Abstract
Of the seven genetic groups, or assemblages, currently recognized in the Giardia duodenalis species complex, only assemblages A and B are associated with human infection, but they also infect other mammals. Recent investigations have suggested the occurrence of genetic exchanges among isolates of G. duodenalis, and the application of assemblage-specific PCR has shown both assemblages A and B in a significant number of human infections. In this work, three real-time quantitative (qPCR) assays were developed to target the G. duodenalis triose phosphate isomerase, glutamate dehydrogenase, and open reading frame C4 sequences. Primers were designed to allow the specific amplification of the DNA of assemblage A or B and to generate products distinguishable by their melting curves or, after qPCR, by their sequences, sizes, or restriction patterns. The assays showed full specificity and detected DNA from a single trophozoite (4 to 8 target copies). We applied these assays, as well as a TaqMan assay that targets the β-giardin gene, to genomic DNA extracted from 30 human stools and to Giardia cysts purified by immunomagnetic capture from the same samples. Simultaneous detection of both assemblages was observed in a large number of DNAs extracted from stools, and experiments on the cysts purified from the same samples showed that this was essentially attributable to mixed infections, as only one assemblage was detected when dilutions of cysts were tested. In a few cases, detection of both assemblages was observed even when single cysts were tested. This result, which suggests the presence of recombinants, needs to be confirmed using more accurate methods for cyst separation and enumeration. The assays described in this study can be used to detect Giardia cysts infectious to humans in samples from animals and in water and food.
Giardia duodenalis (syn. Giardia intestinalis and Giardia lamblia) is the only species within the genus Giardia that infects humans, although it is also found in other mammals, including pets and livestock (1). The infection has a global distribution and, with an estimated 2.8 × 108 cases per year, represents the most common gastrointestinal parasitic infection of humans in developed countries (20). In Asia, Africa, and Latin America, about 200 million people have symptomatic giardiasis, with some 500,000 new cases reported each year (35). Several characteristics of G. duodenalis influence the epidemiology of infection: (i) in humans, the infective dose is about 10 to 100 cysts; (ii) cysts are immediately infectious when excreted in feces and can be transmitted by person-to-person or animal-to-animal contact; (iii) cysts are remarkably stable and can survive for weeks to months in the environment; and (iv) environmental contamination can lead to the contamination of drinking water and food (6, 32).
A considerable amount of data has shown that G. duodenalis should be considered a species complex whose members show little variation in their morphology yet can be assigned to at least seven distinct assemblages (A to G) based on genetic analyses (7, 34). The analysis of more than a thousand human isolates from different geographical locations, examined by PCR amplification of DNA extracted directly from feces, has demonstrated that in almost all cases, only G. duodenalis assemblages A and B are associated with human infections (6). The prevalence of each assemblage varies considerably from country to country; assemblage B seems more common overall, but no strong conclusions can be drawn from current data. The remaining assemblages (C to G) are likely to be host specific, as assemblages C and D have been identified in dogs, cats, coyotes, and wolves; assemblage E in cattle, sheep, goats, pigs, water buffaloes, and muflons; assemblage F in cats; and assemblage G in rats.
The epidemiology of human giardiasis is further complicated by the occurrence of mixed infections and the possibility of genetic exchanges between isolates of assemblage A (10) or even between isolates of assemblages A and B (21, 33). Ideally, genotyping should be performed on single cysts, as this allows a distinction between mixed infections and recombinants. To reach this technically demanding high level of sensitivity and specificity, real-time quantitative PCR (qPCR) appears to be a promising technique.
This work describes the development of new qPCR assays that, through the use of assemblage-specific primers, allow the specific and simultaneous detection of DNAs of assemblages A and B. The application of these assays to DNA extracted from human stools and to cysts purified from the same samples is described.
MATERIALS AND METHODS
Investigated isolates.
Genomic DNAs from reference strains of assemblage A (WB and Bris-162) and assemblage B (Ad28 and GS/M) (24, 27) were used to test the specificities of the qPCR assays. Freshly collected trophozoites of the WB strain, grown under axenic conditions, were concentrated by centrifugation, resuspended in phosphate-buffered saline (PBS), counted under the microscope, and used to test the sensitivities of the qPCR assays.
Human fecal samples positive for Giardia (n = 30) were used in the study. The samples were collected in Italy from both residents (n = 7) and immigrants (n = 4) and from a group of African children of the Saharwi population (19). The samples were from asymptomatic and symptomatic patients of both sexes, with ages from 1 to 59 years. All the samples had been previously genotyped by standard PCR at the triose phosphate isomerase (tpi) gene, the glutamate dehydrogenase (gdh) gene, the open reading frame C4 (orfC4) gene, and the β-giardin gene under the conditions described previously (8, 37). Genotyping results were reported elsewhere (8, 19), except those for the orfC4 gene, which were not published before.
From the same samples, G. duodenalis cysts were recovered from frozen aliquots (about 500 μl) of stools by an immunomagnetic separation method (Dynabeads GC-Combo; Invitrogen, Norway) according to the manufacturer's instructions. The purified cysts were enumerated by immunofluorescence (IF) microscopy using fluorescein isothiocyanate (FITC)-conjugated cyst wall-specific antibodies (Merifluor; Meridian Bioscience, Cincinnati, OH) and further evaluated for integrity of the nuclei by staining them with 4′-6-diamidino-2-phenylindole (DAPI). The percentage of DAPI-positive cysts in the total amount of cysts was estimated. The cysts were stored at 4°C in sterile water until they were used.
Design of assemblage-specific primers.
Giardia duodenalis A and B assemblage-specific primers targeting the tpi, gdh, and orfC4 genes were designed to be used in qPCR assays with a SYBR green detection mode. All available DNA sequences were retrieved from GenBank and aligned using the software SeqMan II (DNAstar, Madison, WI). Over these multiple alignments, primers were designed to match regions (about 20 bp in length) showing the largest number of fixed differences between assemblage A and assemblage B sequences. The primers are listed in Table 1.
TABLE 1.
Primers used in the studya
| Primer | Sequence | Binding site | Amplicon size (bp) | Melting peak (°C) |
|---|---|---|---|---|
| TPI A forward primer | 5′-TCGTCATTGCCCCTTCCGCC-3′ | 116-135 | 77 | 86 |
| TPI B sequence | 5′-.T..TG....T..C...TTT-3′ | |||
| TPI A reverse primer | 3′-CAGTTGAGGATAGCAGCG-5′ | 175-192 | ||
| TPI B sequence | 3′-TGTC...AA.........-5′ | |||
| TPI B forward primer | 5′-GATGAACGCAAGGCCAATAA-3′ | 397-416 | 77 | 80 |
| TPI A sequence | 5′-..C..G...........CCG-3′ | |||
| TPI B reverse primer | 3′-AAGAAGGAGATTGGAGAATC-5′ | 454-473 | ||
| TPI A sequence | 3′-GGC......C.C.....G..-5′ | |||
| GDH A forward primer | 5′-CCGGCAACGTTGCCCAGTTT-3′ | 710-729 | 180 | 83 |
| GDH B sequence | 5′-.T...........T....AC-3′ | |||
| GDH A reverse primer | 3′-TCCGAGTTCAAGGACAAGT-5′ | 871-889 | ||
| GDH B sequence | 3′-G.T...........G....-5′ | |||
| GDH B forward primer | 5′-CGTATTGGCGTCGGCGGT-3′ | 492-510 | 133 | 83 |
| GDH A sequence | 5′-...C..C..........CC-3′ | |||
| GDH B reverse primer | 3′-CTATCAGACCAGAGGCCACA-5′ | 604-623 | ||
| GDH A sequence | 3′-T..........G........G-5′ | |||
| ORFC4 A forward primer | 5′-CTGTAGACAGGGCCCAGGCC-3′ | 135-154 | 103 | 85 |
| ORFC4 B sequence | 5′-......G....AG..G.ATG-3′ | |||
| ORFC4 A reverse primer | 3′-ATTAAGGGCAGGGGACATCAT-5′ | 217-237 | ||
| ORFC4 B sequence | 3′-GGC...T.T.A.....G....-5′ | |||
| ORFC4 B forward primer | 5′-ACTGTCCATTTCTATCTGAG-3′ | 281-300 | 171 | 79 |
| ORFC4 A sequence | 5′-C...C........G..ATCC-3′ | |||
| ORFC4 B reverse primer | 3′-AGGTGGAGGCCAATGGAATCC-5′ | 431-451 | ||
| ORFC4 A sequence | 3′-CT..A.....AG........A-5′ |
The sequence of each assemblage-specific primer is aligned over the corresponding sequence of the other assemblage. The dots indicate identical nucleotides. The 3′ ends of each primer are in boldface. The binding site, amplicon size, and melting peaks are also given.
Assemblage-specific qPCR assays.
Each PCR consisted of 1× LightCycler 480 Probes Master mix (containing FastStart Taq DNA polymerase, a proprietary buffer, deoxynucleotide triphosphates [dNTPs], and MgCl2 at a 3.2 mM final concentration), 300 nM each primer, SYBR green I dye (at a 1:40,000 final concentration), and genomic DNA, trophozoites from in vitro culture, or purified cysts in a final volume of 20 μl. All reactions were performed in triplicate. PCRs were performed in a LightCycler 480 PCR system (Roche Diagnostics GmbH, Germany). The PCR conditions consisted of a 5-min incubation at 95°C, followed by 45 cycles of denaturation (95°C for 12 s), annealing (59°C for 12 s for the tpi and gdh assays or 62°C for 12 s for the orfC4 assay), and extension (72°C for 6 s). Fluorescence data were collected at the end of each cycle as a single acquisition. The melting curve program was performed at the end of each reaction and consisted of 95°C for 5 s, 60°C for 1 min, and heating to 97°C with continuous acquisition (5 acquisitions per degree Celsius). In every run, positive and negative controls were included, and the cycle threshold (CT) values and melting-curve values were recorded. A previously developed TaqMan assay targeting the β-giardin gene was used under the experimental conditions described previously (15).
Post-qPCR analysis.
The reaction products from the tpi qPCR were purified using the QIAquick PCR purification kit (Qiagen, Milan, Italy), and 10 μl of purified products was digested using HincII (Promega, Madison, WI) or AluI (New England BioLabs Inc., Beverly, MA) for 4 h at 37°C in a final reaction volume of 20 μl. The digestion products were separated by electrophoresis on a 3% MetaPhor agarose gel (Cambrex, Karlskoga, Sweden). The reaction products from the gdh qPCR were purified using the QIAquick PCR purification kit (Qiagen, Milan, Italy), and 10 μl of purified products was digested using DdeI (Promega, Madison, WI) for 4 h at 37°C in a final reaction volume of 20 μl. The digestion products were separated by electrophoresis on a 3% MetaPhor agarose gel (Cambrex, Karlskoga, Sweden). Randomly chosen qPCR gdh products were sequenced to confirm their identities.
RESULTS
Overall strategy of qPCR assays.
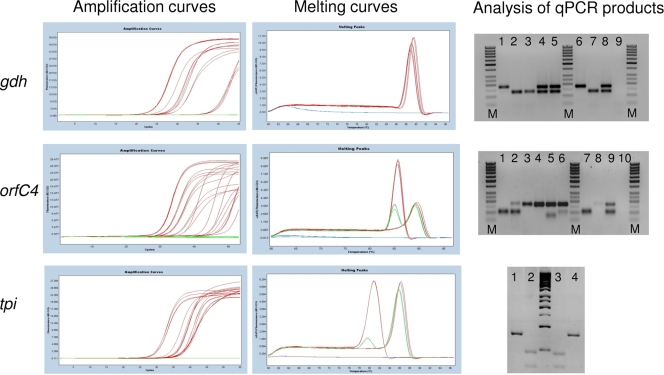
Primers were designed for each locus by taking into account the genetic variability found in all available sequences from assemblage A and B isolates. In particular, to increase their specificity in qPCR, the 3′ ends of each primer were designed to match regions where more than one adjacent substitution was found (Table 1). Next, the assemblage A and B amplification products were checked for their predicted melting curves, using the software Poland (http://www.biophys.uni-duesseldorf.de/local/POLAND/poland.html). This showed that the tpi and orfC4 qPCR amplification products from assemblages A and B had different melting curves, whereas this was not the case for the gdh qPCR (Table 1). Finally, to allow further confirmation of the specificity of the assays, primers were designed to generate amplified products having different sizes (gdh and orfC4) or different restriction patterns (tpi) (Table 1 and Fig. 1). The overall strategy of the qPCR assays is illustrated in Fig. 1.
FIG. 1.
Schematic representation of the amplification curves (left), melting curves (middle), and electrophoretic separation of products (right) obtained using the gdh, orfC4, and tpi qPCR assays. In gel electrophoresis of gdh products, lane M is the 50-bp size ladder; lanes 1 and 6 show the assemblage A product (180 bp); lanes 2, 3, and 7 show the assemblage B product (133 bp); lanes 4, 5, and 8 show the presence of both assemblages A and B; and lane 9 shows the negative control. In gel electrophoresis of orfC4 products, lane M is the 50-bp size ladder; lanes 1 and 7 show the assemblage A product (103 bp); lanes 3, 4, 5, and 8 show the assemblage B product (171 bp); lanes 2, 6, and 9 show the presence of both assemblages A and B; and lane 10 shows the negative control. In gel electrophoresis of tpi amplicons, the product from assemblage A is not cut by AluI (lane 1) but is cut by HincII into two fragments of 47 bp and 31 bp (lane 2), whereas the B product is cut by AluI into two fragments of 45 bp and 32 bp (lane 3) but is not cut by HincII (lane 4).
Specificities and sensitivities of qPCR assays.
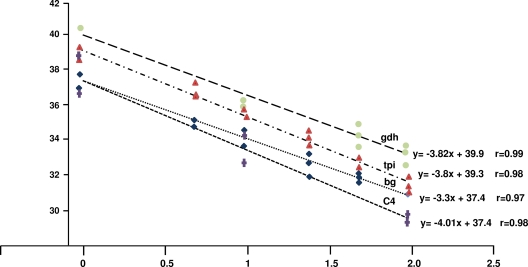
The specificities of the three qPCR assays were first evaluated with genomic DNAs from reference strains of assemblages A and B by testing the abilities of assemblage A-specific primers to amplify assemblage B genomic DNA, and vice versa. No cross-reaction was observed under the experimental conditions used (data not shown). Using 10-fold serial dilutions of genomic DNAs of the WB (assemblage A) and Ad28 (assemblage B) strains, the sensitivities of the assays were estimated to be close to 30 to 60 copies. This was calculated by taking into account the concentration of genomic DNA (measured by spectrophotometry), the genome size (11.7 Mb), and the ploidy (4n) of Giardia. When the sensitivity was estimated using dilutions of freshly collected and enumerated WB trophozoites, a detection level of a single trophozoite (4 to 8 copies, depending on the G1/G2 phase) was obtained using the tpi, gdh, and orfC4 assays. A similar level was also obtained using a published TaqMan assay that targets the β-giardin gene. The standard curves for the four qPCR assays are shown in Fig. 2.
FIG. 2.
Standard curves of the different qPCR assays obtained using serial dilutions of freshly collected trophozoites of the G. duodenalis WB strain using assemblage A-specific primer pairs. The number of trophozoites (in 10 base log units) is indicated on the x axis, while the number of qPCR cycles is indicated on the y axis. The slopes, intercepts, and correlation coefficients are indicated for each target gene.
Application of qPCR assays to DNA extracted from human stools.
A total of 30 DNA extracts from human stool samples, previously genotyped using standard PCR, were analyzed by qPCR.
At the tpi locus, of the 10 isolates typed as assemblage A by standard PCR, 9 could be analyzed by qPCR, and all were classified as A plus B (no DNA was available for the remaining isolate). Similarly, of the 20 isolates typed as assemblage B by standard PCR, 18 could be analyzed by qPCR, and 2 were classified as B, whereas 16 were A plus B (no DNA was available for the remaining two samples). Interestingly, all the fecal samples from African children, which were classified as A or B by standard PCR, were identified as A plus B at this locus. To confirm the detection of both assemblages A and B in those samples, the qPCR products were digested with HincII and AluI, and the digestion products were separated by electrophoresis on agarose gels. This confirmed the genuine amplification of assemblage A (the product was cut by HincII into two fragments of 31 bp and 47 bp, whereas the B product was uncut) and of assemblage B (the product was cut by AluI into two fragments of 32 bp and 45 bp, whereas the A product was uncut). An example is shown in Fig. 1.
At the β-giardin locus, of the 12 isolates classified as assemblage A by standard PCR, 11 could be analyzed by qPCR, and 9 were classified as assemblage A, whereas 2 were classified as A plus B (no DNA was available for the remaining isolate). Similarly, of the 18 isolates classified as assemblage B by standard PCR, 16 could be analyzed by qPCR, and 3 were classified as assemblage B, whereas 13 were classified as A plus B (no DNA was available for the remaining 2 isolates). Of note, of the 15 samples for which coamplification of assemblages A and B at the β-giardin locus was observed, 11 were from African children.
At the gdh locus, of the 11 isolates classified as assemblage A by standard PCR, 10 could be analyzed by qPCR, and all were classified as A plus B (no DNA was available for the remaining isolate). Of the 17 isolates classified by standard PCR as assemblage B, 15 could be analyzed by qPCR, and 12 were classified as assemblage B, whereas 3 were classified as A plus B (no DNA was available for the remaining 2 samples). For 2 samples, no information was available from standard PCR, yet they could be analyzed by qPCR and were classified as assemblage A (1 sample) or assemblage B (1 sample). Again, the simultaneous detection of assemblages A and B at the gdh locus was mainly observed in samples from African children (11 of 13 samples). To confirm the detection of both assemblages A and B in those samples, the qPCR products were subjected to electrophoresis to verify the expected sizes of the amplicons, which differed for assemblage A (180 bp) and assemblage B (133 bp). All amplifications were as expected. An example of this analysis is shown in Fig. 1.
Finally, at the orfC4 locus, of the 13 isolates classified as assemblage A by standard PCR, 12 could be analyzed by qPCR, and 7 were classified as assemblage A, 4 as A plus B, and 1 as assemblage B (no DNA was available for the remaining isolate). Similarly, of the 17 isolates classified as assemblage B by standard PCR, 15 could be analyzed by qPCR, and 8 were classified as assemblage B and 7 as A plus B (no DNA was available for the remaining 2 isolates). Of note, 8 of the 11 fecal samples classified as A plus B at the orfC4 locus were from African children. To confirm that the detection of both assemblages A and B in those samples was not an artifact, the qPCR products were subjected to electrophoresis to verify the expected sizes of the amplicons, which differed for assemblage A (103 bp) and assemblage B (171 bp). All amplification products were confirmed as genuine (Fig. 1 shows an example).
Application of qPCR assays to cysts purified from human stools.
Human stools from the same 30 isolates were processed, using immunomagnetic separation, to purify G. duodenalis cysts. The total number of recovered cysts varied from 100 to 2,000, and the percentage of DAPI-positive cysts varied from 5% to 100%. The purified cysts were used directly in qPCR assays without any prior treatment. The qPCR experiments were first performed using 2 to 4 μl of purified cysts, which therefore contained from 2 to 80 cysts. At the tpi and β-giardin loci, 29 of 30 cyst samples (97%) yielded positive results, whereas 26 (87%) samples were positive at the gdh locus and 21 (70%) at the orfC4 locus (Table 2).
TABLE 2.
Genotyping results obtained for 30 human samples at four loci using regular PCR and qPCR on genomic DNA from stools, on purified cysts, and on single cysts
| Sample | Resulta |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
tpi |
β-giardin |
gdh |
orfC4 |
|||||||||||||
| PCR | qPCR stool | qPCR cysts | qPCR single cysts | PCR | qPCR stool | qPCR cysts | qPCR single cysts | PCR | qPCR stool | qPCR cysts | qPCR single cysts | PCR | qPCR stool | qPCR cysts | qPCR single cysts | |
| Gd84 | B | B | B | ND | A | A+B | B | ND | B | B | B | ND | B | B | B | ND |
| Gd103 | B | ND | B | ND | B | ND | B | ND | B | ND | B | ND | B | ND | B | ND |
| Gd105 | B | A+B | B | ND | B | B | B | ND | B | B | B | ND | B | B | B | ND |
| Gd106 | B | A+B | B | ND | B | B | B | ND | B | B | B | ND | B | B | B | ND |
| Gd109 | B | A+B | B | ND | B | A+B | B | ND | B | B | B | ND | B | B | B | ND |
| Gd113 | B | B | B | ND | B | A+B | B | ND | B | B | B | ND | B | A+B | B | ND |
| Gd142 | B | A+B | B | ND | B | A+B | B | ND | B | B | B | ND | B | A+B | Neg | ND |
| Gd145 | B | A+B | B | ND | B | A+B | B | ND | B | A+B | B | ND | B | B | B | ND |
| Gd147 | B | A+B | B | ND | B | A+B | B | ND | B | B | B | ND | B | B | B | ND |
| Gd189 | B | A+B | B | ND | B | A+B | B | ND | B | B | B | ND | B | A+B | B | ND |
| Gd190 | B | A+B | B | ND | B | A+B | B | ND | B | B | B | ND | B | B | B | ND |
| Gd143 | B | A+B | B | ND | A | A | Neg | ND | Neg | A | Neg | ND | A | A | Neg | ND |
| Gd208 | B | A+B | Neg | ND | A | A | B | ND | A | A+B | B | ND | B | A+B | Neg | ND |
| Gd198 | A | A+B | B | ND | A | A | B | ND | A | A+B | B | ND | A | A | Neg | ND |
| Gd107 | A | ND | A | ND | A | ND | A | ND | A | ND | A | ND | A | ND | A | ND |
| Gd108 | A | A+B | A+B | A | A | A | A | A | A | A+B | A | A | A | A | A | A |
| Gd194 | B | A+B | B | B | A | A | B | B | A | A+B | B | B | A | A | A | B |
| Gd192 | B | A+B | A+B | B | B | A+B | A+B | B | B | B | B | B | B | A+B | B | B |
| Gd204 | B | A+B | A+B | B | B | A+B | B | B | B | A+B | B | B | B | A+B | B | B |
| Gd82 | B | A+B | A+B | B | B | A+B | B | B | B | B | B | B | B | A+B | B | B |
| Gd193 | A | A+B | B | B | A | A | A | Neg | A | A+B | Neg | ND | A | A | Neg | ND |
| Gd141 | A | A+B | A | A | B | B | B | B | B | A+B | B | ND | A | B | Neg | ND |
| Gd200 | A | A+B | A+B | A+B | A | A | B | B | A | A+B | Neg | ND | A | A+B | Neg | ND |
| Gd191 | A | A+B | A+B | Neg | B | A+B | B | B | A | A+B | Neg | ND | A | A | Neg | ND |
| Gd196 | B | A+B | A+B | A+B | B | A+B | B | B | Neg | B | B | B | A | A | Neg | ND |
| Gd111 | B | ND | A+B | A+B | B | ND | B | B | B | ND | B | B | B | ND | B | B |
| Gd140 | B | A+B | A+B | A+B | B | A+B | B | B | B | B | B | B | B | B | B | B |
| Gd197 | A | A+B | A+B | A+B | A | A+B | B | B | A | A+B | A+B | Neg | A | A+B | B | Neg |
| Gd206 | A | A+B | A+B | A+B | A | A | A+B | A | A | A+B | A+B | A | A | A+B | A | A |
| Gd174 | A | A+B | A | A | A | A | A+B | A+B | A | A+B | A | A | A | A+B | A | A |
Nd, not done; Neg, negative.
In 12 of the 30 samples (40%), only one assemblage was detected in all assays (11 samples were assemblage B, and one was assemblage A) (Table 2), independent of the number of cysts used. Compared to the results obtained by standard PCR, almost perfect agreement was found (only one sample was typed as A at the β-giardin locus but was typed as B at the remaining 3 loci). Interestingly, the qPCR of DNA from stools indicated the presence of both assemblages A and B in 8 of the 11 samples at both the tpi and β-giardin loci but only in 3 and 1 at the orfC4 and gdh loci, respectively. In 2 samples, a congruent assignment to assemblage B was obtained at 2 or 3 loci, whereas no results were obtained at the other loci. In four samples, assignment to assemblage B (for Gd82, Gd192, and Gd204) (Table 2) or assemblage A (for Gd108) (Table 2) was supported by 3 assays, whereas the tpi assay indicated these samples were A plus B. However, when the purified cysts from these samples were further diluted to a nominal level of 1 cyst and the qPCR at the tpi locus was repeated, only assemblage B (3 samples) or A (1 sample) was detected. In sample Gd194 (Table 2), a single result at the orfC4 locus supported the presence of assemblage A, whereas this sample was typed as assemblage B at the other three loci. However, when the orfC4 assay was repeated on cysts diluted to a nominal level of 1 cyst, only assemblage B was detected. Therefore, in 19 of the 30 samples (63%), a consistent assignment to one assemblage was obtained.
In eight samples (Gd111, Gd140, Gd174, Gd191, Gd196, Gd197, Gd200, and Gd206), both assemblages A and B were detected at the tpi locus (all samples except Gd174), β-giardin (in samples Gd174 and Gd206), and gdh (in samples Gd197 and Gd206). qPCR experiments performed on diluted cysts showed that only assemblage A was detectable by the β-giardin and gdh assays of sample Gd206; however, qPCR at the tpi locus still detected both assemblages A and B, even for diluted cysts, as did the β-giardin assay of sample Gd174 (Table 2). Therefore, six samples were classified as assemblage B by 3 assays but as A plus B by the tpi assay, one sample (Gd206) was classified as assemblage A by 3 assays but as A plus B by the tpi assay, and one sample (Gd174) was classified as assemblage A by 3 assays but as A plus B by the β-giardin assay. Interestingly, of the six samples for which coamplification of assemblages A and B was observed at the tpi locus, five were from African children. Notably, qPCR on DNA from stools also indicated the presence of both assemblages A and B in seven (tpi), five (gdh), and four (β-giardin and orfC4) of these eight samples, respectively (Table 2).
In samples Gd141 and Gd193, the results of qPCR were not consistent, in that either assemblage A or assemblage B was detected by the different assays (Table 2). Finally, in sample Gd143, only the tpi assay was positive and indicated assemblage B, although this sample was typed as assemblage A by standard PCR and qPCR of DNA from stools (Table 2).
DISCUSSION
Human giardiasis is caused by two genetically very distinct assemblages (A and B) of G. duodenalis, and a number of molecular assays have been developed for their specific detection in stool and environmental samples (7). All those assays are based on the in vitro amplification of specific gene fragments, but the detection formats differ, ranging from the simple analysis of restriction patterns to sequence analysis or to more versatile platforms, like microarrays and qPCR. In particular, qPCR can offer increased specificity and sensitivity, and several assays have been developed for the detection of Giardia in water samples (4, 15) and clinical samples (17).
In this work, we developed three qPCR assays based on the triose phosphate isomerase, glutamate dehydrogenase, and ORFC4 sequences of G. duodenalis. The choice of these markers was driven by the availability of consistent sequence information, by the large amount of genetic polymorphism between assemblage A and B isolates (36), and by the fact that the genes are unlinked, at least in the fully sequenced genome of assemblage A (26). This allowed the design of assemblage A-specific primers that have, over a length of 20 nucleotides, 3 to 8 mismatches when aligned with the corresponding B sequence, and vice versa (Table 1). Under the experimental conditions used, no amplification was observed when assemblage A primers were used on assemblage B genomic DNA, or vice versa. The A-specific and B-specific qPCR products could be easily distinguished, with the exception of the gdh products, by melting-curve analysis (Table 1 and Fig. 1). During the development of these assays, further confirmation of their specificity was obtained by determining the sequence, the size, or the restriction pattern expected for each amplification product at each locus (Table 1 and Fig. 1). The sensitivity, estimated using serial dilutions of freshly collected WB trophozoites, was a single trophozoite (Fig. 2), which, since the unsynchronized trophozoites are likely to be in the G2 phase (3), corresponds to 8 target copies. When these assays were tested on serial dilutions of purified cysts, the sensitivity was close to a single cyst. Thus, qPCR has a sensitivity similar to that reported for a tpi nested-PCR assay of individually isolated Giardia cysts (25).
The qPCR assays are informative with respect to several aspects of current research on Giardia. First, they can be used to get a better estimate of the occurrence of mixed infections in clinical samples, which is likely to be strongly underestimated. Indeed, using tpi assemblage-specific primers in a standard PCR protocol, it has been shown that 31% of the calf isolates can be identified as mixed assemblage A and E infections (13), and a similar proportion of mixed A and B infections was found in human and monkey isolates using the same approach (14, 22). Here, by applying the more sensitive qPCR assays, we have shown that an even larger percentage of human stool samples (37 to 83%, depending on the sensitivities of the different assays) contained DNAs of both assemblages A and B. Importantly, the rationale of our assay should allow us to distinguish between mixed infections and possible recombinants, whereas this is not possible using a standard PCR approach. Indeed, if the simultaneous detection of assemblages A and B is due to the presence of both types of cysts in the host feces (i.e., a mixed infection), then the number of cysts used in qPCR will affect the result to the point that, when dilutions of cysts are tested, only one or the other assemblage is detected. On the other hand, if there is only one type of cyst which contains DNAs of both assemblages (i.e., a recombinant), then the detection will be independent of the number of cysts used, even at the level of a single cyst. Here, we have shown that mixed infections accounted for the vast majority of cases where both assemblages A and B were detected by qPCR. Indeed, when the cysts purified from these samples were further diluted to a nominal level of 1 cyst and retested, the DNA of only one assemblage was still detectable (Table 2). Of note, in 6 of the 29 samples analyzed with the tpi qPCR and in 1 out of 29 samples analyzed with the β-giardin qPCR, amplification products of both assemblages A and B were detected using single cysts as templates (Table 2), suggesting the presence of recombinants. However, the relative inaccuracy of the dilution approach and the fact that cysts tend to clump together indicate the need to use more precise methods, such as micromanipulation or flow cytometry, to ensure that single cysts are indeed analyzed. The occurrence of recombination between assemblages A and B has been recently suggested, although this was tested on in vitro-adapted strains or inferred from a comparative analysis of sequences deposited in GenBank (21, 33). The genome sequencing of the GS strain of assemblage B (11) has not confirmed the presence of A sequences in the genome and suggested that previous results (33) were biased by laboratory contamination. Certainly, in view of the implications of genetic exchanges for the taxonomy and the molecular epidemiology of giardiasis (9, 10, 23), more studies are needed, and they should be based on the analysis of more genetic loci.
The qPCR assays presented in this work can also be used to reevaluate the correlation between the genetic background of the parasite and the clinical manifestations in humans. It is well known that the clinical manifestations of giardiasis are quite variable and range from the absence of symptoms to acute or chronic diarrhea, dehydration, abdominal pain, nausea, vomiting, and weight loss (5). The severity of the disease is likely determined by the interplay between the parasite virulence and the developmental, nutritional, and immunological status of the host. Studies conducted until now have been discordant; some authors have reported an association between assemblage A and symptoms (2, 16, 28, 30, 31), while others have pointed to an association between assemblage B and symptoms (12, 18, 29). Almost all of these studies were based on the analysis of a single gene, and it is interesting that genotyping at the ssu-rRNA and tpi genes supports the assemblage A-symptoms association, whereas genotyping at the β-giardin and gdh genes supports the assemblage B-symptoms association. However, if humans are often infected with both assemblages, as is suggested by the qPCR results presented here (Table 2) and by other studies (14), then this correlation is probably more complex than anticipated.
Finally, the assays can be used on cysts recovered from water samples, where the crucial point is to demonstrate (and quantify) the presence of human pathogens, i.e., assemblages A and B, to infer a possible risk of waterborne transmission. Several qPCR assays have been tested on water samples (4, 15), and particular attention was given to the optimization of DNA extraction methods and the relief of PCR inhibitors from those complex matrices (38). Here, we have shown that cysts purified by immunomagnetic capture, a method that is routinely applied to water samples, can be analyzed directly by qPCR without the need for DNA extraction, but a careful evaluation of these assays is required to demonstrate the applicability of our protocols to complex environmental samples. In perspective, a protocol that combines genotyping by sensitive qPCR assays with methods that allow efficient recovery and enumeration of Giardia cysts will be instrumental in unraveling the intricate epidemiology of giardiasis and the genetics of these parasites.
Acknowledgments
This work was supported by the European MED-VET-NET project, contract FOOD-CT-2004-506122.
We are grateful to Daniele Tonanzi for his excellent technical support.
Footnotes
Published ahead of print on 15 January 2010.
REFERENCES
- 1.Adam, R. D. A. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aydin, A. F., B. A. Besirbellioglu, I. Y. Avci, M. Tanyuksel, E. Araz, and A. Pahsa. 2004. Classification of Giardia duodenalis parasites in Turkey into groups A and B using restriction fragment length polymorphism. Diagn. Microbiol. Infect. Dis. 50:147-151. [DOI] [PubMed] [Google Scholar]
- 3.Bernander, R., J. E. D. Palm, and S. G. Svard. 2001. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell Microbiol. 3:55-62. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand, I., C. Gantzer, T. Chesnot, and J. Schwartzbrod. 2004. Improved specificity for Giardia lamblia cyst quantification in wastewater by development of a real-time PCR method. J. Microbiol. Methods 57:41-53. [DOI] [PubMed] [Google Scholar]
- 5.Buret, A. G. 2008. Pathophysiology of enteric infections with Giardia duodenalis. Parasite 15:261-265. [DOI] [PubMed] [Google Scholar]
- 6.Cacciò, S. M., R. C. Thompson, J. McLauchlin, and H. V. Smith. 2005. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 21:430-437. [DOI] [PubMed] [Google Scholar]
- 7.Cacciò, S. M., and U. Ryan. 2008. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 160:75-80. [DOI] [PubMed] [Google Scholar]
- 8.Cacciò, S. M., R. Beck, M. Lalle, A. Marinculic, and E. Pozio. 2008. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 38:1523-1531. [DOI] [PubMed] [Google Scholar]
- 9.Cacciò, S. M., and H. Sprong. 2010. Giardia duodenalis: genetic recombination and its implications for taxonomy and molecular epidemiology. Exp. Parasitol. 124:107-112. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, M. A., R. D. Adam, M. Worobey, and C. R. Sterling. 2007. Population genetics provides evidence for recombination in Giardia. Curr. Biol. 17:1984-1988. [DOI] [PubMed] [Google Scholar]
- 11.Franzén, O., J. Jerlström-Hultqvist, E. Castro, E. Sherwood, J. Ankarklev, D. S. Reiner, D. Palm, J. O. Andersson, B. Andersson, and S. G. Svärd. 2009. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 5:e1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelanew, T., M. Lalle, A. Hailu, E. Pozio, and S. M. Cacciò. 2007. Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. 102:92-99. [DOI] [PubMed] [Google Scholar]
- 13.Geurden, T., P. Geldhof, B. Levecke, C. Martens, D. Berkvens, S. Casaert, J. Vercruysse, and E. Claerebout. 2007. Mixed Giardia duodenalis assemblage A and E infections in calves. Int. J. Parasitol. 38:259-264. [DOI] [PubMed] [Google Scholar]
- 14.Geurden, T., B. Levecke, S. M. Cacció, A. Visser, G. De Groote, S. Casaert, J. Vercruysse, and E. Claerebout. 2009. Multilocus genotyping of Cryptosporidium and Giardia in non-outbreak related cases of diarrhea in human patients in Belgium. Parasitology 136:1161-1168. [DOI] [PubMed] [Google Scholar]
- 15.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haque, R., S. Roy, M. Kabir, S. E. Stroup, D. Mondal, and E. R. Houpt. 2005. Giardia assemblage A infection and diarrhea in Bangladesh. J. Infect. Dis. 192:2171-2173. [DOI] [PubMed] [Google Scholar]
- 17.Haque, R., S. Roy, A. Siddique, D. Mondal, S. M. Rahman, E. R. Houpt, and W. A. Petry Jr. 2007. Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am. J. Trop. Med. Hyg. 76:713-717. [PubMed] [Google Scholar]
- 18.Homan, W. L., and T. G. Mank. 2001. Human giardiasis: genotype linked differences in clinical symptomatology. Int. J. Parasitol. 31:822-826. [DOI] [PubMed] [Google Scholar]
- 19.Lalle, M., F. Bruschi, B. Castagna, M. Campa, E. Pozio, and S. M. Cacciò. 2009. High genetic polymorphism among Giardia duodenalis isolates from Sahrawi children. Trans. R. Soc. Trop. Med. Hyg. 103:834-838. [DOI] [PubMed] [Google Scholar]
- 20.Lane, S., and D. Lloyd. 2002. Current trends in research into the waterborne parasite Giardia. Crit. Rev. Microbiol. 28:123-147. [DOI] [PubMed] [Google Scholar]
- 21.Lasek-Nesselquist, E., D. M. Welch, R. C. Thompson, R. F. Steuart, and M. L. Sogin. 2009. Genetic exchange within and between assemblages of Giardia duodenalis. J. Eukaryot. Microbiol. 56:504-518. [DOI] [PubMed] [Google Scholar]
- 22.Levecke, B., P. Geldhof, E. Claerebout, P. Dorny, F. Vercammen, S. M. Cacciò, J. Vercruysse, and T. Geurden. 2009. Molecular characterisation of Giardia duodenalis in captive non-human primates reveals mixed assemblage A and B infections and novel polymorphisms. Int. J. Parasitol. 39:1595-1601. [DOI] [PubMed] [Google Scholar]
- 23.Logsdon, J. M., Jr. 2008. Evolutionary genetics: sex happens in Giardia. Curr. Biol. 18:R66-R68. [DOI] [PubMed] [Google Scholar]
- 24.Mayrhofer, G., R. H. Andrews, P. L. Ey, and N. B. Chilton. 1995. Division of Giardia isolates from humans into two genetically distinct assemblages by electrophoretic analysis of enzymes encoded at 27 loci and comparison with Giardia muris. Parasitology 111:11-17. [DOI] [PubMed] [Google Scholar]
- 25.Miller, K. M., and C. R. Sterling. 2007. Sensitivity of nested PCR in the detection of low numbers of Giardia lamblia cysts. Appl. Environ. Microbiol. 73:5949-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison, H. G., A. G. McArthur, F. D. Gillin, S. B. Aley, R. D. Adam, G. J. Olsen, A. A. Best, W. Z. Cande, F. Chen, M. J. Cipriano, B. J. Davids, S. C. Dawson, H. G. Elmendorf, A. B. Hehl, M. E. Holder, S. M. Huse, U. U. Kim, E. Lasek-Nesselquist, G. Manning, A. Nigam, J. E. Nixon, D. Palm, N. E. Passamaneck, A. Prabhu, C. I. Reich, D. S. Reiner, J. Samuelson, S. G. Svard, and M. L. Sogin. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921-1926. [DOI] [PubMed] [Google Scholar]
- 27.Nash, T. E., T. McCutchan, D. Keister, J. B. Dame, J. D. Conrad, and F. D. Gillin. 1985. Restriction-endonuclease analysis of DNA from 15 Giardia isolates obtained from humans and animals. J. Infect. Dis. 152:64-73. [DOI] [PubMed] [Google Scholar]
- 28.Paintlia, A. S., S. Descoteaux, B. Spencer, A. Chakraborti, N. K. Ganguly, R. C. Mahajan, and J. Samuelson. 1998. Giardia lamblia groups A and B among young adults in India. Clin. Infect. Dis. 26:190-191. [DOI] [PubMed] [Google Scholar]
- 29.Pelayo, L., F. A. Nuñez, L. Rojas, E. Furuseth Hansen, B. Gjerde, H. Wilke, B. Mulder, and L. Robertson. 2008. Giardia infections in Cuban children: the genotypes circulating in a rural population. Ann. Trop. Med. Parasitol. 102:585-595. [DOI] [PubMed] [Google Scholar]
- 30.Read, C., J. Walters, I. D. Robertson, and R. C. Thompson. 2002. Correlation between genotype of Giardia duodenalis and diarrhoea. Int. J. Parasitol. 32:229-231. [DOI] [PubMed] [Google Scholar]
- 31.Sahagun, J., A. Clavel, P. Goni, C. Seral, M. T. Llorente, F. J. Castillo, S. Capilla, A. Arias, and R. Gomez-Lus. 2008. Correlation between the presence of symptoms and the Giardia duodenalis genotype. Eur. J. Clin. Microbiol. Infect. Dis. 27:81-83. [DOI] [PubMed] [Google Scholar]
- 32.Smith, H. V., S. M. Cacciò, A. Tait, J. McLauchlin, and R. C. Thompson. 2006. Tools for investigating the environmental transmission of Cryptosporidium and Giardia infections in humans. Trends Parasitol. 22:160-167. [DOI] [PubMed] [Google Scholar]
- 33.Teodorovic, S., J. M. Braverman, and H. G. Elmendorf. 2007. Unusually low levels of genetic variation among Giardia lamblia isolates. Eukaryot. Cell 6:1421-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, R. C., and P. T. Monis. 2004. Variation in Giardia: implications for taxonomy and epidemiology. Adv. Parasitol. 58:69-137. [DOI] [PubMed] [Google Scholar]
- 35.WHO. 1996. The world health report 1996. Fighting disease, fostering development. World Health Organization, Geneva, Switzerland. [PubMed]
- 36.Wielinga, C. M., and R. C. Thompson. 2007. Comparative evaluation of Giardia duodenalis sequence data. Parasitology 134:1795-1821. [DOI] [PubMed] [Google Scholar]
- 37.Yong, T., K. Han, H. Yang, and S. Park. 2002. PCR-RFLP analysis of Giardia intestinalis using a Giardia-specific gene, GLORF-C4. Parasite 9:65-70. [DOI] [PubMed] [Google Scholar]
- 38.Yu, X., M. I. Van Dyke, A. Portt, and P. M. Huck. 2009. Development of a direct DNA extraction protocol for real-time PCR detection of Giardia lamblia from surface water. Ecotoxicology 18:661-668. [DOI] [PubMed] [Google Scholar]