Abstract
The objective of this study was to investigate how changes in soil pH affect the N2O and N2 emissions, denitrification activity, and size of a denitrifier community. We established a field experiment, situated in a grassland area, which consisted of three treatments which were repeatedly amended with a KOH solution (alkaline soil), an H2SO4 solution (acidic soil), or water (natural pH soil) over 10 months. At the site, we determined field N2O and N2 emissions using the 15N gas flux method and collected soil samples for the measurement of potential denitrification activity and quantification of the size of the denitrifying community by quantitative PCR of the narG, napA, nirS, nirK, and nosZ denitrification genes. Overall, our results indicate that soil pH is of importance in determining the nature of denitrification end products. Thus, we found that the N2O/(N2O + N2) ratio increased with decreasing pH due to changes in the total denitrification activity, while no changes in N2O production were observed. Denitrification activity and N2O emissions measured under laboratory conditions were correlated with N fluxes in situ and therefore reflected treatment differences in the field. The size of the denitrifying community was uncoupled from in situ N fluxes, but potential denitrification was correlated with the count of NirS denitrifiers. Significant relationships were observed between nirS, napA, and narG gene copy numbers and the N2O/(N2O + N2) ratio, which are difficult to explain. However, this highlights the need for further studies combining analysis of denitrifier ecology and quantification of denitrification end products for a comprehensive understanding of the regulation of N fluxes by denitrification.
Denitrification is the microbial reduction of NO3− via NO2− to gaseous NO, N2O, and N2, which are then lost into the atmosphere (36). It therefore results in considerable loss of nitrogen, one of the most limiting nutrients for crop production in agriculture (20). Denitrification is also of environmental concern since, together with nitrification, it is the main biological process responsible for N2O emissions (7). N2O is a potent greenhouse gas which has a global warming potential about 320 times greater than that of CO2 and has a lifetime of approximately 120 years (32). In the stratosphere, N2O can also react with O2 to produce NO, which induces the destruction of stratospheric ozone (8). N2O can be released into the atmosphere by incomplete denitrification due to the effect of environmental conditions on the regulation of the different denitrification reductases (14, 41, 51), but it has recently been suggested that it could also be due to lack of nitrous oxide reductase in some denitrifiers (19, 41). Since N2O is an intermediate in the denitrification pathway, both the amount of N2O produced and the N2O/(N2O + N2) ratio are important in understanding and predicting N2O fluxes from soils.
The main environmental factors known to influence the N2O/(N2O + N2) ratio are pH, organic carbon and NO3− availability, water content, and O2 partial pressure (50). Soil pH is one of the most important factors influencing both denitrification and N2O production (43). In general, the denitrification rate increases with increasing pH values (up to the optimum pH) while, in contrast, the N2O/(N2O + N2) ratio decreases (50). This relationship has been characterized in laboratory experiments (9, 45), but it is not clear whether the same relationships exist in the field because of methodological limitations of in situ measurement of N2 emissions (16). Nevertheless, 15N tracing experiments based on the addition of a labeled denitrification substrate to soil offer a useful tool to quantify emissions of both N2O and N2 in situ (47, 49). Soil pH is also an important factor influencing denitrifier community composition (35, 39), which can be an important driver of denitrification activity and N2O emissions (5, 21). A recent study reported a negative relationship between the proportion of bacteria genetically capable of reducing N2O within the total bacterial community and the N2O/(N2O + N2) ratio, with both being strongly correlated with soil pH (38).
The objective of the present study was to explore the effect of changes in soil pH on in situ N2O and N2 emissions, denitrifying enzyme activity (DEA), and potential N2O production. In addition, we also investigated whether differences in N fluxes could be related to changes in the size of the microbial community possessing the different denitrification genes. A field experiment was conducted using replicated grassland plots in which the soil pH was modified by addition of either acid or hydroxide to the soil. A 15N tracer method was used to provide information on N emissions. In addition to measuring potential denitrification activity, the size of the denitrifier community was determined by real-time PCR quantification of the denitrification genes.
MATERIALS AND METHODS
Experimental field.
The field experiment with manipulation of the soil pH was established in a grassland area at Borová Farm near Český Krumlov in South Bohemia, Czech Republic (48°52′N, 14°13′E; altitude, 630 m above sea level). The pasture site had been irregularly and occasionally grazed by cattle over the past 10 years. The soil at the site is classified as Haplic Phaeozem (arenic; World Reference Base system) containing 60 to 80% sand, 14 to 32% silt, and 6 to 14% clay (USDA classification system). The mean annual precipitation in the area is 650 mm, and the annual average temperature is 7°C. After the establishment of the experimental site in July 2007, it was fenced to prevent further access by cattle and the sward was periodically cut.
The experimental field (12 by 18 m) was divided into 24 plots (each 3 by 3 m), 12 of which were used in this study for manipulation of the soil pH as follows: (i) four random plots were amended with a KOH solution to increase the pH, (ii) four random plots were amended with an H2SO4 solution to decrease the pH, and (iii) four random plots were amended with water (control plots with a natural pH). The KOH and H2SO4 solutions were applied three times (11 July 2007, 13 September 2007, and 2 April 2008) before the experiment commenced in May 2008. The KOH (15 liters of a 0.97 M solution for each of the three subsequent amendments) and H2SO4 (15 liters of a 0.73 M solution for each of the three subsequent amendments) solutions were applied uniformly to the whole surface (3 by 3 m) of each plot using a sprinkling can, and the same volume of water was applied to the control plots. For the following assessment of N emissions and soil sampling, only the inner 2.5-by-2.5-m plots were used to ensure a 1-m protective zone between the treatments. The plant cover was a mixture of perennial grasses (mainly Lolium perenne and Phleum pratense), clover (Trifolium hybridum), and other dicotyledonous plants (e.g., Achillea millefolium, Plantago sp., Rumex sp., and Vicia sp.). The application of H2SO4 and KOH solutions led to an approximately 40% reduction in the vegetation cover (based on the covered soil surface). Moreover, clover was suppressed in both cases and Echinochloa crus-galli with Elytrigia repens appeared on the acidic plots while Chenopodium album with Atriplex patula appeared on the alkaline plots.
In situ gas emission measurements.
Gas fluxes were determined using medium-size (basal area, 0.096 m2; volume, 10 dm3), nonvented static chambers consisting of a permanent base (bottom part) and top (lid with a rubber septum for gas sampling) (23). Three days before the measurements were started, the permanent bases were inserted 5 cm into the soil and the vegetation inside the bases was clipped to ca. 5 cm above the soil surface. There were 3 chambers in each plot and a total of 36 chambers (12 chambers for each pH treatment). One day before adding the 15N-labeled tracer, we measured the natural rates of N2O and CO2 emission in each chamber. For this purpose, headspace gas samples were collected using a hypodermic needle and a polypropylene syringe immediately (9:00 a.m.) and 2 h after closing the chambers (11:00 a.m.). Gas samples for N2O analyses were collected using a 15-ml syringe and stored in pre-evacuated (<100 Pa), septum-capped, 12-ml vials (Labco), while gas samples for CO2 analyses were collected using a 5-ml syringe and stored in pre-evacuated (<100 Pa), septum-capped, 3.5-ml vials (Venoject). N2O and CO2 were quantified using 6890N and 6850N gas chromatographs (Agilent Technologies) equipped with μECD and TCD detectors, respectively. Integration of measured peaks and N2O and CO2 quantification were conducted using GC ChemStation software (Rev. B.02.01.; Agilent Technologies).
The 15N gas flux method was used to measure the N2O and N2 emissions, and nitrogen was added as 15N-labeled KNO3 at a 60 atom% excess at a rate of 10 kg N ha−1. The required amount of N was dissolved in 250 ml H2O and dispensed uniformly over the soil surface inside the chamber using a small sprinkling can at 9:00 a.m. of the day after the measurement of natural N2O and CO2 emissions. Fluxes of N gases were determined five times following KNO3 application (2 to 4, 6 to 8, 24 to 26, 48 to 50, and 72 to 74 h). At the time the lid of the chamber was fitted to the base section and after 2 h, headspace gas samples were collected using a 12-ml syringe and needle. The linearity of gas accumulation in the chambers was successfully tested in a preliminary experiment (intensive sample routine every 15 min over 2 h). The gas samples were stored in evacuated (<100 Pa), septum-capped, 12-ml vials (Labco), and the concentration and 15N content of N2O and the 15N content of the N2 in each 12-ml vial were determined by automated isotope ratio mass spectroscopy as described by Stevens et al. (48), using a Europa Scientific 20-20 stable-isotope analyzer interfaced with a Europa Scientific ANCA-TG trace gas preparation system (Europa Scientific, Crewe, United Kingdom) with a Gilson autosampler (Anachem, Luton, United Kingdom). The ion currents (I) at m/z 44, 45, and 46 enabled concentrations and molecular ratios 45R (45I/44I) and 46R (46I/44I) to be calculated for N2O. The sources of N2O were then apportioned into the fraction (dD) derived from the denitrifying pool of enrichment aD and the fraction dN = (1 − dD) derived from the nitrifying pool or pools at natural abundance (2). For N2, the ion currents at m/z 28, 29, and 30 enabled the 29R (29I/28I) and 30R (30I/28I) molecular ratios to be determined. Differences between the molecular ratios in enriched and normal atmospheres were calculated as Δ29R and Δ30R. The flux of N2 was calculated by using data for Δ29R and Δ30R to calculate the enrichment of the denitrifying pool (15XN) and then the N2 flux according to Mulvaney and Boast (31). The total N fluxes during the 74-h period after the application of 15N-labeled KNO3 were calculated by integration, assuming a linear change in flux rates between observation times.
Soil sampling and analyses.
On the day of labeled KNO3 addition, topsoils (0 to 20 cm) were sampled next to all 36 chambers. Three soil samples were taken from a ca. 25-cm distance outside each chamber and combined to produce one composite sample associated with each chamber. Soils were sieved (5 mm) immediately after sampling and stored at field moisture content in plastic bags at 4°C until required (denitrifying activity plus mineral N concentrations and other soil characteristics were determined within 3 days and 2 weeks after the soil sampling, respectively). Subsamples for molecular analyses were stored in Eppendorf tubes at −80°C. Soil pH was measured using a combined electrode (SenTix 61; WTW GmbH, Weilheim, Germany) and pH meter (526/538 pH meter; WTW) in a 10-g/50-ml soil-to-H2O mixture. Before the pH measurements, the soil slurries were shaken on an end-to-end shaker for 5 min and allowed to stand for 2 h. Soil moisture was determined gravimetrically by drying the soil at 105°C for 24 h, and all results were expressed per gram of dry soil. Soil mineral N (NH4+, NO3−) was measured colorimetrically in 1 M KCl extracts using a soil (fresh field moisture)/KCl solution ratio of 40 g/200 ml (55). Total organic carbon (Corg) was determined by wet oxidation with acid dichromate, and total nitrogen content (Ntot) was determined by Kjeldahl digestion (55). Microbial biomass C (Cmic) was determined by the chloroform fumigation extraction method as described by Vance et al. (53), followed by dichromate digestion of extractable C.
DEA measurement.
DEA was determined by the phase I assay of Smith and Tiedje (46), which was slightly modified as described by Šimek and Hopkins (44). Briefly, soil slurries were made by mixing 25-g field-moist soil samples in 120-ml serum bottles with 25 ml of a solution containing 1 mM glucose and 1 mM KNO3. Bottles were capped with rubber stoppers and metal holders, evacuated, and flushed four times with 99.99% He. The slurries were then incubated either with or without acetylene (10%, vol/vol) on an end-to-end shaker at 25°C for measurements of DEA and potential N2O emission, respectively. After 30 and 60 min, 0.5-ml headspace samples were taken with a gas-tight syringe and N2O was immediately quantified using gas chromatography (see above).
DNA extraction and quantification of the denitrifier community.
For each of the 36 samples, DNA was extracted from 250 mg of soil by the method of Martin-Laurent et al. (27). Briefly, samples were homogenized in 1 ml of extraction buffer for 30 s at 1,600 rpm in a Mini-Beadbeater cell disrupter (Mikro-Dismembrator S; B. Braun Biotech International). Soil and cell debris were eliminated by centrifugation (14,000 × g for 5 min at 4°C). After precipitation with ice-cold isopropanol, nucleic acids were purified using both polyvinylpolypyrrolidone and Sepharose 4B spin columns. Soil DNA quality and size were checked by electrophoresis on 1% agarose. DNA was also quantified by spectrophotometry at 260 nm using a BioPhotometer (Eppendorf, Hamburg, Germany).
The size of the denitrifier community was estimated by quantitative PCR (qPCR) of genes encoding the catalytic subunit of the key enzymes of the denitrification pathway. Fragments of the narG, napA, nirK, nirS, and nosZ genes encoding the membrane and periplasmic nitrate reductases, the cd1 and copper nitrite reductases, and the nitrous oxide reductase, respectively, were amplified using primers and thermal cycling conditions described previously (4, 18, 19). The total bacterial community was quantified using 16S rRNA as a molecular marker (25). Reactions were carried out in an ABI Prism 7900 sequence detection system (Applied Biosystems). Quantification was based on the fluorescence intensity of the SYBR green dye, which binds to double-stranded DNA. The 20-μl PCR mixture contained 12.5 μl of SYBR green PCR Master Mix (Absolute QPCR SYBR green Rox ABgen), 1 μM of each primer, 100 ng of T4 gp32 (Qbiogene), and 12.5 ng of DNA. Standard curves were obtained using serial dilutions of linearized plasmids containing cloned narG, napA, nirK, nirS, nosZ, and 16S rRNA genes amplified from denitrifying strains. No-template controls gave null or negligible values. The presence of PCR inhibitors in DNA extracted from soil was estimated by (i) diluting soil DNA extract and (ii) mixing a known amount of standard DNA with soil DNA extract prior to qPCR. No inhibition was detected.
Statistical analyses.
All data obtained were first analyzed using descriptive statistical methods, and the Kolmogorov-Smirnov test was used to check the normality of the data. Differences in means of characteristics among the three pH treatments were tested by hierarchical analysis of variance, where the factor “plot” was nested in the factor “pH treatment,” followed by a post hoc test of comparison (Tukey honestly significant difference [HSD] test). To test possible relationships among N emissions, DEA, abundance of denitrification genes, and soil parameters, Pearson's correlation coefficients (r) were calculated for data from all 36 soil samples and chambers. All statistical tests were performed in the program package STATISTICA (v7.1., StatSoft, Inc., 2005). Significance was accepted at a probability (P) level of <0.05.
RESULTS
Soil pH and other chemical and biological parameters.
Table 1 shows means and standard deviations of selected soil properties and microbial activities in soils from the experimental plots differing in pH management. Manipulation of soil pH via the application of acid or alkali solutions during the 10 months before the gas flux measurements resulted in a significant change in the soil reaction. The pH values were statistically significantly different (Tukey HSD test, P < 0.05) with a difference of 0.85 to 2.15 pH units between the treatments. Several soil properties other than pH were also affected by the addition of acid or alkali solutions. For example, the NH4+ concentration was higher in the acidic soil than in the natural pH and alkaline soils, while the NO3− concentration was higher in the soils with alkaline pH and decreased significantly with decreasing pH (Tukey HSD test, P < 0.05). The microbial biomass determined by the fumigation extraction method (Cmic) was lowest in the acidic soils and highest in the alkaline soils (Tukey HSD test, P < 0.05). Emissions of CO2 and N2O determined before the application of 15N-labeled KNO3 were also affected by the treatments. Thus, significantly higher CO2 flux occurred in the control plots with natural pH soil (182 μg C m−2 h−1) than in the other plots (118 and 122 μg C m−2 h−1 in the acidic and alkaline soils, respectively). In contrast, N2O emissions were similar from the natural pH and alkaline soils (32 to 36 μg N m−2 h−1) but significantly lower from the acidic soils (12 μg N m−2 h−1).
TABLE 1.
Selected properties of soils from experimental plots differing in pH management
| Parameter | Value for indicated type of soila |
||
|---|---|---|---|
| Acidic pH | Natural pH | Alkaline pH | |
| pH | 5.52 ± 0.48 (a)a | 6.82 ± 0.40 (b) | 7.67 ± 0.17 (c) |
| Moisture (g H2O g−1) | 0.23 ± 0.02 (a) | 0.24 ± 0.04 (ab) | 0.26 ± 0.04 (b) |
| NH4+ (μg N g−1) | 2.74 ± 2.65 (a) | 0.56 ± 0.27 (b) | 0.68 ± 0.31 (b) |
| NO3− (μg N g−1) | 1.88 ± 0.82 (a) | 3.77 ± 1.41 (b) | 5.03 ± 2.03 (c) |
| Ntot (mg N g−1) | 3.31 ± 0.71 (a) | 2.90 ± 1.26 (ab) | 2.51 ± 1.21 (b) |
| Corg (mg C g−1) | 20.9 ± 6.5 (a) | 20.5 ± 4.4 (a) | 17.5 ± 8.5 (b) |
| Cmic (μg C g−1) | 587.1 ± 170.6 (a) | 951.8 ± 354.8 (b) | 1,249.7 ± 331.0 (c) |
| CO2 emissionb (μg C m−2 h−1) | 118.3 ± 35.1 (a) | 182.4 ± 23.6 (b) | 122.1 ± 19.6 (a) |
| N2O emissionb (μg N m−2 h−1) | 12.3 ± 3.8 (a) | 31.5 ± 17.4 (b) | 36.1 ± 26.5 (b) |
Mean values ± standard deviations (n = 12) are shown. The different letters in parentheses indicate significant differences between the specific pH treatments (P < 0.05).
Natural emission determined before KNO3 addition.
In situ N2O and N2 emissions after the application of 15N-labeled NO3−.
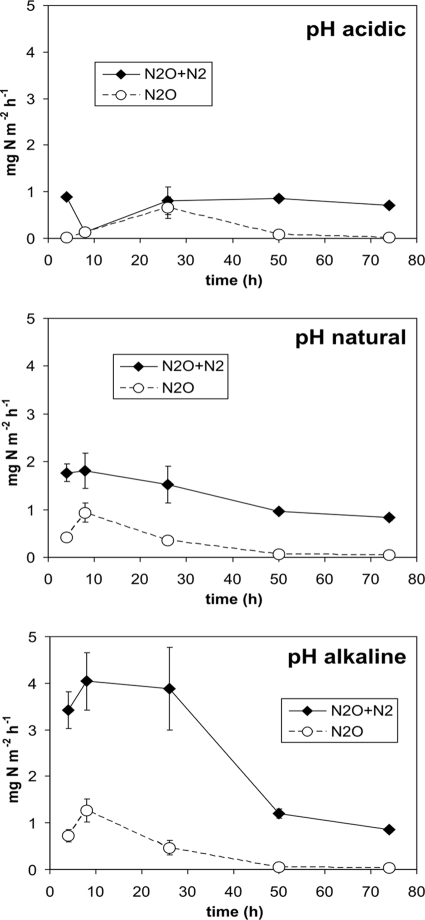
Figure 1 shows the time course of the total N fluxes (N2O + N2) and of the N2O fluxes from the soils with different pH treatment during the 74 h after the 15N-labeled KNO3 amendment. Higher N gas emissions were observed in the alkaline plots than in the control (natural pH) and acidic plots. Different total N fluxes were observed among the three treatments during the first 24 h after the application of KNO3. However, 50 to 74 h after the KNO3 amendment, the total N fluxes were around 1 mg N m−2 h−1 in all treatments. In the control and alkaline plots, the N2O emissions reached a maximum of ca. 1.1 mg N m−2 h−1 within 8 h after KNO3 application and then decreased to ca. 0.4 mg N m−2 h−1. In the acidic soil, the N2O emissions increased up to 26 h before decreasing. In all three pH treatments, N2 was the only gas product at the end of the experimental period.
FIG. 1.
Dynamics of total N (N2O + N2) and N2O in situ fluxes after the addition of 15N-labeled NO3− to acidic, natural pH, and alkaline soils. Mean values ± standard errors of the means are shown (n = 12).
Table 2 shows the fraction of the N2O flux which was derived from the NO3− (denitrification) pool (dD) together with the enrichment (aD) of this pool and also the fraction from the NH4+ (nitrification) pool (dN). Valid calculations could only be done, however, whenever there was a significant N2O flux. Therefore, the values were not determined at 50 and 74 h after the KNO3 addition for all three soils and 4 h after the KNO3 addition for the acidic soil. In general, denitrification was the major process responsible for N2O emissions in all three soils, accounting for ca. 61 to 87%. The exception was the acidic soil 8 h after KNO3 addition, where the N2O emissions were split equally between nitrification and denitrification. The enrichment of the denitrifying pool (aD; on average, 54 atom%) was close to the enrichment of the added KNO3 (60 atom%).
TABLE 2.
Causes of N2O flux in acidic, natural pH, and alkaline soilsa
| Parameter | Soil | Mean value ± SD at following time after KNO3 addition: |
||||
|---|---|---|---|---|---|---|
| 4 h | 8 h | 26 h | 50 h | 74 h | ||
| dD (fraction) | Acidic | NDb | 0.48 ± 0.22 | 0.73 ± 0.22 | ND | ND |
| Natural pH | 0.75 ± 0.15 | 0.83 ± 0.12 | 0.61 ± 0.24 | ND | ND | |
| Alkaline | 0.83 ± 0.14 | 0.87 ± 0.13 | 0.63 ± 0.26 | ND | ND | |
| dN = (1 − dD) (fraction) | Acidic | ND | 0.52 ± 0.22 | 0.27 ± 0.22 | ND | ND |
| Natural pH | 0.25 ± 0.15 | 0.17 ± 0.12 | 0.39 ± 0.24 | ND | ND | |
| Alkaline | 0.17 ± 0.14 | 0.13 ± 0.13 | 0.37 ± 0.26 | ND | ND | |
| aD (enrichment; atom%) | Acidic | ND | 0.56 ± 0.02 | 0.56 ± 0.03 | ND | ND |
| Natural pH | 0.54 ± 0.03 | 0.55 ± 0.03 | 0.50 ± 0.06 | ND | ND | |
| Alkaline | 0.54 ± 0.05 | 0.55 ± 0.04 | 0.51 ± 0.07 | ND | ND | |
The fractions of the N2O flux which were derived from the NO3− (denitrification) pool (dD), from the NH4+ (nitrification) pool (dN), and the enrichment (aD) of the NO3− (denitrifying) pool (atom%) in acidic, natural pH, and alkaline soils are shown (n = 12).
ND, not determined due to low N2O fluxes.
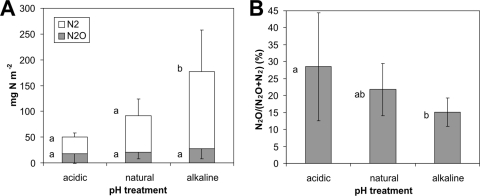
Cumulative fluxes of N2O + N2 over the experimental period (74 h) were 50.7, 91.2, and 177.1 mg N m−2 from the acidic, natural pH, and alkaline soils, respectively (Fig. 2A). The N2 emissions were mostly responsible for the differences in the total N fluxes between the pH treatments, as the N2O emissions were not significantly different (Tukey HSD test, P > 0.24). The large differences in N2 emissions between acidic and control plots were not significant (although marginally: Tukey HSD test, P = 0.055), probably due to the large variation in fluxes between individual chambers (Fig. 2A). Figure 2B shows the relative contribution of N2O to the total cumulative N2O + N2 fluxes expressed as the N2O/(N2O + N2) molar ratio. The N2O/(N2O + N2) ratio increased with decreasing soil pH. The total losses of added NO3-N (10 kg N ha−1) as N2O and N2 (emission factors) were 5.1, 9.1, and 17.7% during the experimental period for the acidic, natural pH, and alkaline soils, respectively. The emission factors for N2O only were 1.8, 2.1, and 2.8% for the acidic, natural pH, and alkaline soils, respectively.
FIG. 2.
In situ cumulative losses of N (separately as N2O and N2) (A) and relative N2O production expressed as the N2O/(N2O + N2) molar ratio (B) over the 74 h after the addition of 15N-labeled KNO3 to acidic, natural pH, and alkaline soils. Mean values and ± standard deviations are shown (n = 12). The different letters next to the bars indicate significant differences between the specific pH treatments (P < 0.05).
Correlation analysis, which included data from all 36 experimental points (both soil samples and chambers for emission measurements), showed significant negative correlations between the N2O/(N2O + N2) ratio and soil pH (r = −0.493, P = 0.002) and NO3− concentration (r = −0.458, P = 0.005) (Table 3). We also found significant correlations between total N fluxes (N2O + N2) and pH (r = 0.540, P < 0.001) but also Ntot, Corg, Cmic, and NO3− concentration (Table 3). There were no other statistically significant correlations with soil properties (Table 3).
TABLE 3.
Pearson's correlation coefficients for relationships between soil parameters (including abundance of denitrification genes and their ratios) and N gas fluxes estimated in situ using 15N and under laboratory conditions using DEA with the respective N2O/(N2O + N2) molar ratio
| Parameter |
P value |
|||
|---|---|---|---|---|
|
In situ |
In laboratory |
|||
| N2O + N2 | N2O/(N2O + N2) ratio | N2O + N2 | N2O/(N2O + N2) ratio | |
| pH | 0.540a | −0.490b | 0.637a | −0.848a |
| Moisture | 0.195 | −0.209 | 0.835a | −0.349c |
| NO3− | 0.353c | −0.458b | 0.825a | −0.578a |
| NH4+ | −0.266 | 0.153 | −0.299 | 0.567a |
| Cmic | 0.465b | −0.269 | 0.940a | −0.599a |
| Ntot | −0.525b | 0.215 | −0.293 | 0.309 |
| Corg | −0.500b | 0.066 | −0.119 | 0.143 |
| 16S rRNA gene copy no. | −0.040 | −0.006 | −0.096 | −0.268 |
| narG gene copy no. | −0.156 | −0.339c | 0.243 | −0.452b |
| napA gene copy no. | 0.122 | −0.130 | 0.175 | −0.470b |
| nirS gene copy no. | −0.012 | −0.415c | 0.441b | −0.602a |
| nirK gene copy no. | 0.080 | −0.005 | −0.121 | −0.186 |
| nosZ gene copy no. | −0.062 | −0.095 | 0.191 | −0.261 |
| narG/16S rRNA ratio | −0.139 | −0.481b | 0.544a | −0.386c |
| napA/16S rRNA ratio | 0.387c | −0.304 | 0.657a | −0.686a |
| nirS/16S rRNA ratio | 0.063 | −0.493b | 0.666a | −0.612a |
| nirK/16S rRNA ratio | 0.101 | −0.022 | −0.120 | −0.043 |
| nosZ/16S rRNA ratio | −0.062 | −0.167 | 0.409c | 0.015 |
| nosZ/nirS ratio | −0.275 | 0.433b | −0.491b | 0.706a |
| nosZ/nirK ratio | −0.066 | −0.124 | 0.349 | 0.005 |
| nosZ/(nirS + nirK) ratio | −0.239 | 0.381c | −0.359c | 0.662a |
P < 0.001 (n = 36).
P < 0.01.
P < 0.05.
DEA.
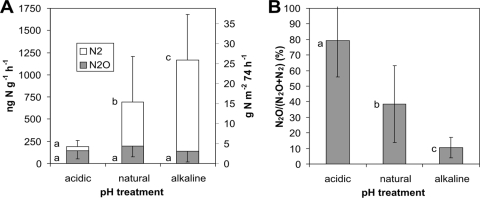
DEA, expressed as the overall production of both denitrification products, N2O and N2, was significantly affected by the pH treatment (189, 692, and 1,169 ng N g−1 h−1 for the acidic, natural pH, and alkaline soils, respectively; Tukey HSD test, P < 0.01; Fig. 3A). Similar to cumulative N emissions in situ, the N2 production was responsible for the differences in DEA between the pH treatments, since N2O production was not affected by pH (Tukey HSD test, P > 0.1). Correlation analysis showed a positive relationship between DEA and pH (Table 3). However, the best predictors of DEA were Cmic (r = 0.940, P < 0.001), soil moisture (r = 0.835, P < 0.001), and NO3− concentration (r = 0.825, P < 0.001). The proportion of N2O produced as the terminal product of denitrification and calculated as the N2O molar ratio (N2O/[N2O + N2]) was also highly dependent on the pH treatment, being 79.2% in acidic soil, 38.5% in natural pH soil, and 10.5% in alkaline soil (Fig. 3B). In contrast to the overall N2O and N2 production (DEA), soil pH was the best predictor of the N2O/(N2O + N2) molar ratio (r = −0.848, P < 0.001) (Table 3). In addition, correlation analysis showed weak but significant relationships between in situ total N fluxes and DEA (r = 0.393, P = 0.018) and between the N2O/(N2O + N2) molar ratio from the field measurements and that from the DEA assay (r = 0.379, P = 0.023) (data not shown).
FIG. 3.
DEA (separately as N2O and N2 production) of soils with different pH treatments (A) and relative N2O production during DEA determination expressed as the N2O/(N2O + N2) molar ratio (B). DEA (A) was also estimated and is expressed as g N m−2 74 h−1 (right vertical axis). Mean values ± standard deviations are shown (n = 12). The different letters next to the bars indicate significant differences between the specific pH treatments (P < 0.05).
Abundance of 16S rRNA and denitrification genes.
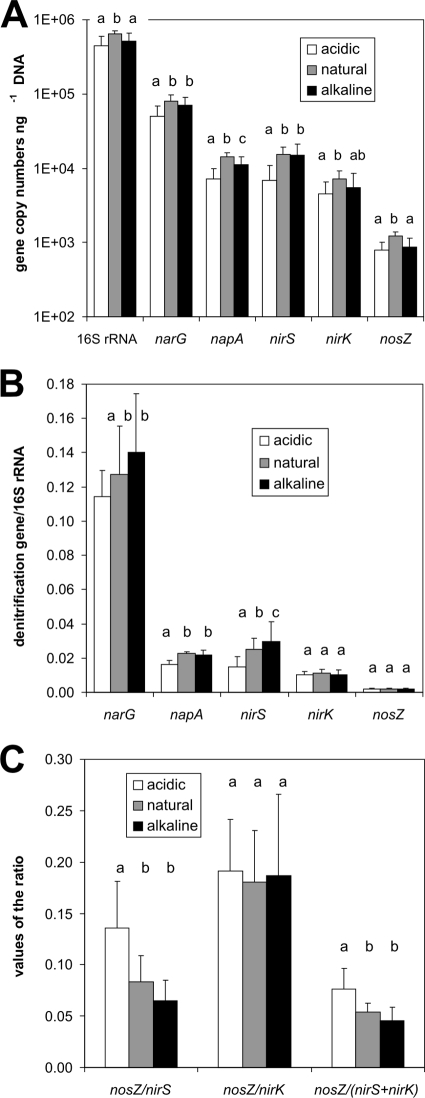
The highest denitrification gene copy numbers were observed in the natural pH plots, with significantly lower gene copy numbers in the acidic soil (Fig. 4A). A decrease compared to the natural pH plots was also observed in the alkaline plots, which was significant only for the napA and nosZ genes. The abundance of total bacteria estimated using 16S rRNA gene copy numbers was also affected by the H2SO4 and KOH amendments (4.4 × 105, 6.4 × 105, and 5.1 × 105 gene copies ng−1 DNA for the acidic, natural pH, and alkaline soils, respectively), similar to the denitrification genes. To better understand how the denitrifier community was influenced by soil pH, we also estimated the relative abundance of denitrifiers potentially capable of performing different steps in the denitrification cascade by calculating the ratios of different denitrification genes to the 16S rRNA gene (e.g., the narG/16S rRNA gene ratio) (Fig. 4B). The nirS/16S rRNA gene ratio was the only one which differed significantly among the three pH treatments (Tukey HSD test, P < 0.05), with the lowest and highest ratios in the acidic and alkaline soils, respectively. Calculation of the nosZ/nirK and nosZ/nirS gene ratios revealed that only the latter was affected by pH, with a significantly higher ratio in the acidic soils (Fig. 4C).
FIG. 4.
Abundances of the 16S rRNA, narG, napA, nirS, nirK, and nosZ genes in soils with different pH treatment expressed as the number of gene copies ng−1 of DNA (A), the specific denitrification gene/16S rRNA gene ratio (B), and nosZ/nirS, nosZ/nirK, and nosZ/(nirS + nirK) ratios (C). Mean values and standard deviations are shown (n = 12). The different letters above the bars indicate significant differences between the specific pH treatments (P < 0.05).
We also performed an analysis of the correlation between N fluxes (in situ N emissions, DEA, and N2O/[N2O + N2] ratio) and the size of the denitrifying community (expressed as absolute values of denitrification gene abundance, as the ratios of different denitrification genes to the 16S rRNA gene, and as ratios of the nosZ gene to genes encoding nitrite reductase) to identify possible linkages between them (Table 3). We found no correlation between in situ total N fluxes and the abundance of the denitrification genes, and only the abundance of the nirS gene was correlated with DEA. The abundance of this gene also correlated with the N2O/(N2O + N2) molar ratio from the field measurements and from the DEA assay. Significant correlations between the N2O/(N2O + N2) ratio and the abundances of the genes narG and napA (napA was correlated only with the relative N2O production determined during the DEA assay) and the nirS/16S rRNA gene, napA/16S rRNA gene, nosZ/nirS, and nosZ/(nirS + nirK) ratios were also observed (Table 3).
DISCUSSION
Manipulation of soil pH in situ is difficult to achieve because of the high buffering capacity of most soils. However, since soils having different natural pHs also differ in many other properties, the effect of soil pH is often investigated by modifying the pH gradually (1, 33), typically by the application of lime. This method of pH manipulation can take years, with repeated lime applications being required. In the present study, we significantly modified the soil pH by short-term application (three times in 10 months) of H2SO4 and KOH solutions. Such additions of KOH or H2SO4 solutions to the soil also represent a source of exchangeable potassium cations and sulfate anions, respectively, which might also have direct or indirect side effects on the microbial community. The shift in soil pH was accompanied by changes in soil functioning, leading to modifications of other soil properties (Table 1). Thus, the relatively high NH4+ concentration and low NO3− concentration in the acidic soil (H2SO4) suggest a decrease in nitrifying activity, as previously reported (12, 34). In contrast, the opposite was found in the alkaline soil, suggesting a higher nitrification rate, which could have stimulated denitrification. The application of the acid and hydroxide to the soil also affected the vegetation cover, which probably caused the observed differences in the CO2 emissions and moisture content between the pH treatments (Table 1). Therefore, the correlations between pH and N fluxes or denitrifying community size observed in our work might not represent a direct causal relationship, as confounding effects cannot be ruled out.
Analysis of the denitrification process in the soils with different pH treatments revealed a significant decrease in N gas production with soil pH for both cumulative N fluxes in situ and DEA (Fig. 2A and 3A), which is in agreement with previous findings of lower denitrification rates in acidic soil (13, 45). Denitrification was the major process responsible for N2O emissions in all three soils (Table 2); the fraction of the N2O flux derived from the denitrifying NO3− pool (dD) was 0.72 (72%) averaged over all treatments at 4, 8, and 26 h after the application of 15N-labeled NO3−. In contrast to the total N fluxes in the field and DEA, we did not find any differences in N2O emissions between the pH treatments for either the cumulative in situ N2O emissions (Fig. 2A) or potential N2O emissions (Fig. 3A). This result was surprising since higher N2O emissions in acidic soils have been reported in several studies (30, 54). For example, Weslien et al. (54) found a strong negative correlation between N2O emissions in situ and soil pH (r = 0.93, P < 0.01) from a Swedish organic soil with a pH range of 3.6 to 5.9. One of the possible reasons for the lack of difference in N2O fluxes between the pH treatments in our study could be the relatively moderate change in pH obtained in the field, which might have been too limited to lead to changes in N2O emissions estimated by the 15N tracer experiment or the potential assay. We also found that the percentage of the applied nitrate (10 kg N ha−1) emitted as N2O in situ was between 1.8 and 2.8%, which is higher than the estimate of 1.25% according to the Intergovernmental Panel on Climate Change (IPCC) (22) but in agreement with recent studies reporting emission rates of up to 2.5% and suggesting that the IPCC methodology underestimates N2O emissions (15). Interestingly, the potential N2O production rates estimated per g N m−2 74 h−1 (Fig. 3A) were ca. 3,500, 1,900, and 1,100 times higher than the in situ natural N2O emissions from the acidic, natural pH, and alkaline soils, respectively (Table 1), and ca. 180, 210, and 40 times higher than the cumulative losses of N2O over 74 h after the amendment with 15N-labeled NO3− (Fig. 2A). The DEA assay optimizes the N production rate under laboratory conditions from the pool of denitrification enzymes present in the soil at the time of sampling and consists of incubating the soil at a temperature of 25°C without oxygen and with the addition of NO3− and a surplus of electron donor (glucose). Incubation of the soil under these nonlimiting denitrifying conditions resulted in the stimulation of the rates observed in our study.
Calculation of the N2O/(N2O + N2) ratio showed a decreasing molar ratio with increasing soil pH and denitrification rates, in agreement with previous studies (43). However, the range of the N2O/(N2O + N2) ratio was different according to the approach used. Thus, it varied between 15.1 and 28.5% and between 10.5 and 79.2% when calculated using the in situ measurement and potential denitrification, respectively (Fig. 2B and 3B). Since NO3− is preferred over N2O as an electron acceptor, the high nitrate concentration in the DEA assay may explain the high N2O/(N2O + N2) ratio. Nevertheless, notwithstanding the differences between the two approaches, the N2O/(N2O + N2) ratio in the field was significantly correlated with the N2O/(N2O + N2) ratio calculated from the DEA assay under laboratory conditions (r = 0.379, P = 0.023). Moreover, the total N fluxes in situ were significantly correlated with potential denitrification (DEA). This is an important finding indicating that although the magnitudes of the fluxes were different, determination of the N2O/(N2O + N2) ratio under laboratory conditions can reflect treatment differences in the field. We found soil pH to be the best predictor of the N2O/(N2O + N2) ratio determined by both DEA and in situ N flux measurements (Table 3). Similarly, simultaneous determination of N2O and N2 emissions from intact soil cores taken from a beech forest ecosystem with differences in pH values showed that pH was the only significant determinant of the N2/N2O ratio (11). In our study, soil pH was also the best predictor of total N fluxes in situ but not of DEA, which was correlated more with Cmic, NO3−, and soil moisture. Overall, our results confirm the role of soil pH in situ in determining the nature of the denitrification end products and process rates. It is, however, important to underline the fact that in our experimental field study many variables, such as pH and NO3−, correlated with each other.
Most recent studies investigating linkages between denitrification activity and denitrifying community ecology have focused on denitrifier diversity and process rates without considering the nature of the denitrification end products (13, 17, 26, 40). However, recent studies have shown that some denitrifiers can have a truncated denitrification pathway and lack the nosZ gene encoding nitrous oxide reductase (24, 37), which results in the emission of N2O as the end product, whatever the environmental conditions. In the present study, we therefore quantified the narG, napA, nirK, nirS, and nosZ denitrification genes to verify whether differences in the denitrification rates and in the N2O/(N2O + N2) ratio could be related to changes in the size of the microbial community possessing the different denitrification genes. In our study, the abundance of the nosZ gene was less then 6% of the total abundance of the nirS and nirK genes. A higher abundance of the nirS and nirK genes than of the nosZ gene has already been observed in several studies (3, 19, 38). However, in a recent study by Hallin et al. (17) using the same primers that we used in our study, similar densities of the nirK gene or nirS and nosZ genes were found, which rules out the possibility that the imbalanced densities of the nirK, nirS, and nosZ genes are due to the fact that the nosZ primers may not be as universal as those for the nirK and nirS genes. Together, these results indicate that, in some environments, a significant fraction of the denitrifiers lacks the nosZ gene and therefore these denitrifiers are not genetically able to reduce N2O, as suggested by bacterial genome analysis (24). Higher copy numbers of all of the denitrification genes and the 16S rRNA gene were observed in the natural pH soil (Fig. 4A), in agreement with the soil respiration data (determined as CO2 emissions, Table 1), indicating that both denitrifiers and total bacteria were affected by the pH treatment. However, the abundance of the denitrification genes was not correlated with total N fluxes in situ and only the abundance of the nirS gene was correlated with DEA (Table 3). A positive correlation between nirS gene abundance and DEA was also reported by Philippot et al. (38) and Chroňáková et al. (6) at sampling sites in the same grassland area while, similar to our results, the abundance of the nosZ gene was uncoupled from the denitrification activity in the studies by Miller et al. (28, 29) and Dandie et al. (10). In contrast, other studies reported significant correlations between the nirK or nosZ gene copy numbers and potential denitrification rates (17, 52). Even though denitrification is a facultative respiratory process and is therefore primarily regulated by nitrate, carbon, and oxygen availability, it is possible that the size of the denitrifying community contributes to the regulation of the process rates when conditions are favorable for denitrification. Such a correlation between the size and activity of the denitrifying community is even more likely when the measured rates result from the activity of all of the denitrification enzymes present in the sample, such as in the DEA assay. However, the discrepancies between the studies cited above underline the complexity of the hierarchical regulation of biochemical processes (42) and indicate that counting of denitrifiers is not enough for a comprehensive understanding of N fluxes by denitrification. In contrast to total N fluxes and DEA, the N2O/(N2O + N2) ratio estimated both in situ and in the laboratory was negatively correlated with the abundance of nirS and to a lesser extent with that of napA (only for the N2O/[N2O + N2] ratio estimated by DEA) and narG (Table 3). Further experiments are required over a broader range of pHs to verify whether there is a relationship between the size of the denitrifying community possessing these genes and the nature of the denitrification end products.
To better understand the relationships among soil pH, denitrification activity, and the denitrifying community, we calculated the proportion of denitrifiers performing the different steps in the denitrification cascade within the total bacterial community (Fig. 4B) and found a positive correlation between the nirS/16S rRNA gene ratio and the soil pH. This correlation, which suggests that NirS denitrifiers are more sensitive to soil pH than the rest of the total bacterial community, has not, however, been observed in previous studies and therefore could represent a spurious relationship (17, 38). Nitrite reductases, which reduce nitrite to gaseous NO, are key enzymes of the denitrification pathway, and bacteria must have this reductase to be recognized as denitrifiers (50). Denitrifying bacteria possess either a cytochrome cd1 nitrite reductase (NirS) or a copper nitrite reductase (NirK) (56), but both enzymes perform the same reaction and are interchangeable. The increase in the nirS/16S rRNA gene ratio with soil pH but not of the nirK/16S rRNA gene ratio supports the findings from recent studies suggesting niche differentiation between denitrifiers possessing the two types of nitrite reductases (17, 38). We found that the ratio of all of the denitrification genes, but not the nirK/16S rRNA gene ratio, was correlated significantly with DEA (Table 3), suggesting that the proportion of denitrifiers within the total bacterial community could be more important for denitrification rates than the size of the denitrifier community itself. Probably due to the significant correlation between the nirS, narG, and napA gene copy numbers and the N2O/(N2O + N2), the nirS/16S rRNA gene, narG/16S rRNA gene, napA/16S rRNA gene, nosZ/nirS, and nosZ/(nirS + nirK) ratios were also correlated with the N2O/(N2O + N2) ratio determined in the laboratory. This putative influence of the proportion of denitrifiers on N2O production might also be interlaced with the influence of the denitrifier community composition (5), which has been shown to be affected by shifts in soil pH (13, 17). In contrast to the results of Philippot et al. (38) suggesting that the proportion of bacteria able to reduce N2O could be of importance in determining the nature of the denitrification end products, we did not find any negative correlation between the proportion of denitrifiers possessing the nosZ gene and the N2O/(N2O + N2) ratio. This indicates that, not surprisingly, the routes for N2O production are numerous and the relative importance of the denitrifier community composition, denitrification enzyme regulation (41), and the proportion of denitrifiers lacking the gene for an N2O reductase remains to be experimentally demonstrated.
To conclude, our results indicate that manipulation of soil pH affected the N2O/(N2O + N2) ratio, which increased with decreasing pH due to changes in the total denitrification activity but not in N2O production. We also showed that denitrification activity and N2O emissions measured under laboratory conditions were correlated with N fluxes in situ and therefore could reflect treatment differences in the field. The size of the denitrifying community was uncoupled from in situ N fluxes, but the potential denitrification was significantly correlated with the number of NirS denitrifiers. We also found a relationship between the narG, napA, and nirS gene copy numbers and the N2O/(N2O + N2) molar ratio which requires further understanding. However, in this study, the proportion of denitrifiers capable of reducing N2O did not seem to have a role in determining the N2O/(N2O + N2) ratio. It is crucial in future studies to continue to bridge the gap between studies of denitrifier ecology and of N fluxes for a comprehensive understanding of the role of denitrifier community ecology in determining not only total denitrification rates but also the nature of the denitrification end products.
Acknowledgments
This research was supported by the Ministère des Affaires Etrangères et Européennes in France (Egide-Barrande), by the Ministry of Education, Youth and Sports in the Czech Republic (Programme Barrande 2-07-26), by research grants AV0Z60660521, MSM 6007665801, and LC 06066, and by the Grant Agency of the Academy of Sciences of the Czech Republic (IAA600660605).
L. Jíšová, V. Šlajchrtová, A. Škeříková, J. Jirout, J. Hynšt, and especially S. Šafarčíková are gratefully acknowledged for field work and laboratory analyses. V. and M. Kamír are thanked for allowing access to the experimental field and their significant support in the field work. Thanks are also due to M. Nicholson from the Agri-Food and Biosciences Institute, Belfast, for gas chromatography and isotope ratio mass spectrometry analyses.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Adams, T. M., and S. N. Adams. 1983. The effects of liming and soil pH on carbon and nitrogen contained in the soil biomass. J. Agric. Sci. 101:553-558. [Google Scholar]
- 2.Arah, J. R. M. 1997. Apportioning nitrous oxide fluxes between nitrification and denitrification using gas-phase mass spectrometry. Soil Biol. Biochem. 29:1295-1299. [Google Scholar]
- 3.Baudoin, E., L. Philippot, D. Chèneby, L. Chapuis-Lardy, N. Fromin, D. Bru, B. Rabary, and A. Brauman. 2009. Direct seeding mulch-based cropping increases both the activity and abundance of denitrifier communities in a tropical soil. Soil Biol. Biochem. 41:1703-1709. [Google Scholar]
- 4.Bru, D., A. Sarr, and L. Philippot. 2007. Relative abundance of the membrane-bound and periplasmic nitrate reductase. Appl. Environ. Microbiol. 73:5971-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavigelli, M. A., and G. P. Robertson. 2000. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81:1402-1414. [Google Scholar]
- 6.Chroňáková, A., V. Radl, J. Čuhel, M. Šimek, D. Elhottová, M. Engel, and M. Schloter. 2009. Overwintering management on upland pasture causes shifts in an abundance of denitrifying microbial communities, their activity and N2O-reducing ability. Soil Biol. Biochem. 41:1132-1138. [Google Scholar]
- 7.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crutzen, P. J., and D. H. Ehhalt. 1977. Effects of nitrogen fertilizers and combustion on the stratospheric ozone layer. Ambio 6:112-117. [Google Scholar]
- 9.Čuhel, J. 2004. B.S. thesis. Faculty of Biological Sciences, University of South Bohemia, České Budějovice, Czech Republic.
- 10.Dandie, C. E., D. L. Burton, B. J. Zebarth, S. L. Henderson, J. T. Trevors, and C. Goyer. 2008. Changes in bacterial denitrifier community abundance over time in an agricultural field and their relationship with denitrification activity. Appl. Environ. Microbiol. 74:5997-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dannenmann, M., K. Butterbach-Bahl, R. Gasche, G. Willibald, and H. Papen. 2008. Dinitrogen emissions and the N2:N2O ratio of a Rendzic Leptosol as influenced by pH and forest thinning. Soil Biol. Biochem. 40:2317-2323. [Google Scholar]
- 12.de Boer, W., and G. A. Kowalchuk. 2001. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol. Biochem. 33:853-866. [Google Scholar]
- 13.Enwall, K., L. Philippot, and S. Hallin. 2005. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 71:8335-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firestone, M. K., R. B. Firestone, and J. M. Tiedje. 1980. Nitrous oxide from soil denitrification: factors controlling its biological production. Science 208:749-751. [DOI] [PubMed] [Google Scholar]
- 15.Giles, J. 2005. Fallout of fertilizers set too low, studies warn. Nature 434:262. [DOI] [PubMed] [Google Scholar]
- 16.Groffman, P. M., M. A. Altabet, J. K. Bohlke, K. Butterbach-Bahl, M. B. David, M. K. Firestone, A. E. Giblin, T. M. Kana, L. P. Nielsen, and M. A. Voytek. 2006. Methods for measuring denitrification: diverse approaches to a difficult problem. Ecol. Appl. 16:2091-2122. [DOI] [PubMed] [Google Scholar]
- 17.Hallin, S., C. M. Jones, M. Schloter, and L. Philippot. 2009. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3:597-605. [DOI] [PubMed] [Google Scholar]
- 18.Henry, S., E. Baudouin, J. C. López-Gutiérrez, F. Martin-Laurent, A. Brauman, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. (Erratum, 61:289-290.) [DOI] [PubMed] [Google Scholar]
- 19.Henry, S., D. Bru, B. Stres, S. Hallet, and L. Philippot. 2006. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofstra, N., and A. F. Bouwman. 2005. Denitrification in agricultural soils: summarizing published data and estimating global annual rates. Nutrient Cycling Agroecosyst. 72:267-278. [Google Scholar]
- 21.Holtan-Hartwig, L., P. Dörsch, and L. R. Bakken. 2000. Comparison of denitrifying communities in organic soils: kinetics of NO3− and N2O reduction. Soil Biol. Biochem. 32:833-843. [Google Scholar]
- 22.Houghton, J. T., L. G. Meira Filho, B. A. Callander, N. Harris, N. Kattenberg, and K. Maskell (ed.). 1996. Climate change 1995: the science of climate change. Cambridge University Press, Cambridge, United Kingdom.
- 23.Hynšt, J., P. Brůček, and M. Šimek. 2007. Nitrous oxide emissions from cattle-impacted pasture soil amended with nitrate and glucose. Biol. Fert. Soils 43:853-859. [Google Scholar]
- 24.Jones, C. M., B. Stres, M. Rosenquist, and S. Hallin. 2008. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25:1955-1966. [DOI] [PubMed] [Google Scholar]
- 25.López-Gutiérrez, J. C., S. Henry, S. Hallet, F. Martin-Laurent, G. Catroux, and L. Philippot. 2004. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57:399-407. [DOI] [PubMed] [Google Scholar]
- 26.Ma, W. K., A. Bedard-Haughn, S. D. Siciliano, and R. E. Farrel. 2008. Relationship between nitrifier and denitrifier community composition and abundance in predicting nitrous oxide emissions from ephemeral wetland soils. Soil Biol. Biochem. 40:1114-1123. [Google Scholar]
- 27.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, M. N., B. J. Zebarth, C. E. Dandie, D. L. Burton, C. Goyer, and J. T. Trevors. 2008. Crop residue influence on denitrification, N2O emissions and denitrifier community abundance in soil. Soil Biol. Biochem. 40:2553-2562. [Google Scholar]
- 29.Miller, M. N., B. J. Zebarth, C. E. Dandie, D. L. Burton, C. Goyer, and J. T. Trevors. 2009. Influence of liquid manure on soil denitrifier abundance, denitrification, and nitrous oxide emissions. Soil Sci. Soc. Am. J. 73:760-768. [Google Scholar]
- 30.Mkhabela, M. S., R. Gordon, D. Burton, A. Madani, and W. Hart. 2006. Effect of lime, dicyandiamide and soil water content on ammonia and nitrous oxide emissions following application of liquid hog manure to a marshland soil. Plant Soil 284:351-361. [Google Scholar]
- 31.Mulvaney, R. L., and C. W. Boast. 1986. Equations for determination of nitrogen-15 labeled dinitrogen and nitrous oxide by mass spectrometry. Soil Sci. Soc. Am. J. 50:360-363. [Google Scholar]
- 32.Nakicenovic, N., and R. Swart (ed.). 2000. Special report on emissions scenarios: a special report of Working Group III of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom.
- 33.Nicol, G. W., S. Leininger, C. Schleper, and J. I. Prosser. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966-2978. [DOI] [PubMed] [Google Scholar]
- 34.Nugroho, R. A., W. F. M. Röling, A. M. Laverman, and H. A. Verhoef. 2007. Low nitrification rates in acid Scots pine forest soils are due to pH-related factors. Microb. Ecol. 53:89-97. [DOI] [PubMed] [Google Scholar]
- 35.Parkin, T., A. J. Sexstone, and J. M. Tiedje. 1985. Adaption of denitrifying populations to low soil pH. Appl. Environ. Microbiol. 49:1053-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul, E. A., and F. E. Clark. 1996. Soil microbiology and biochemistry. Academic Press, San Diego, CA.
- 37.Philippot, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 38.Philippot, L., J. Čuhel, N. P. A. Saby, D. Chèneby, A. Chroňáková, D. Bru, D. Arrouays, F. Martin-Laurent, and M. Šimek. 2009. Mapping field-scale spatial distribution patterns of size and activity of the denitrifier community. Environ. Microbiol. 11:1518-1526. [DOI] [PubMed] [Google Scholar]
- 39.Philippot, L., S. Hallin, and M. Schloter. 2007. Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 96:249-305. [Google Scholar]
- 40.Rich, J. J., and D. D. Myrold. 2004. Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biol. Biochem. 36:1431-1441. [Google Scholar]
- 41.Richardson, D., H. Felgate, N. Watmough, A. Thomson, and E. Baggs. 2009. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key? Trends Biotech. 27:388-397. [DOI] [PubMed] [Google Scholar]
- 42.Röling, W. F. M. 2007. Do microbial numbers count? Quantifying the regulation of biogeochemical fluxes by population size and cellular activity. FEMS Microbiol. Ecol. 62:202-210. [DOI] [PubMed] [Google Scholar]
- 43.Šimek, M., and J. E. Cooper. 2002. The influence of soil pH on denitrification: progress towards the understanding of this interaction over the last 50 years. Eur. J. Soil Sci. 53:345-354. [Google Scholar]
- 44.Šimek, M., and D. W. Hopkins. 1999. Regulation of potential denitrification by soil pH in long-term fertilized arable soil. Biol. Fert. Soils 30:41-47. [Google Scholar]
- 45.Šimek, M., L. Jíšová, and D. W. Hopkins. 2002. What is the so-called optimum pH for denitrification in soil? Soil Biol. Biochem. 34:1227-1234. [Google Scholar]
- 46.Smith, M. S., and J. M. Tiedje. 1979. Phases of denitrification following oxygen depletion in soil. Soil Biol. Biochem. 11:261-267. [Google Scholar]
- 47.Stevens, R. J., and R. J. Laughlin. 1998. Measurement of nitrous oxide and di-nitrogen emissions from agricultural soils. Nutrient Cycling Agroecosyst. 52:131-139. [Google Scholar]
- 48.Stevens, R. J., R. J. Laughlin, G. J. Atkins, and S. J. Prosser. 1993. Automated determination of nitrogen-15-labeled dinitrogen and nitrous oxide by mass-spectrometry. Soil Sci. Soc. Am. J. 57:981-988. [Google Scholar]
- 49.Stevens, R. J., R. J. Laughlin, and J. P. Malone. 1998. Measuring the mole fraction and source of nitrous oxide in the field. Soil Biol. Biochem. 30:541-543. [Google Scholar]
- 50.Tate, R. L. 1995. Soil microbiology. John Wiley, New York, NY.
- 51.Thomsen, J., T. Geest, and R. Cox. 1994. Mass spectrometric studies of the effect of pH on the accumulation of intermediates in denitrification in Paracoccus denitrificans. Appl. Environ. Microbiol. 60:536-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Throbäck, I. N., M. Johansson, M. Rosenquist, M. Pell, M. Hansson, and S. Hallin. 2007. Silver (Ag+) reduces denitrification and induces enrichment of novel nirK genotypes in soil. FEMS Microbiol. Lett. 270:189-194. [DOI] [PubMed] [Google Scholar]
- 53.Vance, E. D., P. C. Brookes, and D. S. Jenkinson. 1987. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19:703-707. [Google Scholar]
- 54.Weslien, P., Å. K. Klemedtsson, G. Börjesson, and L. Klemedtsson. 2009. Strong pH influence on N2O and CH4 fluxes from forested organic soils. Eur. J. Soil Sci. 60:311-320. [Google Scholar]
- 55.Zbíral, J., I. Honsa, and S. Malý. 1997. Soil analyses, part III. Czech Central Institute for Supervising and Testing in Agriculture, Brno, Czech Republic. (In Czech.)
- 56.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]