array independently to distinguish the population of absent genes from that of present genes (see Fig. S1 in the supplemental material). It did not rely on comparison to signals of positive- and negative-control strains, so it was robust to technical processing variation in the array experiment.
For each array, genes with spot signals below the lower 0.5th percentile of the estimated distribution of present genes were called absent, i.e., at P values of <0.005. For C. coli strains tested on multiple arrays, a gene for a given probe was called present if it had a present call on at least one array. For genes with multiple probes, a present call ratio, based on the total number of present calls out of the total number of probes for each gene, was used as a measurement of the divergence of the test strain from the reference strain. The ratio was in the range of 0 to 1, which reflects the gene divergence level (absent to present, respectively) in the test strain compared to the reference strain.
Verification of microarray data.
To assess the microarray performance, gene presence and absence predictions were compared to the genome sequence of two randomly selected C. coli strains, cco4 and cco74 (swine and human origins, respectively). Draft genome sequences were obtained for these two strains by using the Illumina Genome Analyzer II system. For each strain, one lane was used and yielded 5 million and 8 million 36-bp reads, respectively. Reads were first aligned to the RM2228 draft genome with the mapping-and-assembly-with-qualities (MAQ) method, using default parameters (34). A preliminary analysis of the MAQ consensus sequences revealed that many regions of both genomes were too divergent for the reads to be mapped and for a polymorphism to be accurately differentiated from the absence of the region in the sequenced genome. To resolve these undetermined regions, de novo assemblies were performed using Velvet (62). Several hash lengths and coverage cutoffs were used, and the best assembly was selected on the basis of a combination of the N50, contig number, and total contig size statistics. For each of these resulting assemblies, open reading frames were called by using Glimmer, using default settings (8). Then, using BLAST, each MAQ coding consensus sequence that had any unresolved positions (i.e., either absent or too divergent) in the original mapping was searched against the de novo assemblies. When a single hit was found, the corresponding open reading frame was then aligned to the consensus sequence, and whenever possible, the undetermined positions were resolved using the de novo assembled sequence. The resulting enhanced MAQ consensus sequences were used to predict gene presence/absence by counting the percentage of sites absent for each coding sequence. The distribution of the percentages of absent sites appeared clearly bimodal, with a peak at 0% and 100%, corresponding to present and absent genes, respectively (see Fig. S2 in the supplemental material). There were nevertheless a few genes showing intermediate levels of present sites, which appeared to be duplicated and divergent genes for which the de novo assemblies could not be used (more than one hit). In the following analyses, we used a 50% absent-site threshold to delimit present and absent genes.
On the basis of both microarray designs, a presence ratio was calculated, with the number of present calls divided by the number of probes in a particular ORF, and used to predict ORF presence and absence. The presence ratio threshold used to call a gene present was determined using the two control strains sequenced using the Illumina technology and was drawn at 0.5 (see Fig. S3 in the supplemental material). When compared with the Illumina sequencing presence and absence calls, the microarray showed a false-negative rate (FNR) ranging between 3% and 5% (for cco74, 3.3%, and for cco4, 4.8%), while the false-positive rate (FPR) remained between 0% and 3% (for cco74, 0%, and for cco4, 2.6%). Following the Illumina sequencing and assembly procedure, there were only 5 genes for strain cco4 that were absent but were called present by the array. These 5 loci were plasmid genes and included the following: CCOA0022, CCOA0027, CCOA0031, CCOA0151, and CCOA0152. The alignment of the Illumina reads against these genes showed that very small portions of the genes (<100 bp) were indeed conserved but that the vast majority of the locus was absent. Coincidentally, some of the microarray probes were designed in these regions and therefore suggested that the ORFs were present. Given the very recombinant nature of plasmid genes, it is difficult to state if these genes should be considered present or not. If one excludes the plasmid genes, FPRs equal to 0 for both control strains are obtained. Thus, the microarray double design used in this paper yields virtually no false positives, while maintaining a reasonable number of false negatives (<5%).
Accession numbers.
All sequence data arising from the nine additional housekeeping loci have been deposited in the NCBI GenBank database under accession numbers GQ325800 to GQ326546. The Illumina reads were deposited in the NCBI trace archive under accession numbers SRX016174 and SRX016251, and the two assembled sequences are available for download at http://stanhope.vet.cornell.edu/data.html. The microarray data discussed in this paper have been deposited in NCBI's Gene Expression Omnibus (16) and are accessible through GEO Series accession number GSE16787.
RESULTS AND DISCUSSION
Evolutionary history.
A total of 84 isolates were successfully typed by both MLST schemes. A comparison of the data derived from the 7-gene scheme to the Campylobacter MLST database (http://pubmlst.org/campylobacter/) indicated that our 84 isolates comprised 55 STs (Table 2), including 11 STs from swine (11/11; 100% unique), 8 from bovines (8/19; 42% unique), 7 from chickens (7/12; 58% unique), 13 from turkeys (13/16; 81% unique) and 23 from humans (23/26; 88% unique). A total of 15 STs, including 4 isolates from swine, 4 from bovines, 4 from humans, and 3 from turkeys, were new to the MLST database. With the default eBURST setting of 6 out of 7 shared alleles, the vast majority of our isolates could be assigned to the ST 828 complex (71/84) and included isolates from all the different host sources involved in this study. An examination of host species and ST, based on the 7-gene scheme, that incorporates our data along with the previously existing data from the MLST database, indicated that many C. coli STs are present in multiple host species. For example, one of the more commonly represented genotypes, ST 825, has been detected among isolates from human, chicken, turkey, swine, and environmental samples. There were, however, several exceptions to this, including, for example, ST 1104 (n = 6), currently listed only in the pubMLST database from bovines, as well as several other STs nearly exclusively represented in particular host species (e.g., ST 1017, represented in 8/10 isolates from poultry [chicken and turkey], and ST 1096, represented in 5/6 isolates from swine). Furthermore, a Fisher exact test rejected the null hypothesis of independence of ST and host species (P values of <0.001, with chicken and turkey included together as poultry), and therefore, these data support a tendency for particular C. coli genotypes (STs) to be more commonly associated with certain host species.
TABLE 2.
Summary of allelic profiles and resulting STs for both MLST schemesa
| Isolate hostb | Allelic profile (7 loci; pubMLST)c | ST for 7 loci | Allelic profile (9 loci; this study)d | ST for 9 loci | eBURST result for 12/16 shared alleles | Country (state) | Collection datee |
|---|---|---|---|---|---|---|---|
| cco001_S* | 33-38-30-167-104-43-17 | 1467 | 1-1-1-1-1-1-1-1-1 | 1 | USA | 2/27/2001 | |
| cco002_S* | 33-38-30-167-104-44-36 | 3859 | 1-1-1-2-1-2-1-1-2 | 2 | CC9 | USA | 1/24/2001 |
| cco003_S* | 33-39-30-82-104-43-17 | 828 | 1-1-1-3-1-3-1-2-3 | 3 | USA | 2/12/2001 | |
| cco004_S* | 53-321-44-82-104-35-36 | 3860 | 1-1-1-4-2-4-2-1-4 | 4 | USA | 3/1/2001 | |
| cco005_S* | 33-38-30-78-104-44-36 | 3861 | 1-2-1-2-1-1-1-1-2 | 5 | CC9 | USA | 2/6/2001 |
| cco006_S* | 33-38-30-82-104-35-36 | 890 | 1-1-1-4-2-5-1-1-5 | 6 | USA | 10/31/2000 | |
| cco007_S* | 32-39-44-82-104-43-36 | 1061 | 1-3-1-4-3-4-1-2-6 | 7 | USA | 11/30/2000 | |
| cco008_S* | 33-38-30-82-152-173-68 | 1102 | 1-1-2-5-2-1-2-1-2 | 8 | USA | 3/20/2001 | |
| cco009_C* | 33-39-30-82-113-43-17 | 829 | 1-1-1-4-4-6-1-3-6 | 9 | CC5 | USA (AR) | 2/21/2000 |
| cco010_S* | 33-322-30-173-104-35-68 | 3862 | 2-1-3-6-5-7-1-1-7 | 10 | USA | 2/29/2001 | |
| cco011_S* | 53-39-30-82-118-35-36 | 1631 | 1-1-1-5-2-4-1-1-4 | 11 | USA | 2/20/2001 | |
| cco012_S* | 33-38-30-78-104-35-17 | 1113 | 2-2-4-7-1-8-3-1-8 | 12 | USA | 11/28/2000 | |
| cco013_C* | 33-39-30-82-113-47-17 | 825 | 2-1-5-8-1-3-1-2-9 | 13 | CC7 | USA (FL) | 10/30/2000 |
| cco014_C* | 33-39-30-82-104-43-41 | 1017 | 2-4-1-9-1-3-1-2-6 | 14 | CC3 | USA (NC) | 3/6/2000 |
| cco015_C* | 33-39-30-82-113-43-17 | 829 | 1-1-6-4-4-6-1-1-6 | 15 | CC5 | USA (AR) | 8/21/2000 |
| cco016_C* | 33-39-30-82-113-47-17 | 825 | 2-1-5-8-1-3-1-2-9 | 13 | CC7 | USA (AL) | 9/18/2000 |
| cco017_C* | 33-39-30-82-113-43-17 | 829 | 1-1-6-10-4-6-1-3-6 | 16 | CC5 | USA | NA |
| cco018_B* | 33-39-44-82-104-44-17 | 1436 | 3-1-7-11-2-1-1-4-10 | 17 | CC10 | USA (WA) | 4/15/2002 |
| cco019_B* | 33-39-30-78-104-43-17 | 1068 | 2-2-4-4-1-9-3-2-8 | 18 | CC1 | USA (WA) | 4/15/2002 |
| cco020_B* | 33-39-30-78-423-43-17 | 3863 | 2-2-4-4-1-9-3-2-8 | 19 | CC1 | USA (WA) | 4/15/2002 |
| cco023_B* | 33-39-30-82-104-85-68 | 1104 | 1-1-1-4-2-3-2-1-11 | 20 | CC2 | USA (WA) | 4/15/2002 |
| cco024_B* | 33-39-30-78-104-43-17 | 1068 | 2-2-4-4-1-9-3-2-8 | 18 | CC1 | USA (WA) | 4/15/2002 |
| cco025_B* | 33-30-30-78-104-43-17 | 3864 | 2-2-4-4-1-9-3-2-8 | 21 | CC1 | USA (WA) | 6/17/2002 |
| cco027_B* | 33-39-30-78-104-43-17 | 1068 | 4-2-4-4-1-9-3-2-8 | 22 | CC1 | USA (WA) | 6/17/2002 |
| cco037_B* | 33-39-44-82-104-85-17 | 3865 | 5-1-7-11-2-1-1-4-10 | 23 | CC10 | USA (WA) | 10/22/2002 |
| cco049_B* | 33-153-30-78-104-43-17 | 3866 | 2-2-4-4-1-10-3-2-8 | 24 | CC1 | USA (OR) | 7/15/2003 |
| cco051_B* | 33-39-30-78-104-43-17 | 1068 | 2-2-4-4-1-9-3-2-8 | 18 | CC1 | USA (WA) | 12/9/2003 |
| cco052_B* | 33-39-30-78-104-43-17 | 1068 | 2-2-4-4-1-9-3-2-8 | 18 | CC1 | USA (WA) | 12/9/2003 |
| cco054_B* | 33-39-30-78-104-43-17 | 1068 | 2-2-4-4-1-9-3-2-8 | 18 | CC1 | USA (WA) | 12/9/2003 |
| cco055_B* | 33-39-30-82-104-85-68 | 1104 | 1-1-1-12-2-9-2-1-10 | 25 | CC2 | USA (CA) | 7/5/2002 |
| cco060_B* | 33-39-30-82-104-85-68 | 1104 | 5-1-1-12-2-9-2-4-4 | 26 | CC2 | USA (CA) | 4/30/2003 |
| cco061_B* | 33-39-30-82-104-85-68 | 1104 | 1-1-1-11-2-11-2-5-2 | 27 | USA (CA) | 10/17/2003 | |
| cco062_B* | 33-153-30-82-104-85-68 | 2698 | 5-1-1-12-2-1-2-4-10 | 28 | CC2 | USA (CA) | 10/17/2003 |
| cco063_B* | 33-153-30-82-104-85-68 | 2698 | 5-1-1-12-2-9-2-4-4 | 29 | CC2 | USA (CA) | 10/17/2003 |
| cco065_B* | 33-153-30-82-104-85-68 | 2698 | 5-1-1-12-2-9-2-4-4 | 29 | CC2 | USA (CA) | 11/4/2003 |
| cco067_B* | 33-39-30-82-104-85-68 | 1104 | 1-1-1-3-2-3-2-1-4 | 30 | CC2 | USA (CA) | 11/30/2003 |
| cco069_H* | 33-39-30-82-189-43-17 | 1585 | 1-1-8-8-1-3-1-1-6 | 31 | Poland | 2004 | |
| cco070_H* | 33-176-30-115-104-43-17 | 2301 | 2-1-8-13-1-6-1-2-2 | 32 | Poland | 2004 | |
| cco071_H* | 33-110-103-350-188-368-265 | 3867 | 1-5-9-14-6-6-4-6-12 | 33 | Poland | 2004 | |
| cco072_H | 33-39-30-82-113-43-17 | 829 | 2-1-10-11-1-6-1-2-13 | 34 | CC12 | Poland | 2004 |
| cco073_H | 33-39-30-82-113-47-17 | 825 | 1-1-11-15-1-12-1-1-2 | 35 | CC8 | Poland | 2005 |
| cco074_H* | 33-39-30-82-189-47-17 | 1191 | 1-1-11-16-1-6-1-1-2 | 36 | CC8 | Poland | 2005 |
| cco075_H | 33-39-30-78-104-43-17 | 1068 | 2-1-4-4-7-9-5-2-8 | 37 | CC1 | Canada | 1990 |
| cco076_H* | 33-176-30-82-451-43-17 | 3868 | 2-1-10-11-1-3-1-2-6 | 38 | CC12 | Slovenia | 2002 |
| cco077_H* | 33-39-30-79-104-35-17 | 855 | 1-1-4-15-1-13-6-1-14 | 39 | CC6 | Bosnia Herzegovina | 2002 |
| cco078_H | 33-39-30-79-104-35-17 | 855 | 1-1-10-15-1-13-6-1-15 | 40 | CC6 | Slovenia | 2002 |
| cco079_H* | 33-176-30-82-113-43-17 | 1586 | 2-1-10-17-1-3-1-7-16 | 41 | CC12 | Belgium | 1998 |
| cco080_T* | 33-39-65-82-113-47-17 | 894 | 5-1-12-13-1-6-1-8-2 | 42 | USA (CO) | 1981 | |
| cco081_H* | 33-39-261-79-104-64-17 | 3869 | 5-1-1-15-1-6-1-1-17 | 43 | Sweden | 1982 | |
| cco082_H* | 33-39-30-82-113-35-17 | 899 | 1-2-13-11-1-14-1-2-18 | 44 | CC4 | Israel | 1983 |
| cco083_H* | 33-39-30-82-113-47-41 | 889 | 2-6-14-18-8-6-1-1-6 | 45 | Belgium | 1984 | |
| cco085_H | 33-38-30-79-104-35-17 | 2642 | 1-1-15-11-1-9-1-1-6 | 46 | Sweden | 2006 | |
| cco086_H* | 33-38-30-115-113-43-17 | 892 | 2-1-16-11-1-6-1-1-16 | 47 | Canada | 1981 | |
| cco087_H* | 33-39-30-79-452-43-17 | 3870 | 6-1-3-17-9-3-7-1-19 | 48 | UK | 1984 | |
| cco088_H* | 32-38-30-82-152-35-17 | 900 | 1-2-1-11-2-9-2-1-20 | 49 | Canada | 1986 | |
| cco090_H* | 32-42-30-82-104-43-17 | 898 | 1-1-3-8-1-15-2-2-9 | 50 | USA | 1985 | |
| cco091_H* | 33-176-30-79-113-43-17 | 3020 | 5-1-10-11-1-3-1-1-2 | 51 | Switzerland | 1993 | |
| cco092_H* | 33-39-30-328-104-43-17 | 3340 | 1-1-8-15-1-6-1-9-21 | 52 | Switzerland | 1993 | |
| cco093_H* | 33-39-30-79-104-43-41 | 901 | 1-5-17-19-1-15-1-9-16 | 53 | Switzerland | 1993 | |
| cco094_H | 33-39-30-82-113-47-17 | 825 | 1-2-13-11-1-14-1-2-18 | 54 | CC4 | Switzerland | 1993 |
| cco095_H | 33-39-66-79-104-47-41 | 3348 | 1-2-18-20-1-16-4-1-22 | 55 | Switzerland | 1993 | |
| cco096_H | 33-38-30-82-104-35-17 | 1096 | 7-1-1-21-2-3-1-1-2 | 56 | Switzerland | 6/28/1995 | |
| cco097_H | 33-39-30-82-113-47-17 | 825 | 1-2-10-8-1-14-1-2-23 | 57 | CC4 | Switzerland | 12/2/2002 |
| cco098_C | 33-38-30-82-104-43-17 | 854 | 1-1-7-11-1-1-8-1-4 | 58 | CC11 | Switzerland | 2/4/2002 |
| cco099_C | 33-38-30-82-104-43-17 | 854 | 1-1-7-11-1-1-8-1-4 | 58 | CC11 | Switzerland | 7/4/2002 |
| cco100_C* | 33-38-30-82-104-332-17 | 3336 | 1-1-7-22-1-1-8-1-6 | 59 | CC11 | Switzerland | 6/25/2002 |
| cco101_C | 53-38-30-82-118-44-36 | 1147 | 1-1-19-5-2-2-1-1-10 | 60 | Switzerland | 10/4/2002 | |
| cco102_C* | 53-38-30-82-118-44-36 | 1147 | 1-1-19-5-2-2-1-1-10 | 60 | Switzerland | 10/4/2002 | |
| cco103_T* | 33-39-30-140-189-43-41 | 3871 | 2-6-1-23-10-3-9-2-24 | 61 | USA | 6/5/2003 | |
| cco104_T* | 33-39-122-140-113-43-17 | 1050 | 1-7-1-10-4-17-1-1-6 | 62 | USA | 5/23/2002 | |
| cco105_T* | 33-39-122-79-113-43-41 | 1167 | 1-1-6-24-4-3-1-1-6 | 63 | USA | 10/22/2002 | |
| cco106_T* | 33-39-30-82-113-47-17 | 825 | 2-1-12-25-1-3-1-2-25 | 64 | CC7 | USA | 4/7/2001 |
| cco107_T* | 33-39-30-140-104-43-41 | 1067 | 2-5-1-26-1-3-1-2-24 | 65 | CC3 | USA | 4/3/2000 |
| cco108_T* | 33-39-30-82-104-43-41 | 1017 | 2-4-1-27-1-3-1-2-6 | 66 | CC3 | USA | 8/13/2003 |
| cco109_T | 33-39-30-82-104-43-41 | 1017 | 2-4-1-27-1-3-1-2-6 | 66 | CC3 | USA | 11/19/2003 |
| cco110_T* | 33-39-30-82-113-35-17 | 899 | 1-2-13-11-1-14-1-2-18 | 44 | CC4 | USA | 5/23/2002 |
| cco111_T* | 33-39-103-82-104-324-41 | 3872 | 2-6-3-8-1-6-10-2-6 | 67 | USA | 6/6/2002 | |
| cco112_T | 33-39-30-82-211-85-17 | 1082 | 1-1-20-8-1-6-11-1-23 | 68 | USA | 6/26/2002 | |
| cco113_T | 33-39-30-82-104-43-41 | 1017 | 2-4-1-27-1-3-1-2-6 | 66 | CC3 | USA | 7/24/2002 |
| cco114_T* | 241-39-262-82-113-56-17 | 3873 | 2-1-12-11-1-3-1-2-9 | 69 | USA | 5/17/2002 | |
| cco115_T | 33-39-30-79-113-47-17 | 860 | 2-1-6-10-4-6-1-3-6 | 70 | CC5 | USA | 5/29/2003 |
| cco116_T | 33-39-30-82-104-43-41 | 1017 | 2-4-1-27-1-18-1-2-6 | 71 | CC3 | USA | 10/22/2003 |
| cco117_T | 33-39-30-82-113-43-17 | 829 | 1-1-12-4-11-6-1-1-6 | 72 | CC5 | USA | 4/1/2003 |
| RM2228_C | 33-39-30-140-113-43-41 | 1063 | 2-4-1-28-1-3-1-2-6 | 73 | CC3 | USA | 1998 |
NA, not available; CC, clonal complex with a group definition of 12/16.
Isolates marked with * were tested on the microarray. S, swine; C, chicken; T, turkey; B, bovine; and H, human.
Allelic profiles for the 7 pubMLST loci, representing aspA, glnA, gltA, glyA, pgm, tkt, and uncA, respectively.
Allelic profiles for the nine MLST loci new to this study, representing adk, aroE, mdh, nadP, pheS, pgi, recA, carB, and trmU, respectively.
Dates are given as month/day/year.
On the basis of the 16-locus MLST scheme, the 84 test isolates were resolved into 73 different sequence types (Table 2). eBURST analysis of the 16-locus MLST scheme, with the two most stringent settings for group definition (15 shared alleles out of 16 or 14 shared alleles out of 16), identified the same four host-specific groups, ranging in size from 3 to 8 isolates. A clonal complex definition of 13/16 alleles resulted in 11 host-specific groups, and a group definition of 12/16 alleles resulted in 12 host-specific groups (with the exception of CC1, which includes 9 bovine isolates and 1 human isolate) (Table 2). The groups ranged in size from 2 to 10 isolates, with the two largest groups coming from bovines. When the number of shared alleles was dropped to 11, several groups of mixed host composition were observed. Fisher exact tests rejected the null hypothesis of independence of genotype (in this case, clonal complex) and host species for all of these group definitions (P values of <0.001). Thus, with decreasing numbers of shared alleles, more groups, some consisting of more isolates, were identified, but nonetheless, each of these groups was host specific up to the level of 11/16 shared alleles, at which point mixed-host groups became evident. This result, combined with the tendency for STs based on the 7-gene scheme also to be host specific, argues for the possible existence of multiple host-preferred groups; however, n is relatively small for any of these groups, suggesting the need for a much more extensive sample before a more definitive view could be reached.
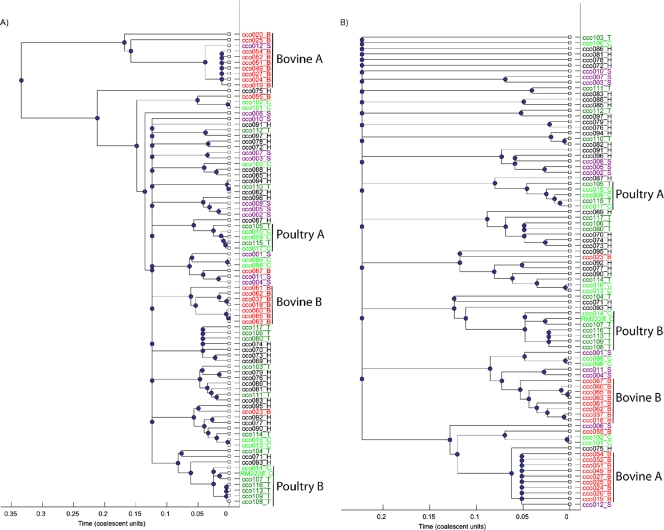
Yet another way to evaluate the evolutionary history and possible host group composition of genetic groups is through ClonalFrame analysis of our 16-gene MLST data. The analysis suggests that the relative impact of recombination versus that of point mutation, expressed as a ratio (r/m), was approximately 1.56 (mean of results from 5 independent runs, summarized in Table 3), and the relative frequency of recombination in comparison to point mutation (ρ/θ) was about 0.25. This estimate of frequency of recombination suggests that recombination is relatively rare compared to some species, such as Streptococcus uberis (ρ/θ, 9.05 [30]), Streptococcus pneumoniae (ρ/θ, 2.1 [22]), Clostridium perfringens (ρ/θ, 3.2 [47]), and Neisseria meningitis (ρ/θ, 1.1 [22]), but roughly similar to that observed for other groups, like lineage I of Listeria monocytogenes (ρ/θ, 0.13 [9]). With some minor exceptions, a 50% majority rule ClonalFrame consensus tree recovered most of the same groups apparent in the eBURST 16-gene analysis (Fig. 1). Phylogenetic analyses using more-traditional approaches, such as neighbor joining (NJ; data not shown), of the 16-gene concatenated alignment also recovered the same ClonalFrane groups highlighted in Fig. 1 (bovine A and B and poultry A and B), with one minor exception: bovine isolate cco067 did not group with other bovine isolates in the NJ tree. The levels of neighbor-joining bootstrap support for the groups indicated in Fig. 1 were as follows: for bovine A, 100%; for bovine B, 90%; for poultry A, 61%; and for poultry B, 80% (relationships between these groups were unresolved). A comparison of the ClonalFrame tree without correction for recombination (Fig. 1A) to that with correction for recombination (Fig. 1B) indicates that the time to the most recent common ancestor (TMRCA) for bovine group A that ignores recombination (Fig. 1A) is about 0.18 coalescent units and that the TMRCA which incorporates recombination (Fig. 1B) is about 0.05. On the other hand, bovine group B has values for TMRCA that are about 0.07 and 0.05 coalescent units for histories which ignore and incorporate recombination, respectively. This indicates that recombination has played a different role in the diversification of these two bovine groups; however, the similar values for TMRCA for the two groups in Fig. 1B suggests (assuming that the mutation rate follows a molecular clock in both lineages) that the two groups are of similar age. There are also two poultry clades, one of which (poultry B) includes within it a turkey-specific subclade. The tree ignoring recombination suggests coalescent times for the poultry A and B clades of about 0.03 and, when recombination is incorporated, about 0.05. Thus, the data suggest that these two poultry groups are of ages similar to one another and to the bovine groups, while the effect of recombination in the diversification of these two clades is more even and overall less significant than that observed for the bovine case. However, recombination does appear to be an important factor in the diversification of the turkey-specific subclade of poultry B; the TMRCA observed for this turkey group when recombination is ignored is less than 0.01, and that observed when recombination is incorporated is about 0.025. This also suggests a relatively recent origin for this turkey group. Multi-isolate, exclusively human clades were not evident; however, a partial human clade was present in both trees, inclusive of three turkey isolates (cco106, cco117, and cco080), with coalescent times without and with correction for recombination at about 0.045 and 0.085 coalescent units, respectively. This suggests a possible older origin for this human/turkey group than for the two poultry and bovine clades. There were no multi-isolate swine clades, but several pairwise associations were scattered throughout both trees.
TABLE 3.
Recombination rates inferred by ClonalFrame analysisa
| Run | ρb | r/mc | ρ/θd |
|---|---|---|---|
| 1 | 40.76 (27.60 58.24) | 1.56 (1.14 2.11) | 0.24 (0.18 0.32) |
| 2 | 37.24 (26.96 52.19) | 1.55 (1.13 2.06) | 0.24 (0.18 0.32) |
| 3 | 40.31(27.17 56.30) | 1.58 (1.14 2.10) | 0.25 (0.18 0.32) |
| 4 | 41.17 (27.46 59.15) | 1.55 (1.12 2.11) | 0.24 (0.18 0.32) |
| 5 | 39.62 (26.14 57.81) | 1.57 (1.14 2.09) | 0.25 (0.18 0.37) |
| All runs combined | 40.63 (26.80 58.20) | 1.56 (1.14 2.10) | 0.25 (0.18 0.32) |
The mean values for the parameters are shown, with the 95% credibility intervals given in parentheses.
Recombination rate.
Relative impact of recombination in comparison to point mutation in the genetic diversification of the lineage.
Relative frequency of occurrence of recombination in comparison to point mutation in the history of the lineage.
FIG. 1.
ClonalFrame trees for the 16-gene MLST data set. (A) Fifty-percent majority rule consensus tree without correction for recombination. (B) Fifty-percent majority rule consensus tree that incorporates recombination in the phylogenetic reconstruction. T, isolates from turkey; C, isolates from chickens; H, isolates from humans; S, isolates from swine; and B, isolates from bovines.
Our sample of isolates does, however, have certain geographic aspects to it that should be considered along with any inferences regarding host preference. For example, our bovine samples come predominately from the states of Washington and California, and the two bovine groups discussed above have a distinct state bias to their composition: bovine group A includes 9 bovine isolates and 1 swine isolate, of which 8 bovine isolates come from Washington State and 1 comes from Oregon. Bovine group B includes 8 bovine isolates, 6 from California and 2 from Washington State. For these bovine isolates, we have more specific information available, and if we look at this from the perspective of town or herds, this geographic tendency tends to break down, with isolates from different herds or towns frequently more closely related on the 16-gene ClonalFrame tree; thus, there tends to be a state clustering but not a town or herd clustering. Interestingly, Miller et al. (39) noted that C. coli samples from bovines tended to be more clonal than those derived from other hosts. They found ST 1068 (7 locus MLST scheme) to comprise 83% of the 63 isolates sampled from bovines in 26 feedlots and 11 different states. We have 7 examples of ST 1068, 6 of them from bovines and 1 from a human. Thus, although our geographic sample of isolates from bovines is limited, our results are consistent with a pattern of particular clones being predominately associated with this host organism on a much broader geographic scale. An examination of the relative importance of geography versus host species in the poultry and human host groups tends to provide mixed support for the significance of geography. For example, the human/turkey ClonalFrame group includes 4 human isolates and 3 turkey isolates; all of the turkey isolates are from the United States, and all the human isolates are from Poland. However, our data set includes two further human isolates from Poland, and these do not group together. Another point arguing against geographic influence involving the human isolates is that we have 7 human isolates in the data set from Switzerland and they appear in various places scattered throughout the ClonalFrame tree. Similarly, we have 5 chicken isolates from Switzerland, and they do not group together or with the human isolates from Switzerland. Overall, the genotyping and ClonalFrame analyses provide tentative support for the development of C. coli host-preferred groups and suggest that recombination has played various roles in their diversification; however, a more diversified geographic sampling involving proximal and distal samples from the same and different hosts would be necessary for definitive exclusion of geography as a contributing factor behind host group formation.
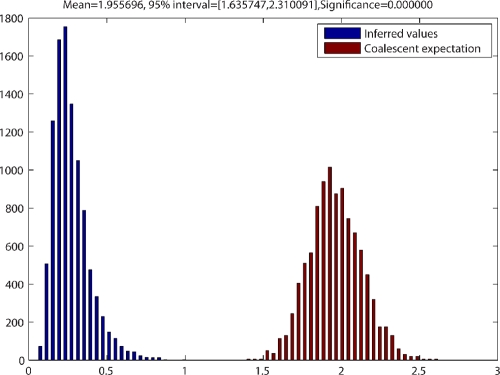
Yet further inferences regarding population history can be derived from the ClonalFrame analyses by implementing the external/internal branch length ratio test, which computes the lengths of the external branches divided by the sum of the lengths of the internal branches, compares it to the expected distribution under the coalescent model, and calculates the statistical significance of the deviation between the observed and expected ratios. In our case, the external-to-internal branch length ratio is significantly smaller than expected (Fig. 2), which indicates that our C. coli evolutionary history is consistent with an expansion of population size or the acquisition of a fitness advantage early in the history of the group (11). This is what one might expect with repeated evolution of multiple host-preferred groups and the presumed population expansion and/or fitness advantage accompanying the development of new host resources. It has been suggested elsewhere (51) that C. coli and C. jejuni may be converging as a consequence of recent changes in gene flow, involving an acceleration of import of C. jejuni alleles by C. coli, and that this could be associated with the development of agricultural practices that brought the two taxa together (however, for an alternative opinion, see references 5 and 60). Part of this overall picture could be the repeated evolution of C. coli host-preferred groups, which may be coincidental with the widespread development of the bovine and poultry industries, reflecting a highly adaptable bacterial species.
FIG. 2.
Distribution obtained by testing the external/internal branch length ratios of trees resulting from ClonalFrame analysis. Shown in blue is the distribution of the sum of the lengths of the external branches (connecting leaves of the tree) divided by the sum of the lengths of the internal branches (connecting internal nodes). Shown in red is the expected distribution of the external-to-internal branch length ratio under the coalescent model. The statistical significance of the deviation between the observed and expected ratios is shown at the top of the figure. The x axis represents the external/internal branch length ratio, and the y axis represents the tree sample frequency.
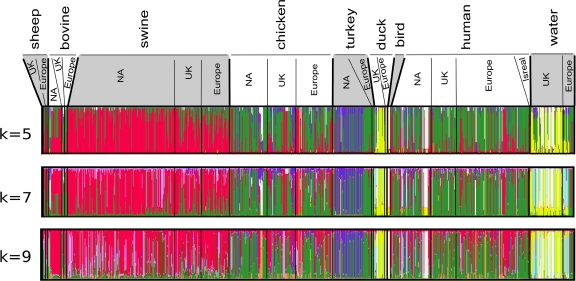
The limitations of our data regarding lack of geographic diversity in the samples, and the relatively limited number of isolates, can to an extent be addressed in analysis of the pubMLST database; however, in our opinion the drawback to this database is the resolution provided by the 7-gene data set. For example, ClonalFrame analysis of just our 84 isolates by use of only the 7-gene MLST data set (data not shown), which incorporates recombination in the evolutionary reconstructions, fails to recover any of the host groups discussed above, with the exception of one bovine group, although the composition of this group is not at all similar to that observed in the 16-gene analysis. In fact, the vast majority of isolates in this 7-gene analysis are entirely unresolved. Nonetheless, it is possible to take advantage of the large number of isolates represented in the pubMLST database, with their relatively broad geographic distribution, and analyze the 7-gene MLST data collectively in ways other than ClonalFrame, employing population genetic approaches. Analysis using STRUCTURE assumes that the observed data are derived from K ancestral subpopulations. The optimal K was determined by doing multiple runs with different K values and choosing that with the highest likelihood score. Our analysis of C. coli by use of this approach suggests that a K value of 9 may be optimal; however, approximately similar results were obtained with K values as low as 5 (Fig. 3). The program, therefore, infers for each site of each sequence its posterior probability of deriving from one of the K ancestral subpopulations and computes the average proportion of genetic material derived from each ancestral subpopulation by each individual. The analysis is summarized and presented in Fig. 3 and illustrates a few important general trends: (i) isolates from swine tend to be a relatively homogeneous genetic group, largely distinct from isolates derived from other hosts; (ii) chicken and human isolates tend to show a great deal of genetic overlap, strongly suggesting that the majority of C. coli human infections arise from chickens; (iii) isolates from ducks and wild birds show distinct similarity to those from environmental water samples, suggesting transmission from wild birds to water or vice versa; (iv) isolates from turkeys have a connection with human infection similar to that observed for isolates from chickens, but with a greater proportion forming a unique genetic group; and (v) geography (at least at the macrogeographic level of North America, the United Kingdom, and Europe) tends to have little bearing on the determination of genetic groups, and host species is a more influential factor. Our surmise above regarding the potential source of human infections is in agreement with recent studies of C. jejuni and C. coli which find chicken meat as the most likely source of human infection for both species of Campylobacter (50, 59). This analysis is also in agreement with other studies suggesting that campylobacteriosis due to C. coli infection is less commonly associated with consumption of pork products (35, 52). Earlier studies have also implicated waterfowl as a possible source of water infection (e.g., reference 40), but generally, this evidence has centered primarily on C. jejuni; our analysis provides a probable similar connection for wild birds and water involving C. coli. The degree to which contaminated water is then a source of human infection remains uncertain, although there are occasional outbreaks which have been linked in this regard (24, 28). A recent study from northwest England suggests that C. coli samples from surface water comprise a population genetically distinct from that described for human cases of disease (52). A population genetic study of C. jejuni involving modeling DNA sequence evolution of MLST genotyping data from over 1,000 clinical isolates in Lancashire, England, concludes that the vast majority (97%) of sporadic disease can be attributed to animals farmed for meat and poultry (59). Our analysis of C. coli supports only minimal direct connection between birds/water and humans, although the connection between birds and water seems quite clear. AMOVA of the 7-gene C. coli pubMLST database, using Arlequin, does not support geography (North America, Europe, and the United Kingdom) as a significant factor in explaining the observed genetic variation but does support host species as a significant factor in explaining genetic variation (Table 4). It should be acknowledged that although the pubMLST database includes a variety of hosts from diverse geographic regions, the sample set is not completely randomized with regard to host species and geography. Some hosts, for example, are better represented in certain geographic areas, and there is a lack of information on the ecological success of isolates from different host/geographic settings. Nonetheless, the results that we obtained with ClonalFrame, STRUCTURE, and AMOVA collectively suggest that the most parsimonious explanation is that isolates have evolved certain host preferences and then spread throughout different geographic areas rather than that certain clones diversify, largely independently, in separate geographic regions.
FIG. 3.
Genetic mixture of different C. coli host groups determined by STRUCTURE analysis. Results for 3 different values of K are presented, with a K value of 9 judged to be optimal. Geographic regions from which the isolates arise are indicated along the top. Each part of the figure consists of vertical bars, one for each isolate. Each bar shows the proportion of nucleotides inherited from different color-coded ancestral populations. NA, North America.
TABLE 4.
AMOVA for 7 gene MLST data for C. coli isolates grouped by host and geographic region
| Structure tested | df | Sum of squares | Variance component | % of variance | F statistica | P |
|---|---|---|---|---|---|---|
| Geographic groups (North America vs UK vs Europe) | ||||||
| Among groups | 2 | 1147.719 | −0.52905 | −1.54 | FCT = −0.01537 | 0.32649 |
| Among populations/within groups | 16 | 6356.643 | 8.55246 | 24.84 | FSC = 0.24467 | 0.00000 |
| Among individuals/within populations | 936 | 24712.207 | 26.40193 | 76.69 | FST = 0.23307 | 0.00000 |
| Host groups (bovine vs swine vs chicken vs turkey vs duck vs wild bird vs human vs water) | ||||||
| Among groups | 7 | 6220.950 | 7.07052 | 19.90 | FCT = 0.19905 | 0.00000 |
| Among populations/within groups | 11 | 1283.412 | 2.04969 | 5.77 | FSC = 0.07204 | 0.00000 |
| Among individuals/within populations | 936 | 24712.207 | 26.40193 | 74.33 | FST = 0.25675 | 0.00000 |
FCT, variance among groups; FSC, variance within groups among populations; FST, variance within populations among individuals.
Core and variable genomes.
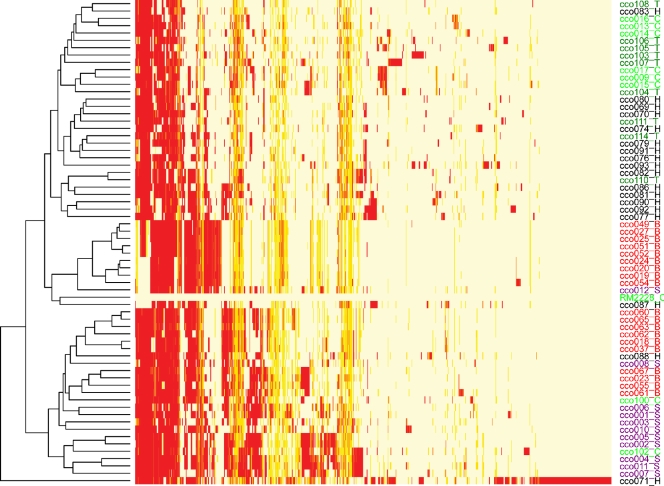
The hybridization profile for each of 65 strains analyzed in this study (Fig. 4) was obtained by hybridizing labeled genomic DNA to a C. coli microarray designed from sequence data for the genome strain RM2228. Divergent and absent genes are expected to show decreased hybridization signals with respect to those obtained with the reference, sequenced strain, while the core C. coli genome consists of genes present and highly conserved in nucleotide sequence across a wide diversity of strains. In the first design, 991 ORFs out of 1,942 tested ORFs were conserved across test strains. This study employed a double design strategy to eliminate potential ambiguous calls. The second design was based on 615 ambiguous ORFs, which showed no hybridization or intermediate levels of hybridization in at least one test strain in the first design. Of this set of 615 ORFs, 98 were misidentified as absent or highly divergent in the first design and were corrected as present in the second design. Of the tested 1,942 genes, 1,089 were common to all strains, representing 56% of the C. coli RM2228 genome; 853 ORFs were variable in at least one of the 65 test strains. However, based on our FNR estimate of 4% (see Materials and Methods, “Verification of microarray data”), we can predict that the microarray will overlook some core genes. For example, assuming a Bernoulli distribution, with the 65 test strains, the probability that a core gene will be found absent in at least one strain is 0.93, while the probability that a core gene will be found in 60 or more strains is 0.95. If one considers that any gene present in at least 60 strains is a core gene, this is consistent with the convention of Lan and Reeves (29), which suggests that genes present in 95% or more of independent strains are considered the core genome, and we obtain a core genome size estimate of 1,473, which is approximately similar to other estimates of core genome size for C. jejuni (15). This result highlights the fact that microarrays will generally underestimate the core genome size, as the number of false negatives quickly sums up, and therefore, microarray approaches should be considered a minimum estimate of the core genome. This phenomenon is illustrated by a core genome size cumulative plot (see Fig. S4 in the supplemental material), where it is apparent that, despite the analysis of a relatively large number of strains, a plateau was not reached. A similar result, pertaining to core genome size estimation from microarray data, was reported recently for Streptococcus thermophilus (46). A list of the C. coli core genome loci appears in Table S2 in the supplemental material.
FIG. 4.
Heat map dendrogram involving the dispensable portion of the C. coli genome across 65 test isolates. Red, absent locus; orange, divergent locus; yellow, present locus. T, isolates from turkey; C, isolates from chickens; H, isolates from humans; S, isolates from swine; and B, isolates from bovines. The y axis dendrogram is a hierarchical clustering of the strains based on gene content.
Dendrogram clustering of the gene presence/absence data indicated that isolates of the same host tended to cluster together (Fig. 4). For example, the heat map dendrogram suggests a poultry group, including 67% (12/18) of the poultry isolates in our analysis, plus a single human isolate (cco083). This group bears only partial similarity in terms of isolate composition to the poultry clades on the ClonalFrame tree, instead including isolates scattered throughout the ClonalFrame tree, suggesting that the similarity in genome composition evident in the heat map clustering is largely independent of common ancestry and instead reflects a tendency for certain genes to be more common in isolates derived from poultry hosts, presumably arising through lateral gene transfer (LGT). A similar line of logic applies to the human isolates, which form two adjacent heat map dendrogram clusters. One of these groups bears some partial resemblance in isolate composition to an association of human isolates in the ClonalFrame analysis, while the other group is composed of human isolates scattered throughout the ClonalFrame tree. There are also two bovine heat map clusters; however, in this case they are not immediately adjacent to one another, and both of these are identical, or nearly so, in composition to the ClonalFrame bovine clades. Finally, in complete contrast to the ClonalFrame analysis, where nearly all the swine isolates were scattered throughout the tree, all but two of the swine isolates cluster together, with the inclusion of a single chicken isolate. Thus, we have a combination of common ancestry in some cases and lateral gene transfer in others, which underlie a tendency for sets of genes to be common to isolates derived from particular hosts. It should be noted, however, that factors other than LGT, such as lineage-specific gene deletion, could result in some of the same clustering as what we observe in Fig. 4.
Groups of genes common to particular host groups were occasionally evident in a more detailed inspection of the microarray data. For example, in the case of the bovine isolates, there was a group of genes common to bovine group A that was largely absent from the rest of the isolates, which included the following 8 loci: haloacid dehalogenase hydrolase (CCO1528), cyclase (CCO1530), dihydroxyhept-2-ene-1,7-dioic acid aldolase (CCO1533), alpha-2,3-sialyltransferase (CCO1538), sulfate adenylyltransferase (CCO1541), 3′(2′),5′-bisphosphate nucleotidase (CCO1543), a glycosyl transferase (CCO1546), and capsular polysaccharide synthesis (CPS) C (CCO1547). Conversely, there was also a group of genes largely absent in bovine group A, generally present in the other isolates, including 9 loci: periplasmic proteins (CCO0110 and CCO0113), peptidyl-prolyl cis-trans isomerase (CCO1240), hypothetical proteins (CCO1241 and CCO1672), arginyl-tRNA synthetase (CCO1244), uroporphyrinogen decarboxylase (CCO1333), a MoaA/NifB/PqqE family protein (CCO1334), and a membrane protein (CCO1335). Of course, this assessment of presence and absence is all relative to the reference microarray strain RM2228, and because of the likelihood of considerable gene diversity in the dispensable portion of the C. coli genome (see, for example, references 31 and 55), this will not reflect a thorough picture of genes that are possibly present or absent in particular host groups. However, the facts that we do detect a few genes that appear characteristic of some groups and that we do get some clustering of host specificity on the heat map dendrogram strongly suggest that there are sets, or combinations of genes, more important to particular types of host adaptation.
A detailed look at the gene presence/absence data for several important pathogenic gene clusters across the different strains of C. coli reveals very different levels of gene conservation across the different gene regions (Fig. 5). The most divergent region was the CPS locus, with only 5 strains having gene composition similar to that observed for the sequenced strain. The majority of the remaining strains did not have just a few genes that were different from RM2228, but instead, most of these strains had an almost entirely different gene composition for this cluster. This suggests enormous gene diversity for the CPS locus in the species C. coli, similar to that reported in comparative genomic hybridization studies involving C. jejuni (41, 45) and for comparative sequence studies of other species of pathogenic bacteria (e.g., Streptococcus pneumoniae [3]). The genes involved in O-linked glycosylation and lipid oligosaccharide synthesis (LOS) were the next most variable gene clusters. No clear pattern of presence and absence within these clusters was correlated with host type. In each of these gene groups, there were a few loci that were consistently represented across all isolates and blocks of genes that were much more variable. The block of genes that were most frequently absent in the LOS locus (CCO1211 to CCO1218) includes loci which share variable levels of low homology with C. jejuni strains and includes several sialyl transferase genes, which have been implicated in Guillain-Barre and Fisher syndromes (61). This block of genes was generally either entirely absent or, more rarely, entirely present across the C. coli strains. Similar to genomic comparisons of C. jejuni strains (15), the N-linked glycosylation gene cluster (pgl) was largely conserved in gene composition across C. coli isolates.
FIG. 5.
Gene presence/absence for several important C. coli pathogenic gene clusters. Red, absent locus; yellow, divergent locus; blue, present locus. LOS, lipooligosaccharide synthesis; pgl, N-linked glycosylation; CPS, capsular polysaccharide synthesis.
The results of this work suggest C. coli may have evolved multiple host-preferred groups, although more-extensive geographic sampling would be required for definitive evaluation of this issue. Comparative genomic hybridization data suggested that there were combinations of genes in the dispensable portion of the genome, more commonly associated with isolates derived from particular hosts, with a history of common ancestry in some cases and lateral gene transfer in others. This suggests that a more thorough understanding of the pan-genome of C. coli could result in the identification of genes of key functional significance to host specific adaptation.
Supplementary Material
Acknowledgments
This work was supported by NIH contract N01-AI-30054 (ZC003-05), awarded to M.J.S.
Footnotes
Published ahead of print on 22 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alfredson, D. A., and V. Korolik. 2007. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 277:123-132. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, R. P., F. E. Nascimento, J. Chau, S. S. Prado, C. W. Tsai, S. A. Lopes, and J. R. Lopes. 2008. Genetic structure and biology of Xylella fastidiosa strains causing disease in citrus and coffee in Brazil. Appl. Environ. Microbiol. 74:3690-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bull, S. A., V. M. Allen, G. Domingue, F. Jorgensen, J. A. Frost, R. Ure, R. Whyte, D. Tinker, J. E. L. Corry, J. Gillard-King, and T. J. Humphrey. 2006. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl. Environ. Microbiol. 72:645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caro-Quintero, A., G. P. Rodriguez-Castano, and K. T. Konstantinidis. 2009. Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species. J. Bacteriol. 191:5824-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champion, O. L., M. W. Gaunt, O. Gundogdu, A. Elmi, A. A. Witney, J. Hinds, N. Dorrell, and B. W. Wren. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U. S. A. 102:16043-16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colles, F. M., K. E. Dingle, A. J. Cody, and M. C. J. Maiden. 2008. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl. Environ. Microbiol. 74:3583-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delcher, A. L., K. A. Bratke, E. C. Powers, and S. L. Salzberg. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Bakker, H., X. Didelot, E. Fortes, K. Nightingale, and M. Wiedmann. 2008. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol. Biol. 8:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didelot, X., M. Barker, D. Falush, and F. G. Priest. 2009. Evolution of pathogenicity in the Bacillus cereus group. Syst. Appl. Microbiol. 32:81-90. [DOI] [PubMed] [Google Scholar]
- 11.Didelot, X., and D. Falush. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingle, K. E., F. M. Colles, D. Falush, and M. C. J. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do, T., K. A. Jolley, M. C. Maiden, S. C. Gilbert, D. Clark, W. G. Wade, and D. Beighton. 2009. Population structure of Streptococcus oralis. Microbiology 155:2593-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falush, D., M. Stephens, and J. K. Pritchard. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7:574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falush, D., M. Stephens, and J. K. Pritchard. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falush, D., M. Torpdahl, X. Didelot, D. F. Conrad, D. J. Wilson, and M. Achtman. 2006. Mismatch induced speciation in Salmonella: model and data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, C., W. P. Hanage, and B. G. Spratt. 2005. Neutral microepidemic evolution of bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 102:1968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.French, N. P., A. Midwinter, B. Holland, J. Collins-Emerson, R. Pattison, F. Colles, and P. Carter. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frost, J. A. 2001. Current epidemiological issues in human campylobacteriosis. J. Appl. Microbiol. 90:85S-95S. [DOI] [PubMed] [Google Scholar]
- 25.Gelman, A., and D. B. Rubin. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7:457-472. [Google Scholar]
- 26.Ghindilis, A. L., M. W. Smith, K. R. Schwarzkopf, K. M. Roth, K. Peyvan, S. B. Munro, M. J. Lodes, A. G. Stover, K. Bernards, K. Dill, and A. McShea. 2007. CombiMatrix oligonucleotide arrays: genotyping and gene expression assays employing electrochemical detection. Biosens. Bioelectron. 22:1853-1860. [DOI] [PubMed] [Google Scholar]
- 27.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, and K. R. Neal. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänninen, M.-L., H. Haajanen, T. Pummi, K. Wermundsen, M. L. Katila, H. Sarkkinen, I. Miettinen, and H. Rautelin. 2003. Detection and typing of Campylobacter jejuni and Campylobacter coli and analysis of indicator organisms in three waterborne outbreaks in Finland. Appl. Environ. Microbiol. 69:1391-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 30.Lang, P., T. Lefébure, W. Wang, R. N. Zadoks, Y. Schukken, and M. J. Stanhope. 2009. Gene content differences across strains of Streptococcus uberis identified using oligonucleotide microarray comparative genomic hybridization. Infect. Genet. Evol. 9:179-188. [DOI] [PubMed] [Google Scholar]
- 31.Lefebure, T., and M. J. Stanhope. 2007. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 8:R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 33.Leonard, E. E., II, L. S. Tompkins, S. Falkow, and I. Nachamkin. 2004. Comparison of Campylobacter jejuni isolates implicated in Guillain-Barre syndrome and strains that cause enteritis by a DNA microarray. Infect. Immun. 72:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, H., J. Ruan, and R. Durbin. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Litrup, E., M. Torpdahl, and E. M. Nielsen. 2007. Multilocus sequence typing performed on Campylobacter coli isolates from humans, broilers, pigs and cattle originating in Denmark. J. Appl. Microbiol. 103:210-218. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy, N. D., F. M. Colles, K. E. Dingle, M. C. Bagnall, G. Manning, M. C. Maiden, and D. Falush. 2007. Host-associated genetic import in Campylobacter jejuni. Emerg. Infect. Dis. 13:267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meinersmann, R. J., R. W. Phillips, K. L. Hiett, and P. Fedorka-Cray. 2005. Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl. Environ. Microbiol. 71:6368-6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletz, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 40.Obiri-Danso, K., and K. Jones. 1999. Distribution and seasonality of microbial indicators and thermophilic campylobacters in two freshwater bathing sites on the River Lune in northwest England. J. Appl. Microbiol. 87:822-832. [DOI] [PubMed] [Google Scholar]
- 41.Parker, C. T., B. Quinones, W. G. Miller, S. T. Horn, and R. E. Mandrell. 2006. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 44:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 43.Poly, F., D. Threadgill, and A. Stintzi. 2004. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J. Bacteriol. 186:4781-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinones, B., M. R. Guilhabert, W. G. Miller, R. E. Mandrell, A. J. Lastovica, and C. T. Parker. 2008. Comparative genomic analysis of clinical strains of Campylobacter jejuni from South Africa. PLoS One 3:e2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen, T. B., M. Danielsen, O. Valina, C. Garrigues, E. Johansen, and M. B. Pedersen. 2008. Streptococcus thermophilus core genome: comparative genome hybridization study of 47 strains. Appl. Environ. Microbiol. 74:4703-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rooney, A. P., J. L. Swezey, R. Friedman, D. W. Hecht, and C. W. Maddox. 2006. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics 172:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosef, O., B. Gondrosen, G. Kapperud, and B. Underdal. 1983. Isolation and characterization of Campylobacter jejuni and Campylobacter coli from domestic and wild mammals in Norway. Appl. Environ. Microbiol. 46:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin: a software for population genetics data analysis. Version 2.000. University of Geneva, Geneva, Switzerland.
- 50.Sheppard, S. K., J. F. Dallas, N. J. Strachan, M. MacRae, N. D. McCarthy, D. J. Wilson, F. J. Gormley, D. Falush, I. D. Ogden, M. C. Maiden, and K. J. Forbes. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheppard, S. K., N. D. McCarthy, D. Falush, and M. C. J. Maiden. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237-239. [DOI] [PubMed] [Google Scholar]
- 52.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, S. James, J. Kempster, M. Painter, V. Edwards-Jones, K. Osborn, M. Regan, Q. Syed, and E. Bolton. 2010. Investigation of food and environmental exposures relating to the epidemiology of Campylobacter coli in humans in northwest England. Appl. Environ. Microbiol. 76:129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taboada, E. N., R. R. Acedillo, C. D. Carrillo, W. A. Findlay, D. T. Medeiros, O. L. Mykytczuk, M. J. Roberts, C. A. Valencia, J. M. Farber, and J. H. E. Nash. 2004. Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J. Clin. Microbiol. 42:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tay, C. Y., H. Mitchell, Q. Dong, K. L. Goh, I. W. Dawes, and R. Lan. 2009. Population structure of Helicobacter pylori among ethnic groups in Malaysia: recent acquisition of the bacterium by the Malay population. BMC Microbiol. 9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. DeBoy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. B. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thakur, S., D. G. White, P. F. McDermott, S. Zhao, B. Kroft, W. Gebreyes, J. Abbott, P. Cullen, L. English, P. Carter, and H. Harbottle. 2009. Genotyping of Campylobacter coli isolated from humans and retail meats using multilocus sequence typing and pulsed-field gel electrophoresis. J. Appl. Microbiol. 106:1722-1733. [DOI] [PubMed] [Google Scholar]
- 57.Volokhov, D., V. Chizhikov, K. Chumakov, and A. Rasooly. 2003. Microarray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 41:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waldenström, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenaar, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, D. J., E. Gabriel, A. J. H. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, P. Fearnhead, C. A. Hart, and P. J. Diggle. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson, D. J., E. Gabriel, A. J. H. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, C. A. Hart, P. J. Diggle, and P. Fearnhead. 2009. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 26:385-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuki, N. 2007. Campylobacter sialyltransferase gene polymorphism directs clinical features of Guillain-Barre syndrome. J. Neurochem. 103(Suppl. 1):150-158. [DOI] [PubMed] [Google Scholar]
- 62.Zerbino, D. R., and E. Birney. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.