Abstract
P pili are extracellular appendages responsible for the targeting of uropathogenic Escherichia coli to the kidney. They are assembled by the chaperone-usher (CU) pathway of pilus biogenesis involving two proteins, the periplasmic chaperone PapD and the outer membrane assembly platform, PapC. Many aspects of the structural biology of the Pap CU pathway have been elucidated, except for the C-terminal domain of the PapC usher, the structure of which is unknown. In this report, we identify a stable and folded fragment of the C-terminal region of the PapC usher and determine its structure using both X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. These structures reveal a β-sandwich fold very similar to that of the plug domain, a domain of PapC obstructing its translocation domain. This structural similarity suggests similar functions in usher-mediated pilus biogenesis, playing out at different stages of the process. This structure paves the way for further functional analysis targeting surfaces common to both the plug and the C-terminal domain of PapC.
Adhesive surface organelles termed pili mediate the adhesion of bacteria to host cells. Pili assembled by the chaperone-usher (CU) pathway form one of five major classes of nonflagellar surface appendages in Gram-negative bacteria, with the P pilus system from uropathogenic Escherichia coli being one of the two best-characterized CU systems. These pili are multisubunit structures consisting of two distinct subassemblies, a rigid rod with a diameter of 6.8 nm and a distal flexible tip fibrillum with a diameter of 2 nm (18, 21). In P pili the helical rod is comprised of more than 1,000 copies of the PapA subunits arranged in a right-handed helical cylinder with 3.3 subunits per turn (3, 8, 14), and the tip fibrillum is comprised of 5 to 10 copies of the PapE subunits (21). Two “adaptor” subunits, PapK and PapF, connect the PapE tip fibrillum to the PapA rod and the PapE tip fibrillum to the distal PapG adhesin (16, 21). The proximal end of the pilus is terminated by the PapH subunit (2, 50). The PapG adhesin mediates the bacterial colonization of the kidney (25, 40) by binding to the globoseries of glycolipids present in the human kidney (25, 40) (Fig. 1A), an event that is critical in pyelonephritis.
FIG. 1.
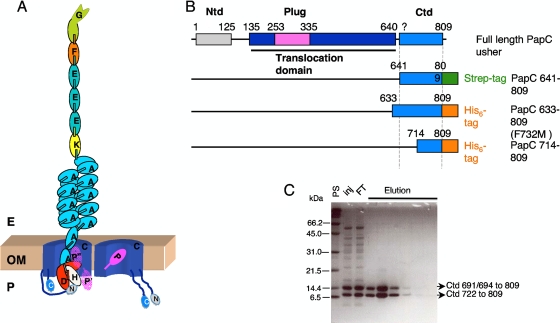
(A) Schematic diagram of a P pilus assembled in the usher translocation platform. Subunits are represented by oval shapes, and N-terminal extensions are represented by short rectangular shapes. The usher homodimer is represented in the outer membrane (OM). In the usher protomer through which the nascent pilus passes, two positions of the plug are indicated by P where the plug is positioned to the side of the transmembrane barrel's lumen and P′ where the plug has swung into the periplasmic space. (B) Domain organization of the PapC usher based on amino acid sequence. The C-terminal domain sequences are indicated in marine blue. The constructs used in this study are schematically represented underneath; all converge to a fragment containing residues 722 to 809, termed the “PapC CTD.” Ntd, N-terminal domain. (C) Identification of a discrete folding unit at the C terminus of PapC. Shown is an SDS-PAGE gel stained with Coomassie blue of the eluted PapC C-terminal fragments obtained with a construct comprising residues 641 to 809 after the first purification step. PS, prestained protein standards; Inj, loaded sample; FT, flowthrough.
The assembly of pili is a coordinated process requiring two proteins: a chaperone and an outer membrane assembly platform, the usher. Pilus subunits are translocated into the periplasm via the general secretory machinery (38, 47). The binding of the PapD chaperone to the nascently translocated subunits facilitates their folding on the chaperone template. The chaperone remains bound to the folded subunits capping their interactive surfaces, thus preventing nonproductive interactions in the periplasm (7). Chaperone-subunit complexes are then targeted to the usher (PapC), where subunits polymerize in an ordered fashion and translocate across the outer membrane through the usher pore (47, 52). Subunit folding and stabilization occur when the chaperone and subunit form a complex through a mechanism termed donor strand complementation (DSC) (9, 41). In this mechanism the C-terminally truncated Ig-like fold of the pilus subunits, which contains only six of the seven β-strands that constitute the canonical Ig fold, is complemented by the donation of a β-strand from the chaperone (9, 41). Chaperone-subunit complexes are then targeted to the outer membrane usher, where the chaperone is released and subunits are noncovalently joined to preceding subunits in the nascent pilus fiber. This polymerization process is made possible by the presence of a disordered N-terminal extension sequence (NTES) in each subunit (except the adhesin) (41), which during pilus assembly displaces the strand donated by the chaperone, thereby substituting for the missing secondary structure in the previously assembled subunit. This mechanism is called donor-strand exchange (DSE) (9, 41, 42, 55). It is believed that this structural reorganization provides the driving force for pilus biogenesis, since no ATP hydrolysis or other type of external energy source is required (17, 56).
DSE occurs at the outer membrane usher, which acts as a catalyst for polymerization (34). Biophysical and cryo-electron microscopy (EM) studies of the FimD usher (a close homolog of PapC) have shown that the usher is a twinned pore in both detergent and lipid bilayers (23, 46). Only one pore is used for secretion, but two pores are required for subunit recruitment (39). For PapC, both monomers and dimers have been described (15, 39). The usher has four functional domains (Fig. 1B): a translocation domain forming a β-barrel with 24 transmembrane β-strands (15, 39), a plug domain in the middle of the translocation domain, and two periplasmic domains, one at each of the N- and C-terminal ends of the usher polypeptide (35, 48). The plug domain has a β-sandwich fold and completely occludes the pore in the inactive usher. Its function, besides gating the channel, seems to be further associated with pilus biogenesis since the deletion of the plug domain abolishes pilus formation in vitro and in vivo (15, 26, 54). The N-terminal domain selectively binds chaperone-subunit complexes (12, 33). The structure of the N-terminal domain of FimD bound to chaperone-subunit complexes indicated that the first 24 residues of FimD are involved in the recognition of chaperone-subunit complexes; the deletion of this region was shown previously to abolish pilus biogenesis (12, 32, 33).
The role of the usher C-terminal domain (CTD) is not well understood. The binding of the chaperone-adhesin complex to the usher C terminus was previously demonstrated in vitro (46), while protease susceptibility in FimD shows that, following targeting to the usher N terminus, the chaperone-adhesin complex forms stable interactions with the FimD C terminus, inducing a conformational change in FimD that may be fundamental in the activation step of pilus biogenesis (29, 30, 43). The structure of the C-terminal domain is unknown and is the only part of the CU pilus biogenesis pathway not yet represented in structural terms. Here we provide evidence for the presence of a discrete folding unit in the PapC CTD and report its structure determined by nuclear magnetic resonance (NMR) spectroscopy and X-ray crystallography.
MATERIALS AND METHODS
Design of constructs for production of the PapC CTD.
In this study, clones encoding residues 633 to 809, residues 641 to 809, and residues 714 to 809 were used. The sequence encoding residues 633 to 809 was cloned into plasmid pTrcPapD (described in reference 49), where the PapD sequence was replaced by PapC with the addition of a C-terminal six-histidine tag and the signal sequence of PapD. The sequences encoding residues 641 to 809 and residues 714 to 809 were cloned into vectors pASK-IBA2 and pASK-IBA32 (IBA bioTAGnology), respectively. By design, cloning into both pASK-IBA2 and pASK-IBA32 adds the signal peptide of the OmpA protein at the N terminus of the construct, ensuring that both constructs are delivered to the periplasm while adding a Strep-Tactin (pASK-IBA2) tag and a six-histidine tag (pASK-IBA32) at the C terminus of the constructs. For phasing using the multiwavelength anomalous-dispersion method, three substitutions to Met were introduced into the fragment at residues 633 to 809, one at a time: F732M, A733M, and W767M. Oligonucleotides used to generate these clones and mutants are listed in Table 1.
TABLE 1.
Oligonucleotides used to generate clones and mutants used in this study
| DNA amplified or PapC CTD mutation | Primer | Sequence (5′-3′) |
|---|---|---|
| DNA amplified | ||
| papC(641-809)a | PapC641_IBA2-f | ATGGTAGGTCTCAGGCCGCGTTACATGCAGGGGGAATGT |
| PapC809_IBA2-r | ATGGTAGGTCTCAGCGCTTTTCTGAGGCGTACAGGGAAGC | |
| papC(714-809)a | PapC714_IBA32-f | ATGGTAGGTCTCAGGCCACAGAAGGTGCCATTGGTTACCG |
| PapC809_IBA32-r | ATGGTAGGTCTCAGCGCTTTTCTGAGGCGTACAGGGAAGC | |
| papC(633-809)b | pTrc5seq_f | GTGGAATTGTGAGCGG |
| PapC 3′6his BamHI_r | GCCGGGATCCATTAGTGGTGATGATGGTGATGTTTCTGAGGCGTAC | |
| papDb | DsigCcter_f | TCCGGGGCAGACGCTGCAACAATTACCGGAAAAGGTGCG |
| DsigCcter_r | CGCACCTTTTCCGGTAATTGTTGCAGCGTCTGCCCCGGA | |
| PapC CTD mutation | ||
| F732Mc | F732M_f | GGAAACGTCTGATGGCAATACTGCG |
| F732M_r | CGCAGTATTGCCATCAGACGTTTCC | |
| A733Mc | A733M_f | CGTCTGTTTATGATACTGCGTCTTG |
| A733M_r | CAAGACGCAGTATCATAAACAGACG | |
| L765Mc | L765M_f | CGACGAAGGCATGGCCTGGCTG |
| L765M_r | CAGCCAGGCCATGCCTTCGTCG |
For PapC residues 641 to 809 and PapC residues714 to 809 in pASK-IBA vectors used in NMR studies (using pDG2 as a template, where the full-length PapC sequence is encoded) (23).
For the construction of plasmid pKDC3, used in X-ray crystallographic studies (using pTrcPapD as the donor of plasmid pTrc99a and pPAP5 as the donor of the PapC CTD sequence, with the addition of a six-histidine tag at the C terminus via the first listed primer pair and the signal sequence from PapD via the second listed primer pair).
For mutagenesis, used in X-ray crystallographic studies (using pKDC3 as a template).
Expression and purification of the PapC CTD.
All proteins were expressed from the clones described above introduced into C600 cells. Expression and purification from the periplasm proceeded as previously described (49). All proteins degraded rapidly to yield a stable domain encompassing residues 722 to 809, termed the “PapC CTD.” For crystallographic studies, the PapC CTD from the construct encoding residues 633 to 809 was purified on a cobalt affinity column (Talon; Clontech) followed by a Q Sepharose column (Source 15Q; GE Healthcare) and dialysis against a solution containing 20 mM MES (morpholineethanesulfonic acid) (pH 6.5) and 20 mM NaCl. For NMR studies, the PapC CTD from the construct encoding residues 641 to 809 was purified on a Strep-Tactin Sepharose affinity column followed by a Q column (Hitrap HP Q; GE Healthcare) and dialyzed against a solution containing 20 mM MES (pH 6.0) and 50 mM NaCl. The PapC CTD from the construct encompassing residues 714 to 809 was purified on a nickel affinity column (Histrap; GE Healthcare) followed by size-exclusion chromatography (Superdex 75; GE Healthcare) in a solution containing 20 mM MES (pH 6.5) and 50 mM NaCl.
Production of labeled proteins.
For the production of the selenomethionine-containing PapC CTD, of the three Met mutants generated from the clone encoding residues 633 to 809, only the F732M variant produced suitable crystals. Following expression in a laboratory strain of Escherichia coli C600 defective in methionine synthesis in minimal medium containing selenomethionine (11), the selenomethionine-labeled PapC CTD was purified as described above for the wild-type protein and was assessed by mass spectrometry to be approximately 65% derivatized with selenomethionine. For the production of the 15N- and 13C-isotope-labeled PapC CTD, the construct at residues 714 to 809 in laboratory strain BL21 was expressed in PG minimal medium and grown with ammonium sulfate-15N2 and d-glucose-13C6 (Cambridge Isotope Laboratories, Inc.) as the sole nitrogen or carbon source, respectively. Purification was performed as described above for unlabeled PapC CTD.
Crystallization and X-ray data collection.
PapC CTD F732M was crystallized by the hanging-drop vapor diffusion method by mixing 2 μl of protein at 15 mg/ml in 20 mM MES (pH 6.5) with 2 μl of mother liquor consisting of 60% saturated ammonium sulfate and 100 mM citric acid (pH 5.0). Crystals were cryoprotected by submergence in 75% saturated sodium malonate (pH 5.7) for 10 s. PapC CTD F732M crystallized in the I4122 space group with cell dimensions of a=b=133.3 Å, c=285.5 Å, and diffracted to a 2.85-Å resolution. These crystals contain seven copies of the PapC CTD in the asymmetric unit. The wild-type PapC CTD was crystallized by mixing 2 μl of protein at 15 mg/ml in 20 mM MES (pH 6.5) with 2 μl of mother liquor consisting of 20% saturated Tacsimate (28) (pH 8.0), made in-house, and cryoprotected in a solution of 60% saturated Tacsimate (pH 8.0) and 10% glycerol. The wild-type PapC CTD crystallized in the space group P42212 with cell dimensions of a=b=100.9 Å, c=89.5 Å, and diffracted to a 2.1-Å resolution. These crystals contained five copies of the PapC CTD in the asymmetric unit.
Three-wavelength multiple-anomalous-dispersion (MAD) diffraction data were collected from a single flash-cooled crystal of PapC CTD F732M (−180°C) at beamline 4.2.2 of the Advanced Light Source. The data were processed with XDS and XSCALE (19). FA values were calculated with XPREP (45). MAD phasing was performed with SHELXE after the location of the anomalous scattering substructure by SHELXD (44). Fourteen primary selenium sites with an occupancy greater than 0.2 and 10 minor sites with an occupancy of between 0.1 to 0.2 were found with SHELXD. High-quality initial phases were obtained after phasing and density modification with SHELXE (see Fig. S1 in the supplemental material), which was assisted by the high solvent content of the crystal (70%). A model of a single PapC CTD was built, duplicated, manually placed for the remaining six molecules, and then rigid-body refined with COOT (13). After further rigid-body refinement and simulated annealing with PHENIX (1), final refinement against the peak wavelength data set was carried out with REFMAC (31). Translation libration screw (TLS) refinement was performed by using 1 TLS group per PapC CTD protomer.
Subsequently, a representative monomer chain was used to perform molecular replacement into the wild-type PapC CTD data set by using the program PHASER (27), which located five protomers in the asymmetric unit. Refinement of wild-type PapC CTD was carried out with PHENIX, with the final model refined using 4 to 5 TLS groups per protomer as selected by the TLSMD server (36, 37). Data collection, phasing, and refinement statistics are summarized in Table S1 in the supplemental material.
NMR spectroscopy.
The majority of the NMR spectra were acquired at either 298 K or 310 K with a Bruker Avance III or UnityINOVA spectrometer (operating at nominal 1H frequencies of 500 MHz, 600 MHz, and 700 MHz). A three-dimensional (3D) 13C-separated nuclear Overhauser effect (NOE) spectroscopy (NOESY) data set was recorded at 950 MHz on the home-built instrument at the University of Oxford. Sequence-specific resonance assignments were obtained by using standard triple-resonance NMR spectroscopy (4). The final set of distance restraints was derived from 3D 15N- and 13C-edited NOESY-HSQC spectra with a mixing time of 125 ms (310 K). {1H}15N heteronuclear NOE data (20) were recorded with 3.0 s of 1H saturation in the latter part of a 3.5-s preparation delay, which was also used without radio frequency pulses for the reference two-dimensional (2D) spectrum without NOE. Analysis of these data indicated that the four extreme N-terminal residues (Val735 to Gly728) and two C-terminal residues (Qln808 and Lys809) are substantially disordered in solution. All NMR spectra were processed by using NMRpipe/NMRDraw (10) and analyzed by using CCPN Analysis, version 2 (51). 1H, 13C, and 15N chemical shifts were referenced indirectly to sodium 2,2-dimethyl-2-silanepentane-5-sulfonate (DSS) by using absolute frequency ratios for the 1H signals (53).
NMR structure calculations.
Interproton distance restraints were derived from 3D 15N-NOESY-HSQC and 13C-NOESY-HSQC spectra. A proportion of the resonances were successfully assigned in a manual fashion without ambiguity. The cross-peaks were grouped into five categories according to their relative peak heights as assessed by Analysis (very strong, strong, medium, weak, and very weak) and were designated with the corresponding interproton distance restraint limits of 1.8 to 2.5 Å, 1.8 to 2.8 Å, 1.8 to 4.0 Å, 1.8 to 5.0 Å, and 1.8 to 6.0 Å, respectively.
All structures for the PapC CTD were calculated by using an ab initio-simulated annealing protocol within the CNS program (6) with PARALLHDGv5.3 force field and PROLSQ nonbonded energy function (24). The protocol adopts a mixture of Cartesian molecular dynamics (MD) and torsion angle dynamics simulated annealing to refine structures starting from randomly generated conformers with good local geometry.
A total of 1,309 unique NOE-derived interproton distance restraints for the PapC CTD were included in the final iterations of the structure calculations. Backbone torsion angle restraints for ϕ and ψ were derived from analyses of 1Hα, 13Cα, 13Cβ, 13C′, and 15NH chemical shift databases as implemented in the DANGLE module of Analysis. The experimental NMR restraints bring the two PapC CTD cysteine side chains within disulfide-bridging distance; later rounds of refinement therefore included the explicit definition of the disulfide bound as part of the molecular topology. Hydrogen bond restraints for backbone amide protons were derived from an assessment of the regular secondary structure elements and the absence of NH-solvent H2O exchange cross-peaks in the 2D and 3D NOESY spectra. A total of 77 dihedral angle and 152 hydrogen bond (76 H-bonds; two distance restraints per H-bond) interatomic distance restraints were used.
Protein structure accession numbers.
The atomic coordinates of the final 10 representative simulated annealing conformers and the list of experimental restraints have been deposited in the RCSB Protein Data Bank (PDB) (accession number 2KT6). The atomic coordinates of the PapC CTD have been deposited in the PDB (accession number 3L48). Chemical shifts for resonance assignments were deposited in the BioMagResBank (accession number BMRB 16684). Structural statistics for the final conformer bundle are reported in Table S2 in the supplemental material.
RESULTS AND DISCUSSION
The folded part of the C-terminal region of PapC extends from residues 722 to 809.
The elucidation of the structure of the PapC usher translocation domain (residues 145 to 639) provided the precise boundaries of this domain (39). In order to investigate the structure of the C-terminal domain of PapC, the region encoding residues 641 to 809 (residue 809 is the C-terminal residue) (Fig. 1B and Table 1) was cloned and expressed, and the corresponding protein was purified by means of a Strep tag fused at the C terminus. During purification, this polypeptide rapidly breaks down into two fragments (Fig. 1C), one comprising residues 691 to 809 and the other comprising residues 722 to 809, as established by N-terminal sequencing (both fragments were purified by using the C-terminal tag and thus contain an intact C terminus). The two fragments can be separated by using anion-exchange chromatography. However, over time, the longer fragment is converted to the shorter one. A new construct encompassing residues 722 to 809 with a six-histidine tag at the C terminus was produced, but this clone expressed very poorly. The region encoding residues 714 to 809 was then cloned, and when the corresponding peptide was expressed, it also converted to the fragment at residues 722 to 809. Thus, we have identified a domain between residues 722 and 809 in the C terminus of the PapC usher that is stable over a long period of time, suggesting that this domain might be properly folded. We refer thereafter to this domain as the C-terminal domain of the PapC usher (PapC CTD).
Crystal structure of the PapC CTD.
The crystal structure of the PapC CTD was solved to a 2.85-Å resolution by using the MAD phasing method performed on a single crystal of the PapC CTD containing a selenomethionine at residue 732 (the wild-type PapC CTD contains a Phe at this position, but it was mutated to Met for phasing purposes) and residue 759. This structure was then used as a search model to determine the 2.1-Å resolution structure of the wild-type PapC CTD (see Materials and Methods and Fig. S1 and Table S1 in the supplemental material). Residues prior to G728 are not represented by electron density and are presumed to be disordered.
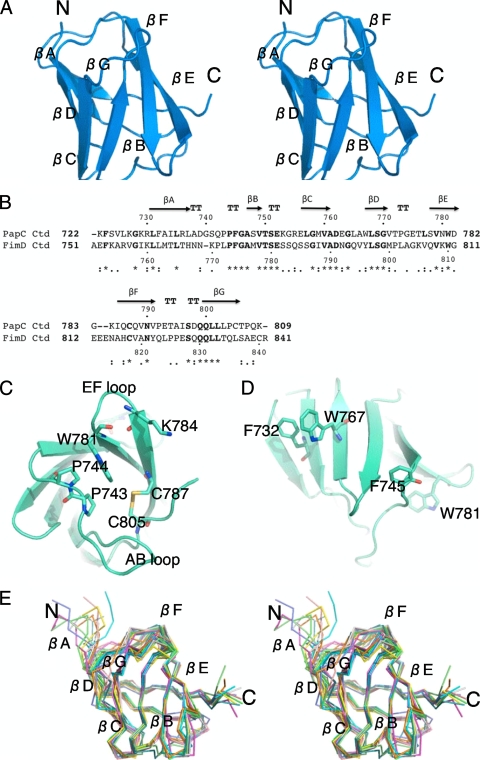
The overall fold of the PapC CTD is that of a small β-sandwich containing seven β-strands (βA through βG) and a central Greek key motif (Fig. 2A), measuring 25 Å on its long axis and 20 Å across. All seven β-strands are connected by well-ordered loops. Conserved residues among PapC CTD homologs (Fig. 2B) are for the most part located in the structure's core. Among them, P743, P744, and W781 make an unusual perpendicular aromatic-aromatic stacking interaction network. Given the high degree of sequence conservation for these residues and by analogy to similar side-chain-side-chain interactions seen in Trp-cage domains (5), this structural motif may be important to the stability of the PapC CTD. It may also contribute to the stability of the relatively long AB and EF loops (Fig. 2C).
FIG. 2.
Crystal and NMR structures of the PapC CTD. (A) Stereo view of the crystal structure of the PapC C-terminal domain in ribbon representation. (B) Sequence alignment of the PapC and FimD CTDs. The secondary structure elements for the PapC CTD are represented on the top of the PapC sequence and are labeled βA to βG. Numbering for both sequences is indicated. “*,” identical; “:,” conserved substitutions; “•,” semiconserved substitutions. “TT” indicates boundaries of β-turns. The sequence alignment was generated by clustalW (22). (C) Details of the residues mentioned in the text related to the crystal structure. This view is a bottom view of the structure shown in A. (D) Details of the residues mentioned in the text related to the NMR structure. This view corresponds to a 180° rotation along the vertical axis compared to the view shown in A. (E) Cα tracing of the NMR structure bundle presented as a best-fit superposition of 10 lowest-energy conformers.
In the crystal, the PapC CTD is essentially ordered to its C terminus, with only K809 and the six-histidine tag unrepresented by electron density. The distal C terminus is stabilized by a disulfide bond between C787 and C805 linking the base of the βF strand and the C-terminal sequence after the βG strand (Fig. 2C). A C787S mutant of full-length PapC was previously shown by So and Thanassi to be expressed at wild-type levels but defective in assembling pili (46). In addition, in an alanine scan limited to charged and polar residues of the PapC CTD within full-length PapC, those same researchers found that the K784A mutation inhibited pilus biogenesis, most likely due to a disruption of the expression of PapC. In the PapC CTD crystal structure, K784 lies on a solvent-exposed turn immediately N terminal to βF and exhibits a complete disordering of its side chain (Fig. 2C).
NMR analysis structure of the PapC CTD.
Full backbone resonance assignments were obtained by using standard triple-resonance NMR methods. While the 2D 15N,1H-HSQC spectrum displays excellent chemical dispersion consistent with a stable H-bonded globular fold and the dispersion of the 1Hα chemical shifts suggests significant β-sheet content (see Fig. S2 in the supplemental material), the dispersion of the aliphatic side-chain 1H chemical shifts is relatively poor. The most upfield chemical shift occurs at ∼0.45 ppm despite the presence of a few aromatic residues (two Trp residues [W767 and W781] and two Phe residues [F732 and F745]) (Fig. 2D) in the protein construct, thus leading to some difficulty in unambiguously resolving the full complement of side-chain proton resonances. Nevertheless, a high proportion of the amino acid side-chain spin systems could be traced in 2D and 3D J-correlated spectra, which is sufficient to provide a basis for the analysis of 3D 15N- and 13C-separated 1H-NOESY spectra to determine the 3D solution structure to at least medium resolution (Table S2). Interestingly, the structure reveals that the four aromatic-residue side chains mentioned above align along one lateral solvent-facing surface of the structure (Fig. 2D), which rationalizes the relatively limited aliphatic side-chain chemical shift dispersion and the relative sparseness of interproton NOE cross-peaks connecting aromatic to aliphatic side-chain groups compared to proteins where a proportion of aromatic side chains contributes to the hydrophobic core.
The NMR structure is essentially the same as the crystal structure but for one notable difference (Fig. 2E): while in the crystal structure, the βG strand is much shorter and makes main-chain-main-chain interactions only with the βA strand, in the NMR structure, it is much longer and divided in two parts, one making main-chain-main-chain H bonding with βA (as in the crystal structure) and the other, at the C terminus, making these interactions with the βF strand. Otherwise, the differences between the two structures (crystal and NMR) are negligible, with a root mean square deviation (RMSD) between backbone atoms of 1.58 Å when the βG strand is omitted from the calculation. The NMR data also provide some dynamic information, and remarkably, as in the crystal structure, all areas of the structure, including loops, are well defined, suggesting little structural flexibility overall.
Structural homology with the plug domain and conservation of residues in the plug and C-terminal domains.
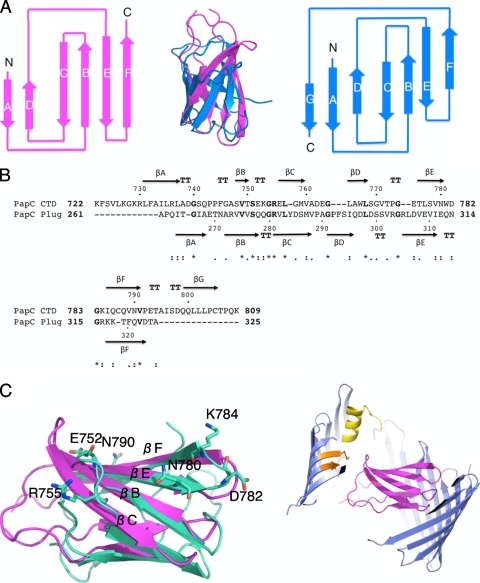
β-Sandwich structures are common among proteins. Indeed, a search for structural homologs of the PapC CTD using a number of available structural homology search engines yielded very similar structures, but the exercise did not shed any biological insight into PapC CTD function. Indeed, one of the highest-scoring structures using 3D-PSSM is the carboxypeptidase regulatory domain (PDB accession number 1UWY), a totally unrelated protein. More striking and perhaps biologically insightful is the homology between the PapC CTD and the plug domain of the same protein. A comparison of the two structures (Fig. 3A) shows that they superpose very well with an RMSD of 2.8 Å for the backbone atoms excluding strand βG of the PapC CTD. Moreover, the topologies of the two proteins (the way the secondary structures are connected) (Fig. 3A) are identical, except for the presence of the C-terminal βG strand in the PapC CTD (Fig. 3B). Thus, the plug and CTD of PapC possess essentially the same fold.
FIG. 3.
Comparison of the structures of the PapC plug and CTD. (A) Topology diagram of the PapC plug domain (magenta) and PapC CTD (marine) and structure alignment (middle). β-Strands are represented by arrows. Strands are labeled βA to βF or βA to βG in the PapC plug or PapC CTD domain, respectively. Aligned structures are shown in a ribbon representation with the same color coding. (B) Sequence alignment of the PapC CTD and PapC plug domain. The secondary structure elements for the PapC CTD and PapC plug domain are represented on the top or bottom of the sequences and are labeled βA to βG or βA to βF, respectively. Numbering for both sequences is indicated. “*,” identical; “:,” conserved substitutions; “•,”: semiconserved substitutions. “TT” indicates boundaries of β-turns. The sequence alignment was generated by clustalW (22). (C) Localization of residues conserved between the PapC CTD and plug. (Left) Surface residues highlighted with “*” and “:” in B are mapped onto the superimposed structures of the PapC CTD (light green) and plug (magenta). Both structures are in cartoon representation. Only residues of the PapC CTD are shown in stick representation, with carbon, oxygen, and nitrogen atoms color coded in light green, red, and blue, respectively. (Right) Structure of the PapC translocation domain showing the plug (magenta) in the same orientation as that at the left. The PapC structure is in a cartoon representation color coded in dark blue for the barrel, magenta for the plug, yellow for the unique helix in the structure, and orange for the trigger hairpin (see the definition of the trigger hairpin described previously by Remaut et al. [39]).
The PapC CTD is indispensable for pilus biogenesis. It is also known to bind the chaperone-adhesin complex, leading to the incorporation of the adhesin at the tip of the pilus (43, 46). Indeed, chaperone-adhesin complex binding to the usher is known to induce conformational changes in the usher, rendering the usher competent for the further polymerization/assembly of subunits (34). This activation process requires the usher CTD. Attempts at mapping the site of the interaction of the PapC CTD and the PapD-PapG chaperone-adhesin complex by NMR yielded very limited indications of ligand-induced chemical shift perturbations for the PapC CTD, presumably because of weak binding and the difficulty in bringing the binding partners to a sufficiently high concentration to achieve a significant population of the bound state under these conditions.
The usher plug was previously shown to be essential for pilus biogenesis both in vivo and in vitro (15, 54). The direct binding of the plug to the chaperone-adhesin complex, however, has never been reported. However, the similarities in fold and structure between the plug and the CTD revealed here suggest functional similarities between them. Since the precise function of the CTD is not yet known, the resolution of the functional significance of these structural similarities in pilus biogenesis will be of major interest. It is noteworthy that most of the surface-exposed polar residues that are conserved between the plug and the CTD (Fig. 3B) locate to the βC, βB, βE, and βF face of the β-sandwich fold (Fig. 3C, left), suggesting that these residues in both the plug and CTD may be important for function. The βC, βB, βE, and βF side of the plug in the PapC translocation domain structure faces the extracellular milieu, and thus, it is likely that the residues in that region of the plug structure will be involved in function only once the plug “hinges” by swinging into the periplasmic space (Fig. 1A) to unplug the channel. Thus, the plug and CTD could collocate in the periplasm to exert similar functions. Although surfaces unique to the CTD (for example, the cluster of aromatic residues mentioned above) could be targeted for mutagenesis, we believe that further work should perhaps focus on probing the role of amino acid residues that are common to the plug and the CTD in order to establish the order of events in which the plug and CTD are involved during the activation and catalysis of pilus polymerization.
Supplementary Material
Acknowledgments
This work was funded by MRC grant 85602 to G.W. and by NIH grants 49950, 29549, and 48689 to S.H. Work by the group of P.C.D. is supported by the MRC (file reference U117574559). A.T.R. was supported by Fundação para a Ciência e Tecnologia grant SFRH/BD/22254/2005.
We acknowledge Acely Garza-Garcia and Diego Esposito (MRC-NIMR) for valuable discussions. We thank Tom Frenkiel and the staff of the MRC Centre for Biomedical NMR and Christina Redfield of the University of Oxford for their assistance in recording the NMR data. We gratefully acknowledge Jay Nix at beamline 4.2.2 at ALS for assistance with X-ray data collection.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Baga, M., M. Norgren, and S. Normark. 1987. Biogenesis of E. coli Pap pili: papH, a minor pilin subunit involved in cell anchoring and length modulation. Cell 49:241-251. [DOI] [PubMed] [Google Scholar]
- 3.Baga, M., S. Normark, J. Hardy, P. O'Hanley, D. Lark, O. Olsson, G. Schoolnik, and S. Falkow. 1984. Nucleotide sequence of the papA gene encoding the Pap pilus subunit of human uropathogenic Escherichia coli. J. Bacteriol. 157:330-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bax, A., and S. Grzesiek. 1993. Methodological advances in protein NMR. Acc. Chem. Res. 16:131-138. [Google Scholar]
- 5.Biedermannova, L., K. E. Riley, K. Berka, P. Hobza, and J. Vondrasek. 2008. Another role of proline: stabilization interactions in proteins and protein complexes concerning proline and tryptophane. Phys. Chem. Chem. Phys. 10:6350-6359. [DOI] [PubMed] [Google Scholar]
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Bullitt, E., C. H. Jones, R. Striker, G. Soto, F. Jacob-Dubuisson, J. Pinkner, M. J. Wick, L. Makowski, and S. J. Hultgren. 1996. Development of pilus organelle subassemblies in vitro depends on chaperone uncapping of a beta zipper. Proc. Natl. Acad. Sci. U. S. A. 93:12890-12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullitt, E., and L. Makowski. 1995. Structural polymorphism of bacterial adhesion pili. Nature 373:164-167. [DOI] [PubMed] [Google Scholar]
- 9.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 10.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. Nmrpipe—a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 11.Doublié, S. 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276:523-530. [PubMed] [Google Scholar]
- 12.Eidam, O., F. S. Dworkowski, R. Glockshuber, M. G. Grutter, and G. Capitani. 2008. Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD. FEBS Lett. 582:651-655. [DOI] [PubMed] [Google Scholar]
- 13.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 14.Gong, M., and L. Makowski. 1992. Helical structure of P pili from Escherichia coli. Evidence from X-ray fiber diffraction and scanning transmission electron microscopy. J. Mol. Biol. 228:735-742. [DOI] [PubMed] [Google Scholar]
- 15.Huang, Y., B. S. Smith, L. X. Chen, R. H. Baxter, and J. Deisenhofer. 2009. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc. Natl. Acad. Sci. U. S. A. 106:7403-7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob-Dubuisson, F., J. Heuser, K. Dodson, S. Normark, and S. Hultgren. 1993. Initiation of assembly and association of the structural elements of a bacterial pilus depend on two specialized tip proteins. EMBO J. 12:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob-Dubuisson, F., R. Striker, and S. J. Hultgren. 1994. Chaperone-assisted self-assembly of pili independent of cellular energy. J. Biol. Chem. 269:12447-12455. [PubMed] [Google Scholar]
- 18.Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. U. S. A. 92:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 20.Kay, L. E., D. A. Torchia, and A. Bax. 1989. Backbone dynamics of proteins as studied by N-15 inverse detected heteronuclear NMR-spectroscopy—application to staphylococcal nuclease. Biochemistry 28:8972-8979. [DOI] [PubMed] [Google Scholar]
- 21.Kuehn, M. J., J. Heuser, S. Normark, and S. J. Hultgren. 1992. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature 356:252-255. [DOI] [PubMed] [Google Scholar]
- 22.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 23.Li, H., L. Qian, Z. Chen, D. Thibault, G. Liu, T. Liu, and D. G. Thanassi. 2004. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 344:1397-1407. [DOI] [PubMed] [Google Scholar]
- 24.Linge, J. P., M. A. Williams, C. A. E. M. Spronk, A. M. J. J. Bonvin, and M. Nilges. 2003. Refinement of protein structures in explicit solvent. Proteins 50:496-506. [DOI] [PubMed] [Google Scholar]
- 25.Lund, B., F. Lindberg, B. I. Marklund, and S. Normark. 1987. The PapG protein is the alpha-D-galactopyranosyl-(1-4)-beta-D-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 84:5898-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mapingire, O. S., N. S. Henderson, G. Duret, D. G. Thanassi, and A. H. Delcour. 2009. Modulating effects of the plug, helix, and N- and C-terminal domains on channel properties of the PapC usher. J. Biol. Chem. 284:36324-36333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McPherson, A., and B. Cudney. 2006. Searching for silver bullets: an alternative strategy for crystallizing macromolecules. J. Struct. Biol. 156:387-406. [DOI] [PubMed] [Google Scholar]
- 29.Munera, D., S. Hultgren, and L. A. Fernandez. 2007. Recognition of the N-terminal lectin domain of FimH adhesin by the usher FimD is required for type 1 pilus biogenesis. Mol. Microbiol. 64:333-346. [DOI] [PubMed] [Google Scholar]
- 30.Munera, D., C. Palomino, and L. A. Fernandez. 2008. Specific residues in the N-terminal domain of FimH stimulate type 1 fimbriae assembly in Escherichia coli following the initial binding of the adhesin to FimD usher. Mol. Microbiol. 69:911-925. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 32.Ng, T. W., L. Akman, M. Osisami, and D. G. Thanassi. 2004. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186:5321-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishiyama, M., R. Horst, O. Eidam, T. Herrmann, O. Ignatov, M. Vetsch, P. Bettendorff, I. Jelesarov, M. G. Grutter, K. Wuthrich, R. Glockshuber, and G. Capitani. 2005. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 24:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiyama, M., T. Ishikawa, H. Rechsteiner, and R. Glockshuber. 2008. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320:376-379. [DOI] [PubMed] [Google Scholar]
- 35.Nishiyama, M., M. Vetsch, C. Puorger, I. Jelesarov, and R. Glockshuber. 2003. Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD. J. Mol. Biol. 330:513-525. [DOI] [PubMed] [Google Scholar]
- 36.Painter, J., and E. A. Merritt. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439-450. [DOI] [PubMed] [Google Scholar]
- 37.Painter, J., and E. A. Merritt. 2006. Web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39:109-111. [Google Scholar]
- 38.Pugsley, A. P. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Remaut, H., C. Tang, N. S. Henderson, J. S. Pinkner, T. Wang, S. J. Hultgren, D. G. Thanassi, G. Waksman, and H. Li. 2008. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133:640-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts, J. A., B. I. Marklund, D. Ilver, D. Haslam, M. B. Kaack, G. Baskin, M. Louis, R. Mollby, J. Winberg, and S. Normark. 1994. The Gal(alpha 1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. U. S. A. 91:11889-11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauer, F. G., K. Futterer, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 42.Sauer, F. G., J. S. Pinkner, G. Waksman, and S. J. Hultgren. 2002. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111:543-551. [DOI] [PubMed] [Google Scholar]
- 43.Saulino, E. T., D. G. Thanassi, J. S. Pinkner, and S. J. Hultgren. 1998. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 17:2177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheldrick, G. M. 2008. A short history of SHELX. Acta Crystallogr. A 64:112-122. [DOI] [PubMed] [Google Scholar]
- 45.Sheldrick, G. M. 2003. XPREP. Bruker-Nonius, Inc., Madison, WI.
- 46.So, S. S., and D. G. Thanassi. 2006. Analysis of the requirements for pilus biogenesis at the outer membrane usher and the function of the usher C-terminus. Mol. Microbiol. 60:364-375. [DOI] [PubMed] [Google Scholar]
- 47.Thanassi, D. G., E. T. Saulino, and S. J. Hultgren. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1:223-231. [DOI] [PubMed] [Google Scholar]
- 48.Thanassi, D. G., C. Stathopoulos, K. Dodson, D. Geiger, and S. J. Hultgren. 2002. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 184:6260-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verger, D., E. Bullitt, S. J. Hultgren, and G. Waksman. 2007. Crystal structure of the P pilus rod subunit PapA. PLoS Pathog. 3:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verger, D., E. Miller, H. Remaut, G. Waksman, and S. Hultgren. 2006. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 7:1228-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vranken, W. F., W. Boucher, T. J. Stevens, R. H. Fogh, A. Pajon, P. Llinas, E. L. Ulrich, J. L. Markley, J. Ionides, and E. D. Laue. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687-696. [DOI] [PubMed] [Google Scholar]
- 52.Waksman, G., and S. J. Hultgren. 2009. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7:765-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wishart, D. S., C. G. Bigam, J. Yao, F. Abildgaard, H. J. Dyson, E. Oldfield, J. L. Markley, and B. D. Sykes. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6:135-140. [DOI] [PubMed] [Google Scholar]
- 54.Yu, X., G. R. Visweswaran, Z. Duck, S. Marupakula, S. MacIntyre, S. D. Knight, and A. V. Zavialov. 2009. Caf1A usher possesses a Caf1 subunit-like domain that is crucial for Caf1 fibre secretion. Biochem. J. 418:541-551. [DOI] [PubMed] [Google Scholar]
- 55.Zavialov, A. V., J. Berglund, A. F. Pudney, L. J. Fooks, T. M. Ibrahim, S. MacIntyre, and S. D. Knight. 2003. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113:587-596. [DOI] [PubMed] [Google Scholar]
- 56.Zavialov, A. V., V. M. Tischenko, L. J. Fooks, B. O. Brandsdal, J. Aqvist, V. P. Zav'yalov, S. Macintyre, and S. D. Knight. 2005. Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem. J. 389:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.