Abstract
Phase variation of the outer membrane protein Ag43 encoded by agn43 in Escherichia coli is controlled by an epigenetic mechanism. Sequestration of the regulatory region from Dam-dependent methylation has to be established and maintained throughout a generation to obtain and maintain the OFF phase. This work shows that hemimethylated DNA, which is formed by the passage of the DNA replication fork in an ON-phase cell, can be sequestered from methylation by OxyR binding, which is thus a key event for the switch from ON to OFF. No evidence was found that the protein SeqA, which also binds to the region, is involved in sequestration. To facilitate the dissection of this process further, a novel approach was introduced that does not alter the sequence of the regulatory region or the cellular concentration of Dam or OxyR, which consists of inserting auxiliary OxyR binding sites upstream of the regulatory region. Using this strategy, it was shown that the ON-to-OFF switch frequency can be modulated without changing the OFF-to-ON frequency. The data support a model in which in an ON-phase cell, the subcellular OxyR availability at the replication fork as it passes through the agn43 regulatory region is key for initiating an ON-to-OFF switch. In contrast, this availability is not a determining factor for the switch from OFF to ON. This finding shows that different variables affect these two stochastic events. This provides new insight into the events determining the stochastic nature of epigenetic phase variation.
DNA methylation contributes to controlling cellular processes, including chromosome structure, DNA repair, and gene regulation in both prokaryotes and eukaryotes. In the bacterium Escherichia coli, these are conferred by the methylation of adenine by the deoxyadenosine methyltransferase (Dam) target sequence GATC in conjunction with a variety of other proteins. Some Dam-dependent processes rely on the temporary or persistent sequestration of specific GATC sites from Dam-dependent methylation (11, 31).
Temporary sequestration is evident at the origin of replication, oriC. As a result of sequestration, the hemimethylated state, i.e., when only one strand of the double helix is methylated, is protected from methylation for up to one-third of a generation (10). This temporary sequestration is essential for the correct timing of the initiation of DNA replication (43). Non-oriC GATC sites can also be temporarily sequestered, but in general, this occurs for shorter time periods, with remethylation happening within minutes (32, 43). In contrast, the persistent sequestration of specific non-oriC GATC sequences in the genome can be maintained for an entire generation, and this sequestered state can be inherited by daughter cells (11). This is the basis of methylation-dependent epigenetic regulation, as exemplified by Dam-dependent phase variation in E. coli.
Proteins that are required for both temporary and persistent sequestration have been identified. The temporary sequestration of oriC requires the proteins DnaA and SeqA, which bind at and sequester hemimethylated oriC (1, 32). SeqA also binds at non-oriC sequences that contain multiple hemimethylated GATC sequences that are appropriately spaced. Consistent with this finding, SeqA foci have been observed to form at the replication fork where hemimethylated DNA (HM-DNA) is formed (9, 43).
Phase variation is a heritable yet reversible form of gene expression where a reversible switch occurs between an ON (expressing) phase and an OFF (nonexpressing) phase. Dam-dependent phase variation systems require persistent but reversible sequestration. Regulatory DNA binding proteins that are required for this sequestration have been identified as being Lrp and OxyR at pap and agn43, respectively (7, 18). A nonmethylated GATC sequence that is typically associated with Dam-dependent phase variation can be obtained by first sequestering hemimethylated DNA that is formed after DNA replication. Persistent sequestration and a second round of DNA replication will generate nonmethylated sequences. The regulatory process is reversible, and sequestration has to be alleviated to facilitate a switch in the gene expression phase from off to on (11). For agn43 these changes occur at a frequency of about 1 in a thousand cells per generation (18).
The agn43 gene (also known as flu) encodes the outer membrane protein Ag43 (reviewed in references 40 and 41). The relationship between sequestration from Dam, OxyR, and agn43 phase variation is as follows. OxyR is a repressor of agn43 (18, 22, 41, 44). Three Dam target (GATC) sequences are contained within the binding site for OxyR at agn43, and methylation on both strands at this sequence prevents OxyR binding. Thus, methylation alleviates OxyR-mediated repression and allows the ON phase (18, 46). Conversely, OxyR binding to this region can occur when the GATC sequences are unmethylated and results in the OFF phase. Furthermore, this binding blocks the access of Dam to the GATC sequences affecting stable sequestration, which yields the heritability of the off expression phase. The oxidation state of OxyR does not affect this regulation (45). The regulatory region of agn43 contains a SeqA binding site in addition to and overlapping with the OxyR binding site, and SeqA binds to hemimethylated agn43 DNA in vitro (15).
This model for the ON and the OFF phases and the roles of Dam and of OxyR are well supported, but less is understood about the transitions between the DNA methylation states and the expression phase. Here we address the molecular basis of sequestration further to gain insight into how sequestration at agn43 is initially established and then maintained at a high frequency. Our data indicate that the overlap of the SeqA and OxyR binding sites at agn43 has no significance and that SeqA does not play a role in the sequestration of the agn43 region from Dam. Further insight was obtained by applying a novel approach. Introducing auxiliary OxyR binding sites as far upstream of the agn43 transcription start site as 466 bp was sufficient to introduce a bias to the OFF phase. Together with in vitro analyses of OxyR-nucleoprotein interactions presented here, this suggests that OxyR availability at the replication fork is a key determinant for initiating a switch specifically in the ON-to-OFF direction but not for the OFF-to-ON switch. The implications of these findings are discussed in the context of contrasts between SeqA- and OxyR-dependent sequestrations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) broth and on LB agar (33). Media were supplemented with the following antibiotics as necessary: ampicillin (100 μg ml−1), chloramphenicol (34 μg ml−1), kanamycin (30 μg ml−1), and spectinomycin (50 μg ml−1). Fifty microliters of a 10-mg ml−1 catalase solution (Sigma-Aldrich) was applied onto the surface of plates to facilitate the growth of oxyR mutant strains.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype or descriptiona | Reference |
|---|---|---|
| Strains | ||
| AAEC100 | MG1655 (ΔlacZYA) | 4 |
| MV472 | MC4100 oxyR::(Spec) containing pMV108 | 46 |
| MV537 | BL21 containing pMV161 | This study |
| MV1115 | AAEC100 att::pMV244 [agn(−466-+275)′-lacZ] | This study |
| MV1116 | AAEC100 att::pMV245 [agn(−112-+275)′-lacZ] | This study |
| MV1153 | AAEC100 att::pMV253 [(OxyR BS)1agn(−466-+275)′-lacZ] | This study |
| MV1154 | AAEC100 att::pMV254 [(OxyR BS)1agn(−112-+275)′-lacZ] | This study |
| MV1276 | AAEC100 att::pMV280 [(OxyR BS)2agn(−112-+275)′-lacZ] | This study |
| MV1277 | AAEC100 att::pMV278 [(OxyR BS)2agn(−466-+275)′-lacZ] | This study |
| MV1190 | AAEC100 att::pMV257 [(OxyR BS)3agn(−112-+275)′-lacZ] | This study |
| MV1279 | AAEC100 att::pMV279 [(OxyR BS)3agn(−466-+275)′-lacZ] | This study |
| MV1278 | AAEC100 att::pMV281 [(OxyR BS)4agn(−112-+275)′-lacZ] | This study |
| MV1280 | AAEC100 att::pMV293 [(SeqA BS) agn(−39-+275)′-lacZ] | This study |
| MV1305 | AAEC100 att::pMV301 [(SeqA BS) agn(−466-+275)′-lacZ] | This study |
| MV1311 | AAEC100 att::pMV302 [inverted (OxyR BS)-agn(−466-+275)′-lacZ] | This study |
| MV1314 | AAEC100 att::pMV305 [inverted (OxyR BS)3agn(−466-+275)′-lacZ] | This study |
| Plasmids | ||
| pKD3 | cat (Cmr) flanked with FRT sites | 16 |
| pAH125 | CRIM vector lacZ transcriptional fusion | 19 |
| pMV108 | pZE24 with a fragment encoding OxyR(C199S) | 15 |
| pMV140 | pUC19 with 741-bp (agn43 GATC I-II-III mutant) fragment | 46 |
| pMV161 | pET2a (Novagen) with an NdeI-XhoI fragment encoding SeqA | This study |
| pMV243 | CRIM Cmr; pAH125 with ahpC replaced by cat flanked by FRT | This study |
| pMV244 | pMV243 with SalI-EcoRI insert agn(−466-+275) | This study |
| pMV245 | pMV243 with SalI-EcoRI insert agn(−112-+275) | This study |
| pMV253 | pMV244 with 1 OxyR binding siteb inserted at SalI site | This study |
| pMV254 | pMV245 with 1 OxyR binding siteb inserted at SalI site | This study |
| pMV257 | pMV245 with 3 OxyR binding sitesb inserted at SalI site | This study |
| pMV278 | pMV244 with 2 OxyR binding sitesb inserted at SalI site | This study |
| pMV279 | pMV244 with 3 OxyR binding sitesb inserted at SalI site | This study |
| pMV280 | pMV245 with 2 OxyR binding sitesb inserted at SalI site | This study |
| pMV281 | pMV245 with 4 OxyR binding sitesb inserted at SalI site | This study |
| pMV293 | pMV243 with agn(−39-+275)′-lacZ and SeqA BS site (GATC6) at −39 | This study |
| pMV301 | pMV244 with SeqA binding site (GATC6) inserted at SalI site (position −466) | This study |
| pMV302 | pMV253 with inversion of [(OxyR BS)-agn(−466-+275)-lacZ] | This study |
| pMV305 | pMV279 with inverted [(OxyR BS)3-agn(−466-+275)-lacZ] |
nt numbering is that of agn43K-12 relative to the +1 transcription start site (46). BS, binding site.
See the text.
Genetic manipulations.
All genetic manipulations were carried out according to standard protocols (33) unless otherwise noted. Oligonucleotides used this study are listed in Table 2.
TABLE 2.
Oligonucleotides used in this work
| Oligonucleotide | Sequence (5′-3′)a | Function |
|---|---|---|
| oMV102 | GGAATTCCCGTCATGTGATTCCATAC | R, agn(+275)b |
| oMV329 | AATGCCTCATGATGTGTAGGCTGGAGCTGCTTC | F, cat/FRT |
| oMV330 | AATTGGCGGCCGCCATGGGAATTAGCCATGGTCC | R, cat/FRT |
| oMV396 | CGCGTCGACTCCTGCACAGGCACAGAAT | F, agn(−466) |
| oMV397 | CGCGTCGACTCTTCTGAACCTGTCGTGAC | F, agn(−112) |
| oMV419 | CATACTAGTCGACTCAATAATAGAATAAAACG | R, agn(−17); OxyR BS |
| oMV420 | GGCAACTCGAGTACTGACAAAGGTTATTAC | F, agn(+64); OxyR BS |
| oMV659 | GTCGACGTCGGCTTGAGAAAGACCTG | F, oriC (nt 3923331)c |
| oMV687 | GTCGACTATTGCACGGACAAGAGAG | R, agn(−39) |
Restriction sites are in boldface type.
agn sequences are from E. coli K-12 MG1655; the coordinate is 5′ relative to the +1 transcription start site (46). F and R indicate forward and reverse orientations, respectively.
According to MG1655 genome numbering.
Strains containing transcriptional lacZ reporter fusions integrated into the chromosome at the att site were made based on the CRIM system (19). Reporter fusions were made in pMV243, which is a Cmr Kans derivative of pAH125. pMV243 was made by amplifying the Cm resistance marker flanked by FLP recombination target (FRT) sites from pKD3 with oMV329 and oMV330, and this was cloned into the BspHI and NotI sites of pAH125. Thus, in pMV243, ahp was replaced with cat, which can be excised by using Flip recombinase. pMV243 derivates were integrated into the chromosome as described previously (19).
Regulatory regions of agn43 of E. coli K-12 were amplified with either primer oMV396 or oMV397, in combination with oMV102, and cloned as SalI and EcoRI fragments in pMV243, yielding CRIM derivatives pMV244 and pMV245, respectively. Strains with these plasmids integrated into the chromosome at the att site are MV1115 and MV1116, respectively (Table 1).
Derivatives of pMV244 and pMV245 with additional OxyR and SeqA binding sites.
A sequence with an OxyR binding site was generated by amplifying the agn43 binding site with point mutations in all three GATC sequences (46) from pMV140 using oMV419 and oMV420 and cloned as an SalI fragment into pMV244 or pMV245. Reporter plasmids were integrated into the chromosome, and genomic DNA was analyzed by using PCR to confirm that insertions were stably maintained.
A SeqA binding region consisting of oriC (nucleotides [nt] 3923331 to 3923428) containing 5 GATC sequences and a GATC sequence from a previous BamHI cloning site was obtained by amplification with oMV659 and oMV687 from pMV224, which contains a larger oriC fragment inserted at position −39 of the agn43 regulatory region (our unpublished data). This fragment was cloned into the SalI site of pMV244, resulting in pMV301. The spacing of the GATC sequences in this fragments allows for SeqA binding (9). The SeqA binding region was also placed at position −39 of agn43 (nt 3923331 to 3923428), using the same template with oMV659 and oMV102. This construct was cloned into pMV243, resulting in pMV293. Reporter fusions were integrated into the chromosome, resulting in MV1280 and MV1305, respectively.
Inverting the orientation of the reporter fusion.
The orientation of the reporter fusion relative to the direction of the DNA replication fork was reversed by the inversion of the agn-lacZ fragment in selected CRIM derivative plasmids. Plasmids were digested with NheI and SphI, which flank the agn-lacZ sequence, end filled with Klenow enzyme, and ligated. Clones with inverted inserts were identified by restriction enzyme digestion, and plasmids were integrated into the chromosome. The resulting plasmids and strains are listed in Table 1.
Analysis of agn′-lacZ expression.
β-Galactosidase activity was assayed on logarithmic-phase cultures grown in M9 minimal medium with glucose as a carbon source (34). Each assay was done at least in triplicate with a minimum of two independent biological samples. Activity is given in Miller units (MU) (34). The switch frequency is the fraction of cells per generation that change expression phases. The switch frequency is calculated according to the equation (M/N)/g, where M is the number of cells that changed, N is the total number of cells, and g is the number of generations, as described previously (5).
Protein purification.
OxyR(C199S) (29) was purified from MV472 as described previously (15), with the following modifications. Purification on a HiTrap heparin column (Amersham Biosciences) was performed with a 0.1 to 0.8 M KCl gradient (elution step) and was followed by gel filtration on a Superdex 200 column (Amersham Biosciences). The purified OxyR sample was dialyzed against buffer Z (50 mM HEPES [pH 8.0], 5 mM MgCl2, 0.5 mM EDTA [pH 8.0], 10% [vol/vol] glycerol).
SeqA was expressed and purified with a C-terminal His6 tag. The fusion protein has full functionality both in vitro and in vivo (17, 39). The seqA gene was cloned into pET21a, resulting in pMV161 (Tables 1 and 2). Expression was induced in MV537 with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.5 for 3 h at 37°C. Purification was based on the pellet retrieval method (17) with the following modifications. The cell pellet was resuspended in buffer A (25 mM HEPES-KOH [pH 7.5], 10 mM magnesium acetate, 4 mM β-mercaptoethanol, 20% sucrose) supplemented with protease inhibitor cocktail (Complete mini-EDTA-free protease inhibitor cocktail tablets; Roche Diagnostics) and lysed by using a French press (Spectronic). After centrifugation the cell debris was resuspended in buffer B (1 M NaCl, 25 mM HEPES-KOH [pH 7.5], 10 mM magnesium acetate, 4 mM β-mercaptoethanol, 20% sucrose, 0.1% Igepal) and lysed by sonication. Cell debris was removed by centrifugation (10,000 × g at 4°C). SeqA-His was purified from the supernatant by using a HiTrap HP column (GE Healthcare Life Sciences) and a 0.25 to 500 mM imidazole elution gradient. Purified SeqA was dialyzed against buffer M (300 mM ammonium sulfate, 20 mM HEPES-KOH [pH 8.0], 10 mM magnesium acetate, 1 mM EDTA, 4 mM dithiothreitol [DTT], 15% [vol/vol] glycerol). All column chromatography was carried out with an Äkta Purifier 100 chromatography system (GE Healthcare Life Sciences). Protein samples were analyzed by SDS-PAGE, Coomassie blue staining, and an EPI Chemi II Darkroom imaging system with LabWorks image acquisition and analysis software (UVP, Inc.). The protein concentration was determined by a Bradford assay (7a) (Bio-Rad) with bovine serum albumin (BSA) as a standard.
EMSA and immunoblotting.
An electrophoretic mobility shift assay (EMSA) was performed with purified proteins and 6-carboxyfluorescein (FAM)-labeled DNA. To obtain a 66-bp FAM-labeled, hemimethylated DNA (HM-DNA) probe of the agn43K-12 regulatory region, two oligonucleotides comprising the region at nt 9231 to 9295 (GenBank accession number AE000291) (18) were annealed, of which the bottom strand contained N-6-methyl deoxyadenosine residues at the three GATC sites and FAM at its 5′ end (Integrated DNA Technologies, Inc.), and the top strand contained no methylated adenines. The affinity of SeqA for the hemimethylated agn43 regulatory sequence with either the top or the bottom strand methylated is the same (15).
Binding reactions with purified proteins were carried out for the specified time at 30°C in 20-μl mixtures containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 10% (vol/vol) glycerol, 5 mg/ml BSA, and poly(dI-dC) as a nonspecific competitor. This buffer is close to optimal for SeqA binding and still allows OxyR binding and DNA methylation to occur.
Protein-DNA complexes were subjected to electrophoresis in a 5% nondenaturing polyacrylamide gel in a high-ionic-strength buffer (50 mM Tris base, 380 mM glycine, 1.5 mM EDTA). The gels were analyzed by using a Molecular Imager FX apparatus (Bio-Rad), and bands were quantified with QuantityOne software (Bio-Rad).
To identify proteins present in nucleoprotein complexes (6), the proteins were transferred from EMSA gels onto a Hybond-P polyvinylidene difluoride (PVDF) membrane (GE Healthcare Life Sciences) in a denaturing transfer buffer (20 mM Tris [pH 8.0], 150 mM glycine, 0.5% SDS, 20% methanol) by using a Trans-Blot semidry system (Bio-Rad). Membranes were probed as described previously for Western blots (33), with total serum containing anti-OxyR or anti-His-SeqA polyclonal antibodies (Eurogentec S.A., Belgium). Enhanced chemiluminescence (ECL) (Amersham Biosciences) was used to detect the peroxidase-linked secondary anti-rabbit IgG antibody (Amersham). Bands were visualized by using X-Ray film. TTBS buffer (20 mM Tris, 120 mM NaCl, 0.05% Tween 20 [pH 7.4]) with 2% SDS was used to strip the membranes when additional probing was required.
RESULTS
In vitro competition for HM-DNA indicates that OxyR but not SeqA binding can mediate a switch to the off phase.
Hemimethylated agn43 DNA is a substrate for Dam and a binding template for OxyR and SeqA (15). The gene expression state is a direct result of the outcome of the competition of these proteins for this DNA, with methylation leading to the reestablishment of an on phase and sequestration initiating a switch to the off phase. To identify factors affecting the competition, the preferred protein-DNA interactions were analyzed in vitro. An EMSA was performed with SeqA, OxyR, and Dam, with the hemimethylated agn43 regulatory region as a probe. The DNA probe includes the agn43 OxyR binding site and the three Dam target sequences. There are no additional SeqA binding sites within 500 bp of the agn43 regulatory region either in the native sequence context in E. coli K-12 or for the agn′-lacZ reporter fusion. Thus, the probe contains the only sequence for SeqA binding in the agn43 regulatory region. OxyR is a global regulator that is oxidized in response to oxidative stress (38). However, the reduced form is the predominant form under standard growth conditions and is necessary and sufficient for agn43 phase variation (18, 45). OxyR(C199S) (29) has all the properties of reduced OxyR and was used in all experiments. OxyR(C199S) from here on is referred to as OxyR. Proteins present in a nucleocomplex were identified by immunoblotting of the EMSA gels.
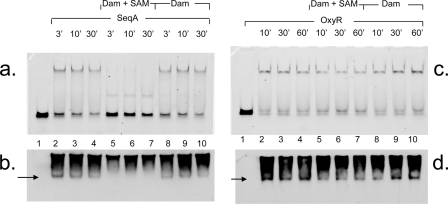
First, we assessed whether the methylation of agn43 GATC sequences of HM-DNA in complex with either SeqA or OxyR can occur. Nucleoprotein complexes were allowed to form with 7 pmol of either SeqA or OxyR, followed by the addition of Dam in the presence or absence of the methyl group donor S-adenosylmethionine (SAM) (Fig. 1). Note that 7 pmol of OxyR in this assay correlates to 350 nM and that 7 pmol of SeqA correlates to 500 nM. Cellular concentrations of OxyR and SeqA have been estimated to be 170 to 340 nM (S. Ishihama, personal communication) and 1.5 μM (37), respectively, but this does not necessarily represent soluble protein that is not already in a nucleoprotein complex. The addition of Dam to the SeqA/HM-DNA complex resulted in a rapid disappearance of the preexisting complex (Fig. 1a and b, lanes 5 to 7), with less than 15% of the complex remaining after a 3-min incubation with Dam and SAM. This displacement is dependent on SAM (Fig. 1a and b, lanes 8 to 10). These results suggest that SeqA dissociates rapidly from the DNA to allow the access of Dam and the methylation of HM-DNA. This conversion to fully methylated DNA prevents SeqA from rebinding (15).
FIG. 1.
Dam methylation rapidly disrupts existing SeqA/HM-DNA complexes, but OxyR/HM-DNA complexes are more stable. (a and c) Results of EMSA after the addition of 7 pmol SeqA (a, lanes 2 to 10) or OxyR (c, lanes 2 to 10) with 0.1 pmol agn43 HM-DNA. Lane 1, DNA only. Complexes were allowed to form for 10 min, after which no protein (lanes 2 to 4), 0.5 pmol Dam supplemented with 64 nM SAM (lanes 5 to 7), or Dam without SAM (lanes 8 to 10) was added. (b and d) After incubation for the times indicated, complexes were analyzed by EMSA gel. The protein identity in the complex was confirmed by Western blot analysis with anti-SeqA (b) or anti-OxyR (d). Arrows in b and d indicate the positions of DNA-protein complexes, and smears represent excess free protein.
In contrast, the OxyR/HM-DNA complex was very stable in the presence of Dam and SAM, with the majority of DNA still present in the nucleocomplex after an hour (Fig. 1c and d, lanes 5 to 7). This suggests that this OxyR binding to HM-DNA is stable even in the presence of Dam and therefore could be maintained for a generation and thereby could initiate a switch to the off phase in an on-phase cell.
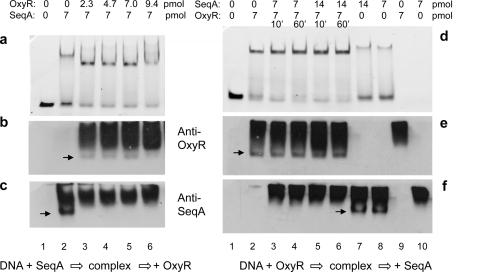
The generation of new DNA binding sites as part of the DNA replication process can affect the stability of existing complexes. We examined the stabilities of SeqA and OxyR DNA complexes after the addition of (unlabeled) agn43 HM-DNA in vitro (Fig. 2a and b). In essence, this examines the dissociation of the protein from the DNA. SeqA (Fig. 2a) was displaced from the labeled HM-DNA complex by a 1.5-fold excess of HM-DNA within 10 min. This is consistent with a high rate of dissociation and the above-described observation that the SeqA/HM-DNA complex is not stable in the presence of Dam and SAM.
FIG. 2.
Complexes of OxyR or SeqA with agn43 HM-DNA are displaced rapidly by an excess of agn43 HM-DNA. Shown are data from EMSA analysis of 0.2 pmol HM-DNA (a and b) or 0.4 pmol unmethylated agn43 DNA as a comparison (c). Samples have no protein (lanes 1) or an addition of 9 pmol SeqA (a) or 9 pmol OxyR (b and c), and complexes were allowed to form and then incubated for a further 10 min with no addition (lanes 2) or with the addition of non-FAM-labeled agn43 HM-DNA at 0.3 pmol (lanes 3), 0.6 pmol (lanes 4), 1.2 pmol (lanes 5), and 2.8 pmol (c, lane 6).
Similar to SeqA, OxyR (Fig. 2b) was rapidly displaced from HM-DNA by excess (unlabeled) agn43 HM-DNA. This was despite the fact that, and in contrast to the SeqA/HM-DNA complex, the OxyR/HM-DNA complex was stable in the presence of Dam and SAM (Fig. 1c). OxyR in complex with agn43 unmethylated DNA (UM-DNA) was more stable in the presence of an excess of HM-DNA (Fig. 2c). This finding is consistent with the previously reported higher affinity of OxyR for agn43 UM-DNA than HM-DNA (15).
Consistent with the findings described above, OxyR can also replace SeqA in complex with HM-DNA. OxyR was added to preformed SeqA/HM-DNA complexes, and complexes were analyzed after 10 min (Fig. 3a to c). The results show that OxyR replaced SeqA to form an OxyR/HM-DNA complex (Fig. 3a to c, lanes 2 to 6). Conversely and consistent with this finding, the addition of SeqA did not affect OxyR in complex with HM-DNA, even at a 2-fold molar excess or with a 60-min incubation (Fig. 3d to f, lanes 2 to 6). These in vitro analyses indicate that SeqA bound to agn43 DNA is displaced rapidly by both Dam and OxyR and thus suggest that SeqA binding at agn43 HM-DNA will not bias the competition between Dam and OxyR for this substrate. By extrapolation to the situation in the cell, this suggests that SeqA does not play a role in establishing or maintaining the DNA methylation patterns that are essential for agn43 phase variation.
FIG. 3.
OxyR complexes are preferentially formed with agn43 HM-DNA. Shown are EMSA gels (a and d) and Western blots of these EMSA gels probed with OxyR antiserum (b and e) or SeqA antiserum (c and f). In b, c, d and f, arrows indicate DNA-protein complexes, and smears represent excess free protein. (a to c) A total of 0.1 pmol HM-DNA (lanes 1 to 10) was incubated with no protein (lane 1) or for 10 min with 7 pmol SeqA (lanes 2 to 6) with the subsequent addition of OxyR at the indicated amount and an additional 10 min of incubation (lanes 3 to 6). (d to f) A total of 0.1 pmol HM-DNA was incubated with OxyR followed by the addition of SeqA (lanes 3 to 6) at 7 or 14 pmol, as indicated, for 10 min (lanes 3 and 5) or 60 min (lanes 4 and 6) or incubated with SeqA only (lanes 7 and 8). OxyR (lane 9) and SeqA (lane 10) were incubated in the absence of HM-DNA.
SeqA affects agn43 phase variation only with enhanced binding at the promoter sequence.
In a cell, SeqA binds to two or more GATC sequences on the same face of the DNA as a primary binding site but also forms higher-order structures that involve protein bound at distantly located sites (12, 43). These non-oriC foci could be significant by directly or indirectly facilitating the sequestration that is important for gene regulation. The following strategy was developed to address this possibility.
A SeqA binding site that contains six GATC sequences (GATC6) but no other known protein binding sites was inserted at position −466 of agn43 (MV1305). This binding site would increase the local concentration of SeqA, but six GATC sites are not sufficient to mediate oriC-like sequestration (3). The introduction of GATC6 did not affect phase variation or the level of expression of agn′-lacZ. The same GATC6 sequence was also placed at position −39 in the regulatory region (MV1280). In this strain the spacing between the GATC6 sequence and the three agn43 GATC sequences is such that the cooperative binding of SeqA is predicted to occur at this GATC(6 + 3) sequence (8). Colonies of MV1280 were grades of blue on M9-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates, and progeny colonies had similar variable levels of Lac expression. Thus, the addition of these sites disrupted the heritable nature of agn43 phase variation. As expected and consistent with the variable colony phenotypes, transcription levels in cultures derived from individual colonies ranged from 150 to 545 MU. This is lower than the agn promoter maximum of 1,300 MU (46), which suggests that the enhanced SeqA binding blocked access to the agn43 promoter region of Dam and/or RNA polymerase. Importantly, SeqA-dependent changes in agn43 expression were obtained only by introducing SeqA binding in immediate proximity of the agn43 promoter region.
Introduction of auxiliary OxyR binding sites up to 466 bp from agn43 affects phase variation.
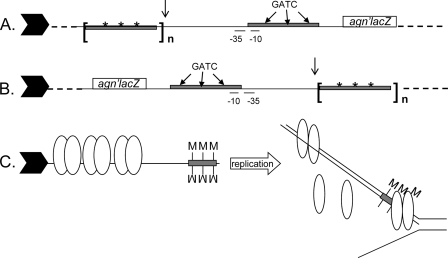
OxyR is important for maintaining and establishing the sequestration that is essential for phase variation and imposes heritability on the gene expression phase. Increasing the cellular level of OxyR above that in wild-type cells leads to a locked OFF-phase population with stable, fully sequestered agn43 DNA (18, 46). To further analyze this effect without altering the global concentration, a novel approach was taken. OxyR binding sites were introduced as auxiliary binding sites for agn43. Each site has the same binding affinity for reduced OxyR as that for the unmethylated agn43 sequence, but they do contain GATC sequences (46). Clones with one to four of these fragments at 112 or at 466 bp upstream of the agn′-lacZ transcription start site (Fig. 4A) were isolated, and the phase variation of agn′-lacZ was examined. Note that the global cellular Dam and OxyR concentrations as well as the agn43 regulatory sequence were identical to those of the wild type in all these isolates.
FIG. 4.
Schematic of important elements of the reporter fusion of agn′-lacZ showing the relative positions of the additional OxyR binding sites and the orientation in the chromosome (not to scale). Indicated are the promoter elements (−10 and −35) and the Dam target sequences (GATC). Gray rectangles indicate OxyR tetramer binding regions, asterisks indicate mutated GATC sequences, and n indicates the number of OxyR binding sites. The line arrow indicates the insertion position of the −112 or −466 element in different isolates (Table 3). The filled block arrow indicates the direction of the passage of the replication fork. (A) Orientation relative to the replication fork for isolates except MV1311 and MV1314. (B) In MV1311 and MV1314 the sequence elements between the dashed lines were inverted on the chromosome. (C) Schematic model depicting how three upstream auxiliary OxyR binding sites can bias phase variation to the off phase. Symbols are described above; each oval represents an OxyR dimer, M designates a methyl group, and the open arrow indicates DNA replication. Only half of the replication fork is shown.
Phase variation rates for isolates with just one or two OxyR binding sites were similar to the that of the wild type regardless of the insertion site (Table 3). However, with three extra sites, a bias to the OFF phase was effected at both insertion sites. A locked OFF phenotype was effected with three and four binding sites inserted at positions −466 and −112, respectively (MV1279 and MV1278, respectively). The reason for the difference in the magnitude of the effect is not clear but could be due to a different occupation level of the binding sites for unknown reasons. There was no correlation between the phase of the helix of the new site(s) and the agn43 operator sequence. The transformation of MV1279 with pTP166(dam), which results in the overproduction of Dam, reverted the Lac− phenotype to LacZ+ colonies, as would occur with the wild-type operator sequence (46), and shows that this induced OFF phase is also reversible and involves Dam-dependent sequestration at agn43. Importantly, this experimental strategy for the first time allowed the phase variation of agn43 to be locked OFF without altering either the agn43 OxyR binding site affinity, the methylation target sequences, or the cellular concentration of the regulatory factors.
TABLE 3.
Phase variation of agn43 is biased to the off phase as a result of introducing auxiliary OxyR binding sitesa
| Strain | No. of auxiliary OxyR BS and insertion site | Switch frequency |
|
|---|---|---|---|
| Off to on | On to off | ||
| MV1116 | agn′(−112)-lacZ | 2.3 × 10−3 | 1.9 × 10−3 |
| MV1154 | +1 OxyR BS at −112 | 3.4 × 10−3 | 4.6 × 10−3 |
| MV1276 | +2 OxyR BS at −112 | 5.7 × 10−3 | 3.7 × 10−3 |
| MV1190 | +3 OxyR BS at −112 | 3.4 × 10−3 | 25.2 × 10−3 |
| MV1278 | +4 OxyR BS at −112 | NA | NA |
| MV1115 | agn′(−466)-lacZ | 3.7 × 10−3 | 0.7 × 10−3 |
| MV1153 | +1 OxyR BS at −466 | 2.9 × 10−3 | 2.0 × 10−3 |
| MV1277 | +2 OxyR BS at −466 | 4.3 × 10−3 | 1.1 × 10−3 |
| MV1279 | +3 OxyR BS at −466 | NA | NA |
| MV1311 | Inverted [agn′(−466)-lacZ + 1 OxyR BS] | 4.9 × 10−3 | 0.7 × 10−3 |
| MV1314 | Inverted [agn′(−466)-lacZ + 3 OxyR BS] | 3.7 × 10−3 | 13.8 × 10−3 |
Representative data are given.
b NA, not applicable (locked off).
In these isolates the orientation of the agn′-lacZ region on the chromosome is such that the DNA replication fork will pass through the additional OxyR binding sites before passing through the native agn43 regulatory region. We therefore postulated that in this experiment, the bias to the OFF phase may have been obtained by an increase in the local subcellular concentration of free OxyR as a result of the displacement of OxyR by the passage of the replication fork. In the isolates with auxiliary binding sites, this displacement would occur prior to the generation of hemimethylated DNA in an ON-phase cell. If this model is correct, an inverse orientation of the OxyR-containing sequence relative to agn43 on the chromosome should decrease the effect on phase variation rates as a result of the auxiliary OxyR binding sites (Fig. 4B and C). Indeed, compared to the locked OFF phenotype of MV1279 as a result of three auxiliary binding sites, isolate MV1314 with an inverted region now showed a phase variation of agn′-lacZ but with a directional bias of a high ON-to-OFF rate and a normal OFF-to-ON rate. As expected, the inversion did not affect the switch frequency with a single additional binding site (MV1311), since the single binding site also had no measurable effect in the original orientation (Table 3). The results are consistent with the interpretation that the additional binding sites increase the OxyR availability and thus create a higher probability that OxyR can outcompete Dam for the regulatory region than in MV115 with no additional sites. This results in an increase of the ON-to-OFF rate, which in MV1314 causes an increase in the ON-to-OFF switch frequency and in MV1279 causes the persistent sequestration and maintenance of the OFF phase and, thus, a locked OFF phase.
DISCUSSION
The methylation state of three GATC Dam target sequences at the agn43 regulatory region establishes the heritability of the expression phase (46). The ON phase switches to the OFF phase in 1 out of 1,000 cells per generation (18), indicating that at this rate, the sequestration of the region is newly established. When DNA replication of the fully methylated sequence in an on-phase cell generates HM-DNA, an opportunity arises to change the expression phase: Dam access is required to maintain the ON phase, whereas OxyR binding can be the first step to switch to the OFF phase. agn43 HM-DNA in the cell is a substrate for methylation by Dam and a binding substrate for SeqA (15, 21) and OxyR (15).
No role in sequestration has been identified for SeqA binding to natural or synthetic regions with a low number of GATC sequences (2, 3, 20). At agn43, however, a prolonged sequestration like that at oriC should not be required to mediate an effect on phase variation, since it is the competition between OxyR and Dam for the HM-DNA that ultimately determines the regulatory phase. The outcome of this competition could be influenced by SeqA if the SeqA/HM-agnDNA complex is more stable in the presence of one or the other (15). SeqA binds at agn43 HM-DNA in vitro (15), but the significance of this was not known. A seqA mutation affects phase variation through an indirect effect on the general level of genomic DNA methylation (15). The overproduction of SeqA did not affect phase variation, but this result is difficult to interpret since this SeqA exists largely in inclusion bodies (M. W. van der Woude, unpublished data). Therefore, we examined protein-DNA interactions in vitro to address events as they may occur following the passage of the replication fork at methylated agn43 DNA in an on-phase cell.
The in vitro assays used here to address the stability of nucleoprotein complexes showed that the SeqA/HM-DNA complex is not maintained in the presence of Dam and SAM (Fig. 1), excess unlabeled SeqA DNA binding substrate (Fig. 3A), or OxyR (Fig. 2). SeqA also cannot displace OxyR in complex with HM-DNA (Fig. 2). These data indicate that SeqA binding at agn43 does not result in a measurable sequestration of the agn43 regulatory region from either Dam or OxyR. Thus, it is unlikely that SeqA binding at agn43 will bias the competition between OxyR and Dam for HM-DNA.
Non-oriC sequence SeqA-DNA complexes were previously proposed to be recruited to preexisting SeqA foci (17, 35). The sequence context of agn43 varies among E. coli isolates, which raised the question of whether this may influence regulation (46). However, a SeqA binding site introduced at position −466 from the agn43 transcription start site did not affect agn43 phase variation. In contrast, phase variation was altered and heritability was compromised when this sequence was placed directly adjacent to the agn43 promoter, where the combination of existing and novel GATC sequences results in an improved binding site (2). This may alter regulation directly (2, 43) or indirectly through sequestration from the methylation of the agn43 promoter (18, 46). Significantly, this approach also failed to identify a role for SeqA binding at the natural agn43 GATC3 cluster. Taken together, these data do not support a role of the temporary binding of SeqA in establishing OxyR-dependent persistent sequestration of the agn43 region.
The important new insight obtained from the in vitro analyses is that OxyR binding to HM-DNA may be sufficient to destine a cell to switch from the ON phase to the OFF phase. The OxyR/HM-DNA complex was stable in the presence of Dam-SAM (Fig. 1c and d). Thus, OxyR binding sequesters the agn43 GATC sequences from Dam when it is bound to the hemimethylated region as well as the unmethylated region (15). Furthermore, OxyR titration as a result of genome replication is unlikely to be significant for phase variation even though the OxyR/HM-DNA complex was not stable in the presence of competitor OxyR binding sites (Fig. 2b). This is unlikely to be biologically relevant since there are few binding sites for the reduced form of OxyR in the chromosome (47, 48), and with an estimated 150 monomers of OxyR in the cell (125 nM dimer) (A. Ishihama, personal communication), new OxyR binding sites formed as a result of DNA replication can be occupied without titration from occupied sites like agn43. Taken together, these results suggest that OxyR dissociates from HM-DNA at a high rate but also that (re)association occurs rapidly and prevents Dam access and subsequent methylation. The relative stability of the complex in the presence of Dam-SAM suggests that OxyR-DNA interactions are not the only factor determining the access of Dam and that the complex Dam-SAM-DNA interactions and methylation kinetics that are still being elucidated are also a contributing factor (13, 25, 26, 50). Importantly, our findings regarding stability in the presence of Dam-SAM support a model in which OxyR binding to HM-DNA in replicating ON-phase cells initiates a switch to the OFF phase.
Further analysis will be required to determine the exact contribution of each interaction in this competition for DNA, as is also ongoing for the Dam- and Lrp-dependent phase variation of pap (36). Differences between the systems will exist. In addition to different protein-DNA interactions between Lrp and OxyR, the GATC flanking sequences that play a role in pap methylation kinetics (14) are not present in agn43.
Here it is shown that the sequestration process that is essential for Dam-dependent phase variation can be modulated without changing the cellular levels of regulatory proteins or the sequence of the regulatory region of agn43 (18, 23, 44, 46). Specifically, by introducing OxyR binding sites upstream as far as 466 bp of the regulatory region, the phase variation of agn43 was altered with a bias to the OFF phase (Table 3). One model to explain this effect is that the auxiliary OxyR binding sites allow novel interactions between distant OxyR tetramers and stabilize the interaction at agn43. There is, however, no precedence for this type of interaction for OxyR, even though the interaction between OxyR multimers bound to DNA remains to be fully resolved (28). There is also no apparent correlation with the face of the helix, the OxyR binding sites, and the observed effects. Therefore, a different model is favored.
We propose that the bias to the OFF phase is a result of introducing an increase in the OxyR subcellular concentration at the regulatory region merely as a result of the additional binding sites. This correlation is consistent with the locked OFF phenotype that is seen for an isolate with an increased total cellular level of OxyR due to the presence of an OxyR-producing plasmid (18, 23, 44, 46). The effect of this artificial system of auxiliary OxyR binding sites can be compared to that occurring naturally for lac regulation. LacI binds at low-affinity secondary sites, which creates a LacI sink (42). This is required to maintain an effective local LacI concentration at the high-affinity binding site that mediates LacI-dependent repression. In contrast, the auxiliary OxyR binding sites have an affinity for OxyR that is the same as that for the (unmethylated) binding site that mediates repression. However, the cellular concentration of OxyR is high enough to allow the occupation of all sites. Thus, we propose that the auxiliary binding sites function as a synthetic OxyR sink that increases the OxyR concentration locally and thereby biases the competition between OxyR and Dam to favor OxyR binding. This in turn facilitates a switch to the OFF phase. The data are consistent with our conclusion from the in vitro analyses that the formation of an OxyR/HM-DNA nucleoprotein complex will initiate a switch to the OFF phase.
The passage of the DNA replication fork may facilitate the dissociation of OxyR from the auxiliary binding sites (Fig. 4C). Thus, OxyR will be made available in these isolates before HM-agnDNA is formed. The effect of these auxiliary OxyR sites could therefore change if the orientation relative to the passage of the replication fork was inverted. In that case the HM-DNA is formed before the OxyR is dislodged and becomes available (Fig. 4B and C), increasing the chance that Dam accesses the DNA before OxyR and resulting in a maintenance of the on phase. Indeed, the result of inverting the relative position of the reporter construct and auxiliary sites on the chromosome was a decreased directional bias to the OFF phase (Table 3), which is consistent with this model. In contrast, the OxyR-sink-dependent changes did not affect the OFF-to-ON switch (Table 3). This suggests that in progeny cells from an OFF parent cell in which OxyR is bound at the agn43 region to maintain an OFF phase in both of them, OxyR availability is not a determining factor. The event(s) that affect the stochastic OFF-to-ON switch remain to be determined, but Dam availability at the replication fork is a variable for consideration.
In a deterministic model for agn43 phase variation proposed previously by Lim and van Oudenaarden (30), a transient intermediate state is incorporated in the switch from the on to the off phase with HM-DNA that is proposed to be either ON or ON with a lower expression level (“partial ON”). In our model the “partial ON” state that is proposed as a result of unoccupied hemimethylated DNA will not persist to yield a subpopulation of cells. Our analyses suggest that OxyR bound to hemimethylated DNA is stable enough to be maintained and repress transcription throughout a cell cycle. This is consistent with an OFF phase or perhaps due to the low affinity of a “partial ON” phase but not an ON phase. Furthermore, the insight obtained here that the subcellular OxyR concentration at agn43 is a key feature suggests that at one of the two daughter chromosomes of a replicating OFF-phase genome, OxyR rebinding will occur and the OFF state will be inherited due to the availability of the OxyR tetramer. In the second daughter cell, the partial expression state (lower than that in an on cell) that was proposed in the model described previously by Lim and van Oudenaarden for cells with no OxyR-bound and unmethylated DNA could indeed occur, since only one OxyR tetramer (two dimers in solution) would be readily available for rebinding, consistent with the finding that the transcription level is decreased from an unmethylated agn43 promoter compared to that from a methylated one, even in the absence of OxyR (18, 23, 44, 46). However, this too would probably not persist since the DNA is available as both a Dam template and an OxyR template. The work here may inform further experiments or directly contribute to refining the deterministic model and the development of novel stochastic models for this regulation.
Further application of the “artificial sink” approach described here can yield new insights into regulation in general and into this epigenetic regulation in particular. It can form the basis to acquire quantitative data for modeling, which is of particular interest for stochastic regulation, including agn43 and pap Dam-dependent phase variation (24, 27, 30, 31, 49). Pursuing novel insights into the pap and agn43 systems may ultimately allow us to define “molecular rules” that govern Dam-dependent epigenetic regulation.
Acknowledgments
This work was supported by grant BB/C502849/1 from the BBSRC.
We thank the Technology Facility at the University of York, Nisha Gandhi and Sophie Foppolo for technical support, R. Verstraten and M. Bouwman for assistance, S. Broadbent for critical reading of the manuscript, and A. Ishihama for sharing unpublished data.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Bach, T., Morigen, and K. Skarstad. 2008. The initiator protein DnaA contributes to keeping new origins inactivated by promoting the presence of hemimethylated DNA. J. Mol. Biol. 384:1076-1085. [DOI] [PubMed] [Google Scholar]
- 2.Bach, T., and K. Skarstad. 2005. An oriC-like distribution of GATC sites mediates full sequestration of non-origin sequences in Escherichia coli. J. Mol. Biol. 350:7-11. [DOI] [PubMed] [Google Scholar]
- 3.Bach, T., and K. Skarstad. 2004. Re-replication from non-sequesterable origins generates three-nucleoid cells which divide asymmetrically. Mol. Microbiol. 51:1589-1600. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisentstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 5.Blyn, L. B., B. A. Braaten, C. A. White-Ziegler, D. A. Rolfson, and D. A. Low. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowen, B., J. Steinberg, U. K. Laemmli, and H. Weintraub. 1980. The detection of DNA-binding proteins by protein blotting. Nucleic Acids Res. 8:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, B. A., J. V. Platko, M. W. van der Woude, B. H. Simons, F. K. de Graaf, J. M. Calvo, and D. A. Low. 1992. Leucine-responsive regulatory protein (Lrp) controls the expression of both the pap and fan pili operons in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:4250-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brendler, T., and S. Austin. 1999. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 18:2304-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brendler, T., J. Sawitzke, K. Sergueev, and S. Austin. 2000. A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J. 19:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 11.Casadesus, J., and D. Low. 2006. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70:830-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung, Y. S., T. Brendler, S. Austin, and A. Guarne. 2009. Structural insights into the cooperative binding of SeqA to a tandem GATC repeat. Nucleic Acids Res. 37:3143-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffin, S. R., and N. O. Reich. 2009. Escherichia coli DNA adenine methyltransferase: the structural basis of processive catalysis and indirect read-out. J. Biol. Chem. 284:18390-18400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin, S. R., and N. O. Reich. 2008. Modulation of Escherichia coli DNA methyltransferase activity by biologically derived GATC-flanking sequences. J. Biol. Chem. 283:20106-20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Correnti, J., V. Munster, T. Chan, and M. van der Woude. 2002. Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol. Microbiol. 44:521-532. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fossum, S., S. Soreide, and K. Skarstad. 2003. Lack of SeqA focus formation, specific DNA binding and proper protein multimerization in the Escherichia coli sequestration mutant seqA2. Mol. Microbiol. 47:619-632. [DOI] [PubMed] [Google Scholar]
- 18.Haagmans, W., and M. van der Woude. 2000. Phase variation of Ag43 in E. coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877-887. [DOI] [PubMed] [Google Scholar]
- 19.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, J. S., S. Kang, H. Lee, H. K. Kim, and D. S. Hwang. 2003. Sequential binding of SeqA to paired hemi-methylated GATC sequences mediates formation of higher order complexes. J. Biol. Chem. 278:34983-34989. [DOI] [PubMed] [Google Scholar]
- 21.Han, J. S., S. Kang, S. H. Kim, M. J. Ko, and D. S. Hwang. 2004. Binding of SeqA protein to hemi-methylated GATC sequences enhances their interaction and aggregation properties. J. Biol. Chem. 279:30236-30243. [DOI] [PubMed] [Google Scholar]
- 22.Henderson, I. R., M. Meehan, and P. Owen. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol. Lett. 149:115-200. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernday, A., B. Braaten, and D. Low. 2004. The intricate workings of a bacterial epigenetic switch. Adv. Exp. Med. Biol. 547:83-89. [DOI] [PubMed] [Google Scholar]
- 25.Horton, J. R., K. Liebert, M. Bekes, A. Jeltsch, and X. Cheng. 2006. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J. Mol. Biol. 358:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton, J. R., K. Liebert, S. Hattman, A. Jeltsch, and X. Cheng. 2005. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of Dam methyltransferase. Cell 121:349-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarboe, L. R., D. Beckwith, and J. C. Liao. 2004. Stochastic modeling of the phase-variable pap operon regulation in uropathogenic Escherichia coli. Biotechnol. Bioeng. 88:189-203. [DOI] [PubMed] [Google Scholar]
- 28.Knapp, G. S., J. W. Tsai, and J. C. Hu. 2009. The oligomerization of OxyR in Escherichia coli. Protein Sci. 18:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kullik, I., M. B. Toledano, L. A. Tartaglis, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim, H., and A. van Oudenaarden. 2007. A multistep epigenetic switch enables the stable inheritance of DNA methylation states. Nat. Genet. 39:269-275. [DOI] [PubMed] [Google Scholar]
- 31.Low, D. A., and J. Casadesus. 2008. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 11:106-112. [DOI] [PubMed] [Google Scholar]
- 32.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Morigen, I. Odsbu, and K. Skarstad. 2009. Growth rate dependent numbers of SeqA structures organize the multiple replication forks in rapidly growing Escherichia coli. Genes Cells 14:643-657. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, S. N., and N. O. Reich. 2008. Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J. Mol. Biol. 383:92-105. [DOI] [PubMed] [Google Scholar]
- 37.Slater, S., S. Wold, M. Lu, E. Boye, K. Skarstad, and N. Kleckner. 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82:927-936. [DOI] [PubMed] [Google Scholar]
- 38.Storz, G., L. A. Tartaglia, and B. A. Ames. 1990. Transcriptional regulation of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189-194. [DOI] [PubMed] [Google Scholar]
- 39.Taghbalout, A., A. Landoulsi, R. Kern, M. Yamazoe, S. Hiraga, B. Holland, M. Kohiyama, and A. Malki. 2000. Competition between the replication initiator DnaA and the sequestration factor SeqA for binding to the hemimethylated chromosomal origin of E. coli in vitro. Genes Cells 5:873-884. [DOI] [PubMed] [Google Scholar]
- 40.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van der Woude, M. W., and I. R. Henderson. 2008. Regulation and function of Ag43 (Flu). Annu. Rev. Microbiol. 62:153-169. [DOI] [PubMed] [Google Scholar]
- 42.Vilar, J. M., and L. Saiz. 2005. DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev. 15:136-144. [DOI] [PubMed] [Google Scholar]
- 43.Waldminghaus, T., and K. Skarstad. 2009. The Escherichia coli SeqA protein. Plasmid 61:141-150. [DOI] [PubMed] [Google Scholar]
- 44.Waldron, D. E., P. Owen, and C. J. Dorman. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44:509-520. [DOI] [PubMed] [Google Scholar]
- 45.Wallecha, A., J. Correnti, V. Munster, and M. van der Woude. 2003. Phase variation of Ag43 is independent of the oxidation state of OxyR. J. Bacteriol. 185:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallecha, A., V. Munster, J. Correnti, T. Chan, and M. van der Woude. 2002. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184:3338-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng, M., X. Wang, B. Doan, K. A. Lewis, T. D. Schneider, and G. Storz. 2001. Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J. Bacteriol. 183:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, B., D. Beckwith, L. R. Jarboe, and J. C. Liao. 2005. Markov chain modeling of pyelonephritis-associated pili expression in uropathogenic Escherichia coli. Biophys. J. 88:2541-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zinoviev, V. V., A. A. Evdokimov, E. G. Malygin, B. Sclavi, M. Buckle, and S. Hattman. 2007. Differential methylation kinetics of individual target site strands by T4Dam DNA methyltransferase. Biol. Chem. 388:1199-1207. [DOI] [PubMed] [Google Scholar]