Abstract
The Ibis T5000 is a novel diagnostic platform that couples PCR and mass spectrometry. In this study, we developed an assay that can identify all known pathogenic Vibrio species and field-tested it using natural water samples from both freshwater lakes and the Georgian coastal zone of the Black Sea. Of the 278 total water samples screened, 9 different Vibrio species were detected, 114 (41%) samples were positive for V. cholerae, and 5 (0.8%) samples were positive for the cholera toxin A gene (ctxA). All ctxA-positive samples were from two freshwater lakes, and no ctxA-positive samples from any of the Black Sea sites were detected.
The genus Vibrio, within the family Vibrionaceae, is a diverse group of Gram-negative bacteria found exclusively in the aquatic environment. Important pathogenic members include Vibrio cholerae, the causative agent of cholera, and Vibrio parahaemolyticus and Vibrio vulnificus, which have been implicated in diarrhea, septicemia, and wound infections (5). The Ibis T5000 uses electrospray ionization-mass spectrometry to analyze the products of broad-range PCR (PCR-electrospray ionization-mass spectrometry [PCR/ESI-MS]) and is designed to rapidly detect and identify emerging pathogens and biothreat agents without prior knowledge of a pathogen's nucleic acid sequence (4, 10).
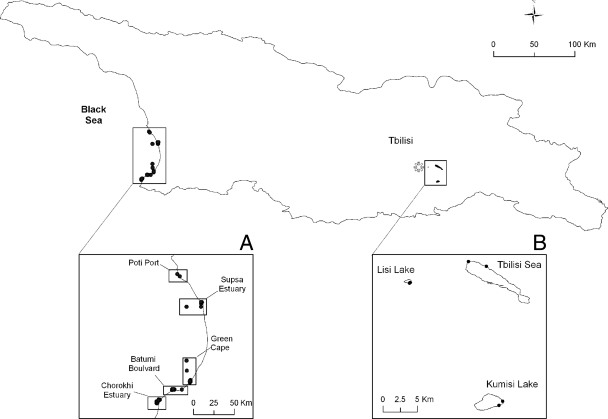
Forty-two well-characterized bacterial strains were used for the validation of the Vibrio PCR/ESI-MS assay (Table 1). Four sampling sites, the Chorokhi estuary, Bulvari, Green Cape, and the Supsa estuary along the Georgian coastal zone of the Black Sea, and three freshwater lakes, Kumisi Lake, Lisi Lake, and the Tbilisi Sea near the city of Tbilisi, Georgia (Fig. 1), were sampled monthly from July 2006 to October 2007 and biweekly during the summer (July to September). One hundred liters of surface water was collected and processed by following methods described previously (8).
TABLE 1.
Bacterial strains used in this study
| Species and/or straina | Description | Source (yr of isolation)b |
|---|---|---|
| Vibrio strains | ||
| V. cholerae O1 | ||
| 026 | El Tor Ogawa ctxAB+ | Kenya (1985) |
| MJ-1236 | El Tor Inaba ctxAB+ | Matlab, Bangladesh (1994) |
| B33 | El Tor Ogawa ctxAB+ | Beira, Mozambique (2004) |
| BX 330286 | El Tor Inaba ctxAB+ | Australia; water (1986) |
| 12129(1) | El Tor Inaba ctxAB mutant | Australia; water (1985) |
| CIRS101 | El Tor Inaba ctxAB+ | Dhaka, Bangladesh (2002) |
| 098 | Classical ctxAB+ | Indonesia (1991) |
| TM 11079-80 | El Tor Ogawa ctxAB mutant | Brazil; sewage (1980) |
| FMU 87295/0 | Classical ctxAB+ | Mexico; clinical isolate (1983) |
| UMSM N-16961 | El Tor Inaba ctxAB+ | Bangladesh (1975) |
| O395 | Classical Ogawa ctxAB+ | India (1965) |
| V. cholerae O139 | ||
| MO10 | ctxAB+ | India (1992) |
| MO45 | ctxAB+ | India (1985) |
| CO-393 | ctxAB+ | India (1993) |
| EM0158 | ctxAB+ | Mathbaria, Bangladesh; pond water (2004) |
| V. cholerae non-O1, non-O139 | ||
| TMA 21 | ctxAB mutant | Brazil; seawater (1982) |
| TMA 135 | ctxAB mutant | Brazil; seawater (1981) |
| CB98-179 | ctxAB mutant | Chesapeake Bay, Maryland, USA; water |
| UM4056 | ctxAB mutant | Mud Lake, Louisiana, USA; water |
| CT 5369-93 | ctxAB mutant | Brazil; sewage (1993) |
| V. vulnificus | ||
| 324 (CDC B9629) | Florida, USA; clinical isolate, ATCC 27562 | |
| 332 (CDC A6546)* | Alaska, USA; clinical isolate, ATCC 33816 | |
| V. parahaemolyticus | ||
| EB 101 | Japan; clinical isolate, type strain ATCC 17802 | |
| EB 102 | Japan; clinical isolate, ATCC 17803 (1953) | |
| V. mimicus | ||
| VM223 | ||
| MB-451 | Bangladesh | |
| V. metschnikovii LMG 11664 | Fowl; ATCC 700040 | |
| V. albensis VL426 | Maidstone, Kent, UK; water | |
| V. furnissii 9119-82 | Japan; human feces, CIP 102972 | |
| V. hollisae | Maryland, USA; human feces, CIP 101886 | |
| V. logei 584 | Gut of Arctic mussel; ATCC 29985 | |
| V. mediterranei 50 | Spain; coastal sediment, CIP 103203 | |
| V. cincinnatiensis | Ohio, USA; human clinical samples, CIP 104173 | |
| V. coralliilyticus YB | Tanzania; diseased coral, ATCC BAA-450 | |
| V. orientalis 717 | China; seawater, CIP 102891 (1983) | |
| Other bacteria | ||
| Aeromonas caviae ATCC 15468 | ATCC | |
| Aeromonas hydrophila ATCC 7966 | ATCC | |
| Aeromonas jandaei | Brazil (1996) | |
| Aeromonas salmonicida subsp. masoucida ATCC 27013 | ATCC | |
| Aeromonas sobria ATCC 9071 | ATCC | |
| Aeromonas veronii ATCC 35624 | ATCC | |
| Photobacterium damselae subsp. damselae ATCC 33539 | ATCC |
*, confirmed as V. mimicus by 16S rRNA sequence analysis and biochemical characterization.
ATCC, American Type Culture Collection; CIP, Collection de l'Institut Pasteur.
FIG. 1.
Map of Georgia with sampling sites. (A) Georgian coast of the Black Sea showing the sites Poti, Supsa, Green Cape, and Batumi. (B) Map of the three freshwater lakes, with two sampling sites at each lake. (Adapted from reference 6 with permission of the publisher.)
An 8-primer-pair assay with breadth of coverage across the entire family Vibrionaceae was developed (see Fig. S1 in the supplemental material). All primers used in the assay are listed in Table 2. All PCRs were performed in 96-well microtiter plates as described previously (1). A Bruker Daltonics microTOF (Billerica, MA) mass spectrometer was used for analyzing the purified DNA as described previously (3, 7). The presence of V. cholerae was confirmed by using species-specific intergenic spacer region (ISR) PCR primers (2). For detection of the cholera toxin A gene (ctxA), PCR was performed as described by Rivera et al. (9). For confirmation of the Vibrio population data obtained with the PCR/ESI-MS assay, an environmental clone library was constructed from one of the samples (collection date, 12 August 2006) from the Chorokhi estuary using the universal 16S rRNA primer 27F (5′-AGAGTTTGATYMTGGCTCAG-3′) and the Vibrio-specific primer 680R (5′-GAAATTCTACCCCCCTCTACAG-3′). The resulting PCR product was cloned and sequenced, and sequences with identities of ≥99% were considered matches.
TABLE 2.
PCR primers used in the Vibrio PCR/ESI-MS assay
| Primer pair no. | Orientationa | Sequence (5′-3′) | Target gene | Breadth of coverage |
|---|---|---|---|---|
| 1098 | F | TCCGCGGAGTTGACTGGGT | RNase P | All Vibrionaceae |
| R | TGACTTTCCTCCCCCTTATCAGTCTCC | |||
| 2001 | F | TGAGTGCCAACATATCAGTGCTGAAGA | fur | All Vibrionaceae |
| R | TCCGCCTTCAAAATGGTGGCGAGT | |||
| 2011 | F | TAAAGCCCGTGAAATGACTCGTCGTAAAGG | gyrB | All Vibrionaceae |
| R | TGAGTCACCCTCCACAATGTATAGTTCAGA | |||
| 2993 | F | TTCCCACCGATATCATGGCTTACCACGG | ompU | Vibrio and Listonella |
| R | TCGGTCAGCAAAACGGTAGCTTGC | |||
| 2927 | F | TCAATGAACGACCAACAAGTGATTGATG | gapA | All Vibrionaceae |
| R | TCCTTTATGCAACTTGGTATCAACAGGAAT | |||
| 3003 | F | TCAGCATATGCACATGGAACACCTC | ctxB | V. cholerae specific |
| R | TGCCGTATACGAAAATATCTTATCATTTAGCGT | |||
| 2012 | F | TACGCTGACGGAATCAACCAAAGCGG | ompU | V. cholerae specific |
| R | TGCTTCAGCACGGCCACCAACTTCTAG | |||
| 2323 | F | TGCCAAGAGGACAGAGTGAGTACTTTGA | ctxA | V. cholerae specific |
| R | TAACAAATCCCGTCTGAGTTCCTCTTGCA |
F, forward; R, reverse.
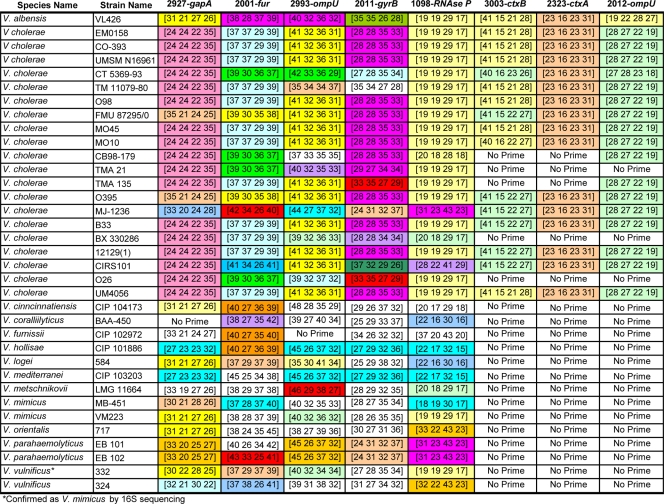
Base composition data were produced for all 35 Vibrio isolates (20 strains of V. cholerae and 15 non-cholera Vibrio species) (Fig. 2). Based on unique DNA base composition signatures, all the Vibrio strains were correctly identified at the species level. Reactions were negative for Photobacterium damselae and for all species of Aeromonas tested (data not shown). To determine the ability of the assay to detect mixtures of multiple species of Vibrio, we spiked equal amounts of one to four different Vibrio species in phosphate-buffered saline. In all cases, the assay successfully identified the mixture components correctly (see Fig. S2 in the supplemental material).
FIG. 2.
Base compositions of eight selected amplicons from Vibrio ribosomal and housekeeping genes. Within each column, base compositions that are common to multiple types are similarly colored. The numbers in the columns indicate the numbers of each base (A, G, C, and T) in the PCR amplicons generated for each gene target. Base compositions with a white background are unique to a particular type. No Prime, no PCR amplification product was expected.
The limits of detection for PCR/ESI-MS were tested using serial dilutions of the following three distinct Vibrio species: V. cholerae, V. vulnificus, and V. alginolyticus. Twofold serial dilutions of purified DNA were made, with a final concentration ranging from 0.05 ng to 50 ng per reaction mixture. Each sample was diluted 1:100 with genome dilution buffer and analyzed by PCR/ESI-MS. To confirm reproducibility, samples were run in duplicate. All three test strains showed detection at approximately 4 to 8 genome copies per PCR (see Table S1 in the supplemental material).
To test the ability of the assay to detect and identify Vibrio spp. from natural aquatic samples, we applied it to a subset of total community DNA samples collected in 2006 from freshwater lakes and sites along the Georgian coast of the Black Sea. Six different Vibrio species were detected and identified from 19 natural water samples collected from both freshwater and seawater sites spanning the seasons summer (July/August) to winter (November/December) in 2006 (Table 3). There was a high prevalence of V. cholerae DNA detection for several of the sites in Georgia, evidenced by 13 out of the 19 samples testing positive for V. cholerae. All but one of these detections was confirmed by standard PCR targeting the V. cholerae-specific 16S-23S rRNA ISR. As expected, Vibrio counts, as determined by plate counting on plates with thiosulfate, citrate, bile salts, and sucrose (TCBS plates), were high in the summer months and decreased significantly during the winter months. For the Chorokhi estuary samples, counts fell from 3,500 CFU/100 ml in July to as low as 7 CFU/100 ml in December. Conversely, molecular analysis of the water samples using the PCR/ESI-MS assay suggested the presence of large numbers of vibrios late into the winter (Table 3), suggesting the possibility that these organisms were in the viable but nonculturable state.
TABLE 3.
PCR/ESI-MS detection and quantification of Vibrio species in selected Georgian natural water samples
| Sampling site | Date of sampling (day-mo-yr) | Sample type | Total Vibrio sp. count (CFU/ 100 ml) | PCR/ESI-MS detection (no. of genome copies/reaction mixture)a | V. cholerae-specific ISR PCRc |
|---|---|---|---|---|---|
| Chorokhi | 27-Jul-06 | Enriched | 3,500 | V. cholerae, V. parahaemolyticus, V. vulnificus | Pos |
| Bulvari | 27-Jul-06 | Enriched | 3,000 | V. parahaemolyticus, V. vulnificus | ND |
| Chorokhib | 10-Aug-06 | Enriched | 1,320 | V. cholerae, V. parahaemolyticus, V. vulnificus | ND |
| Chorokhi | 27-Aug-06 | Enriched | 1,600 | V. cholerae, V. parahaemolyticus, V. vulnificus | Pos |
| Supsa | 27-Aug-06 | Enriched | 275 | V. cholerae, V. parahaemolyticus, V. vulnificus | Pos |
| Kumisi Lake | 6-Sept-06 | Enriched | V. cholerae | Pos | |
| Tbilisi Sea | 9-Sept-06 | Concentrated | V. cholerae (38) | Pos (weak) | |
| Supsa | 20-Sept-06 | Enriched | 300 | V. cholerae, V. vulnificus | Pos |
| Green Cape | 20-Sept-06 | Enriched | 1,400 | V. parahaemolyticus, V. vulnificus | ND |
| Chorokhi | 20-Sept-06 | Enriched | 400 | V. cholerae, V. parahaemolyticus | Pos |
| Green Cape | 25-Oct-06 | Concentrated | 200 | V. cholerae (6,998) | Pos |
| Chorokhi | 24-Oct-06 | Concentrated | 600 | V. vulnificus (67) | ND |
| Tbilisi Sea | 30-Oct-06 | Concentrated | V. cholerae (5,583) | Pos | |
| Lisi Lake | 7-Nov-06 | Enriched | V. cholerae | Pos | |
| Tbilisi Lake | 7-Nov-06 | Concentrated | V. cholerae (3,266) | Pos | |
| Lisi Lake | 7-Nov-06 | Enriched | V. mimicus | ND | |
| Chorokhi | 2-Dec-06 | Concentrated | 7 | V. alginolyticus (172), V. vulnificus (178) | ND |
| Bulvari | 2-Dec-06 | Enriched | 88 | V. metschnikovii | ND |
| Supsa | 2-Dec-06 | Enriched | 60 | V. cholerae, V. parahaemolyticus | Neg |
Quantitation not shown for enriched samples.
Chosen for 16S rRNA environmental clone library.
ND, not done; Pos, positive; Neg, negative.
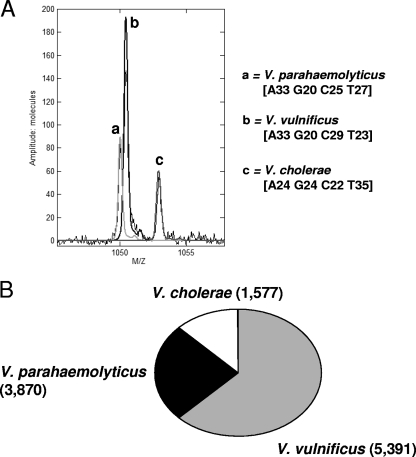
For further confirmation of the PCR/ESI-MS results, we constructed a 16S rRNA clone library for the Chorokhi estuary sample collected on 10 August 2006 (Fig. 3). Vibrio population analysis using the PCR/ESI-MS assay revealed the presence of V. cholerae, V. parahaemolyticus, and V. vulnificus in the sample (Fig. 3A). Sequence identification by clone library analysis revealed the presence of the same Vibrio species, and their relative abundances were similar to those indicated by the PCR/ESI-MS analysis (Fig. 3B).
FIG. 3.
Detection of multiple pathogenic Vibrio species in a sample collected in August 2006 from the Chorokhi estuary site on the coast of the Black Sea. (A) Mass spectra of PCR amplicons derived from the RNase P housekeeping gene from total community DNA extracted from the sample showing detections of V. cholerae, V. parahaemolyticus, and V. vulnificus. (B) Relative proportion of DNA sequences matching Vibrio species obtained from an environmental 16S rRNA clone library using primers 24F and 680R, as described in Materials and Methods. For comparison, the genomic quantifications (numbers of genome copies per reaction mixture) derived from the PCR/ESI-MS assay are given in parentheses.
For the 278 total water samples screened, 9 different Vibrio species were detected, with 114 (41%) samples positive for V. cholerae and 5 (0.8%) positive for the ctxA gene. All ctxA-positive samples were from two freshwater lakes, either Kumisi Lake or Lisi Lake, located near the capital city of Tbilisi, Georgia. No ctxA-positive samples were detected from any of the Black Sea sites. All ctxA-positive samples, except for one, were confirmed by using ctxA-specific standard PCR.
To determine the specificity of the assay to identify vibrios isolated from Georgian natural waters, we characterized 10 Vibrio isolates, comprising five different species isolated from both freshwater lakes and the Black Sea. All 10 isolates were correctly identified with high confidence using the PCR/ESI-MS assay (see Table S2 in the supplemental material). Importantly, the results show that Vibrio mimicus can be correctly identified and distinguished from the closely related species V. cholerae.
Our objectives in this study were to develop an assay based on PCR/ESI-MS using the Ibis T5000 platform for the rapid detection and identification of all pathogenic vibrios and then to field-test it using natural water. This study marks the first application of this technology to Vibrio spp. and the first use of the Ibis T5000 platform for the direct detection of bacterial pathogens from natural aquatic environments.
Supplementary Material
Acknowledgments
This research was supported, in part, by the U.S. Defense Threat Reduction Agency (DTRA) Cooperative Threat Reduction Program (project GG-13). C. Grim was supported by an IC Postdoctoral Research Fellowship (NGA grant HM15820612010).
Opinions, interpretations, conclusions, and recommendations stated herein are ours and are not necessarily endorsed by the U.S. Army.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baldwin, C. D., G. B. Howe, R. Sampath, L. B. Blyn, H. Matthews, V. Harpin, T. A. Hall, J. J. Drader, S. A. Hofstadler, M. W. Eshoo, K. Rudnick, K. Studarus, D. Moore, S. Abbott, J. M. Janda, and C. A. Whitehouse. 2009. Usefulness of multilocus polymerase chain reaction followed by electrospray ionization mass spectrometry to identify a diverse panel of bacterial isolates. Diagn. Microbiol. Infect. Dis. 63:403-408. [DOI] [PubMed] [Google Scholar]
- 2.Chun, J., A. Huq, and R. R. Colwell. 1999. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl. Environ. Microbiol. 65:2202-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ecker, D. J., J. Drader, J. Gutierrez, A. Gutierrez, J. Hannis, A. Schink, R. Sampath, J. A. Ecker, L. B. Blyn, M. W. Eshoo, T. A. Hall, M. Tobarmosquera, Y. Jiang, K. Sannes-Lowery, L. Cummins, B. Libby, D. J. Walcott, C. Massire, R. Ranken, S. M. Manalili, C. Ivy, R. Melton, H. Levene, V. Harpin, F. Li, N. White, M. Pear, V. Samant, D. Knize, D. Robbins, K. Rudnick, F. Hajjar, and S. A. Hofstadler. 2006. The Ibis T5000 universal biosensor—an automated platform for pathogen identification and strain typing. JALA 11:341-351. [Google Scholar]
- 4.Ecker, D. J., R. Sampath, C. Massire, L. B. Blyn, T. A. Hall, M. W. Eshoo, and S. A. Hofstadler. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6:553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farmer, J. J. I., J. M. Janda, and K. M. Birkhead. 2003. Vibrio, p. 706-718. In P. R. Murray (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 6.Grim, C. J., E. Jaiani, C. A. Whitehouse, N. Janelidze, T. Kokashvili, M. Tediashvili, R. R. Colwell, and A. Huq. 2009. Detection of toxigenic Vibrio cholerae O1 in freshwater lakes of the former Soviet Republic of Georgia. Environ. Microbiol. Rep. [Epub ahead of print.] doi: 10.1111/j.1758-2229.2009.00073.x. [DOI] [PubMed]
- 7.Hofstadler, S. A., K. A. Sannes-Lowery, and J. C. Hannis. 2005. Analysis of nucleic acids by FTICR MS. Mass Spectrom. Rev. 24:265-285. [DOI] [PubMed] [Google Scholar]
- 8.Huq, A., R. R. Colwell, R. Rahman, A. Ali, M. A. Chowdhury, S. Parveen, D. A. Sack, and E. Russek-Cohen. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera, I. N., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampath, R., T. A. Hall, C. Massire, F. Li, L. B. Blyn, M. W. Eshoo, S. A. Hofstadler, and D. J. Ecker. 2007. Rapid identification of emerging infectious agents using PCR and electrospray ionization mass spectrometry. Ann. N. Y. Acad. Sci. 1102:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.