Abstract
The human pathogen L. monocytogenes is a facultatively intracellular bacterium that survives and replicates in the cytosol of many mammalian cells. The listerial metabolism, especially under intracellular conditions, is still poorly understood. Recent studies analyzed the carbon metabolism of L. monocytogenes by the 13C isotopologue perturbation method in a defined minimal medium containing [U-13C6]glucose. It was shown that these bacteria produce oxaloacetate mainly by carboxylation of pyruvate due to an incomplete tricarboxylic acid cycle. Here, we report that a pycA insertion mutant defective in pyruvate carboxylase (PYC) still grows, albeit at a reduced rate, in brain heart infusion (BHI) medium but is unable to multiply in a defined minimal medium with glucose or glycerol as a carbon source. Aspartate and glutamate of the pycA mutant, in contrast to the wild-type strain, remain unlabeled when [U-13C6]glucose is added to BHI, indicating that the PYC-catalyzed carboxylation of pyruvate is the predominant reaction leading to oxaloacetate in L. monocytogenes. The pycA mutant is also unable to replicate in mammalian cells and exhibits high virulence attenuation in the mouse sepsis model.
Listeria monocytogenes is a human pathogen that can cause systemic infections, especially in immunocompromised people, with symptoms such as septicemia, (encephalo)meningitis, placentitis, and stillbirth. These Gram-positive bacteria are able to enter the cytosol of many mammalian cells after being taken up via normal or induced phagocytosis by professional phagocytes, mainly macrophages and dendritic cells, and nonphagocytic cells, such as epithelial cells, fibroblasts, and endothelial cells (1, 8, 13). While the virulence genes and their regulation (4, 21), as well as the encoded virulence factors (20, 22), necessary for the various steps of the intracellular replication cycle of L. monocytogenes have been extensively studied in the past few decades, there is still little information concerning the metabolic capacities and the metabolic adaptation processes (10) that enable these bacteria to efficiently replicate in the cytosol of their host cells.
The information on listerial metabolism obtained from the genome sequence (7) suggests that these heterotrophic bacteria are capable of utilizing a variety of carbohydrates as carbon sources, since a large number of genes encoding phosphoenolpyruvate (PEP)-phosphotransferase systems (PTS) were identified. Furthermore, all genes encoding the enzymes necessary for the catabolism of glycerol and dihydroxyacetone are present in the L. monocytogenes genome (7, 11). This genomic information is in accord with data from previous and more recent physiological studies (11, 17, 24).
Most genes encoding the enzymes for the major catabolic pathways, namely, glycolysis, the citrate cycle, and the pentose phosphate cycle, are present in L. monocytogenes. The citrate cycle, however, seems to be interrupted, since the genes encoding 2-oxoglutarate dehydrogenase have not been identified in all L. monocytogenes strains sequenced so far, including EGD-e (7), or in Listeria innocua strain Clip 11262. This enzymatic gap in the citrate cycle was recently confirmed by 13C isotopologue perturbation studies using uniformly 13C-labeled glucose. The results showed that two C4 amino acids, aspartate and threonine, are generated in L. monocytogenes, predominantly from building blocks comprising one or three 13C atoms, respectively (2). These data suggested that oxaloacetate, the direct or indirect precursor of both amino acids, is generated by an anaplerotic reaction assembling precursors composed of one and three carbon atoms, respectively. This can be afforded by the carboxylation of pyruvate catalyzed by the ATP-dependent pyruvate carboxylase (PYC) encoded by pycA.
The genes encoding the enzymes for most anabolic pathways, but not those for the biosynthesis of thiamine (vitamin B1), riboflavin (vitamin B2), biotin, and thiotic acid (lipoate), were also identified in L. monocytogenes. However, these bacteria grow efficiently in a mineral salt medium containing a suitable carbon source (e.g., glucose) and these four cofactors only when the amino acids cysteine, methionine, glutamine, arginine, valine, isoleucine, and leucine are also added (17). According to Tsai and Hodgson, strain 10403S requires only methionine and cysteine (24). The missing sulfate reductase in L. monocytogenes readily explains the strict requirement for cysteine/methionine as a sulfur source, while the missing nitrate reductase may be the reason for the stimulatory growth effect of glutamine and arginine as reduced nitrogen sources. However, the need for the three branched-chain amino acids (BCAA) valine, isoleucine, and leucine for efficient growth of L. monocytogenes EGD-e (references 17 and 24 and our unpublished results) is less obvious, since L. monocytogenes has the complete genetic set for synthesis of the BCAA, indicating the role of metabolic intermediates in listerial growth.
The central precursor for the biosynthesis of the BCAA is pyruvate, which is channeled into their biosynthetic pathways either directly, via oxidative decarboxylation of pyruvate to acetyl-coenzyme A (CoA), or more indirectly via oxaloacetate (generated by pyruvate carboxylation) to aspartate and further to threonine. Thus, biosynthesis of the BCAA may compete with the PYC-mediated generation of oxaloacetate for the common substrate pyruvate. These data suggest that PYC may play an important role in the carbon metabolism of L. monocytogenes.
To more precisely determine the significance of this anaplerotic enzyme for listerial metabolism and pathogenesis, we generated a mutant of L. monocytogenes EGD-e defective in pycA, the gene encoding PYC, and studied the replication of this mutant under different extra- and intracellular growth conditions. The results show that PYC indeed plays a crucial role in the intracellular replication of L. monocytogenes and hence in the infection process.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Escherichia coli strain DH5α was used for cloning, and pLSV101 was used as a construction vector for mutagenesis (16). E. coli strains were cultivated in Luria-Bertani (LB) medium at 37°C. The L. monocytogenes wild-type strain EGD-e and the mutant strains were grown under aerobic conditions in brain heart infusion (BHI) or in chemically defined minimal medium (MM) (17) supplemented with different sugars or other carbon sources at 37°C or 42°C. When necessary, the media were supplemented with erythromycin (Sigma, St. Louis, MO) to final concentrations of 5 μg/ml for L. monocytogenes or 300 μg/ml for E. coli. Fresh stock solutions of carbohydrates (glucose or glycerol) or other supplements (adenosine, oxaloacetate, or Casamino Acids [CAA]) were filter sterilized and added to the culture medium at the required final concentrations. To determine growth curves, aliquots were removed at regular intervals, and the optical density (OD) was determined using a spectrophotometer. For shift experiments, overnight cultures of the strains were diluted in fresh BHI, grown to an OD at 600 nm (OD600) of 0.5, and washed once in sterile phosphate-buffered saline (PBS). The sedimented cells were resuspended in MM containing the appropriate supplement(s), and growth was subsequently monitored at 37°C.
For mouse infection experiments, L. monocytogenes strains were grown to the mid-logarithmic phase (OD600 = 1.0) at 37°C in BHI medium, washed two times with endotoxin-free isotonic saline (0.9% NaCl), resuspended in 20% (vol/vol) glycerol in 0.9% NaCl, and stored at −80°C.
General techniques.
PCR amplifications, cloning procedures, isolation of chromosomal DNA, and DNA manipulations were carried out according to standard procedures (18). Cycle sequencing was performed using the CEQ Dye Terminator Cycle Sequencing Quick Start kit (Beckman Coulter), and sequencing reactions were run on an XL2000 Beckman Coulter sequencer. The Listeria home page of the Institut Pasteur (http://genolist.pasteur.fr/ListiList/) and the NCBI database (http://www.ncbi.nlm.nih.gov/) were used for sequence comparisons. All oligonucleotides used for PCR amplification were synthesized by Metabion (Martinsried, Germany).
Disruption of pycA and lmo1073 in L. monocytogenes.
The insertion mutants were constructed by using L. monocytogenes Sv1/2a EGD-e as the parental strain, using a protocol described previously (16), by inserting a pLSV101 recombinant plasmid harboring specific fragments of the pycA or lmo1073 gene into the corresponding gene of L. monocytogenes EGD-e (12, 26). Internal (N-terminal) fragments of 286 bp (pycA) and 239 bp (lmo1073) were PCR amplified from chromosomal DNA derived from L. monocytogenes EGD-e by using the following primer pairs: pycA-BamHI (AAAAAAGGATCCGTAGCAAACCGGC), pycA-EcoRI (AACGAAAAGAATTCCTTCTTGCTCA), lmo1073-BamHI (TGGTTTAAAGGATCCGCCGTAGTT), and lmo1073-EcoRI (TCTGAGCGAATTCTGGATAATCATC) (the restriction endonuclease sites are underlined; boldface indicates deviation from the original sequence). The purified PCR products were digested with the corresponding restriction enzymes and cloned via the restriction sites into pLSV101 to yield the mutagenesis plasmids pLSV101-pycA and pLSV101-lmo1073. These plasmids were transformed by electroporation into L. monocytogenes EGD-e, and insertion mutants (EGD-e/pycA::pLSV101 and EGD-e/lmo1073::pLSV101) were selected by growth on erythromycin at 42°C. Insertional mutagenesis of pycA and lmo1073 was confirmed by PCR and sequencing. The revertant of the pycA insertion mutant (EGD-e/Rev) was obtained by subcultivating the insertion mutant at 30°C without erythromycin. Precise excision of the single plasmid insertion was confirmed by PCR analysis and sequencing.
[U-13C6]glucose incorporation studies.
[U-13C6]glucose was purchased from Campro Scientific (Berlin, Germany). Strains EGD-e and EGD-e/pycA::pLSV101 were grown overnight in BHI at 37°C and 42°C, respectively. The cells were sedimented and washed with PBS. BHI was supplemented with 2.5 mg/ml [U-13C6]glucose, and Erlenmeyer flasks with 100 ml BHI with [U-13C6]glucose were then inoculated with an aliquot of the cell suspensions to an OD600 of 0.1. The cultures were grown at 37°C until OD600 = 0.5 and OD600 = 1.0. Sodium azide was then added to a final concentration of 10 mM. The cells were centrifuged and washed three times with PBS.
Protein hydrolysis and amino acid derivatization.
Bacterial cells (an approximately 10-mg washed wet pellet) were suspended in 0.5 ml of 6 M hydrochloric acid. The mixture was heated at 105°C for 24 h under an inert atmosphere. The hydrolysate was placed on a column of Dowex 50W × 8 (H+ form; 200 to 400 mesh; 5 by 10 mm). The column was washed twice with 750 μl of water and was developed with 1 ml of 2 M ammonium hydroxide. An aliquot of the eluate was dried under a stream of nitrogen, and the residue was dissolved in 50 μl of dry acetonitrile. A total of 50 μl of N-(tert-butyldimethyl-silyl)-N-methyl-trifluoroacetamide containing 1% tert-butyl-dimethyl-silylchloride (Sigma) was added. The mixture was kept at 70°C for 30 min. The resulting mixture of tert-butyl-dimethylsilyl derivatives (TBDMS) of amino acids was used for gas chromatography/mass spectrometry (GC/MS) analysis without further work-up.
Mass spectrometry.
GC/MS analysis was performed on a GCMS-QP2010 Plus Gas Chromatograph/Mass Spectrometer (Shimadzu, Duisburg, Germany) equipped with a fused silica capillary column (Equity TM-5; 30 m by 0.25 mm; 0.25-μm film thickness; Supelco, Bellafonte, PA) working with electron impact ionization at 70 eV. Of a solution containing TBDMS amino acids, 1 μl was injected in 1:10 split mode at an interface temperature of 260°C and a helium inlet pressure of 70 kPa. The column was developed at 150°C for 3 min and then with a temperature gradient of 10°C min−1 to a final temperature of 260°C that was held for 3 min. Data were collected using GCMSsolution version 2.50 SU3 software (Shimadzu). Selected ion-monitoring experiments were carried out. Samples were analyzed at least three times, and data were processed as described previously (3). The excess of multiple labeled isotopologues was calculated according to the formula [2 × (M + 2) + 3 × (M + 3) +… + n × (M + n)]/n, where n is the number of carbon atoms in a given fragment derived from the original amino acid.
[U-14C]aspartate and [3H]oxaloacetate uptake assays.
L. monocytogenes strains EGD-e and EGD-e/pycA::pLSV101 were grown overnight in BHI at 37°C and 42°C, respectively, and subsequently grown in BHI to an OD600 of 0.6. Each culture was then harvested by centrifugation at 5,000 rpm for 3 min at 4°C. The pellet was washed three times in transport buffer (50 mM Tris-HCl, pH 7.2, and 20 mM MgCl2) and resuspended in the same buffer. Labeled l-[U-14C]aspartate (0.1 mCi/ml; Hartmann Analytic, Braunschweig, Germany) or [3H]oxaloacetate was added in a final concentration of 10 mM and incubated at 37°C. Aliquots were retrieved at different time points and filtered rapidly under vacuum through a 0.45-μm-pore-size cellulose nitrate filter (Sartorius, Göttingen, Germany). The filters were washed with 9 ml cold 0.9% NaCl and dried for 20 min at 42°C. Radioactivity was determined using a Perkin-Elmer 1214 Rackbeta liquid scintillation counter. Additionally, the CFU of each sample were determined, and the glucose uptake was calculated for each strain.
Transcriptome analyses.
Transcriptome analyses were performed using whole-genome DNA microarrays as described by Marr et al. (14). Two independently isolated RNA samples from the wild-type L. monocytogenes EGD-e and the pycA insertion mutant grown in BHI to an OD600 of 1.0, were used for the analysis. Each RNA pair was reverse transcribed and hybridized to two microarray slides with dye swap. Another two microarray slides were hybridized using the same principle. In total, four microarray slides to generate 16 replicate expression values were used for further analysis. cDNA labeling and hybridization were performed as described previously (14). The slides were scanned using ScanArray HT and analyzed using Scan-Array express software (Perkin-Elmer, Boston, MA). Spots were flagged and eliminated from analysis when the signal-to-noise ratio was less than 3 or in obvious instances of high background or stray fluorescent signals. The LOWESS method of normalization (27) was performed on the background-corrected median intensity of the spots. The normalized ratios were analyzed further with Microsoft Excel (Microsoft, Redmond, WA) and SAM (significance analysis of microarrays) software for statistical significance (http://www-stat.stanford.edu/∼tibs/SAM/). As described previously (16), genes whose expression values were >2.0 or <0.5 were considered to be differentially regulated. The data discussed in this work are listed in Table 1.
TABLE 1.
Transcriptome analysis of EGD-e/pycA::pLSV101
| Genea | Encoded protein | Relative transcript level |
|---|---|---|
| Downregulated | ||
| lmo0048 | Similar to Staphylococcus two-component sensor histidine kinase AgrB | 0.02 |
| lmo0049 | Unknown | 0.02 |
| lmo0050 | Similar to sensor histidine kinase (AgrC from Staphylococcus) | 0.02 |
| lmo0051 | Similar to 2-component response regulator protein (AgrA from Staphylococcus) | 0.02 |
| lmo0208 | Conserved hypothetical protein | 0.47 |
| lmo0210 (ldh) | Similar to l-lactate dehydrogenase | 0.19 |
| lmo0238 (cysE) | Similar to serine O-acetyltransferase | 0.31 |
| lmo0239 (cysS) | Cysteinyl-tRNA synthetase | 0.33 |
| lmo0240 | Highly similar to B. subtilis YazC protein | 0.35 |
| lmo0241 | Similar to conserved hypothetical proteins like B. subtilis YacO protein | 0.34 |
| lmo0242 | Similar to B. subtilis YacP protein | 0.40 |
| lmo0243 (sigH) | RNA polymerase σ30 factor | 0.44 |
| lmo0283 | Similar to ABC transporter permease protein | 0.34 |
| lmo0284 | Similar to ABC transporter (ATP-binding protein) | 0.38 |
| lmo0285 | Putative lipoprotein | 0.41 |
| lmo0286 | Similar to aminotransferase | 0.34 |
| lmo0302 | Unknown | 0.35 |
| lmo0352 | Highly similar to regulatory proteins (DeoR family) | 0.44 |
| lmo0477 | Putative secreted protein | 0.02 |
| lmo0478 | Putative secreted protein | 0.02 |
| lmo0479 | Putative secreted protein | 0.03 |
| lmo0560d | Similar to NADP-specific glutamate dehydrogenase | 0.30 |
| lmo0641 | Similar to heavy metal-transporting ATPase | 0.25 |
| lmo0642 | Unknown | 0.25 |
| lmo0778 | Unknown | 0.13 |
| lmo0780 | Unknown | 0.44 |
| lmo0788 | Unknown | 0.31 |
| lmo0795 | Conserved hypothetical protein | 0.45 |
| lmo1014 (gbuA)b | Highly similar to glycine betaine ABC transporter (ATP-binding protein) | 0.44 |
| lmo1015 (gbuB)b | Highly similar to glycine betaine ABC transporters (permease) | 0.43 |
| lmo1056 | Unknown | 0.52 |
| lmo1073b | Similar to metal binding protein (ABC transporter) | 0.14 |
| lmo1120 | Unknown | 0.53 |
| lmo1121 | Unknown | 0.55 |
| lmo1227 | Similar to uracil-DNA glycosylase | 0.42 |
| lmo1298 (glnR)e | Similar to glutamine synthetase repressor | 0.15 |
| lmo1299 (glnAe | Highly similar to glutamine synthetases | 0.21 |
| lmo1516d | Similar to ammonium transporter NrgA | 0.10 |
| lmo1517d | Similar to nitrogen regulatory PII protein | 0.16 |
| lmo1634 | Similar to alcohol-acetaldehyde dehydrogenase | 0.11 |
| lmo1983 (ilvD) | Similar to dihydroxy-acid dehydratase | 0.39 |
| lmo1984 (ilvB)b | Similar to acetolactate synthase (acetohydroxy-acid synthase) (large subunit) | 0.49 |
| lmo1985 (ilvN)b | Similar to acetolactate synthase (acetohydroxy-acid synthase) (small subunit) | 0.41 |
| lmo2064 | Similar to large conductance mechanosensitive channel protein | 0.34 |
| lmo2192d | Similar to oligopeptide ABC transporter (ATP-binding protein) | 0.40 |
| lmo2193d | Similar to oligopeptide ABC transporter (ATP-binding protein) | 0.48 |
| lmo2195d | Similar to oligopeptide ABC transporter (permease) | 0.45 |
| lmo2360 | Transmembrane protein | 0.49 |
| lmo2374 | Similar to aspartate kinase | 0.49 |
| lmo2407 | Unknown | 0.53 |
| lmo2410e | Unknown | 0.26 |
| lmo2457 (tpi)d | Highly similar to triose phosphate isomerase | 0.42 |
| lmo2458 (pgk)d | Highly similar to phosphoglycerate kinase | 0.38 |
| lmo2459 (gap)d | Highly similar to glyceraldehyde 3-phosphate dehydrogenase | 0.34 |
| lmo2460d | Similar to B. subtilis CggR hypothetical transcriptional regulator | 0.18 |
| lmo2587 | Conserved hypothetical protein | 0.41 |
| lmo2646 | Unknown | 0.02 |
| lmo2648 | sImilar to phosphotriesterase | 0.02 |
| lmo2753e | Unknown | 0.43 |
| lmo2761 | Similar to β-glucosidase | 0.03 |
| lmo2762 | Similar to PTS cellobiose-specific enzyme IIB | 0.10 |
| lmo2763 | Similar to PTS cellobiose-specific enzyme IIC | 0.12 |
| lmo2764 | Similar to xylose operon regulatory protein and to glucose kinase | 0.12 |
| lmo2768 | Hypothetical membrane protein | 0.45 |
| lmo2785 (kat)e | Catalase | 0.46 |
| lmo2857 | Hypothetical protein | 0.32 |
| Upregulated | ||
| lmo0109b | Similar to transcriptional regulatory proteins, AraC family | 5.14 |
| lmo0110c | Similar to lipase | 3.83 |
| lmo0130d | Similar to 5-nucleotidase, putative peptidoglycan bound protein (LPXTG motif) | 14.16 |
| lmo0135d | Similar to oligopeptide ABC transport system substrate-binding proteins | 3.40 |
| lmo0211 (ctc) | Similar to B. subtilis general stress protein | 2.32 |
| lmo0267 | Similar to other proteins | 1.85 |
| lmo0391b | Unknown | 2.25 |
| lmo0392b | Highly similar to B. subtilis YqfA protein | 2.09 |
| lmo0422b | Similar to unknown protein | 1.95 |
| lmo0514 | Similar to internalin protein, putative peptidoglycan bound protein (LPXTG motif) | 2.26 |
| lmo0530 | Unknown | 2.30 |
| lmo0556b | Similar to phosphoglycerate mutase | 2.31 |
| lmo0715 | Unknown | 2.09 |
| lmo0769 | Similar to α-1,6-mannanase | 3.81 |
| lmo0863 | Unknown | 2.57 |
| lmo0864 | Unknown | 3.56 |
| lmo0865 | Similar to phosphomannomutase | 2.52 |
| lmo0971 (dltD) | DltD protein for d-alanine esterification of lipoteichoic acid and wall teichoic acid | 2.22 |
| lmo0972 (dltC) | d-Alanyl carrier protein | 1.94 |
| lmo0973 (dltB) | DltB protein for d-alanine esterification of lipoteichoic acid and wall teichoic acid | 2.08 |
| lmo0974 (dltA)c | d-Alanine-activating enzyme (Dae), d-alanine-d-alanyl carrier protein ligase (Dcl) | 2.11 |
| lmo1042b | Similar to molybdopterin biosynthesis protein MoeA | 2.60 |
| lmo1043 | Similar to molybdopterin-guanine dinucleotide biosynthesis MobB | 2.56 |
| lmo1044b | Similar to molybdopterin converting factor, subunit 2 | 2.89 |
| lmo1045b | Similar to molybdopterin converting factor, subunit 1 | 2.77 |
| lmo1046b | Similar to molybdenum cofactor biosynthesis protein C | 3.07 |
| lmo1047 | Similar to molybdenum cofactor biosynthesis protein A | 2.37 |
| lmo1048b | Similar to molybdenum cofactor biosynthesis protein B | 1.95 |
| lmo1049 | Similar to molybdopterin biosynthesis protein MoeB | 2.92 |
| lmo1097 | Similar to integrases | 2.81 |
| lmo1142 | Similar to Salmonella enterica PduS protein | 3.73 |
| lmo1143 | Similar to S. enterica PduT protein | 3.56 |
| lmo1144 | Similar to S. enterica PduU protein | 2.29 |
| lmo1145 | Similar to S. enterica PduV protein | 2.37 |
| lmo1146 | Unknown | 2.44 |
| lmo1150 | Regulatory protein similar to S. enterica serovar Typhimurium PocR protein | 7.31 |
| lmo1172b | Similar to two-component response regulator | 10.10 |
| lmo1254d | Similar to α,α-phosphotrehalase | 7.38 |
| lmo1255d | Similar to PTS system trehalose-specific enzyme IIBC | 9.42 |
| lmo1306 | Highly similar to B. subtilis YneF protein | 2.52 |
| lmo1339 | Similar to glucose kinase | 1.99 |
| lmo1349b | Similar to glycine dehydrogenase (decarboxylating) subunit 1 | 4.21 |
| lmo1388 (tcsA)d | CD4+ T-cell-stimulating antigen, lipoprotein | 2.41 |
| lmo1396 | Similar to phosphatidylglycerophosphate synthase | 1.94 |
| lmo1416 | Unknown | 2.83 |
| lmo1425 (opuCD) | Similar to betaine/carnitine/choline ABC transporter | 1.90 |
| lmo1521 | Similar to N-acetylmuramoyl-l-alanine amidase | 2.45 |
| lmo1538b | Similar to glycerol kinase | 4.35 |
| lmo1539b | Similar to glycerol uptake facilitator | 5.31 |
| lmo1648 | Unknown | 2.65 |
| lmo1690 | Similar to hypothetical proteins | 4.01 |
| lmo1879 (cspD)b | Similar to cold shock protein | 11.39 |
| lmo1883b | Similar to chitinases | 8.00 |
| lmo1900 (panD) | Similar to aspartate 1-decarboxylases | 3.86 |
| lmo1901 (panC) | Similar to panthotenate synthetases | 3.68 |
| lmo1902 (panB) | Similar to ketopantoate hydroxymethyltransferases | 5.94 |
| lmo1903 | Similar to thioredoxin | 2.47 |
| lmo1954 (drm) | Similar to phosphopentomutase | 3.92 |
| lmo1955b | Similar to integrase/recombinase | 3.04 |
| lmo1970 | Similar to putative phosphotriesterase-related proteins | 2.78 |
| lmo1972 | Similar to pentitol PTS system enzyme IIB component | 4.53 |
| lmo1974 | Similar to transcription regulators (GntR family) | 2.87 |
| lmo1999b | Weakly similar to glucosamine-fructose-6-phosphate aminotransferase | 2.14 |
| lmo2007 | Weakly similar to putative sugar-binding lipoproteins | 2.10 |
| lmo2084d | Unknown | 2.66 |
| lmo2104 | Unknown | 2.42 |
| lmo2257 | Hypothetical CDS | 16.96 |
| lmo2293d | Protein gp10 (bacteriophage A118) | 6.05 |
| lmo2295d | Protein gp8 (bacteriophage A118) | 7.76 |
| lmo2296d | Similar to coat protein (bacteriophage SPP1) | 13.00 |
| lmo2298d | Protein gp4 (bacteriophage A118) | 12.60 |
| lmo2300 | Putative terminase large subunit (bacteriophage A118) | 6.06 |
| lmo2304 | Protein gp65 (bacteriophage A118) | 3.61 |
| lmo2305 | Unknown | 2.63 |
| lmo2306 | Similar to phage protein | 3.61 |
| lmo2307 | Hypothetical protein | 4.35 |
| lmo2308 | Similar to single-stranded DNA-binding protein | 3.72 |
| lmo2309 | Unknown | 2.58 |
| lmo2310 | Unknown | 3.71 |
| lmo2311 | Unknown | 3.97 |
| lmo2312 | Unknown | 4.29 |
| lmo2315 | Similar to protein gp51 (bacteriophage A118) | 2.92 |
| lmo2322 | Protein gp44 (bacteriophage A118) | 3.20 |
| lmo2335 (fruA) | Highly similar to PTS fructose-specific enzyme IIABC component | 54.79 |
| lmo2336 (fruB) | Fructose-1-phosphate kinase | 98.99 |
| lmo2337 | Similar to regulatory protein DeoR family | 112.00 |
| lmo2343 | Similar to nitrilotriacetate monooxygenase | 2.74 |
| lmo2344 | Similar to B. subtilis YtnI protein | 2.97 |
| lmo2345 | Conserved hypothetical protein | 2.56 |
| lmo2437d | Unknown | 10.25 |
| lmo2522 | Similar to hypothetical cell wall binding protein from B. subtilis | 3.30 |
| lmo2539 (glyA) | Highly similar to glycine hydroxymethyltransferase | 2.20 |
| lmo2590b | Similar to ATP binding proteins | 4.55 |
| lmo2684c | Similar to cellobiose phosphotransferase enzyme IIC component | 18.73 |
| lmo2685c | Similar to cellobiose phosphotransferase enzyme IIA component | 17.18 |
| lmo2708 | Similar to PTS system, cellobiose-specific enzyme IIC | 17.05 |
| lmo2714 | Peptidoglycan-anchored protein (LPXTG motif) | 2.10 |
| lmo2742d | Unknown | 6.51 |
| lmo2743 | Similar to transaldolase | 6.06 |
| lmo2771b | Similar to β-glucosidase | 135.36 |
| lmo2772b | Similar to β-glucoside-specific enzyme IIABC | 207.92 |
| lmo2773b | Similar to transcription antiterminator | 178.03 |
| lmo2786 (bvrC) | Unknown | 2.93 |
| lmo2787 (bvrB)b | β-Glucoside-specific phosphotransferase enzyme IIABC component | 10.06 |
| lmo2788 (bvrA)b | Transcription antiterminator | 4.51 |
| lmo2797b | Similar to phosphotransferase system mannitol-specific enzyme IIA | 5.61 |
| lmo2798b | Similar to phosphatase | 6.56 |
| lmo2851 | Similar to AraC-type regulatory protein | 4.98 |
Genes were identified by microarray analyses as down- or upregulated in EGD-e/pycA::pLSV101 relative to L. monocytogenes EGD-e grown in BHI medium.
Gene or operon whose transcription was shown previously to be controlled by CcpA and Hpr-SerP (16).
Gene or operon whose transcription was shown previously to be controlled by CcpA only (16).
Gene or operon whose transcription was shown previously to be controlled by Hpr-SerP only (16).
Cell infection assays.
Human colon epithelial cells (Caco-2; ACC 169) and mouse monocytes/macrophages (J774A.1; ACC 170) were received from the German Collection of Microorganisms and Cell Cultures (DMSZ) and cultured at 37°C and 5% CO2 in RPMI 1640 medium supplemented with 2 mM l-glutamine (Gibco, Eggenstein, Germany) and 10% heat-inactivated fetal calf serum (FCS) (Biochrom KG, Berlin, Germany). A total of 1 × 105 Caco-2 or J774A.1 cells per well were seeded in 24-well culture plates 24 h prior to infection. The cells were washed twice with phosphate-buffered saline (PBS) supplemented with Mg2+ Ca2+ (PBS-Mg2+ Ca2+) and infected at a multiplicity of infection (MOI) of 10 bacteria per cell for 1 h (Caco-2) or an MOI of 1 for 45 min (J774A.1). The cells were washed with PBS-Mg2+ Ca2+ at time zero (t = 0 h) and incubated with medium containing 50 μg/ml gentamicin, which was replaced with medium containing 10 μg/ml gentamicin after 1 h. Cells were washed and lysed at appropriate time points using cold distilled water, and viable bacterial counts of intracellular bacteria were determined by plating serial dilutions on BHI agar plates.
Infection of mice.
Six- to 12-week-old female C57BL/6 mice were purchased from Harlan Winkelmann GmbH, Borchen, Germany. The animals were housed under specific-pathogen-free conditions at the Biocenter of the University of Würzburg. All animal experiments were approved by the government of Unterfranken and were performed according to the German animal protection guidelines. Groups of five mice were intravenously infected for immunization with 5 × 103 bacteria unless otherwise noted and resuspended in 0.1 ml endotoxin-free 0.9% NaCl. Three days postinfection, the spleens and livers of the mice were harvested for determination of the number of L. monocytogenes bacteria in the lysates of the infected spleens and livers. Counts of viable intracellular bacteria were determined by plating serial dilutions of mechanically lysed cell suspensions on BHI agar.
Microarray data accession number.
The complete microarray data set associated with this study has been deposited in NCBI's Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and is accessible through accession number GSE19014.
RESULTS
Construction and characterization of a pycA insertion mutant of L. monocytogenes EGD-e.
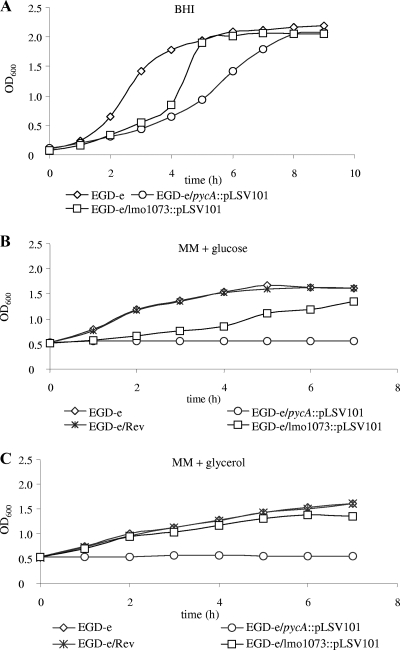
The PYC-mediated synthesis of oxaloacetate appears to be an important anaplerotic reaction in L. monocytogenes leading to C4 metabolites, and hence, the enzyme may play a crucial role in L. monocytogenes carbon metabolism. The present study aimed to analyze the significance of PYC in the listerial metabolism under extra- and intracellular growth conditions and the effect of PYC deficiency on in vivo virulence. For this goal, a pycA insertion mutant (EGD-e/pycA::pLSV101) was constructed. Attempts to construct an in-frame deletion of pycA using a conditionally replicating plasmid (12) failed, probably because the pycA mutant was overgrown by the wild-type strain (see below) in the absence of a selective marker. The pycA insertion mutant grew at a 2- to 3-fold-lower rate than the wild-type strain, EGD-e, when cultured in BHI broth (Fig. 1A) but was completely unable to multiply in a defined minimal medium (17) with glucose or glycerol as a carbon source (Fig. 1B and C).
FIG. 1.
(A) Growth of L. monocytogenes EGD-e, EGD-e/ pycA::pLSV101, and EGD-e/lmo1073::pLSV101 in BHI. (B and C) Growth of L. monocytogenes EGD-e, EGD-e/pycA::pLSV101, the revertant of the pycA insertion mutant (EGD-e/Rev), and EGD-e/lmo1073::pLSV101 in MM supplemented with 50 mM glucose (MM + glucose) or 50 mM glycerol (MM + glycerol). The strains were first grown in BHI medium to an OD600 of 0.5. After centrifugation, the cells were washed twice with MM and then resuspended in MM with the appropriate carbon source. All growth curves are representative of three replicates.
To ensure that the observed phenotype of the pycA mutant was exclusively caused by the inactivation of the pycA gene, we isolated a revertant strain, EGD-e/Rev, that had the pSLV101-pycA insertion precisely excised, as confirmed by DNA sequencing. Spontaneous excision of the insertion occurs in the absence of selection pressure at a rate of approximately 10−7 and restores in most cases the wild-type genotype without mutations (5, 12). Repeatedly isolated revertants grew in all the culture media used at rates similar to that of the wild-type strain (Fig. 1B), ruling out an additional mutation(s) that might be responsible for the failure of the pycA mutant to grow in the defined minimal medium. Since pycA is constitutively expressed under all culture conditions (11), we assumed that the use of the revertant strain as a control was superior to trans complementation by a recombinant pycA-carrying plasmid, which, due to the higher pycA copy number, would probably lead to elevated PYC activity in the complemented pycA mutant with unpredictable consequences for metabolism. Indeed, complementation of the pycA mutant with two recombinant plasmids (pAD123-pycA and pHSP9-pycA) could not be demonstrated by either of these recombinant plasmids when the strains were grown in liquid medium. However, growth of the EGD-e wild-type strain carrying the recombinant pycA plasmids was strikingly inhibited, demonstrating adverse effects on carbon metabolism by both recombinant plasmids, possibly due to an unbalanced PycA concentration or involvement of noncoding RNA (data not shown).
The pycA gene (lmo1072) is the first gene of a transcription unit that contains three additional genes (lmo1073 to lmo1075) located downstream. The insertion of pLSV101 into pycA may therefore cause a polar effect on the expression of these genes. Microarray analysis indeed showed a significantly smaller amount of transcript of the first downstream gene, lmo1073, compared to the wild-type strain (Table 1, note e), while the transcription of the gene lmo1071, located upstream, was unaffected by the insertion in pycA. To test whether the lower expression of lmo1073, encoding a putative metal binding protein of an ABC transporter, may be responsible for the growth defect of the pycA insertion in the defined growth medium, we constructed an insertion mutant of lmo1073 using the same strategy as for pycA. The resulting mutant, EGD-e/lmo1073::pLSV101, grew in BHI at a rate similar to that of the wild-type strain and showed only slight growth inhibition compared to the wild-type strain in the above-described defined minimal medium containing glucose or glycerol (Fig. 1A to C).
These results rule out the possibility that the transcriptional impairment of lmo1073 or of the genes located further downstream, lmo1074 and lmo1075, due to the pLSV101 insertion in pycA is responsible for the growth defect of the pycA insertion mutant, but they suggest that this failure is caused entirely by the inactivation of PYC. We therefore conclude that the PYC-mediated anaplerotic reaction resulting in the production of oxaloacetate is essential during growth of L. monocytogenes in a culture medium where carbon and energy metabolism depends entirely on the carbon source added.
Oxaloacetate cannot be produced in the pycA mutant by alternative reactions.
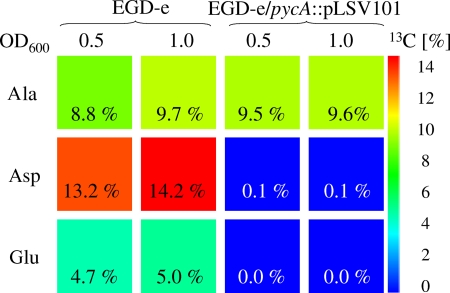
As the pycA mutant was still able to grow in BHI, it might be argued that oxaloacetate is produced in the pycA mutant by an alternative metabolic reaction(s) catalyzed by yet-undefined enzymes that are expressed and/or activated in BHI, thus replacing PYC. To test this hypothesis, the wild-type strain and the pycA mutant were cultured in BHI supplemented with uniformly 13C-labeled glucose ([U-13C6]glucose), and 13C incorporation into protein-derived aspartate, glutamate, and alanine was analyzed by mass spectrometry as described previously (3). As shown in Fig. 2, incorporation of 13C was not detectable in aspartate and glutamate derived from EGD-e/pycA::pLSV101 that was harvested in two different growth phases, namely, at OD600 = 0.5 and 1.0. However, high 13C incorporation into both amino acids of proteins of the wild-type strain cultivated under the same conditions was observed. Oxaloacetate is the direct precursor of the biosynthesis of aspartate and is also needed for the production of oxoglutarate (via the citrate cycle) as the precursor of glutamate. 13C incorporation from [U-13C6]glucose into alanine, used as a control, was similar in the pycA mutant and the wild-type strain (Fig. 2), indicating that the production of the glycolytic intermediate pyruvate, the precursor of alanine, is unaffected by the pycA mutation. The clear difference of 13C enrichment in aspartate and its precursor, oxaloacetate, between the wild-type and the mutant strain shows that the PYC-catalyzed anaplerotic reaction is indeed the predominant reaction leading to oxaloacetate production in L. monocytogenes.
FIG. 2.
13C excess (percent) from [U-13C6]glucose of multiply labeled isotopologues of alanine (Ala), aspartate (Asp), and glutamate (Glu) derived from bacterial protein. The color map indicates 13C excess on a linear scale.
Growth of the pycA mutant in defined minimal medium cannot be restored by supplementation with oxaloacetate or aspartate.
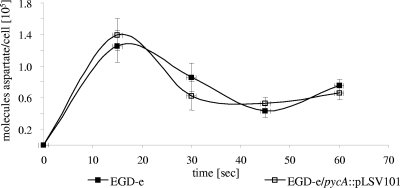
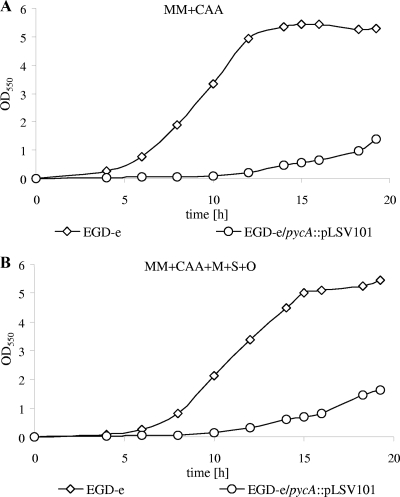
Several growth and transport experiments were performed with the wild-type strain and the pycA mutant. Addition of oxaloacetate or aspartate at concentrations between 1 and 10 mM to the glucose- or glycerol-containing minimal medium could not restore growth of the pycA mutant. External oxaloacetate could not be transported into the listerial cell, as shown by the lack of uptake of 3H-labeled oxaloacetate by either the wild-type strain or the pycA mutant (data not shown). This was expected, as the L. monocytogenes genome apparently lacks a gene(s) for oxaloacetate (and other C4 dicarboxylate) permease(s) (7). In contrast, 14C-labeled aspartate is taken up by the pycA mutant at a rate similar to that by the wild-type strain (Fig. 3) but is apparently not efficiently converted into oxaloacetate. The addition of 1% CAA, which contain all amino acids except tryptophan, or of CAA and malate, succinate, and oxaloacetate to the minimal medium only partially restored the growth of the pycA mutant (Fig. 4A and B). This indicates that the lack not only of synthesis of the amino acids belonging to the aspartate family, but also of other reactions involving oxaloacetate (e.g., the conversion to other C4 dicarboxylates) may be responsible for the poor growth of the pycA mutant under these conditions.
FIG. 3.
[U-14C]aspartate uptake rate in wild-type L. monocytogenes EGD-e and EGD-e/pycA::pLSV101. The strains were pregrown in BHI to an OD600 of 0.6. The y axis indicates the number of aspartate molecules taken up per bacterial cell. The aspartate uptake measurements were performed in triplicate, and the error bars indicate the standard deviations from the means.
FIG. 4.
Strains EGD-e and EGD-e/pycA::pLSV101 were grown overnight in BHI and diluted 1:400 in MM containing 1% CAA (A) and 1% CAA, 1 g/liter malate, 1 g/liter succinate, and 1 g/liter oxaloacetate (B). Bacterial growth was monitored at OD550 for 20 h. The average values of three independent experiments are shown.
Comparative transcript profiles of the pycA mutant and the wild-type strain.
The supply of carbon substrates present in BHI is obviously richer than that in the defined medium and allows rather efficient growth of the pycA mutant. To find out which metabolic pathway(s) may enable the pycA mutant to grow in BHI, comparative transcript profiling of the pycA mutant and the EGD-e wild-type strain was performed. For this purpose, RNAs of the two strains, grown in BHI to an OD600 of 1.0, where both strains were still in the logarithmic growth phase (Fig. 1), was used. In total, 174 genes were found to be differentially expressed (Table 1), 108 were significantly upregulated (>2-fold), and 66 were downregulated (<0.5-fold) in the pycA mutant in comparison to strain EGD-e.
Among the downregulated genes, the most pronounced were those encoding the two-component AgrAB(C) system and the major transcription regulator of glycolysis, CggR. Concomitantly with the reduced level of CggR, the gap operon encoding enzymes of the glycolysis pathway, especially tpi, pgk, and gap, was also downregulated (downregulation of eno and pgm was less reproducible), suggesting reduced glucose catabolism in the pycA mutant.
In line with this assumption, most of the differentially regulated genes of the pycA mutant marked in Table 1 are under positive or negative glucose control by CcpA-dependent and -independent mechanisms in L. monocytogenes and Bacillus subtilis (6, 14). Exceptions among the downregulated genes include the aspartate kinase gene, an operon for a cellobiose-specific PTS, the agrABC operon (lmo0048 to lmo0051), and lmo0477 to lmo0479. The function of the L. monocytogenes-specific genes is presently unknown. Among the upregulated genes whose expression does not seem to be under direct glucose control are several genes with unknown functions, lmo2335 to lmo2337 (fruRAB), encoding a PTSFru; panBCD (lmo1900 to lmo1902), involved in panthotenate biosynthesis; lmo0769 and lmo0865, encoding α-1,6-mannanase and phosphomannomutase, respectively; and several genes involved in cell wall biosynthesis, such as the dlt operon, lmo1521, and lmo2522. The upregulation of these genes may point to a special need for the encoded pathways and enzymes when the pycA mutant grows in BHI.
Many of the glucose-controlled genes whose transcription is significantly upregulated in the pycA mutant encode reactions and pathways involved in carbon metabolism. These genes include lmo0135, encoding a component of a putative oligopeptide ABC transporter; lmo1538 and lmo1539, encoding a glycerol kinase and a glycerol uptake facilitator essential for glycerol metabolism; the operons lmo1042 to lmo1049 and lmo1144 to lmo1150, involved in vitamin B12 synthesis and the vitamin B12-dependent propanediol utilization (Pdu) pathway, respectively; and lmo1349, encoding a glycine dehydrogenase. High upregulation is observed for the operons encoding several PTS permeases, lmo1255, lmo2683 to lmo2685, lmo2771 to lmo2773, and lmo2786 to lmo2788 (bvrCBA). In L. monocytogenes, the expression of these PTS operons appears to be regulated by PTS regulation domain-containing antiterminators and activators (22a) that may be activated by low glucose concentrations (23).
The pycA mutant is taken up by mammalian cells but is unable to replicate in these host cells, and its virulence is attenuated in the mouse.
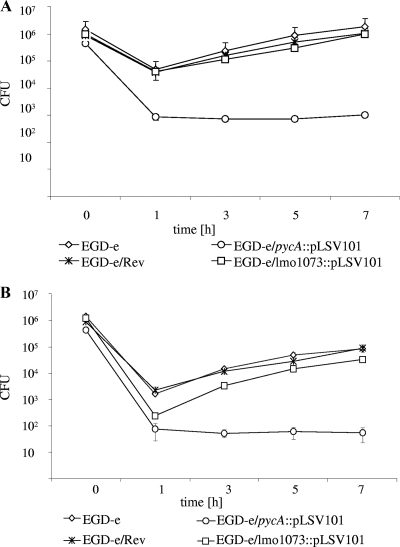
To test whether the pycA mutant encounters within mammalian host cells growth conditions similar to those in rich medium (BHI) or in the nutrient-limited defined culture media, we determined the intracellular replication of the pycA mutant in the macrophage-like J774A.1 cells and in the epitheloid Caco-2 cells (Fig. 5 A and B). The pycA mutant was taken up by both cell types with efficiencies that were about 5-fold lower in the macrophages and almost 100-fold lower in the Caco-2 cells than those of the EGD-e wild-type strain. However, subsequent replication of the pycA mutant in the host cells did not occur, indicating that the host cell is unable to compensate for the lack of PYC-mediated oxaloacetate production of the pycA mutant. The pycA revertant strain EGD-e/Rev was again able to replicate in the cytosol with efficiency similar to that of the wild-type strain. Mutant EGD-e/lmo1073::pLSV101, used as a control, could still replicate in both host cell types at only a slightly lower rate (Fig. 5A and B). These data clearly show that the PYC-catalyzed production of oxaloacetate is also indispensable for the intracellular replication of L. monocytogenes.
FIG. 5.
Effect of the pLSV101 insertion in the pycA gene on the intracellular replication of L. monocytogenes. J774.A1 (A) and Caco-2 (B) cells were infected with either EGD-e, the pycA insertion mutant (EGD-e/pycA::pLSV101), or the revertant of the pycA insertion mutant (EGD-e/Rev), and the numbers of bacteria recovered at various time points (t = 0 h, t = 1 h, t = 3 h, t = 5 h, and t = 7 h) after infection were determined as described in Materials and Methods. Three independent infections were performed for each strain, and the error bars indicate the standard deviations from the means.
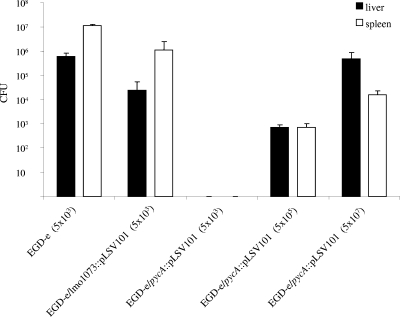
To determine whether and to what extent the virulence of the pycA mutant is attenuated in the mouse sepsis model, 5 × 103 EGD-e/pycA::pLSV101 cells were injected into the tail veins of C57BL/6 mice. As controls, an equal number of EGD-e and mutant EGD-e/lmo1073::pLSV101 bacteria were injected in parallel into C57BL/6 mice. Counts of viable bacteria were determined in the livers and spleens 3 days after intravenous infection. As shown in Fig. 6, the wild-type and EGD-e/lmo1073::pLSV101 strains multiplied up to 106 and 107 viable counts in the livers and spleens, respectively, 3 days postinfection, as reported previously (15), whereas no counts of viable EGD-e/pycA::pLSV101 bacteria could be detected in either the livers or spleens of the infected mice. This result shows that the virulence of the pycA-deficient mutant is highly attenuated, and it is apparently unable to reach these organs. Mice infected with up to 5 × 107 EGD-e/pycA::pLSV101 cells showed still lower counts of viable bacteria in livers and spleens than mice infected with a number of wild-type bacteria 4 orders of magnitude lower. Even with this high dose of intravenously injected pycA mutant bacteria, the infected mice not only survived, but showed no sign of disease.
FIG. 6.
Viable bacterial counts in spleens and livers of C57BL/6 mice 72 h after intravenous infection with 5 × 103 CFU of L. monocytogenes EGD-e and the lmo1073 insertion mutant (EGD-e/lmo1073::pLSV101) or with 5 × 103, 5 × 105, and 5 × 107 CFU of EGD-e/pycA::pLSV101. Each bar represents the infection experiments on a group of five animals, and the error bars indicate the standard deviations from the means.
DISCUSSION
Oxaloacetate plays a central role in the bacterial carbon metabolism (19). It acts as a direct precursor for the biosyntheses of aspartate/asparagine, lysine, threonine, and β-alanine (pantothenate) and indirectly for the biosyntheses of arginine, histidine, purine, nicotinate, and nicotinamide. As an acceptor for acetyl-CoA, it coinitiates the citrate cycle and hence is essential for the generation of additional important intermediates, such as succinyl-CoA and 2-oxoglutarate, the precursor of glutamate/glutamine, proline, and arginine. In most heterotrophic bacteria, including pathogens, the generation of oxaloacetate is safeguarded by the presence of several reactions. In addition to the (complete) citrate cycle, several anaplerotic reactions can produce oxaloacetate, including pyruvate and PEP carboxylases, as well as the often reversible PEP carboxykinase.
For example, B. subtilis, phylogenetically related to L. monocytogenes, possesses a complete citrate cycle and, in addition, PYC and a bifunctional PEP carboxykinase, which ensures the robustness of mutations around the pyruvate node (28). In contrast, L. monocytogenes is characterized by an interrupted citrate cycle that is unable to generate oxaloacetate (2), and only a pycA gene (lmo1072) encoding PYC, but no pckA gene for a PEP carboxykinase, was identified in the L. monocytogenes genome (7).
As shown in this study, PYC-mediated pyruvate carboxylation is the predominant reaction providing oxaloacetate when L. monocytogenes grows on a single carbon substrate that serves as the source for energy and all catabolic intermediates. This is a rather unusual situation even among intracellular bacterial pathogens, all of which, with the exception of Chlamydia, produce oxaloacetate via a complete citrate cycle and, in addition, by PYC and/or PEP carboxylase.
No major alternative reaction leading to oxaloacetate besides pyruvate carboxylation seems to exist in L. monocytogenes.
The pycA mutant characterized in this study was obtained by the insertion of an erythromycin resistance (Emr) plasmid into the pycA gene. Our data show that the growth failure of this mutant is caused by the inactivated PYC and not by polar effects on the genes located up- or downstream. Accidental mutations in the pycA mutant affecting its growth can also be excluded, since a revertant strain with the insertion precisely excised grew in glucose-containing defined medium at a rate comparable to that of the wild-type strain.
The lack of 13C incorporation into aspartate and glutamate in the pycA mutant growing in BHI in the presence of uniformly labeled [13C]glucose shows that there is no induction of an alternative oxaloacetate-generating reaction in the mutant. The recently reported predominant generation of 13C3-labeled aspartate in L. monocytogenes growing in J774 macrophages in the presence of uniformly labeled [13C]glucose is also in line with this conclusion (3).
The failure to restore the growth of the pycA mutant by addition of oxaloacetate to the defined minimal medium is apparently due to the lack of a transporter for oxaloacetate. The L. monocytogenes genome sequence (7) indeed lacks the genes for either a DctA homologue, known to transport C4 dicarboxylates in several Gram-positive and Gram-negative bacteria (9), or a CitM homologue, which, besides citrate, can also transport C4 dicarboxylates (7, 25).
Aspartate, on the other hand, is efficiently taken up by the pycA mutant, probably by the gene products of lmo0847 and lmo1740, encoding putative GltK-homologous ABC transporters for glutamate/aspartate. Nevertheless, even an excess of aspartate cannot restore the growth defect of the pycA mutant in minimal medium. Aspartate can be converted to oxaloacetate by either aspartate aminotransferase or aspartate oxidase. The genes for both enzymes, aspB (lmo1897) and nadB (lmo2023), have been identified in the L. monocytogenes genome (7). Aspartate aminotransferase uses mainly 2-oxoglutarate as an acceptor of the amino group. The cellular concentration of 2-oxoglutarate in the pycA mutant, however, is very low, as indicated by the lack of 13C incorporation into glutamate in the presence of [U-13C6]glucose. Aspartate oxidase, on the other hand, converts aspartate into oxaloacetate and ammonia in the presence of oxygen but generates hydrogen superoxide in the reaction, which might be too toxic for the listerial cell when produced in larger amounts. Thus, the generation of sufficient amounts of oxaloacetate from aspartate is apparently not possible in the pycA mutant.
Since even the addition of Casamino Acids, which provide all amino acids of the aspartate and glutamate families, cannot restore growth of the pycA mutant in the defined minimal medium, the shortage of oxaloacetate in the pycA mutant apparently leads to still other metabolic bottlenecks. One of them might be the insufficient production of other C4 dicarboxylic acids, such as succinyl-CoA, which is needed for several metabolic reactions (19).
Metabolic adaptations of the mutant possibly encompass the lack of PYC when growing in BHI.
The pycA mutant is able to grow (albeit more slowly than the wild-type strain) in BHI, a poorly defined rich culture medium. The transcript profiles of the two strains reveal significant changes in the metabolism of the pycA mutant compared to the wild-type strain. However, these data do not offer ready explanations for the ability of the mutant to grow in BHI but rather indicate that this ability is due to complex metabolic adaptation processes.
As shown in Table 1, the glucose metabolism in the pycA mutant is apparently reduced, as indicated by the downregulation of the glycolysis genes and the differential expression of many genes and operons that are activated or repressed by CcpA-dependent and -independent controls mediated by the cellular glucose concentrations in B. subtilis and L. monocytogenes (6, 16). This partial relief of these glucose-mediated gene control mechanisms could be a direct consequence of the pycA mutation, but the possibility that it is simply caused by the slower growth of the mutant in BHI cannot be excluded.
Upregulated genes and operons that seem to be differently controlled include the fruRBA operon (lmo2335 to lmo2337), the pdu genes (lmo1142 to lmo1150), and the panBCD genes (lmo1900 to lmo1902), as well as lmo0769 and lmo0865, encoding α-1,6-mannanase and phosphomannomutase (7). These genes are involved in glucose-independent carbon metabolism, and their upregulation suggests that the pycA mutant may utilize carbohydrates, such as fructose and/or mannose. These carbohydrates may derive from glycoproteins present in BHI.
The upregulation of the panBCD genes further suggests increased pantothenate production essential for the formation of coenzyme A. This cofactor is required for the synthesis of acetyl-CoA via oxidative decarboxylation of pyruvate by pyruvate dehydrogenase, a reaction that may be favored in the absence of PYC. An increased amount of acetyl-CoA may enhance the production of 2-oxoglutarate at low oxaloacetate concentrations, thereby stimulating the conversion of aspartate to oxaloacetate by aspartate aminotransferase (see above).
The upregulation of the genes encoding the enzymes for the complex, B12-dependent propandiol utilization (Pdu) suggests that the pycA mutant possibly utilizes still other carbon substrates to overcome the lack of oxaloacetate production.
The impact of PYC on virulence.
Our data show that the pycA mutant is taken up by macrophages at a rate similar to that of the wild-type strain. In contrast, the internalin-triggered internalization of the mutant by Caco-2 cells is about 100-fold lower than that of the wild-type strain. The internalization is carried out in RPMI medium, where the pycA mutant is unable to grow. The decreased internalization rate could therefore be due to altered surface structures of the pycA mutant. Alternatively, the internalin-triggered phagocytosis may require metabolically active listeriae, while the uptake by professional phagocytes may occur with metabolically inactive bacteria. In both host cell types, however, the pycA mutant is unable to replicate after being taken up, indicating that the intracellular milieu of the host cells—other than BHI—cannot compensate for the metabolic defect of the pycA mutation. As expected from the inability of the pycA mutant to replicate within mammalian cells, its virulence is also highly attenuated in the mouse sepsis model.
Due to the important role that PYC plays in the intracellular listerial metabolism and in systemic infections, the enzyme may represent an interesting metabolic target for the screening of antilisterial drugs.
Acknowledgments
Victor Weidmann is acknowledged for performing mouse infection assays. We thank Sascha Stoll for helpful discussions and for critically reading the manuscript and Stephanie Grubmüller for help in the preparation of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB479-B1, Go-168/27-3, and EI-384/6-1) and its priority program SPP1316 (EI-384/6 and FU-375/5), the Network of Excellence/EuroPathoGenomics, and the Fonds der chemischen Industrie.
Footnotes
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 2.Eisenreich, W., J. Slaghuis, R. Laupitz, J. Bussemer, J. Stritzker, C. Schwarz, R. Schwarz, T. Dandekar, W. Goebel, and A. Bacher. 2006. 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U. S. A. 103:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eylert, E., J. Schär, S. Mertins, R. Stoll, A. Bacher, W. Goebel, and W. Eisenreich. 2008. Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69:1008-1017. [DOI] [PubMed] [Google Scholar]
- 4.Freitag, N. E. 2006. From hot dogs to host cells: how the bacterial pathogen Listeria monocytogenes regulates virulence gene expression. Future Microbiol. 1:89-101. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs, T. M., J. Klumpp, and K. Przybilla. 2006. Insertion-duplication mutagenesis of Salmonella enterica and related species using a novel thermosensitive vector. Plasmid 55:39-49. [DOI] [PubMed] [Google Scholar]
- 6.Fujita, Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73:245-259. [DOI] [PubMed] [Google Scholar]
- 7.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 8.Hamon, M., H. Bierne, and P. Cossart. 2006. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4:423-434. [DOI] [PubMed] [Google Scholar]
- 9.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 10.Joseph, B., and W. Goebel. 2007. Life of Listeria monocytogenes in the host cells' cytosol. Microbes Infect. 9:1188-1195. [DOI] [PubMed] [Google Scholar]
- 11.Joseph, B., S. Mertins, R. Stoll, J. Schar, K. R. Umesha, Q. Luo, S. Muller-Altrock, and W. Goebel. 2008. Glycerol-metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190:5412-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph, B., K. Przybilla, C. Stühler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lecuit, M. 2005. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin. Microbiol. Infect. 11:430-436. [DOI] [PubMed] [Google Scholar]
- 14.Marr, A. K., B. Joseph, S. Mertins, R. Ecke, S. Müller-Altrock, and W. Goebel. 2006. Overexpression of PrfA leads to growth inhibition of Listeria monocytogenes in glucose-containing culture media by interfering with glucose uptake. J. Bacteriol. 188:3887-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauder, N., R. Ecke, S. Mertins, D. I. Loeffler, G. Seidel, M. Sprehe, W. Hillen, W. Goebel, and S. Müller-Altrock. 2006. Species-specific differences in the activity of PrfA, the key regulator of listerial virulence genes. J. Bacteriol. 188:7941-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertins, S., B. Joseph, M. Goetz, R. Ecke, G. Seidel, M. Sprehe, W. Hillen, W. Goebel, and S. Müller-Altrock. 2007. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J. Bacteriol. 189:473-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Premaratne, R. J., W. J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765-794. [DOI] [PubMed] [Google Scholar]
- 20.Schnupf, P., and D. A. Portnoy. 2007. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 9:1176-1187. [DOI] [PubMed] [Google Scholar]
- 21.Scortti, M., H. J. Monzo, L. Lacharme-Lora, D. A. Lewis, and J. A. Vazquez-Boland. 2007. The PrfA virulence regulon. Microbes Infect. 9:1196-1207. [DOI] [PubMed] [Google Scholar]
- 22.Seveau, S., T. N. Tham, B. Payrastre, A. D. Hoppe, J. A. Swanson, and P. Cossart. 2007. A FRET analysis to unravel the role of cholesterol in Rac1 and PI 3-kinase activation in the InlB/Met signalling pathway. Cell. Microbiol. 9:790-803. [DOI] [PubMed] [Google Scholar]
- 22a.Stoll, R., and W. Goebel. 7 January 2010. Identification of the major PEP-phosphotransferase systems (PTS) for glucose, mannose and cellobiose of Listeria monocytogenes and their significance for extra- and intracellular growth. Microbiology [Epub ahead of print.] doi: 10.1099/mic.0.034934-0. [DOI] [PubMed]
- 23.Stülke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD-a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 24.Tsai, H. N., and D. A. Hodgson. 2003. Development of a synthetic minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 69:6943-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warner, J. B., and J. S. Lolkema. 2002. Growth of Bacillus subtilis on citrate and isocitrate is supported by the Mg2+-citrate transporter CitM. Microbiology 148:3405-3412. [DOI] [PubMed] [Google Scholar]
- 26.Wuenscher, M. D., S. Kohler, W. Goebel, and T. Chakraborty. 1991. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol. Gen. Genet. 228:177-182. [DOI] [PubMed] [Google Scholar]
- 27.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamboni, N., H. Maaheimo, T. Szyperski, H. P. Hohmann, and U. Sauer. 2004. The phosphoenolpyruvate carboxykinase also catalyzes C3 carboxylation at the interface of glycolysis and the TCA cycle of Bacillus subtilis. Metab. Eng. 6:277-284. [DOI] [PubMed] [Google Scholar]