Abstract
Pseudomonas aeruginosa produces three different types of bacteriocins: the soluble S-pyocins and the bacteriophage-like F- and R-pyocins. R-pyocins kill susceptible bacteria of the same or closely related species with high efficiency. Five different types of R-pyocins (R1- to R5-pyocins) have been described based on their killing spectra and tail fiber protein sequences. We analyzed the distribution of R-pyocin genes in a collection of clinical P. aeruginosa isolates. We found similar percentages of isolates not containing R-pyocins (28%) and isolates containing genes encoding R1-pyocins (25%), R2-pyocins (17%), and R5-pyocins (29%). The R-pyocin-deficient isolates were susceptible to R1-, R2-, and R5-pyocins, while most R2- and R5- pyocin producers were resistant. Determination of the O serotypes revealed that the R-pyocin-susceptible isolates belonged to serotypes O1, O3, and O6, while the R-pyocin-resistant isolates were serotype O10, O11, and O12 isolates. We hypothesized that O-serotype-specific lipopolysaccharide (LPS) packaging densities may account for the distinct accessibilities of R-pyocins to their receptors at the cell surface. Using genetically defined LPS mutants, we showed that the l-Rha residue and two distinct d-Glc residues of the outer core are part of the receptor sites for R1-, R2-, and R5-pyocins, respectively. To illustrate R-pyocin-mediated intraspecies biological warfare, we monitored the population dynamics of two different R-pyocin-producing P. aeruginosa clones of sequential respiratory isolates obtained from a colonized patient. The results of this study highlight the potential role of R-pyocins in shaping bacterial populations during host colonization and support use of these molecules as specific and potent bactericidal agents.
Many bacterial species produce bacteriocins, which are proteineous compounds that are able to kill cells of members of the same or closely related species (21). Pyocins, the bacteriocins produced by Pseudomonas aeruginosa, can be classified into three different families: the soluble S-pyocins and the high-molecular-weight F- and R-pyocins. S-pyocins (18) are similar to colicins of Escherichia coli and cause cell death through their endonuclease or pore-forming activities (16). In contrast, the flexible F-pyocins and the rod-shaped R-pyocins are genetically and morphologically related to lambda and P2 bacteriophages, respectively (17). However, unlike bacteriophages, these pyocins lack a phage head structure, do not contain DNA, and are therefore not replicative. R-pyocins kill susceptible bacteria by binding to the cell surface, contracting their sheath, and inserting their core structure through the cell envelope, which results in target cell lysis due to depolarization of the cytoplasmic membrane (26). R-pyocins kill with high efficiency (one pyocin molecule kills one bacterial cell), while for the flexible F-pyocins 100 to 200 molecules are required to kill one cell (16).
The expression of R-, F-, and S-pyocins is positively and negatively regulated by the PrtN and PrtR proteins, respectively, which are activated when a bacterial cell is exposed to DNA-damaging agents (14).
Recently, there has been interest in R-pyocins as specific bactericidal agents. Their specificity, mediated by the tail fiber structure, has been exploited to construct novel R-pyocins with different target spectra (29). The therapeutic efficacy of engineered R-pyocins has been demonstrated in vivo in a mouse model of P. aeruginosa peritonitis (24) and exogenously by using food products contaminated by E. coli O157:H7 (23).
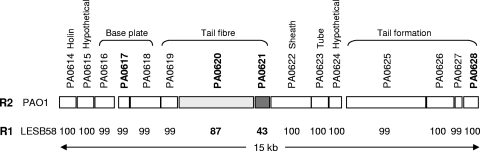
Five different types of R-pyocins have been described based on their killing activities (10) and, more recently, based on a comparison of the amino acid sequences of the tail fiber protein Prf15 (PA0621 of PAO1) (29). While the amino acid sequences of the tail fiber proteins of the R2-, R3-, and R4-pyocins are nearly identical, the amino acid sequences of the R1- and R5-pyocins differ considerably in the C-terminal region of the Prf15 protein (29).
It has been proposed that the lipopolysaccharide (LPS) core contains the R-pyocin-specific recognition sites (15). Purified LPS molecules have also been shown to bind pyocin molecules. The pyocin-LPS interaction has been exploited as an epidemiological typing method to characterize clinical P. aeruginosa strains (3, 5). The precise molecular determinants responsible for the specific R-pyocin-LPS interactions, however, have not been characterized. In this study, we examined the R-pyocin profiles of P. aeruginosa isolates obtained from tracheal aspirates of intubated patients hospitalized in European intensive care units. We compared these profiles with the O serotypes, as defined by the variable B-band oligosaccharide chain of the LPS. Based on the data obtained, we propose an R-pyocin type- and O-serotype-specific killing profile. We also suggest structural determinants required for R-pyocin type-specific pyocin recognition. Our data suggest that LPS plays an essential role both as a protective shield and as a receptor for R-pyocins.
MATERIALS AND METHODS
Bacterial strains and media.
PAO1 (serotype O5), PA14 (serotype O10), and PAK (serotype O6) were used as reference strains (Table 1). We collected clinical isolates daily from 61 colonized intubated patients hospitalized in 17 European centers (11; T. Köhler, R. Guanella, J. Carlet, and C. van Delden, submitted for publication). We obtained approval for this multicenter European study (ANB 006#2001, ClinicalTrials.govID#NCT00610623) from the appropriate local ethics committees and national agencies, as well as written consent from all patients or their legal representatives according to legal and ethical requirements. The first isolate from each patient was used in this study. Isolates were grown in Luria-Bertani (LB) medium at 37°C with agitation (240 rpm).
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant characteristics | Source and/or reference |
|---|---|---|
| Strains | ||
| PAK | R1-pyocin producer, LPS A+ LPS B+ | J. Lam |
| PAO1 | R2-pyocin producer | Laboratory collection |
| PA14 | R2-pyocin producer | 6 |
| 15108/-1 | R5-pyocin producer, clinical isolate | 11 |
| PAKΔR1 | ΔPA0620-PA0621 in R1-pyocin region | This study |
| PA14ΔR2 | ΔPA0620-PA0621 in R2-pyocin region | This study |
| 15108/-1ΔR5 | ΔPA0620-PA0621 in R5-pyocin region | This study |
| PAKwbpP | LPS A+ LPS B− | J. Lam (1) |
| PAKwbpO | LPS A+ LPS B− | J. Lam (1) |
| PAKwbpL | LPS A− LPS B− | J. Lam (1) |
| PAKrmlC | LPS A− LPS B−l-Rha− | J. Lam (20) |
| PAO1wbpM | LPS A+ LPS B− | J. Lam (2) |
| PAO1wbpL | LPS A− LPS B− | J. Lam (22) |
| PAO1rmlC | LPS A− LPS B−l-Rha− | J. Lam (20) |
| PAO1rmd | LPS A− LPS B+ | J. Lam (22) |
| AK1012 | PAO1algC, LPS A− LPS B−, core deficient | 9 |
| Plasmids | ||
| pEX18T | Suicide vector, Tetr | 8 |
| pRK2013 | RK2, helper plasmid | 4 |
| pΔR2T42 | R2 pyocin region of PA14 harboring a 2,285-bp deletion encompassing PA0620 and PA0621 | This study |
| Primers | ||
| PA0620 R5-F | 5′-CGATACAGGACTCATCGTGCAG-3′ | This study |
| PA0620 R5-R | 5′-ACTCGGTAGCGACAATCACACA-3′ | This study |
| PA0621-L | 5′-CCTCCTTGCGAGAGGTTTATGA-3′ | This study |
| PA0621-R | 5′-ATCTCGATGGCGAACAACTAGG-3′ | This study |
| PA0617-L | 5′-CGGCCTGGCTCATCTTAAACA-3′ | This study |
| PA0617-R | 5′-ACTCGCACGGCAGACAATCC-3′ | This study |
| PA0628-L | 5′-CTTCGACGTTACCTTCGACGAC-3′ | This study |
| PA0628-R | 5′-CTCTCCACGGCTGGTATAGAGG-3′ | This study |
| PA0620 R1-F | 5′-GCGCACATGCAGACCATATCTA-3′ | This study |
| PA0620 R1-R | 5′-GGTATCGTGGTTATGCCCTGAG-3′ | This study |
| PA0619-FD | 5′-CGCTTCGTGAGCTTAACGATC-3′ | This study |
| PA0622-RD | 5′-GCAGGCCAGGTAGATAGACGAG-3′ | This study |
PCR assay.
Primers used for amplification of pyocin genes were generated using the primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and are listed in Table 1. Bacterial cell lysates were used as DNA templates for all PCR amplifications. The PCR conditions were as follows: denaturation at 95°C for 2 min, followed by 27 cycles of 95°C for 20 s, 57°C for 30 s, and 72°C for 1 min and a final extension at 72°C for 4 min. R-pyocin genes were detected using multiplex PCR assay mixtures containing primer pairs for PA0617, PA0621, and PA0628 (Fig. 1). The primer sequences used for specific detection of R1-, R2- (R3- and R4-), and R5-pyocin genes (Table 1) were based on the DNA sequences of the tail fiber gene (29) corresponding to open reading frame (ORF) PA0620 of the PAO1 genome (25).
FIG. 1.
Amino acid sequence analysis of R-pyocins encoded by operons of strains PAO1 and LESB58. Amino acid sequences were retrieved and compared by BLAST using the Pseudomonas website (http://pseudomonas.com/). The numbers indicate the percentages of amino acid identity between PAO1 (R2-pyocin) and LESB58 (R1-pyocin) proteins. Genes used in pyocin screening are indicated by bold type. Functions of individual proteins are indicated.
Construction of R-pyocin mutants.
Mutants with the tail fiber gene and the adjacent chaperone gene (PA0620 and PA0621 of PAO1) of the R-pyocin operon deleted were constructed by homologous recombination using plasmid pΔR2T42; a 4,068-bp PCR fragment was generated from PA14 cell lysates using primers PA0617L and PAO621-R. This fragment was digested with HindIII and BamHI, and a 2-kb DNA fragment was ligated into HindIII-BamHI-cleaved suicide vector pEX18T (8) to obtain plasmid pΔR2T4. A second, 6,773-bp PCR fragment was amplified using primers PA0621-L and PA0628-R and digested with KpnI and BglII. The resulting 3-kb DNA fragment was cloned into KpnI and BamHI-cleaved plasmid pΔR2T4 to obtain the final construct, pΔR2T42. This plasmid has a 2,285-bp deletion between the BamHI site in PA0620 and the BglII site in PA0621 (PA14 sequence). To generate R-pyocin-deficient mutants, plasmid pΔR2T42 was subsequently transferred by triparental mating, using pRK2013 as a helper plasmid, into P. aeruginosa recipient strains. Single recombinants were first selected on LB medium containing tetracycline (75 mg/liter) and ampicillin (50 mg/liter), to counterselect against E. coli. Individual colonies were then grown in LB broth without antibiotics for 18 h, and dilutions were spread on LB medium supplemented with 5% sucrose. Three clones were streaked on LB-sucrose medium and then screened by PCR for the presence of the expected DNA deletion using primers PA0619-FD and PA0622-RD.
Bacteriophage and pyocin susceptibility assays.
Strains were grown for 20 h in 2 ml LB medium, and cultures were centrifuged at 14,000 rpm for 5 min. To kill residual bacterial cells, chloroform (5%) was added to each supernatant. The resulting supernatant was used as a source of R-pyocins. The indicator strains used for pyocin susceptibility assays were grown overnight (16 to 18 h) in 2 ml LB medium and diluted to obtain an optical density at 600 nm (OD600) of 0.64 in 0.9% NaCl. Three microliters of each suspension was added to 3 ml top agar (0.4% LB agar) and poured onto fresh LB medium plates. In the spot assay, 4 μl of supernatant was deposited on the top agar, and the plates were incubated for 18 h at 30°C unless otherwise stated. Plaque formation was considered to be the result of lysis by bacteriophages, and clear lysis zones were considered to be results of R-pyocin activity. Susceptibility assays with PAK (R1-pyocin), PA14 (R2-pyocin), and 15108/-1 (R5-pyocin) supernatants, whose results are shown in Table 2, were performed in parallel with susceptibility assays with supernatants from the corresponding R-pyocin deletion strains PAKΔR1, PA14ΔR2, and 15108/-1ΔR5. When supernatants from the mutant strains produced zones of lysis on lawns of the test isolate, which occurred in less than 5% of the cases, lytic activity was considered not to result from R-pyocin activity and was scored as negative.
TABLE 2.
R-pyocin production and susceptibility of 47 clinical isolates
| Isolate | R-pyocin type | SNP genotype | Serotypea | Lysis byb: |
||
|---|---|---|---|---|---|---|
| R1-pyocin | R2-pyocin | R5-pyocin | ||||
| 03101 | A42A | O1 | + | + | + | |
| 05101 | 6C22 | O1 | + | + | + | |
| 13104 | 0C1A | O1 | + | + | + | |
| 13112 | C40A | O1 | + | + | + | |
| 15109 | F429 | O1 | + | + | + | |
| 27101 | EC4A | O1 | + | + | + | |
| 13120 | 6592 | O3 | + | + | + | |
| 15104 | F62A | O3 | + | + | + | |
| 24101 | 2C25 | O3 | + | + | + | |
| 06101 | 279A | O6 | + | + | + | |
| 06103 | CC2A | O6 | + | + | + | |
| 13113 | 0F9E | O6 | + | − | + | |
| 27102 | 841E | NT | + | + | − | |
| 13109 | 1 | F429 | O1 | − | + | + |
| 01101 | 1 | 1BAE | O6 | − | + | + |
| 06104 | 1 | AF9A | O6 | − | − | + |
| 10107 | 1 | 2C2A | O6 | − | + | + |
| 13106 | 1 | 4F8A | O6 | − | + | + |
| 13108 | 1 | 1BAE | O6 | − | + | + |
| 13111 | 1 | 85AA | O6 | − | + | − |
| 13128 | 1 | 062A | O6 | − | ± | − |
| 15101 | 1 | 0C2E | O6 | − | + | + |
| 16101 | 1 | 0C2E | O6 | − | + | + |
| 19105 | 1 | 7C2E | O6 | − | + | + |
| 26103 | 1 | 4F92 | O6 | − | + | − |
| PAK | 1 | 55AA | O6 | − | + | + |
| PAO1 | 2 | 0002 | O5 | − | − | − |
| 27103 | 2 | 0812 | O5 | − | − | − |
| PA14 | 2 | D421 | O10 | − | − | − |
| 06105 | 2 | D421 | O10 | − | − | − |
| 10103 | 2 | D421 | O10 | − | − | − |
| 13124 | 2 | D421 | O10 | − | − | ± |
| 30101 | 2 | D421 | O10 | − | − | − |
| 13122 | 2 | F661 | O10 | − | − | ± |
| 13119 | 2 | 239A | O12 | − | − | − |
| 13118-S1 | 2 | 239A | O15 | − | − | + |
| 08102 | 5 | 2C1A | O4 | + | + | − |
| 10102 | 5 | 2F92 | O4 | + | + | − |
| 21106 | 5 | 4F82 | O4 | − | − | − |
| 03103 | 5 | 0F92 | O11 | ± | − | − |
| 15102 | 5 | E429 | O11 | − | − | − |
| 19101 | 5 | F469 | O11 | − | − | − |
| 08101 | 5 | F469 | O11 | − | − | − |
| 13117 | 5 | F469 | O11 | − | − | − |
| 22101 | 5 | F469 | O11 | − | − | − |
| 26101 | 5 | F469 | O11 | − | − | − |
| 19102 | 5 | 6D92 | O12 | − | − | − |
| 21102 | 5 | 6D92 | O12 | − | − | − |
| 15108 | 5 | E429 | O15 | + | ± | − |
| 28101 | 5 | 261A | NT | − | − | − |
LPS O-serotype lysis was determined with supernatants from strains PAK (R1-pyocin), PA14 (R2-pyocin), and 15108-1 (R5-pyocin), and the results were compared to those for the corresponding R-pyocin deletion mutant. NT, not typeable.
+, lysis; −, no lysis; ±, weak or fuzzy lysis zone.
Serotyping.
Isolates were serotyped by the slide agglutination method using commercial antisera (Bio-Rad) against all 20 serotypes (Habs classification) of P. aeruginosa. Only a small percentage (6%) of isolates were nontypeable.
Genotyping.
Isolates were genotyped using the Clondiag P. aeruginosa array according to the manufacturer's instructions (CLONDIAG, Jena, Germany) (28). The genotypes of most of the isolates have been described previously (11, 28).
RESULTS
Detection of R-pyocin genes.
The genes for synthesis of R- and F-pyocins are located in two adjacent DNA regions, inserted between the trpE and trpG genes in the PAO1 genome (http://pseudomonas.com/). To obtain information on the sequence conservation of R-pyocin genes, we first compared the 15 R-pyocin protein sequences (PA0614 to PA0628) of PAO1, which produces R2-pyocin, with the R-pyocin protein sequences of PA14 and the Liverpool epidemic clone LESB58 using information for their available genome sequences (http://pseudomonas.com/). The PA14 sequences were >99% identical to those of PAO1, and PA14 was thus considered a strain that also produces an R2-pyocin. The protein sequences encoded by 15 ORFs of LESB58 were also identical to the protein sequences of PAO1 and PA14, with the notable exception of the tail fiber protein sequence encoded by PA0620 (87% identity) (Fig. 1). Comparison with the recently reported tail fiber protein sequences of the R1- to R5-pyocins (29) revealed that the amino acid sequence encoded by PA0620 from LESB58 was identical to the amino acid sequence of the R1-pyocin tail fiber protein. An ORF corresponding to the chaperone protein PA0621 ORF, however, was not annotated for strain LESB58 (http://pseudomonas.com/). A search for ORFs in the intergenic region between the PA0620 and PA0622 ORFs using ORF-finder revealed an ORF encoding a putative protein consisting of 146 amino acids that showed 43% amino acid identity with the PA0621 protein of PAO1 (Fig. 1). This sequence analysis suggested that the PA0620 and PA0621 ORFs encoding the tail fiber and cognate chaperone proteins solely define the R-pyocin type, while the sequences of the other ORFs of the operon seem to be conserved among the different R-pyocin types.
To assess the distribution of R-pyocin genes in clinical isolates, we used the initial colonizing isolates obtained from tracheal aspirates of 61 intubated patients hospitalized in 17 European centers (11; Köhler et al., submitted). Using a multiplex PCR assay, we then screened the 61 isolates for three pyocin genes, PA0617, PA0628, and either PA0620 or PA0621, based on the DNA sequences of PAO1 (R2-pyocin) (Fig. 1). Fourteen isolates (23%) showed no signal for any of these ORFs, suggesting that the entire R-pyocin operon is absent. The remaining isolates all contained amplicons of the expected sizes for ORFs PA0617 and PA0628, but only 13 isolates contained an amplicon for PA0621 (see Table S1 in the supplemental material). The 13 PA0621-positive isolates were classified as R2-pyocin isolates, and the 34 negative isolates were expected to contain a PA0621 gene of a different R-pyocin type. Since the amino acid sequences of the tail fiber (PA0620) and chaperone (PA0621) proteins of the R2-, R3-, and R4-pyocins differ by only a few amino acid substitutions, we considered these three pyocins members of the same subtype, which we refer to as the R2-pyocin type below.
Based on the DNA sequences of the R1- and R5-pyocin genes (29), we designed two new primer sets located in the variable 3′-terminal region of PA0620 (Table 1). Among the 34 R2-pyocin-negative isolates, we identified 13 isolates carrying the R1-pyocin gene and 21 isolates carrying the R5-pyocin gene. Thus, the 61 isolates examined could be placed in four different groups: one group consisting of R-pyocin-negative isolates and three groups consisting of isolates containing either R1-, R2-, or R5-pyocin genes (see Table S1 in the supplemental material).
Correlation between R-pyocins and O serotypes.
As R-pyocins bind to LPS (15), we next determined the O serotypes of the clinical isolates by the slide agglutination method. Using the P. aeruginosa Clondiag array (28), we previously identified 34 different genotypes (single-nucleotide polymorphism [SNP] types) for the 61 initial isolates (see Table S1 in the supplemental material) (11; Köhler et al. submitted). To avoid redundancy, we decided to use only one isolate per SNP type and per center for further analysis, which resulted in a final collection of 47 isolates (Table 2). The most frequent serotypes for these isolates were serotypes O6 (15 isolates from eight centers), O11 (seven isolates from seven centers), O1 (seven isolates from five centers), and O10 (six isolates from four centers) (Table 2). Remarkably, five of seven serotype O11 isolates and five of six serotype O10 isolates had the F469 and D421 genotypes, respectively (P < 0.01, Fischer's exact test). In contrast, almost all serotype O1 and O6 isolates had distinct SNP types. For the remaining serotypes (serotypes O3, O4, O5, and O15), the number of isolates was too small to determine a possible association between genotype and serotype.
We then investigated a possible association between O serotype and R-pyocin genes. Fourteen isolates in which R-pyocin genes were not detectable by the PCR assay belonged to either serotype O1 (all 6 isolates tested; P = 0.003), O3 (all 3 isolates tested; P = 0.029), or O6 (3 of 15 isolates tested) (Table 2). Remarkably, 12 of 13 R1-pyocin-producing isolates belonged to serotype O6 (12 of 15 serotype O6 isolates tested; P < 0.001). The 13 R2-pyocin-producing isolates belonged to either serotype O5 (both O5 isolates tested; P = 0.019), O10 (all 6 O10 isolates tested; P = 0.019), O12, or O15, while the 21 R5-pyocin-producing isolates belonged to serotype O4 (all 3 serotype O4 isolates tested; P = 0.009), serotype O11 (all 7 serotype O11 isolates tested; P = 0.009), or serotype O12 or O15 (Table 2; see Table S1 in the supplemental material). Thus, in our clinical collection, seven serotypes (serotypes O1, O3, O4, O6, O10, O11, and O12) showed a statistically significant association with a particular R-pyocin type. In agreement with these results, reference strains PAO1 (serotype O5) and PA14 (serotype O10) both harbor the R2-pyocin gene, while strain PAK (serotype O6) carries an R1-pyocin gene (Table 2).
Pyocin susceptibility profile of clinical isolates.
We next determined the susceptibilities of the 47 clinical isolates to the different R-pyocin types using the spot assay. We used PAK, PA14, and clinical isolate 15108/-1 as producers of R1-, R2-, and R5-pyocins, respectively. To ensure that the observed lysis zones were due to R-pyocins and not to other lytic activities (F-pyocin, S-pyocin, or bacteriophage), we first constructed a deletion in the R-pyocin region in each of the three strains. The mutants, constructed by homologous recombination (see Materials and Methods), had an unmarked 2.2-kb deletion encompassing the ORFs encoding the tail fiber protein PA0620 and the cognate chaperone PA0621. Supernatants from the mutants did not exhibit lytic activity when they were spotted on a clinical isolate (13112/-1) that was susceptible to all three types of R-pyocins produced by the wild-type strains. Supernatants from these deletion mutants were used as negative controls in subsequent experiments.
With the exception of isolates 13113 and 27102, all of the isolates deficient in R-pyocin genes were found to be susceptible to R1-, R2-, and R5-pyocins (Table 2). All 12 isolates harboring an R1-pyocin gene were resistant to the R1-pyocin produced by PAK (P < 0.001); however, 11 isolates were susceptible to R2-pyocin (P = 0.017) and 9 isolates were susceptible to R5-pyocin (P = 0.085) (Table 2).
All isolates harboring an R2-pyocin gene were resistant to both R1- and R2-pyocins (P < 0.001), while three of the eight isolates were weakly susceptible to R5-pyocin. With the exception of two serotype O4 strains and one serotype O15 strain, all R5-pyocin-producing isolates were resistant to the three types of R-pyocins (P = 0.002). These data clearly demonstrate that there is an association between the O serotype and the susceptibility to R-pyocins in clinical isolates.
LPS determinants as receptors for R-pyocins.
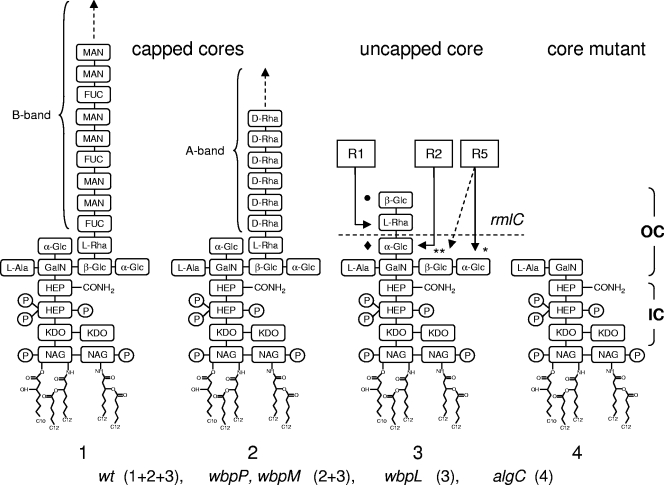
R-pyocins kill susceptible bacterial cells by inserting their core structure through the cell envelope and therefore have to establish close contact with the cell surface. Previous studies have suggested that the core of the LPS might be the receptor for R-pyocins; however, the putative receptor sites have not been characterized in detail (15). Using the spot assay, we tested the R-pyocin susceptibilities of defined isogenic LPS mutants previously constructed using reference strains PAK (serotype O6) and PAO1 (serotype O5) (kindly provided by J. Lam, University of Guelph, Guelph, Canada). As described above, strain PAK was susceptible to R2- and R5-pyocins but resistant to its own R1-pyocin (Table 3). In contrast, the LPS B-band-deficient wbpP mutant (Table 1) was susceptible to its own R1-pyocin (Table 3), suggesting that removal of the long B-band LPS molecules makes the binding site accessible to R1-pyocin. We then tested the wbpL mutant (Table 1) of PAK deficient in both LPS B-band and LPS A-band synthesis. This mutant was susceptible to R1- and R5-pyocins but resistant to R2-pyocin. At first glance, this suggests that the A-band LPS could be involved in R2-pyocin recognition (Fig. 2). However, the rmlC mutant (Table 1) deficient in both A- and B-band LPS and also lacking the l-Rha residue of the core was resistant to R1-pyocin but susceptible to R2- and R5-pyocins. This suggests that the l-Rha residue is the receptor of the R1-pyocin and that the α-Glc residue (filled diamond in structure 3 in Fig. 2) is the receptor for the R2-pyocin. The second α-Glc residue (asterisk in structure 3 in Fig. 2), which is accessible in the capped (structures 1 and 2 in Fig. 2) and uncapped (structure 3 in Fig. 2) core structures, is likely to be the receptor for R5-pyocin.
TABLE 3.
R-pyocin susceptibility of LPS mutants of PAK (serotype O6) and PAO1 (serotype O5)
| Strain | Relevant characteristics | Lysis bya: |
||
|---|---|---|---|---|
| R1-pyocin | R2-pyocin | R5-pyocin | ||
| PAK | LPS A+ LPS B+ | − | + | + |
| PAKwbpP | LPS A+ LPS B− | + | + | + |
| PAKwbpL | LPS A− LPS B− | + | − | + |
| PAKrmlC | LPS A− LPS B−l-Rha− | − | + | + |
| PAO1 | LPS A+ LPS B+ | − | − | − |
| PAO1rmd | LPS A− LPS B+ | − | − | − |
| PAO1wbpM | LPS A+ LPS B− | + | + | + |
| PAO1wbpL | LPS A− LPS B− | + | − | + |
| PAO1rmlC | LPS A− LPS B−l-Rha− | − | + | + |
| PAO1algC | LPS A− LPS B−, core mutant | − | − | − |
+, lysis; −, no lysis.
FIG. 2.
LPS structures of P. aeruginosa PAO1 (serotype O5) and mutant derivatives. Structures 1 and 2 show glycoforms capped by a serotype O5 B-band polysaccharide chain and an A-band (d-rhamnose trisaccharide) chain, respectively. Structure 3 shows the uncapped glycoform. The LPS of a wild-type PAO1 strain (wt) is composed of structures 1, 2, and 3 (1+2+3). wbpP (PAK) and wbpM (PAO1) mutants lack the B-band LPS, while wbpL mutants of PAK and PAO1 are deficient in both A- and B-band LPS synthesis. The algC mutant of PAO1 (structure 4) lacks all glucose (Glc) residues of the outer core (OC) LPS, while it contains an intact inner core (IC) LPS. The sugar residues involved in R-pyocin recognition are indicated by arrows. Involvement of the terminal α-Glc residue (one asterisk) in R5-pyocin recognition (solid arrow) is favored over involvement of the β-Glc residue (two asterisks, dashed arrow). The rmlC mutant lacks the terminal l-Rha residue (dashed line). MAN, mannose; FUC, fucose; GalN, N-galactosamine; HEP, heptose; NAG, N-acetylglucosamine; KDO, 2-keto-3-deoxyoctulosonic acid.
To confirm these findings, we tested five LPS mutants derived from serotype O5 strain PAO1. Remarkably, while the rmd mutant deficient in the A-band LPS was resistant to all three types of pyocins, the wbpM mutant deficient in B-band LPS synthesis was susceptible (Table 3). This suggests again that the long B-band oligosaccharide chains prevent access of the R-pyocins to their receptor sites. As observed for the wbpL mutant of PAK, the PAO1 wbpL mutant was resistant to R2-pyocin. R-pyocins of wbpL mutants should exist only in the uncapped core form (structure 3 in Fig. 2), in which the α-Glc residue (filled diamond in structure 3 in Fig. 2), normally exposed in structures 1 and 2, is blocked by the l-Rha sugar. When this l-Rha residue was not present (e.g., in the rmlC mutant), the strain was susceptible to R2-pyocin (Table 3). Finally, the algC mutant of PAO1 (AK1012), which lacks all of the glucose residues of the outer core but has the inner core structures (structure 4 in Fig. 2), was completely resistant to all three types of R-pyocins, confirming that the glucose residues are involved in R2- and R5-pyocin recognition.
In summary, these results suggest that the long B-band LPS molecules, which determine the serotype of P. aeruginosa strains, may prevent access of R-pyocins to their receptor sites. Moreover, the l-Rha residue in the outer core appears to be the receptor for R1-pyocin, and two distinct Glc residues are part of the receptor sites for the R2- and R5-pyocins. Thus, the LPS has a dual role as both a shield and a receptor for R-pyocin molecules.
R-pyocins and biological warfare.
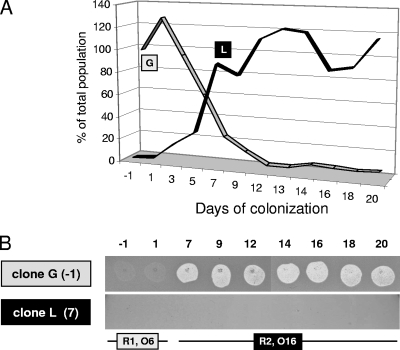
Whether R-pyocins play a role as biological weapons in a clinical context has not been clearly established. Among the 61 intubated patients, we identified several patients colonized with bacteria having two different genotypes. In one of these patients, we previously followed the evolution of the two P. aeruginosa populations using genomic DNA preparations isolated from daily tracheal aspirates (11). In this patient the initially dominant clone, clone G, had been outcompeted by the second clone, clone L, after about 10 days of cocolonization (Fig. 3A). We determined the R-pyocin types and O serotypes of the two clones. Clone G (isolate obtained on the first day of colonization [day −1] produced R1-pyocin and belonged to serotype O6 (isolate 15101 [Table 2 and Fig. 3B]), while clone L (isolate obtained on day 7) produced R2-pyocin and belonged to serotype O16 (Fig. 3B). We tested the lytic activities of the supernatants from nine isolates obtained from this patient, two belonging to clone G (obtained on days −1 and 1) and seven belonging to clone L (obtained on days 7 to 20). When spotted on a lawn of bacteria of the initially dominant clone G (isolate obtained on day −1), lytic activity was observed for all supernatants from clone L isolates (obtained on days 7 to 20) (Fig. 3B). In contrast, when supernatants of clone G or clone L isolates were spotted on a lawn of clone L bacteria (isolate obtained on day 7), no lysis was observed. Therefore, whereas clone G was susceptible to the R2-pyocin produced by clone L, clone L was resistant not only to its own pyocin, as expected, but also to the R1-pyocin produced by clone G (Fig. 3B). The consistency of these in vitro data with the coevolution of the two clones observed directly in the patient's lungs (Fig. 3A) suggests that biological warfare by R-pyocins might have been responsible for shaping the population dynamics in the patient.
FIG. 3.
Population dynamics and biological warfare. (A) Relative proportions of two P. aeruginosa clones (G and L) during host colonization calculated previously by quantitative reverse transcription-PCR using genomic DNA extracts of tracheal aspirates (11). (B) Pyocin susceptibilities of the isolate of clone G obtained on day −1 (−1) and of the isolate of clone L obtained on day 7 (7). Four microliters of culture supernatant from nine colonizing isolates was spotted on top agar plates, and the results were determined after incubation for 18 h at 37°C.
DISCUSSION
In the present study we determined the R-pyocin types and susceptibilities of P. aeruginosa isolates collected from 61 intubated patients throughout Europe. Our results reveal a surprising association between O serotypes and R-pyocin production. Pyocin and LPS biosynthesis genes are not genetically linked; thus, coevolution of these two types of genes seems unlikely. As observed for S-pyocin producers, each R-pyocin-producing isolate was resistant to at least its own R-pyocin. While in the case of S-pyocins the immunity is provided by a special chaperone protein, whose gene is genetically linked to the cognate S-pyocin gene (16), there does not seem to be such an immunity protein for R-pyocins. Indeed, since R-pyocins cause cell lysis after attachment to the cell surface of susceptible bacteria, self-protection can be guaranteed only by extracellular factors. This shielding function could be provided by the long-chain oligosaccharides of the LPS. Note that the average length of an O-chain LPS in P. aeruginosa is 30 to 40 nm (12), while the average length of the main core of a pyocin molecule is 100 nm. This size of R-pyocin particles was the same for the R1-, R2-, and R5-pyocins according to our electron microscope observations (data not shown).
Furthermore, we also demonstrated that there is an association between the O serotype and the R-pyocin susceptibility pattern. Indeed, isolates that were susceptible to all three R-pyocins tested had a serotype O1, O3, or O6 B-band LPS, while the serotype O5, O10, O11, and O12 isolates were completely resistant. This correlation may reflect the packing density of the B-band LPS side chains, which is dictated by the physicochemical properties of the sugar constituents. Indeed, only a low percentage of core LPS units are capped (substituted) with A-band or B-band LPS (structures 1 and 2 in Fig. 2). We suggest that serotype O1, O3, and O6 isolates have a low proportion of capped LPS molecules (loose packing). This would provide comparatively unhindered access of R-pyocin molecules to their surface receptors. Interestingly, freeze substitution electron microscopy studies with P. aeruginosa cells have shown that PAO1 (serotype O5, R-pyocin resistant) has a higher proportion of capped LPS than PAK (serotype O6, R2- and R5-pyocin susceptible) (12). We would therefore expect that serotype O10, O11, and O12 strains, all of which are resistant to the three different R-pyocins tested, would also have dense LPS packing. Indeed, when PA14 (serotype O10) was grown at 45°C, a temperature which prevents expression of B-band LPS but not expression of A-band LPS in PAO1 (13), the strain was susceptible to the R5-pyocin (data not shown). However, we cannot exclude other explanations, including R-pyocin type-specific interactions with the LPS O-side chains, which probably have different electrostatic properties depending on the sugar composition and additional chemical modifications.
The different types of R-pyocins recognize different receptor sites that were proposed to be located in the LPS core (15). Our data obtained using LPS-specific mutants of PAO1 and PAK are in agreement with this proposal. Based on biochemical analysis of spontaneous LPS mutants of serotype O3 strain PAC1, Meadow and Wells proposed that R1-pyocin requires the l-Rha sugar of the core LPS for lytic activity (15). This sugar is the acceptor for both A-band and B-band LPS (19). In the wbpP (LPS A+ LPS B−) and wbpL (LPS A− LPS B−) mutants of PAK, this l-Rha residue is accessible, and this could explain the susceptibility of these mutants to R1-pyocin. In the rmlC mutants of both PAO1 and PAK this l-Rha residue is not present, which results in resistance to R1-pyocin. PAO1 has a β-Glc residue (filled circle in structure 3 in Fig. 2) in the uncapped glycoform, which should prevent access to the adjacent l-Rha. This modification probably is not stoichiometric and leaves sufficient l-Rha residues unmodified to confer susceptibility to R1-pyocin.
Our results also suggest that the α-Glc residues (filled diamond in structure 3 in Fig. 2) must be part of the recognition site for the R2-pyocin, while the terminal α-Glc residue (asterisk in structure 3 in Fig. 2) is part of the R5-pyocin receptor site. Unlike the β-Glc sugar (two asterisks in structure 3 in Fig. 2), the terminal α-Glc residue is accessible in all capped and uncapped forms and is therefore more likely to be the receptor of R5-pyocin. Whether the receptor sites consist solely of these LPS core sugar residues or require other structures (for instance, outer membrane proteins) remains to be determined.
In our clinical collection, we found similar percentages of R-pyocin-deficient strains (28%) and strains producing R1-pyocins (25%), R2-pyocins (17%), and R5-pyocins (29%). This could have resulted from the fact that most patients were colonized by a single genotype, limiting encounters and possible warfare between different clonal populations. However, in one patient colonized by two different genotypes, we observed that an initially dominant clone was outcompeted by a second clone that produced an R-pyocin to which the initial clone was susceptible. The killing behavior of these two clones was in agreement with the R-pyocin production and susceptibility profiles established during this study. The population dynamics in this patient therefore illustrate that biological warfare by R-pyocins may play an important role in shaping the structure of P. aeruginosa populations during host colonization.
Interestingly, the observed in vitro killing of strain PAK (R1-type pyocin, serotype O6) by PA14 (R2-type pyocin, serotype O10) during growth in liquid culture (7) is in agreement with the susceptibility profile established in this study. Pyocins also modulate bacterial population dynamics in biofilms, particularly under anaerobic growth conditions, which were shown to induce R- and F-pyocin genes, as well as S-pyocin genes (27).
Thus, pyocins, particularly the potent R-pyocins, could provide a novel approach for specific targeting of otherwise difficult-to-treat infections caused by multi-drug-resistant strains of P. aeruginosa.
Supplementary Material
Acknowledgments
We thank H. Koch (Kenta AG, Bern, Switzerland) and L. Vettoretti (CHU, Besançon, France) for performing serotyping. We are grateful to E. Anderson and J. Lam for the gift of the P. aeruginosa LPS mutant strains and to P. Cossen for help with the electron microscope.
This work was supported by grant 320000-108106 from the Swiss National Science Foundation and grant RC0732 from Vaincre la Mucoviscidose to C.V.D.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Belanger, M., L. L. Burrows, and J. S. Lam. 1999. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145:3505-3521. [DOI] [PubMed] [Google Scholar]
- 2.Burrows, L. L., D. F. Charter, and J. S. Lam. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481-495. [DOI] [PubMed] [Google Scholar]
- 3.Farmer, J. J., III, and L. G. Herman. 1969. Epidemiological fingerprinting of Pseudomonas aeruginosa by the production of and sensitivity of pyocin and bacteriophage. Appl. Microbiol. 18:760-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fyfe, J. A., G. Harris, and J. R. Govan. 1984. Revised pyocin typing method for Pseudomonas aeruginosa. J. Clin. Microbiol. 20:47-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He, J., R. L. Baldini, E. Deziel, M. Saucier, Q. Zhang, N. T. Liberati, D. Lee, J. Urbach, H. M. Goodman, and L. G. Rahme. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U. S. A. 101:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heo, Y. J., I. Y. Chung, K. B. Choi, and Y. H. Cho. 2007. R-type pyocin is required for competitive growth advantage between Pseudomonas aeruginosa strains. J. Microbiol. Biotechnol. 17:180-185. [PubMed] [Google Scholar]
- 8.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 9.Jarrell, K., and A. M. Kropinski. 1977. The chemical composition of the lipopolysaccharide from Pseudomonas aeruginosa strain PAO and a spontaneously derived rough mutant. Microbios 19:103-116. [PubMed] [Google Scholar]
- 10.Kageyama, M. 1975. Bacteriocins and bacteriophages in Pseudomonas aeruginosa, p. 291-305. In T. Mitsuhashi and H. Hashimoto (ed.), Microbial drug resistance. University of Tokyo Press, Tokyo, Japan.
- 11.Köhler, T., A. Buckling, and C. Van Delden. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U. S. A. 106:6339-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam, J. S., L. L. Graham, J. Lightfoot, T. Dasgupta, and T. J. Beveridge. 1992. Ultrastructural examination of the lipopolysaccharides of Pseudomonas aeruginosa strains and their isogenic rough mutants by freeze-substitution. J. Bacteriol. 174:7159-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makin, S. A., and T. J. Beveridge. 1996. Pseudomonas aeruginosa PAO1 ceases to express serotype-specific lipopolysaccharide at 45°C. J. Bacteriol. 178:3350-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsui, H., Y. Sano, H. Ishihara, and T. Shinomiya. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J. Bacteriol. 175:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meadow, P. M., and P. L. Wells. 1978. Receptor sites for R-type pyocins and bacteriophage E79 in the core part of the lipopolysaccharide of Pseudomonas aeruginosa PAC1. J. Gen. Microbiol. 108:339-343. [Google Scholar]
- 16.Michel-Briand, Y., and C. Baysse. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499-510. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama, K., K. Takashima, H. Ishihara, T. Shinomiya, M. Kageyama, S. Kanaya, M. Ohnishi, T. Murata, H. Mori, and T. Hayashi. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213-231. [DOI] [PubMed] [Google Scholar]
- 18.Parret, A. H., and R. De Mot. 2002. Bacteria killing their own kind: novel bacteriocins of Pseudomonas and other gamma-proteobacteria. Trends Microbiol. 10:107-112. [DOI] [PubMed] [Google Scholar]
- 19.Poon, K. K., E. L. Westman, E. Vinogradov, S. Jin, and J. S. Lam. 2008. Functional characterization of MigA and WapR: putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 190:1857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahim, R., L. L. Burrows, M. A. Monteiro, M. B. Perry, and J. S. Lam. 2000. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 146:2803-2814. [DOI] [PubMed] [Google Scholar]
- 21.Riley, M. A., and J. E. Wertz. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117-137. [DOI] [PubMed] [Google Scholar]
- 22.Rocchetta, H. L., L. L. Burrows, J. C. Pacan, and J. S. Lam. 1998. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 28:1103-1119. [DOI] [PubMed] [Google Scholar]
- 23.Scholl, D., M. Cooley, S. R. Williams, D. Gebhart, D. Martin, A. Bates, and R. Mandrell. 2009. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob. Agents Chemother. 53:3074-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scholl, D., and D. W. Martin, Jr. 2008. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a murine peritonitis model. Antimicrob. Agents Chemother. 52:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 26.Uratani, Y., and T. Hoshino. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157:632-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waite, R. D., and M. A. Curtis. 2009. Pseudomonas aeruginosa PAO1 pyocin production affects population dynamics within mixed-culture biofilms. J. Bacteriol. 191:1349-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiehlmann, L., G. Wagner, N. Cramer, B. Siebert, P. Gudowius, G. Morales, T. Kohler, C. Van Delden, C. Weinel, P. Slickers, and B. Tummler. 2007. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8101-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams, S. R., D. Gebhart, D. W. Martin, and D. Scholl. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74:3868-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.