Abstract
Porphyromonas gingivalis, a Gram-negative oral anaerobe, is strongly associated with chronic adult periodontitis, and it utilizes FimA fimbriae to persistently colonize and evade host defenses in the periodontal crevice. The FimA-related gene cluster (the fim gene cluster) is positively regulated by the FimS-FimR two-component system. In this study, comparative analyses between fimbriate type strain ATCC 33277 and fimbria-deficient strain W83 revealed differences in their fimS loci, which encode FimS histidine kinase. Using a reciprocal gene exchange system, we established that FimS from W83 is malfunctional. Complementation analysis with chimeric fimS constructs revealed that W83 FimS has a defective kinase domain due to a truncated conserved G3 box motif that provides an ATP-binding pocket. The introduction of the functional fimS from 33277 restored the production, but not polymerization, of endogenous FimA subunits in W83. Further analyses with a fimA-exchanged W83 isogenic strain showed that even the fimbria-deficient W83 retains the ability to polymerize FimA from 33277, indicating the assembly of mature FimA by a primary structure-dependent mechanism. It also was shown that the substantial expression of 33277-type FimA fimbriae in the W83 derivative requires the introduction and expression of the functional 33277 fimS. These findings indicate that FimSR is the unique and universal regulatory system that activates the fim gene cluster in a fimA genotype-independent manner.
Robust epidemiological evidence indicates that Porphyromonas gingivalis, a Gram-negative oral anaerobe, is strongly associated with chronic adult periodontitis (37). Of the various virulence factors produced by this bacterium, fimbriae have attracted attention as determinants of persistent colonization in the primary ecological niche, the periodontal crevice. Recent studies have demonstrated that the major structural subunit protein of fimbriae, FimA (fimbrilin), mediates the invasion of gingival epithelial cells by P. gingivalis (27). The FimA-coding gene, fimA, was classified into several genotypes (2), and their correlations with the severity of periodontal diseases also were investigated (2, 8, 22). A recent study has strengthened links between fimbriae and P. gingivalis evasion of host immune system by showing that FimA fimbriae inhibit Toll-like receptor 2 (TLR2)-mediated proinflammatory and antimicrobial responses by host cells (12).
Bacteria use His-Asp phosphorelay systems and, most simply, two-component regulatory systems (TCS) to respond to environmental changes (38). Many studies revealed that TCS regulate the expression of virulence genes in pathogenic bacteria (9, 36, 41). In P. gingivalis, the first TCS, the FimS histidine kinase and FimR response regulator, was discovered in a screen for fimbria-deficient transposon mutants (14). Gene complementation revealed that response regulator FimR is essential for the production of FimA (30). The target gene of FimR was identified by a combination of microarray analyses and chromatin immunoprecipitation (ChIP) assays (31). The comparative expression analysis of fimR wild-type and mutant strains indicated that FimR-responsive genes with the highest fold change values were clustered around the fimA locus, suggesting that FimSR has a dedicated role in the expression of the fim gene cluster. Subsequent ChIP and electromobility shift assays (EMSA) revealed that FimR binds to the promoter region of fimX, the first gene of the fim gene cluster. Transcript analyses of insertion mutants of fimR, and fim cluster genes fimX, pgmA, and fimA, supported a regulation cascade model in which an environmental signal (as-yet unknown) triggers the FimS-FimR phosphorelay. Activated FimR binds to the promoter region of fimX to upregulate the transcription of fimX and pgmA, the first two genes in the cluster. The fimX gene product acts in trans to promote the transcription of downstream gene fimA.
Two strains of P. gingivalis are used frequently by the research community. In early studies, the virulence of strain W83 was noted in mouse abscess models (11), and therefore this strain was chosen for genome sequencing (29). However, it was already shown that W83 cell extracts did not react with anti-FimA monomers or anti-fimbria antibodies (19, 39), suggesting that the strain was fimbria deficient. Alternatively, type strain ATCC 33277 was studied with respect to fimbria production (13). A comparative whole-genome analysis of the two strains was carried out (4), and recently the genome sequence of ATCC 33277 was published (26). These genome-wide studies revealed that both the fim gene cluster and fimSR genes are conserved in the W83 genome, and that was apparently paradoxical to the afimbriate phenotype of W83. It still was possible that the strain does produce FimA fimbrilin and fimbriae, albeit at a low level.
In this study, we used a reciprocal gene exchange system to establish that strain W83 is fimbria deficient. The expression of fimA and related genes are not activated at the transcriptional level, mainly due to a defective FimS histidine kinase with a truncation in a conserved motif required for ATP binding. The introduction of the functional fimS restored the production, but not the polymerization, of endogenous FimA subunits in W83. Further analyses with a fimA-exchanged W83 derivative showed that even afimbriate W83 retains the ability to polymerize 33277-type FimA, indicating the autoassembly of mature FimA by a primary structure-dependent mechanism.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains and plasmids used in this study are shown in Table 1. Escherichia coli strains were grown either on Luria-Bertani (LB) agar plates or in LB broth at 37°C with appropriate antibiotics when necessary. P. gingivalis strains were maintained on blood agar plates (BAPHK) as described previously (31). To prepare RNA and protein samples, cells were grown in supplemented trypticase soy broth (sTSBHK) and harvested at late log or early stationary phase (31). Anaerobic growth conditions (10% CO2, 5% H2, 85% N2) were created by using either AnaeroBox HARD (Hirasawa Works, Tokyo, Japan) or Anoxomat WS9000 (Mart Microbiology BV, Lichtenvoorde, The Netherlands). Procedures for the disruption of fimS with the ermF-ermAM cassette (Emr) by allelic exchange were as described previously (31). The cassette was inserted into the SphI site at the center of fimS so as not to disrupt the C-terminal coding region that is essential for fimS-independent fimR expression. To generate a fimA-exchanged W83, W83fA327, a 1,968-bp chimeric DNA fragment first was constructed by a PCR-based overlap extension method with the primers listed in Table 1. Three fragments were amplified as templates: a 415-bp fragment containing the C-terminal coding region of pgmA and the 5′ common sequence of fimA was from the W83 genome with primers a and b; a 1,175-bp fragment ranging from the start codon of fimA to the downstream intergenic region (IGR) was from the 33277 genome with primers c and d; and a 435-bp fragment covering the IGR and 5′ fimB was from the W83 genome with primers e and f. Consequently, the sequences of the overlapped PCR products with these three templates and the primers a and f were identical to those of the W83 loci, except that only the open reading frame (ORF) of fimA was the same as that of 33277. A SmaI site was designed at the center of both primers d and e (the IGR between fimA and fimB). The chimeric PCR fragment was cloned into pCR4Blunt-TOPO (Invitrogen, Carlsbad, CA) and linearized with SmaI to ligate a PCR-amplified 1,095-bp ermF cassette. The final construct (3,063 bp) was amplified by PCR with primers a and f, directly sequenced for confirmation, and then introduced into W83 by electroporation. Genomic DNAs from erythromycin-resistant transformants were analyzed by PCR and direct sequencing.
TABLE 1.
Strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequencesa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Host for general cloning/construction of plasmids | Invitrogen |
| S17-1 | Host for mobilizing pT-COW to P. gingivalis, Tpr | 10 |
| P. gingivalis strains | ||
| ATCC 33277 | Fimbriate strain with type I fimA | ATCC (26) |
| AG21-5 | 33277 with Tn4351 insertion in fimR | 43 |
| AGFS1 | 33277 with Emr insertion in fimS | This study |
| W83 | Afimbriate strain with type IV fimA | 29 |
| WFS1 | W83 with Emr insertion in fimS | This study |
| W83fA327 | W83 with fimA exchanged for 33277 type | This study |
| Plasmids | ||
| pTCB | pT-COW derivative, Ampr, tetQ | 10, 30 |
| pTCBex | pTCB derivative with the promoter pfR | 25, 30 |
| pTCBex33277fimS | pTCBex carrying 33277 fimS ORF | This study |
| pTCBexW83fimS | pTCBex carrying W83 fimS ORF | This study |
| pTCBexChimera-1 | pTCBex carrying fimS Chimera-1 | This study |
| pTCBexChimera-2 | pTCBex carrying fimS Chimera-2 | This study |
| RT and real-time PCR primers | ||
| 16S rRNA | GTCAATGGGCGAGAGCCTGAA/AGTGTCAGTCGCAGTATGGCAA (383) | This study |
| 33277 fimA specific | CTTGTAACAAAGACAACGAGGCAG/GCAGGTGCAACGTAATTACGGCTC (700) | This study |
| W83 fimA specific | CTTGTAACAAAGACAACGAGGCAG/GACGCCTCCAATTCGTATGTTTCT (700) | This study |
| fimA | ATGAAAAAAACAAAGTTTTTCTTG/ACGGGTTCTGCCTCGTTGTC (83) | This study |
| fimS | GACTGCGAGAAATGAATGACACCA/ACCTTTGTCTGATACAGACACTTT (500) | This study |
| fimR | ATGATTAGTATCGTACTCGTGGAT/ATCATTGATCTGAGCTGTTTTGCA (336) | This study |
| fimSR | GACTGCGAGAAATGAATGACACCA/ATCATTGATCTGAGCTGTTTTGCA (1,052) | This study |
| fimX | GCTTTATTGGGGTACGACGA/TCCAAGTTCCATCTTTTGCC (342) | This study |
| pgmA | TACCCCCATTATTCGTTCCA/CGAGAGCAATGTGGCATAGA (826) | This study |
| fimX-pgmA | GCTTTATTGGGGTACGACGA/CGAGAGCAATGTGGCATAGA (1,532) | This study |
| glk (PG1737) | TGGCAATTTGGTTATAAGTTCGT/ACCGGCAAATCCATCGTGT (415) | This study |
| 33277 fimS specific | ATGCAATCGACACAAACAATGATAATC/CTGAGAGCATAAAGACACATCCAAGA (97) | This study |
| W83 fimS specific | AGTAGACAAATACAAGACACTATTATG/CTGAGAGCATAAAGACACATCCAAGA (88) | This study |
| 5′ RLM-RACE primers | ||
| Adapter outer primer (Ambion) | GCTGATGGCGATGAATGAACACTG | This study |
| Adapter inner primer | GAACACTGCGTTTGCTGGCTTTGATG | This study |
| GSP-RT | TCTCTTTGGCTGAGAGCATAAAGAC | This study |
| GSP1 | CTGAGAGCATAAAGACACATCCAAGA | This study |
| GSP2 | CATCCAAGAAATGTCAGGATAGTT | This study |
| GSP3 | AGAAATGTCAGGATAGTTAGTCGCGTA | This study |
| Primers for construction of fimS expression plasmids | ||
| 33277 fimS | GAGGATCCATGCAATCGACACAAACAATGATAATC/TGCTCGAGTTACGCTGAGGCAAGGACAAAAGAG) (1,912) | This study |
| W83 fimS | CGGATCCATGTTTTTAACAAAAAACTATTA/CCTCGAGTCAATGCATAACTTGCTTTTGTT (1,880) | This study |
| Introduction of a missense mutation in 33277 fimS | TGGAAGCTCCTAAACAATGTTTGGT/ACCAAACATTGTTTAGGAGCTTCCA | This study |
| fimA-exchanged construct to generate W83fA327 | ||
| W83pgmA-33277fimA5′ (primers a/b) | AACGCTACGAAACGTGGACGATAGCAGA/CTGCCTCGTTGTCTTTGTTACAAGCTGT (416) | This study |
| 33277fimA5′-fimAB IGR (primers c/d) | ACAGCTTGTAACAAAGACAACGAGGCAG/GGAGCGAACCCCGGGCCCTGTATTCCGA (1,175) | This study |
| fimAB IGR-W83fimB (primers e/f) | TCGGAATACAGGGCCCGGGGTTCGCTCC/TGGCTGTTTCTTCCAGCATGTGATCGTC (435) | This study |
| ermF cassette | GATAGCTTCCGCTATTGCTTTTTTGCTCAT/CTACGAAGGATGAAATTTTTCAGGGACAAC (1,095) | This study |
Sequences are 5′ to 3′ and forward/reverse (amplicon sizes are in bp). Sequences corresponding to restriction sites are underlined.
Preparation of RNA, reverse transcription, and real-time PCR.
Total RNA was extracted from 2 to 3 ml culture using RiboPure-Bacteria (Ambion, Austin, TX) and further treated with Turbo DNA-free (Ambion) according to the manufacturer's instructions. Reverse transcription (RT) of the purified RNA was performed as follows: a 13.5-μl mixture containing 3 μg of RNA, 2 μl of random decamers (Ambion), and 4 μl of deoxynucleoside triphosphates (dNTPs) (2.5 mM each) was heat denatured at 65°C for 5 min and then placed on ice. The mixture was combined with 4 μl of 5× first-strand buffer, 1 μl of 0.1 M dithiothreitol (DTT), 1 μl of RNase OUT (Invitrogen), and 2 μl of Superscript III reverse transcriptase (200 U/μl; Invitrogen) to make a 21-μl reaction mixture, and it was incubated for annealing at room temperature for 5 min, followed by incubation for cDNA synthesis at 55°C for 1 h. Usually, 300 ng of cDNA or 400 ng of RNA (for negative-control purposes) was used for subsequent semiquantitative RT-PCR (50 μl of total reaction volume with gene-specific primers). Real-time PCR was performed using a 7900HT fast real-time PCR system (Applied Biosystems, Foster City, CA). Reaction mixtures containing 100 ng of cDNA and gene-specific primers were prepared with SYBR green real-time PCR Master Mix (Toyobo, Osaka, Japan) according to the manufacturer's protocol. The primers used are listed in Table 1. The thermal cycle settings used were the following: initial denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 56°C for 15 s, and extension at 72°C for 1 min. The expression of target genes was normalized to the endogenous 16S rRNA in each strain. The relative quantification of target gene expression was performed by using the comparative cycle threshold (CT) method (20) (part number 4364016, Rev. B, relative quantitation using comparative CT, getting started guide, 2005; Applied Biosystems). Strain 33277 was chosen as the calibrator. The relative target gene expression was given by 2−ΔΔCT, where ΔΔCT = ΔCT(W83 sample) − ΔCT(33277 sample) and ΔCT = CT − CT(16S rRNA). The results are presented as the means ± standard deviations from triplicate reactions with three independent cDNA sources.
5′ RNA ligase-mediated (RLM)-RACE.
Six micrograms of total RNA was mixed with 0.6 μg of 5′ rapid amplification of cDNA ends (RACE) adapter (Ambion), 20 U of T4 RNA ligase (Ambion), 2 μl of 10× reaction buffer, and distilled water (dH2O) to make a 20-μl reaction mixture. After incubation at 37°C for 1 h, 4 μl of the adapter-ligated RNA reaction mixture was combined with a 20 pmol gene-specific primer, GSP-RT (Table 1), 10 nmol of each dNTP, and dH2O to yield a 13-μl mixture. The mixture was denatured at 65°C for 5 min followed by annealing at 50°C for 5 min, and then it was combined with 4 μl of 5× first-strand buffer, 1 μl of 0.1 M DTT, 1 μl of RNase OUT (Invitrogen), and 1 μl of Superscript III reverse transcriptase (Invitrogen) to make a 20-μl reaction mixture, which was incubated at 55°C for 1 h. Primers used for subsequent outer PCRs (adapter outer primer and GSP-RT) and inner PCRs (adapter inner primer and GSP1, GSP2, or GSP3) are listed in Table 1. Forty microliters of each nested PCR mixture contained 1 U of KOD FX (Toyobo) and 2 μl of either RT (for outer PCR) or outer PCR (for inner PCR) reaction mixture. The following reaction conditions were used: for outer PCR, denaturing at 94°C for 2 min followed by 30 cycles of 98°C for 10 s, 55°C for 30 s, and 68°C for 30 s; for inner PCR, denaturing at 94°C for 2 min, followed by 40 cycles of 98°C for 10 s, 59°C for 30 s, and 68°C for 30 s. Ten microliters of each reaction mixture was subjected to 3.5% agarose gel electrophoresis (AGE).
Construction of expression plasmids.
The Escherichia coli-P. gingivalis shuttle plasmid pTCB is a derivative of pT-COW (10), which was constructed by exchanging the 1.4-kbp AvaI-HindIII fragment with a 0.18-kbp polylinker sequence that was excised from pBluescript II KS (Stratagene, La Jolla, CA) with BssHII. The expression plasmid pTCBex was constructed by exchanging the XbaI-BamHI fragment of the pTCB polylinker with a fragment containing the fimR promoter (pfR), which was amplified by PCR with primers 5′-TCACTAGTGTGGAAAACATTGGAAAGG-3′ and 5′-ACGGATCCTATTGTGTAATCTTTACGC-3′ to generate a SpeI site at the 5′ end and a BamHI site at the 3′ end. To construct pTCBex33277fimS and pTCBexW83fimS, wild-type fimS ORFs from strains 33277 and W83 were PCR amplified with the primers listed in Table 1, and the products with a BamHI site at the 5′ end and an XhoI site at the 3′ end were ligated in frame to the BamHI-XhoI double-digested pTCBex. pTCBexChimera-1 was constructed by replacing the HindIII-XhoI fragment of pTCBexW83fimS with that from pTCBex33277fimS. To construct pTCBexChimera-2, a missense mutation was introduced in the 33277 fimS ORF by a PCR-based overlap extension method with the primers listed in Table 1. The HindIII-XhoI fragment of pTCBexW83fimS then was replaced with that of the PCR product containing the mutation. All of the plasmids constructed in this study were sequenced to confirm their accuracy.
Complementation of fimS variants in trans.
All of the pTCBexfimS variants were introduced into P. gingivalis by conjugation with E. coli S17-1. The E. coli strains transformed with pTCBexfimS were grown to an optical density at 600 nm (OD600) of 0.1 in LB broth, and the recipient P. gingivalis strains were grown to an OD600 of 0.5 in sTSBHK. One milliliter of P. gingivalis and 0.5 ml of S17-1 cultures were combined in a 2-ml plastic tube and centrifuged at 4,000 × g for 2 min. The cell pellet was suspended in 20 μl of sTSBHK, placed within a diameter of 1 cm (not streaked) on BAPHK, and incubated under aerobic conditions for 12 h at 37°C. The mating mix then was streaked on BAPHK supplemented with 1 μg/ml of tetracycline and incubated under anaerobic conditions for 1 week at 37°C. Black-pigmented colonies were isolated as candidate transconjugants and analyzed by Western blotting and RT-PCR for the expression of FimA and fimS, respectively.
Fractionation of P. gingivalis cells.
P. gingivalis strains cultured in sTSBHK were harvested at late log or early stationary phase by centrifugation at 10,000 × g for 20 min at 4°C. Proteins in the culture supernatant were precipitated with 10% trichloroacetic acid and redissolved in HEPES buffer (10 mM HEPES, pH 7.4, 1 mM leupeptin [Peptide Institute Inc., Osaka, Japan], 1 mM N-α-p-tosyl-l-lysine chloromethyl ketone [TLCK, Sigma-Aldrich Co., St. Louis, MO]). Cells were washed once with HEPES buffer supplemented with 150 mM NaCl and resuspended in a half-culture volume of HEPES buffer. The cells were disrupted by sonication-cooling cycles for 15 min at 4°C and centrifuged at 4,000 × g for 10 min at 4°C to eliminate unbroken cells. The cleared supernatant was collected as the whole-cell lysate and subjected to ultracentrifugation at 100,000 × g for 60 min in a Beckman TLA-100.3 rotor. The supernatant was collected as the cytoplasm/periplasm fraction and the precipitate as the whole-cell envelope (total membrane) fraction. The envelope precipitate was resuspended in the original volume of HEPES buffer containing 1% Triton X-100 and 20 mM MgCl2 to extract inner membrane proteins. The suspension was ultracentrifuged at 100,000 × g for 60 min, the supernatant was collected as the inner membrane fraction, and the precipitate was isolated as the outer membrane fraction.
Antibodies and Western blot analysis.
The preparation of rabbit polyclonal antibodies against type I FimA was described previously (45). Rabbit anti-W83 FimA (type IV) polyclonal antibody was raised against a synthetic peptide, PGGTASDNLVSAGT (Sigma-Aldrich Japan Co., Ishikari, Japan), and affinity purified on a peptide-immobilized antigen column. Anti-FimR antibody was raised and purified as described previously (30). Rabbit anti-PgmA antibody was raised against FLAG-tagged recombinant PgmA that was overexpressed in E. coli DH5α by using pFLAG-MAC expression plasmid (Sigma-Aldrich Co.). For Western blot analyses, cell numbers were equalized by using an equal volume of cell suspension with the turbidity of each cell culture adjusted to 1.0 at 600 nm. Alternatively, concentrations of protein samples were determined either by measuring the OD280 or with the Bio-Rad protein assay and bovine gamma globulin (Bio-Rad Laboratories, Hercules, CA) as a standard. Whole-cell suspensions in phosphate-buffered saline (PBS) containing 1 mM TLCK and 1 mM leupeptin or fractionated protein samples were mixed with 2× SDS sample buffer (0.1 M Tris-HCl [pH 6.8], 4% SDS, 3.1% DTT, 20% glycerol, 0.01% bromophenol blue) and incubated for 15 min at appropriate temperatures before being loaded onto 12.5% polyacrylamide gels. After SDS-PAGE, proteins were transferred electrophoretically to Trans-Blot polyvinyl difluoride (PVDF) membranes (Bio-Rad Laboratories) and probed with primary antibodies, followed by reaction with peroxidase-conjugated anti-rabbit immunoglobulins (DakoCytomation, Glostrup, Denmark). Signals were detected using Amersham ECL Plus Western blotting detection reagents (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) according to the manufacturer's instructions and were quantified with the Spot Denso program of AlphaImager EC (Alpha Innotech Co., San Leandro, CA).
Mass spectrometric analysis.
Protein bands excised from the Coomassie brilliant blue (CBB)-stained gels were destained with 25 mM NH4HCO3 buffer containing 30% CH3CN and dehydrated with 100% CH3CN, reduced with 10 mM DTT in 25 mM NH4HCO3 for 1 h at 56°C, and subsequently alkylated with 55 mM iodoacetamide in 25 mM NH4HCO3 for 45 min in the dark. Samples were dehydrated and digested with 10 ng/μl sequencing-grade trypsin (Promega Co., Madison, WI) in 25 mM NH4HCO3 overnight at 37°C. Peptides were extracted with 5% trifluoroacetic acid in 50% CH3CN for 1 h, spotted onto a matrix-assisted laser desorption ionization (MALDI) target plate to combine with CHCA matrix (Sigma-Aldrich Co.), and then analyzed with a 4800 MALDI-time of flight (TOF)/TOF analyzer (Applied Biosystems/MDS SCIEX) and Mascot software (Matrix Science Co., Tokyo, Japan) with the significance criteria of the program (P < 0.05).
N-terminal amino acid sequencing.
The outer membrane fraction from WFS1 cells harboring pTCBex33277fimS was further fractionated by SDS-PAGE with 7.5 to 15% polyacrylamide gradient gels and then electrotransferred onto a Trans-Blot PVDF membrane (Bio-Rad Laboratories). The single band corresponding to the W83 FimA polypeptide was visualized by staining with CBB and excised from the membrane. Approximately 1.5 μg of W83 FimA polypeptide was subjected to N-terminal peptide sequence analysis by the gas-phase Edman degradation method with Procise 492 cLC (Applied Biosystems).
Transmission electron microscopy (TEM).
P. gingivalis cells were grown in sTSBHK to late logarithmic phase and placed on a high-resolution carbon substrate (Ohkenshoji Co., Tokyo, Japan), followed by negative staining with 1% ammonium molybdate for 1 min. The samples were examined with a transmission electron microscope (LEO LIBRA 120 [Carl Zeiss SMT AG, Oberkochen, Germany] or JEM-1210 [JEOL, Tokyo, Japan]).
RESULTS
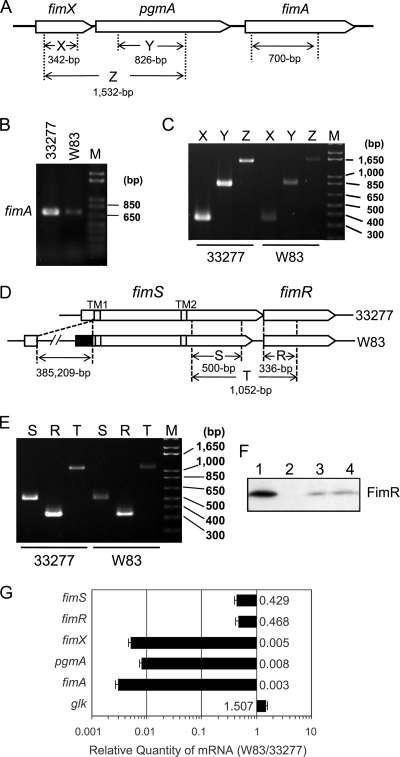
Decreased expression of the fim gene cluster and the fimSR operon in W83.
Previous studies have established that the FimSR TCS plays an essential role in promoting the expression of fimA in the fimbriate strain ATCC 33277 (30). The response regulator FimR directly activates the expression of fimX, which in turn plays a direct positive role in upregulating the expression of the downstream genes pgmA and fimA (31). As an initial step of this study, the expression of fimA, fimX, and pgmA, as well as their transcript organization, in strains 33277 and W83 were examined by RT-PCR using primer sets depicted in Fig. 1A. RNA samples were extracted from cells grown in a standard broth (sTSBHK) and harvested at late log or early stationary phase, because we showed in a previous study that strain 33277 produces FimA fimbriae throughout the growth cycle under the standard broth culture conditions, and also the FimSR TCS was shown to be operational throughout the growth cycle under the same conditions (31). As shown in Fig. 1B, the transcription of fimA in W83 was significantly less than that in 33277. For fimX and pgmA expression (Fig. 1C), a primer set covering both genes amplified cDNA fragments of the predicted size in both ATCC 33277 and W83, indicating that fimX and pgmA were cotranscribed as a monocistronic transcript. This result is consistent with the previous observation that an insertional mutation of the fimX locus abolished the transcription of pgmA (31). However, the level of the expression of the operon in W83 was significantly less than that in 33277, demonstrating that the FimR-FimX-PgmA-FimA regulation cascade is defective in W83, which is consistent with its afimbriate phenotype. We further compared the transcription of fimS-fimR between the two strains by RT-PCR, because W83 is markedly different from 33277 in the 5′ upstream noncoding and coding region of fimS due to the insertion of a 385,209-bp fragment (BROP [http://genome.brop.org/]; The Institute for Genomic Research [TIGR]) (Fig. 1D) (14), and such an alteration in the promoter region could negatively affect the expression of fimS in W83. As shown in Fig. 1E, the results were as anticipated, in that the expression of both fimS and fimR was less in W83 than in 33277. These experiments also revealed that fimS and fimR were cotranscribed as an operon in both strains. In the previous study, we found promoter activity in the C-terminal coding region of fimS (pfR) that is dedicated to the transcription of fimR in a fimS expression-independent manner (30). Taking this result together with those obtained in the present study, we conclude that transcript organization around the fimSR loci is polycistronic. Western blot analysis (Fig. 1F) confirmed the expression of fimR gene products in strains ATCC 33277 and W83; however, a decreased but detectable amount of FimR was produced in W83, equivalent to that produced by a fimS knockout (ko) mutant of 33277 that was driven by the promoter region pfR.
FIG. 1.
Reduced expression of fimSR and fim gene cluster in W83. (A) The structure of fimX-pgmA and fimA loci and the design of amplicons to be generated by RT-PCR. (B) Semiquantitative RT-PCR of fimA. After 22 PCR cycles with 300 ng of cDNA or 400 ng of RNA (negative control; data not shown) as a template, 10 μl out of 50 μl of each reaction mixture was run on a 1.5% agarose gel. (C) Semiquantitative RT-PCR (25 cycles) of the fimX-pgmA operon with reaction conditions similar to those described for panel B. (D) Schematic diagrams of fimSR loci in 33277 and W83. TM1 and TM2 indicate predicted transmembrane regions. In W83, the N-terminal cytoplasmic domain-coding region, derived from the 385-kbp insertional fragment, is depicted in black. Amplicons to be generated by RT-PCR also are indicated. (E) Semiquantitative RT-PCR of fimSR with 300 ng cDNA or 400 ng RNA (negative control; data not shown) as a template. After 25 cycles of the PCR, 10 μl out of 50 μl of each reaction mixture was run on a 2% agarose gel. M, DNA ladder marker. (F) Production of FimR protein in 33277 (lane 1), the fimR mutant (lane 2, AG21-5), the fimS mutant (lane 3, AGFS1), and W83 (lane 4). Total protein samples from each strain, adjusted to equal cell numbers, were fractionated by SDS-PAGE, transferred to a PVDF membrane, and probed with anti-FimR antibody. (G) Relative quantitation of fimSR and fimA-related gene expression in strains W83 and ATCC 33277. Reaction mixtures containing 100 ng of cDNA from either W83 or 33277 total RNA were subjected to real-time PCR as described in Materials and Methods. The expression of target genes was normalized to the endogenous 16S rRNA in each strain. The results are presented as ratios of target mRNA levels in W83 to those in 33277 by the calculation of 2−ΔΔCT, as described in Materials and Methods and Table S1 in the supplemental material. The error bar indicates standard deviations.
Moderate decrease of fimSR expression and a significant decrease of fim gene cluster expression in W83.
To quantify the expression levels of fimSR and their target fim gene cluster components in strains W83 and 33277 more, we carried out relative quantitation assays by the comparative CT method (ΔΔCT) (20) using real-time PCR. As shown in Fig. 1G, mRNA levels of fimSR in W83 were estimated to be 40 to 50% of those in 33277. On the other hand, the expression levels of fimX, pgmA, and fimA in W83 all were less than 1% of those in 33277 (0.495, 0.757, and 0.312%, respectively), indicating a significant decrease of the expression of fim gene cluster components compared to the moderate decrease of fimSR expression in W83. In this experiment, the expression of a glucose kinase gene (glk) also was analyzed as the internal control gene, and the level in W83 was estimated to be 1.5-fold of that in 33277. This result is consistent with a previous study by Naito et al. showing that the expression level of glk in W83 was approximately 2-fold that in 33277 (26), supporting the reliability of the data in this study. These results indicate that merely decreased expression of the TCS itself is not sufficient to explain the significant reduction in the transcriptional activation of the fim gene cluster in W83, and they suggest that there are additional defects in the regulation of FimA production that result in the distinct fimbrial phenotypes of the two strains.
Transcriptional start site of fimS is disrupted in W83.
As described above, due to the insertion of a 385-kbp DNA fragment into the fimS locus in W83, the start codon assigned by TIGR resulted in an extended N-terminal cytoplasmic region of W83 FimS (Fig. 1D). To confirm the transcriptional start site (TS) of fimS experimentally, 5′ RLM-RACE was performed (Fig. 2). The specific amplification of TS-containing fragments was verified by using a set of three different gene-specific primers (GSP1 to GSP3) as depicted in Fig. 2A. The nested PCR products obtained by using a 5′ adapter primer and 3′ GSPs comprise a 29-bp adapter-derived sequence attached to 5′ ends of fimS cDNA. As shown in Fig. 2B, all GSP primers amplified adapter-ligated cDNA fragments from 33277 of length sufficient to contain the previously predicted start codon (Fig. 2B, left) (14), indicating that in 33277 fimS RNA actually is translated from the start codon. In addition, shorter fragments of approximately 80 and 60 bp also were amplified by GSP1 and GSP2 primers, respectively; however, they apparently were too short to code for the N-terminal cytoplasmic region and transmembrane region 1 (TM1; I15 to K34). The additional sequencing of the extra shorter amplicons from 33277 by GSP1 and GSP2 primers revealed deletions of internal sequences, presumably due to the secondary structure specific to 33277 fimS cDNA (data not shown). W83 fimS TS-containing fragments also were amplified using the same sets of primers, and the amplicon generated from any GSP was about 30 bp shorter than that from 33277 (Fig. 2B, right), indicating that the TS of W83 fimS is located about 30 bp downstream of that of 33277 fimS. The amplicons by GSP3 primer were further analyzed by direct sequencing and indicated A−18 as a TS candidate for 33277 fimS and G35 for W83 fimS (Fig. 2A, boxed). Consequently, the predicted alternative start codon for W83 fimS is A55, and translation starts at M19. In this case, the predicted cytoplasmic region is truncated at the N terminus. Therefore, it is apparent that the original transcriptional start site of fimS is disrupted in W83 by the insertion of the large fragment.
FIG. 2.
Comparison of fimS transcriptional start sites (TS) from 33277 and W83 by 5′ RLM-RACE. (A) Alignments of 33277 (327) and W83 fimS sequences around the FimS N-terminal coding regions. Originally predicted amino acid-coding sequences are shown in uppercase letters, and sequences from the inserted fragment in W83 are shown in italics. Arrows indicate three gene-specific reverse primers used for 5′ RACE (Table 1). (B) Results of nested PCR of 5′ RACE. Reaction mixtures were separated on a 3.5% agarose gel. The PCR products shown in lanes 1, 2, and 3 in both panels were amplified by using reverse primers GSP1, GSP2, and GSP3, respectively. The fragments indicated by arrowheads are TS-containing PCR products with the 29-bp 5′ adapter sequence. The products by GSP3 primer from both strains (lane 3) also were extracted and sequenced to determine TSs, which are boxed in Fig. 2A (A−18 for 33277 and G35 for W83).
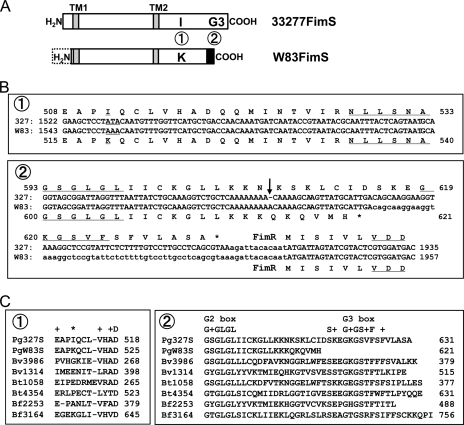
Different structures of FimS kinase domains in strains ATCC 33277 and W83.
Although the production of FimR in W83 was modest (Fig. 1F), the protein still was detectable and should be functional because its amino acid sequence is almost identical to that of 33277, except for the alteration of one residue from serine to threonine (S209T; data not shown). The reduced transcription of the fim gene cluster also may be a consequence of a nonfunctional FimS histidine kinase in W83. Indeed, a comparison of fimS genes revealed two more regions that differ between 33277 and W83 (14), and these differences cause distinct primary structures in the C-terminal histidine kinase domain (Fig. 3A). The alteration of a single nucleotide located upstream of the N box coding region results in a missense mutation from isoleucine to lysine (ATA to AAA) (Fig. 3B, sequence box 1). The other is the insertion of an A downstream of the G2 box coding region that generates a frameshift mutation that shortens the C-terminal region of the kinase (Fig. 3B, sequence box 2). Such differences in the primary structure could affect the functionality of W83 FimS as a histidine kinase. It especially should be noted that, by multiple alignment with several histidine kinases from other Bacteroidetes, the truncated C-terminal region of W83 FimS results in the loss of a predicted G3 box, GKGSVF (Fig. 3C, sequence box 2), a newly discovered conserved motif responsible for providing an ATP-binding pocket (7).
FIG. 3.
Mutations found in the kinase domain-coding region of W83 fimS. (A) Two regions in the kinase domain of W83 FimS differ from that of 33277 FimS. 1, a single-amino-acid substitution from isoleucine to lysine; 2, truncation of the C-terminal region containing the G3 box. (B) Alignments of fimS nucleotide and deduced amino acid sequences around the FimS kinase domain. 1, a missense mutation (ATA [I511] to AAA [K518]) is located upstream of the N box (NLLSNA) coding region; 2, an insertion of A (indicated by an arrow) is found downstream of the G2 box (GSGLGL) coding region of W83. This insertion caused a frameshift mutation and truncated the C-terminal coding region that contains the predicted G3 box (GKGSVF). The VDD motif in FimR is conserved in the receiver domains of response regulators. (C) Alignments of amino acid sequences around the distinct regions, 1 and 2, of 33277 and W83 FimS (Pg327S and PgW83S, respectively) with histidine kinases from other Bacteroidetes. Bv, Bacteroides vulgatus ATCC 8482; Bt, Bacteroides thetaiotaomicron VPI-5482; and Bf, Bacteroides fragilis NCTC9343. Numbers followed by Bv, Bt, and Bf represent gene locus numbers of histidine kinases in each organism. Note that the predicted G3 box (G+GS+F) in region 2 is highly conserved beyond species, whereas the amino acid indicated by an asterisk in region 1 is not.
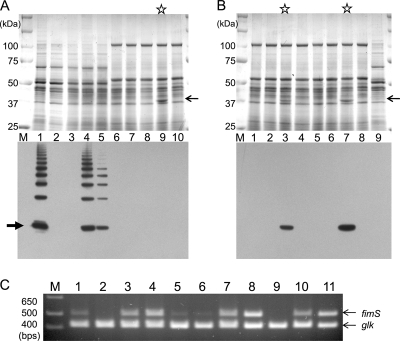
Reciprocal exchange of fimS genes establishes that W83 FimS is functionally deficient.
We developed a gene exchange system to compare the functionality of FimS variants from the different strains. The fimS genes from each strain were subcloned into a pT-COW-based expression vector, pTCBex (25, 30), and introduced into fimS insertion mutants of 33277 and W83 (AGFS1 and WFS1, respectively). The expression of the cloned genes was driven by the same promoter (pfR) on pTCBex, and the functionality of the FimS variants was evaluated by comparing the induced levels of FimA in the same genetic background, i.e., in either ATCC 33277 or W83 with disrupted fimS. To compare the production of FimA induced by the complementation of fimS variants, whole-cell samples were subjected to Western blot analyses with anti-type I FimA antibody (Fig. 4A). In strain AGFS1 complemented with 33277-type fimS (designated 33277 fimS), FimA was induced to wild-type ATCC 33277 levels (Fig. 4A, lower, lanes 1 and 4). However, fimS from strain W83 (W83 fimS) induced a much lower level of FimA in the ATCC 33277 genetic background (Fig. 4A, lower, lane 5; less than 24% of the 33277 fimS-complemented level shown in lane 4), clearly indicating the reduced functionality of the W83 FimS kinase. On the other hand, after the transfer of 33277 fimS into WFS1 (fimS-disrupted W83), a FimA signal was not detected in Western blots probed with anti-type I FimA antibody (Fig. 4A, lower, lane 9). However, the inspection of the CBB-stained gels revealed that the 33277 fimS-complemented WFS1 apparently produced a substantial level of a protein with the size of around 40 kDa (Fig. 4A, upper, lane 9; indicated by an arrow). We assumed that this protein was type IV FimA induced by 33277 FimS and excised the corresponding 40-kDa band from the gel to identify the protein by a mass analysis. The destained band was digested with trypsin, and the extracted sample was subjected to MALDI-TOF/TOF analysis. As anticipated, analysis showed that the protein was identical to W83 FimA (Mowse score 84; data not shown). Thus, we reasoned that the failure to detect a signal by Western blotting was due to the poor cross-reactivity of the anti-type I FimA antibody, and we therefore raised anti-type IV FimA antibody against a W83 FimA-specific peptide (PGGTASDNLVSAGT) to use in additional Western analyses. As expected, the 40-kDa protein induced by the 33277 fimS complementation of WFS1 reacted exclusively with the anti-type IV FimA antibody (Fig. 4B, lower, lane 7). Interestingly, in contrast to the results in the ATCC 33277 background (Fig. 4A, lower, lanes 4 and 5), the FimA signal in the W83 background was detected as a clear single band, not an oligomer ladder, and was detectable only after 33277 fimS complementation (Fig. 4B, lower, lanes 7 and 8). The expression of the introduced fimS constructs were examined by RT-PCR, which confirmed that both types of fimS were successfully transcribed in each fimS ko strain (Fig. 4C). These results provide robust evidence of the malfunctionality of W83-type FimS. The lack of FimA in both wild-type W83 and WFS1 with W83 fimS complementation (Fig. 4B, lanes 1 and 8) establishes that this malfunctionality is the major reason for fimbria deficiency in W83. Additional supportive evidence indicating the malfunctionality of W83 FimS was obtained by introducing the 33277 fimS construct into wild-type W83. In this case, the production of W83 FimA was lower than that in the 33277 fimS-complemented WFS1 (Fig. 4B, lower, lanes 3 and 7), suggesting competitive interference by the endogenous W83 FimS with the introduced functional 33277 FimS. Furthermore, the overexpression of the W83 fimS construct in wild-type W83 (Fig. 4C, lane 8) did not induce detectable levels of FimA (Fig. 4B, lower panel, lane 4), indicating that the decreased expression of the endogenous fimSR operon, as shown in Fig. 1G, is not the major reason for the fimbria-less phenotype of this strain.
FIG. 4.
Phenotype of fimS-exchanged strains. (A) CBB-stained SDS-12.5% PAGE gel (upper) and Western blot (lower) with anti-type I FimA antibody. Total protein samples were denatured at 80°C for 20 min before SDS-PAGE. Lane 1, 33277; 2, AGFS1; 3, AGFS1(pTCBex); 4, AGFS1(pTCBex33277fimS); 5, AGFS1(pTCBexW83fimS); 6, W83; 7, WFS1; 8, WFS1(pTCBex); 9, WFS1(pTCBex33277fimS); 10, WFS1(pTCBexW83fimS). The CBB-stained single band of W83 FimA in lane 9 is indicated by a thin arrow. A thick arrow indicates the signals from 41-kDa monomers. (B) CBB-stained SDS-12.5% PAGE gel (upper) and Western blot with anti-W83 FimA antibody (lower). Total protein samples were denatured at 80°C for 20 min before being subjected to SDS-PAGE. Lane 1, W83; 2, W83(pTCBex); 3, W83(pTCBex33277fimS); 4, W83(pTCBexW83fimS); 5, WFS1; 6, WFS1(pTCBex); 7, WFS1(pTCBex33277fimS); 8, WFS1(pTCBexW83fimS); 9, 33277. Stained bands corresponding to W83 FimA in lanes 3 and 7 are indicated by an arrow. (C) RT-PCR confirming the expression of fimS in the complemented fimS knockout mutants. The 500-bp fimS cDNAs were amplified with the primer set shown in Fig. 1D and Table 1. After 25 cycles of PCR, an equal volume of each reaction mixture was fractionated by 3% AGE. The glucose kinase gene (glk; 415 bp) was coamplified in each reaction as an internal control. Lane 1, 33277; 2, AGFS1(pTCBex); 3, AGFS1(pTCBex33277fimS); 4, AGFS1(pTCBexW83fimS); 5, W83; 6, W83(pTCBex); 7, W83(pTCBex33277fimS); 8, W83(pTCBexW83fimS); 9, WFS1(pTCBex); 10, WFS1(pTCBex33277fimS); 11, WFS1(pTCBexW83fimS).
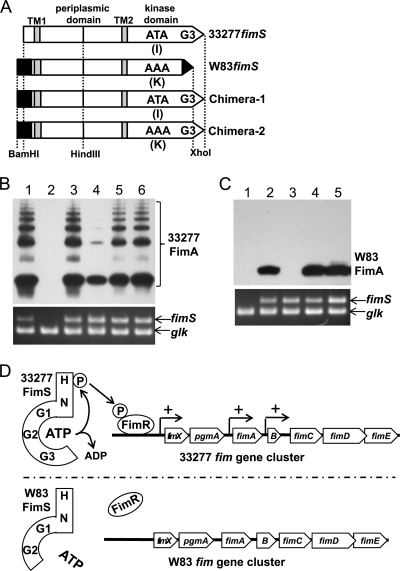
W83-33277 FimS chimeras demonstrate that truncation of the G3 box is responsible for the malfunctionality of W83 FimS.
Given the findings that FimS from W83 is malfunctional and that its structure differs from that of ATCC 33277 in three regions (Fig. 2 and 3), we investigated further which regions negatively affected the function of W83 FimS. A chimeric fimS was constructed in which the kinase domain of W83 FimS was exchanged with that of 33277 FimS (Chimera-1 in Fig. 5A) and expressed in trans in the fimS-disrupted strains. The 5′ region of the ORF, as well as that of W83, was intentionally extended according to the original assignment by TIGR. However, it was confirmed again by 5′ RACE that the transcription started from the same site as that shown in Fig. 2 (data not shown). The functionality of the chimera was evaluated by the ability to induce the production of endogenous FimA as described above. As shown in Fig. 5B, complementation by the chimeric fimS in the fimS ko 33277 background (AGFS1) restored substantial expression of FimA that was similar to that of the 33277 wild-type and 33277 fimS-complemented AGFS1 (lane 5). The same result was obtained in a similar analysis with the fimS ko W83 background (WFS1) (Fig. 5C, lane 4). These results indicated that mutations in the kinase domain-coding region are responsible for the malfunctionality of W83 FimS and also suggested that the difference in the N-terminal cytoplasmic region before TM1 does not affect the function of FimS, at least with regard to the induction of FimA expression. Further complementation analysis with a second type of fimS chimera (Chimera-2 in Fig. 5A) that codes for a W83 FimS variant fused with the G3 box from 33277 FimS again showed the full restoration of FimA expression in both genetic backgrounds (Fig. 5B, lane 6, and C, lane 5). These results were as expected from the distinct conservation of sequences (Fig. 3C) and clearly indicated that this truncation of the G3 box-containing C-terminal region (region 2 in Fig. 3) is responsible for the malfunctionality of W83 FimS, and not the I-to-K amino acid substitution due to the missense mutation (region 1 in Fig. 3).
FIG. 5.
Truncation of G3 box is responsible for functional deficiency of W83 FimS. (A) Design of W83-33277 fimS chimeras. Chimera-1 consists of an N-terminal cytoplasmic domain-coding region from W83 and C-terminal kinase domain-coding region from 33277 possessing the G3 box. Chimera-2 is a derivative of Chimera-1 with the same missense mutation, AAA, as that in W83 fimS. (B) Upper panel, a Western blot with anti-type I FimA antibodies showing induced levels of 33277 FimA by the complementation of the fimS knockout ATCC 33277 with fimS variants. Samples were heated at 80°C for 20 min prior to SDS-12.5% PAGE to observe the polymerization of FimA. Lower panel, expression of introduced fimS revealed by RT-PCR. The coamplified glucose kinase gene (glk) was an internal control. Lane 1, 33277; 2, AGFS1(pTCBex); 3, AGFS1(pTCBex33277fimS); 4, AGFS1(pTCBexW83fimS); 5, AGFS1(pTCBexChimera-1); 6, AGFS1(pTCBexChimera-2). (C) Upper panel, a Western blot with anti-type IV FimA antibodies showing induced levels of W83 FimA by the complementation of the fimS knockout W83 with fimS variants. Samples were boiled for 20 min before SDS-PAGE (12.5%) to maximize signal intensities for detection. Lower panel, expression of introduced fimS revealed by RT-PCR. Lane 1, WFS1(pTCBex); 2, WFS1(pTCBex33277fimS); 3, WFS1(pTCBexW83fimS); 4, WFS1(pTCBexChimera-1); 5, WFS1(pTCBexChimera-2). (D) Summarizing model. The G3 box, in addition to G1 and G2 boxes, in the C terminus of the 33277 FimS kinase domain forms a pocket to bind ATP, which autophosphorylates the histidine residue of the H box. The phosphoryl group then is transferred to the cognate response regulator, FimR, to activate its function as a positive regulator of the fim gene cluster. The truncation of the G3 box in W83 FimS renders it defective in ATP capture, leading to the disruption of the subsequent phosphorelay.
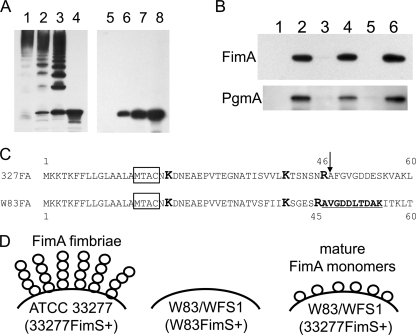
33277 FimS-induced W83 type IV FimA is localized in the outer membrane fraction as a mature polypeptide.
As shown in Fig. 4 and 5, heating at 80°C is an optimal partially denaturing condition to detect the normally polymerized structure of type I FimA fimbriae as a ladder-like signal in Western blots. It is not known, however, whether such a condition is applicable to other FimA genotypes. Therefore, cells expressing W83 FimA were treated at lower temperatures and subjected to similar Western analyses (Fig. 6A). The sample from ATCC 33277 control cells showed signals of high-molecular-weight FimA polymer at 37°C, with released oligomers increasing from 50 to 80°C, until only a single band signal of a 41-kDa monomer was obtained after treatment at 100°C (lanes 1 to 4). In contrast, no signals of higher-molecular-weight polymers or oligomers were observed in the W83 FimA-expressing cell at all temperatures examined (lanes 5 to 8). This result raised the possibility that W83 FimA is not polymerized but remains in or attached to the cell as a monomer, and this prompted further examination of its cellular localization. Cells were disrupted and fractionated according to an established protocol (24) and then subjected to Western analyses with anti-W83 FimA antibody. At the same time, the fractions also were probed with anti-PgmA to validate the fractionation (15). The result clearly indicated that the W83 FimA was localized almost exclusively in the outer membrane fraction (Fig. 6B, lane 6).
FIG. 6.
33277 FimS-induced W83 FimA is a mature polypeptide and is localized on the cell surface. (A) Whole cells were heat treated at 37°C (lanes 1 and 5), 50°C (lanes 2 and 6), 80°C (lanes 3 and 7), and 100°C (lanes 4 and 8) and fractionated by SDS-PAGE, and Western blots were probed with strain-specific anti-FimA antibody. Left, ATCC 33277; right, WFS1(pTCBex33277fimS). (B) Localization of W83 FimA in WFS1. WFS1(pTCBex33277fimS) cells were fractioned and analyzed by Western blotting. Lane 1, culture supernatant; 2, whole cell; 3, cytoplasm/periplasm; 4, envelope; 5, inner membrane; 6, outer membrane. The upper panel shows a membrane probed with anti-W83 FimA antibody. The same samples were transferred onto an equivalent membrane and probed with anti-recombinant PgmA antibody to validate the fractionation (lower panel). (C) Alignment of N-terminal sequences of 33277 and W83 FimA proteins. Boxed sequences (MTAC) are the lipoprotein sorting signals, and lysine (K) and arginine (R) residues of gingipain cleavage sites are highlighted in boldface and a larger size (35). An arrow indicates the first residue (A47) of mature 33277 FimA. The first 10 residues of W83 FimA from outer membrane fraction, determined by an N-terminal peptide sequence analysis, are underlined and highlighted in boldface. (D) Schematic representations showing the distinct phenotype of FimA fimbriae in the P. gingivalis strains studied. Strain W83 lacks FimA fimbriae due to the deficiency in the positive regulation of the fim gene cluster, as depicted in Fig. 5D. In W83 or the fimS knockout derivative (WFS1) expressing 33277 FimS in trans, W83 FimA propeptides are transported to the cell surface and cleaved by RGP to mature monomers. The monomers remain at the cell surface and are not polymerized.
The N-terminal amino acid sequence of W83 FimA also was analyzed to examine its posttranslational processing (the extent of maturation). The outer membrane fraction from W83 FimA-expressing cells was fractionated further by SDS-PAGE and transferred to a PVDF membrane. After being stained with CBB, a single band corresponding to the W83 FimA was excised and subjected to sequence analysis by Edman degradation (17). Ten cycles of analysis revealed the first 10 amino acids, AVGDDLTDAK, indicating that A46 is the first residue of W83 FimA that is localized in the outer membrane fraction (Fig. 6C). The alignment of the N-terminal sequence of FimA from ATCC 33277 with that from W83 revealed that A46 of W83 FimA corresponds to A47 of ATCC 33277 FimA, corresponding to the N terminus of the mature polypeptide after digestion with Arg-gingipain (RGP) on the cell surface (35). These results indicate that in W83, type IV FimA is localized on the cell surface as a mature monomer (Fig. 6D).
To obtain morphological evidence that W83 FimA, if expressed, is not polymerized in the W83 parent or W83 fimS KO background, transmission electron micrographs (TEM) were taken of negatively stained cells. Fibrous structures were observed abundantly at the cell surfaces of the 33277 parent and the derivatives expressing functional FimS and modestly at those of the W83 fimS-complemented 33277 fimS mutant strain, but not of W83 parent or W83 fimS mutant strains even when its endogenous FimA, i.e., W83 FimA, was produced (data not shown). These results are consistent with those of the Western blots shown in Fig. 4B and 6A and clearly indicate that FimA subunits from W83 are not assembled as fimbriae regardless of expression level.
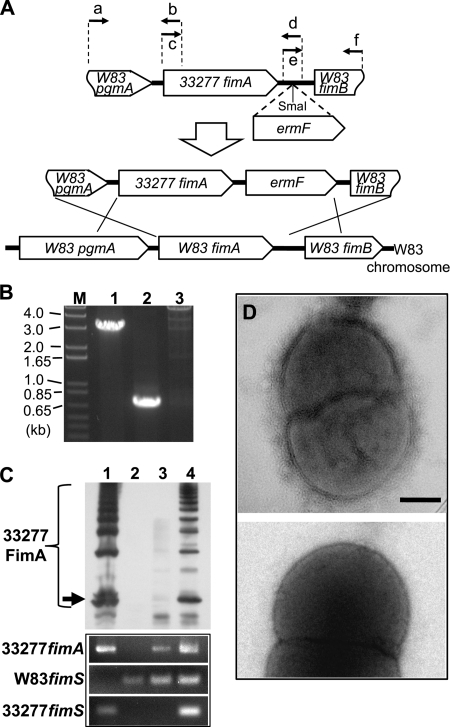
W83 retains the ability to assemble 33277 FimA.
The data shown above raised the question of why strain W83 still was not able to assemble fimbriae even if endogenous type IV FimA was abundantly produced after the introduction of functional fimS constructs. We assumed that two possibilities accounted for this phenotype: either W83 is deficient in assembling FimA subunits regardless of genotype, or assembly is dependent on the FimA genotype. To examine these hypotheses, we generated an additional W83 derivative in which the fimA locus was exchanged for type I 33277 fimA (Fig. 7A). An ermF selection marker cassette was inserted at the center of the intergenic region between fimA and fimB, because previous studies have shown that transcripts of fimA and the downstream genes fimB to fimE are separate, and the disruption of fimB to fimE does not abolish FimA polymerization (31, 32, 44). Genomic DNAs from the erythromycin-resistant transformants were analyzed to confirm their allelic exchange by double crossover recombination (Fig. 7B). One of the confirmed clones, W83fA327, was examined further by Western blotting for its FimA phenotype after the introduction of either an empty expression vector (pTCBex) or the vector carrying 33277 fimS. Interestingly, a trace level of a ladder-like signal of FimA was detected from W83fA327 carrying an empty vector (Fig. 7C, lane 3), and the level was drastically elevated by the introduction and expression of 33277 fimS (Fig. 7C, lane 4). These results clearly demonstrate that even the fimbria-deficient W83 still retains the ability to polymerize 33277-type FimA subunits, and that the substantial production level of the FimA fimbriae is achieved by the expression of functional FimS. We further examined the fimbriation of the W83 derivative harboring both 33277 fimA and 33277 fimS by TEM, and we found that the cells produced many fibrillar structures from the cell surface (Fig. 7D, upper). This result indicates that W83 retains all components and functions required for assembling 33277-type FimA subunits to fimbriae.
FIG. 7.
W83 retains the ability to assemble 33277-type FimA fimbriae. (A) Construction of fimA-exchanged W83 variant W83fA327. A chimeric DNA fragment comprised of the C-terminal coding region of W83 pgmA, full-length 33277 fimA, the N-terminal coding region of W83 fimB, and intergenic sequences from W83 was generated by a PCR-based overlap extension, as described in Materials and Methods. After the insertion of an ermF cassette, the final construct was introduced into W83 by electroporation. (B) Confirmation of allelic exchange by genomic DNA PCR. Lane 1, amplicons with primers a and f (1,968 bp + 1,095 bp of ermF = 3,063 bp; this amplicon also was sequenced); lane 2, amplicons with specific primers for 33277 fimA (700 bp); lane 3, no 700-bp amplicons with W83 fimA-specific primers. (C) Complementation of the fimA-exchanged W83 with the functional 33277 fimS gene is sufficient for the assembly of FimA fimbriae. The upper panel shows a Western blot of 80°C heat-treated whole-cell proteins from 33277 (lane 1), W83 (lane 2), W83fA327 harboring pTCBex (lane 3), and W83fA327 harboring pTCBex33277fimS (lane 4) probed with anti-33277 FimA antibody. Ladder signals are from polymerized FimA. An arrow indicates the signals from 41-kDa monomers. Lower panels represent the expression profile of each strain by RT-PCR. (D) Morphological observation of fimbrial assembly by transmission electron microscopy. The upper panel shows fimbriate W83fA327 harboring pTCBex33277fimS. The lower panel shows a fimbria-less W83 derivative expressing W83 fimA and 33277 fimS (i.e., strain WFS1 harboring pTCBex33277fimS). Scale bar, 200 nm.
DISCUSSION
In the present study, we discovered two deficiencies in the FimA biogenesis pathway of strain W83 that account for its afimbriate phenotype. First, FimA production is drastically reduced because of a mutated FimS histidine kinase. Early in this study, we showed that the N-terminal coding region of W83 fimS contained a 385-kbp insertion that resulted in the decreased expression of fimSR and a potential disruption of the FimS N-terminal cytoplasmic domain. It became apparent from later analyses that this decreased expression of fimSR was not responsible for the inability of FimS to induce FimA production, because elevating the expression level of W83 fimS in wild-type W83 still did not increase FimA production to detectable levels. However, exchanging the kinase domain of W83 FimS with that of the functional 33277 FimS restored FimA production to 33277 levels. The further examination of the W83 FimS-coding DNA sequence revealed a frameshift mutation that resulted in a loss of the C-terminal region containing a G3 box ATP-binding motif. As anticipated, only switching the C-terminal region of W83 FimS to that of 33277 FimS, which contained an intact G3 box, was sufficient to restore the full production of FimA. Losing a G3 box would negatively affect the ATP-binding ability of W83 FimS, causally decreasing, if not completely inhibiting, its ability to autophosphorylate the conserved histidine residue in the H box and subsequent phosphorelay to the cognate FimR response regulator (Fig. 5D). Nonphosphorylated FimR should not function in the positive regulation of the fim gene cluster. To substantiate this model, these phosphorylation assays will be the focus of future work.
The nucleotide sequencing of fimSR from another afimbriate strain, W50, revealed that these genomic loci were identical to those of W83 (data not shown). It is already known that fimA from both strains are identical (GenBank accession no. D38373 for W50, AE015924 and gi:34398108 for W83); therefore, we predict that the same fimS mutations that result in the afimbriate phenotype of W83 are responsible for the same phenotype in strain W50.
The second reason for the afimbriate phenotype of W83 is that even when FimA production was induced by introducing a functional FimS, FimA protein was retained in the outer membrane fraction and not polymerized to form fimbriae. The N-terminal amino acid sequences of the nonpolymerized subunits revealed that they are the mature polypeptide, the result of processing by Arg-gingipain (RGP) at the cell surface (35). Therefore, W83 retains almost all of the components and mechanisms required to express, translocate, and process FimA to the mature polypeptide. Furthermore, W83 was shown to be able to polymerize 33277-type FimA to form fimbriae. A CLUSTALW analysis showed only 46.4% primary sequence identity between mature FimA from W83 and that from 33277, and the prediction of the secondary structures by the new joint method (1) revealed many regions with structural differences throughout the mature W83 FimA polypeptide (data not shown). These results demonstrate that the distinct primary structure of FimA itself leads to the inability to assemble FimA fimbriae. W83 FimA could lose a functional interface domain that is required for polymerization, or, even if this domain is conserved, overall changes in higher structures could interfere with subunit polymerization.
In E. coli, the mature structural subunits of both P and type 1 pili are assembled by the chaperon/usher pathway in which the N-terminal extension motif (Nte) of the subunits is required for donor-strand exchange (DSE) in the periplasm (34). In P. gingivalis, PgmA, with its gene location adjacent to the fim gene cluster and gene product localized in the outer membrane fraction, could be a candidate for the usher in the FimA fimbriation. However, several lines of evidence show inconsistency with this model. First, PgmA does not share homology with known ushers. Second, a pgmA ko mutant still polymerizes FimA subunits and produces fimbriae (16 and K. Nishikawa, unpublished data). Third, disruptions of fimC, fimD, or fimE in the downstream portion of the fim cluster still exhibit a fimbria-positive phenotype (32, 44), i.e., so far no gene within the cluster has been found to code for known usher-like proteins. Fourth, the N-terminal region of polymerizable 33277 FimA does not contain the Nte motif. Fifth, in P. gingivalis immature FimA is retained in the periplasm prior to processing to the mature protein by RGP at the cell surface (35). Thus, our findings, as well as previous studies, suggest that FimA subunits are assembled by either a chaperon/usher-independent pathway or a chaperon/usher-like pathway with functionally, but not structurally, equivalent components. A simple working model deduced from the present findings with ATCC 33277 is that, after the FimA propeptide is transported to the cell surface and cleaved by RGP, the mature polypeptide autopolymerizes in an amino acid sequence-dependent manner. This model is one of the plausible hypotheses, but we do not rule out other possibilities, including that the polymerization of FimA subunits depends on an interaction with other component(s) expressed by both strains. If this is the case, the structural alteration would prevent W83 FimA from associating with minor components or unknown factors that are essential for assembly. Pertinent issues to be addressed in a future study include the identification of functional domains in the FimA subunit that are critical for polymerization. Nevertheless, it should be emphasized that, whatever the strain or genotype of fimA, the expression of a functional FimS is essential for the substantial expression of FimA in P. gingivalis.
The complementation of fimS ko 33277 by W83 fimS still induced a modest level of FimA (Fig. 4A, lane 5, and 5B, lane 4), indicating that the enzymatic activity of W83 FimS is not lost completely. However, when the same W83 fimS construct was introduced and expressed in the W83 background, no signal for FimA was detected (Fig. 4B, lanes 4 and 8, and Fig. 5C, lane 3). We clearly showed by the generation of AGFS1 that the disruption of the fimS locus is sufficient to inhibit the expression of fimA, and also that the full production of FimA is recoverable only by the complementation with functional fimS genes. A possible explanation for the inhibition of W83 FimA production in the W83 background is that W83 FimA is unstable or more susceptible to degradation than 33277 FimA due to its different amino acid sequence.
Previously, we suggested that FimS is constitutively activated under standard laboratory growth conditions, since strain ATCC 33277 showed the constitutive and progressive production of FimA from the early growth phase (31). It was reported recently that FimA production by strain 33277 was decreased under nutrient-limited culture conditions (21). This result implied that an environmental cue for FimS activation is a component of rich media, so that cells would be continually exposed to signal input and FimS kept active until the component was depleted. Our genetic analyses provide preliminary evidence of how these environmental signals are perceived. In complementation assays, we showed evidence of the competitive interference of the functional 33277 FimS by the defective endogenous W83 FimS (Fig. 4B, lanes 3 and 7). Although there is no experimental evidence to date, it is possible that FimS functions as a dimer, like many known histidine kinases. If this is the case, a heterodimer containing FimS from 33277 and W83 would contain only the functional kinase domain from 33277, while a W83 FimS homodimer would contain only malfunctional kinase domains. In spite of variations in their kinase domains, amino acid sequences of the FimS periplasmic regions in strains 33277, W83, and W50 are identical, indicative of how critical these sequences are to FimS function. Thus, it is possible that the competitive interference observed between FimS proteins of W83 and 33277 is the result of both copies having identical and functional sensor domains that respond to the same environmental signal. Supporting this notion is the existence of a TPR motif in the center of the periplasmic domain. Such motifs are known to provide interfaces for protein-protein interactions, and also bind to various types of compounds, including peptides and polyunsaturated fatty acids (5).
The characteristic symptoms of adult periodontitis, i.e., persistent chronicity with the recurrence of acute inflammatory episodes, could be explained, in part, by the ability of P. gingivalis to invade human gingival cells (3). Several studies established a role of FimA fimbriae in host cell invasion by P. gingivalis (18, 28, 33, 40, 42), and the observation that recombinant FimA-coated beads mediated the invasion of epithelial cells (27) suggests that even unpolymerized FimA subunits have invasive potential, and that strains with an abundant FimA monomers on their cell surfaces also can invade host gingival epithelial cells, as long as the FimSR TCS is functional. As found in the present study, defective W83 FimS drastically reduced FimA production, suggesting that W83 is deficient in FimA-dependent niche colonization, epithelial cell invasion, and the evasion of innate immune responses mediated by TLR2 (12). Future studies using animal models will test this prediction. The present study also raises questions regarding FimA production, the fimA genotype, and their relationship to periodontitis. Different fimA genotypes have been associated with periodontal disease activity in Japanese patients (2). However, recent work indicates that with more diverse populations, additional genotyping of, for example, regulatory genes may provide new epidemiological correlations and insights (8, 22, 23).
In summary, the fimS exchange experiments carried out in this study elucidated several of the steps involved in fimbria biogenesis in P. gingivalis. First, FimSR is the unique and universal regulatory system that activates the fim gene cluster in a fimA genotype-independent manner, because switching the G3 box-truncated, defective W83 FimS to the functional 33277 type was sufficient to induce the significant expression of endogenous fimA even in afimbriate strain W83. With regard to controlling the pathogenicity of P. gingivalis in the periodontal niche, we propose advantages in targeting this TCS over vaccination against FimA with its variable antigenicity, as shown in Fig. 4A. Second, in spite of minor differences in amino acid sequences between W83 and 33277 proteins, all of the regulatory components, i.e., FimR, FimX, and PgmA, are functional in W83, except for FimS. Third, the assembly of fimbriae is dependent on the primary structure of FimA. Furthermore, this is the first report establishing a direct link between the functionality of a histidine kinase and the expression of a major virulence factor of P. gingivalis, an important oral pathogen. Also, to our knowledge, this is the first example of a disruption in a two-component phosphorelay mechanism by a naturally occurring truncation of a G3 box. The complete genome sequences of the two P. gingivalis strains studied here now are available for comparison and, as we have demonstrated, comparative analyses of genes of interest and their products will enhance our understanding of the molecular mechanisms underlying their different phenotypic characteristics.
Supplementary Material
Acknowledgments
We thank T. Hirose of the Center for Instrumental Analysis, Hokkaido University, for help with N-terminal amino acid sequencing. Anti-type I FimA antiserum was provided by F. Yoshimura.
This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI 19592139 to K.N.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and by NIH grant DE015931 to M.D.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akiyama, Y., K. Onizuka, T. Noguchi, and M. Ando. 1998. Parallel protein information analysis (PAPIA) system running on a 64-node PC cluster, p. 131-140. In Proc. 9th Genome Informatics Workshop (GIW'98), Universal Academy Press, Tokyo, Japan. [PubMed]
- 2.Amano, A., I. Nakagawa, K. Kataoka, I. Morisaki, and S. Hamada. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontal patients. J. Clin. Microbiol. 37:1426-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 4.Chen, T., Y. Hosogi, K. Nishikawa, K. Abby, R. D. Fleischmann, J. Walling, and M. J. Duncan. 2004. Comparative whole genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 186:5473-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, A. K., P. T. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.Dutta, R., and M. Inouye. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25:24-28. [DOI] [PubMed] [Google Scholar]
- 8.Enersen, M., I. Olsen, Ø. Kvalheim, and D. A. Caugant. 2008. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J. Clin. Microbiol. 46:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Véscovi, E., F. C. Soncini, and E. A. Groisman. 1994. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res. Microbiol. 145:473-480. [DOI] [PubMed] [Google Scholar]
- 10.Gardner, R. G., J. B. Russell, D. B. Wilson, G. R. Wang, and N. B. Shoemaker. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed B-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B14. Appl. Environ. Microbiol. 62:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genco, C. A., C. W. Cutler, D. Kapczynski, H. Maloney, and R. R. Arnold. 1991. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajishengallis, G., M. Wang, S. Liang, M. Triantafilou, and K. Triantafilou. 2008. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc. Natl. Acad. Sci. USA 105:13532-13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamada, N., K. Watanabe, C. Sasakawa, M. Yoshikawa, F. Yoshimura, and T. Umemoto. 1994. Construction and characterization of a fimA mutant of Porphyromonas gingivalis. Infect. Immun. 62:1696-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, J., K. Nishikawa, R. Hirano, T. Noguchi, and F. Yoshimura. 2000. Identification of a two-component signal transduction system involved in fimbriation of Porphyromonas gingivalis. Microbiol. Immunol. 44:279-282. [DOI] [PubMed] [Google Scholar]
- 15.Hongo, H., E. Osano, M. Ozeki, T. Onoe, K. Watanabe, O. Honda, H. Nakamura, H. Tani, and F. Yoshimura. 1999. Characterization of an outer membrane protein gene, pgmA, and its gene product from Porphyromonas gingivalis. Microbiol. Immunol. 43:937-946. [DOI] [PubMed] [Google Scholar]
- 16.Hongo, H., H. Takano, and M. Morita. 2007. Dense fimbrial meshwork enhances Porphyromonas gingivalis adhesiveness: a scanning electron microscopic study. J. Periodont. Res. 42:114-118. [DOI] [PubMed] [Google Scholar]
- 17.Hunkapiller, M. W., and L. E. Hood. 1983. Microsequencing of proteins by gas phase Edman degradation, p. 23-36. In T. Y. Liu, S. Sakakibara, A. N. Schechter, K. Yugi, H. Yajima, and K. Yasunobu (ed.), Frontiers in biochemical and biophysical studies of proteins and membranes. Elsevier, New York, NY.
- 18.Kato, T., S. Kawai, K. Nakano, H. Inaba, M. Kuboniwa, I. Nakagawa, K. Tsuda, H. Omori, T. Ooshima, T. Yoshimori, and A. Amano. 2007. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell. Microbiol. 9:753-765. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. Y., H. T. Sojar, G. S. Bedi, and R. J. Genco. 1991. Porphyromonas (Bacteroides) gingivalis fimbrillin: size, amino-terminal sequences, and antigenic heterogeneity. Infect. Immun. 59:383-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 21.Masuda, T., Y. Murakami, T. Noguchi, and F. Yoshimura. 2006. Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl. Environ. Microbiol. 72:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Missailidis, C. G., J. E. Umeda, C. Ota-Tsuzuki, D. Anzai, and M. P. Mayer. 2004. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol. Immunol. 19:224-229. [DOI] [PubMed] [Google Scholar]
- 23.Miura, M., T. Hamachi, O. Fujise, and K. Maeda. 2005. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J. Periodont. Res. 40:147-152. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, Y., M. Imai, H. Nakamura, and F. Yoshimura. 2002. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 110:157-162. [DOI] [PubMed] [Google Scholar]
- 25.Nagano, K., Y. Murakami, K. Nishikawa, J. Sakakibara, K. Shimozato, and F. Yoshimura. 2007. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 56:1536-1548. [DOI] [PubMed] [Google Scholar]
- 26.Naito, M., H. Hirakawa, A. Yamashita, N. Ohara, M. Shoji, H. Yukitake, K. Nakayama, H. Toh, F. Yoshimura, S. Kuhara, M. Hattori, T. Hayashi, and K. Nakayama. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa, I., A. Amano, M. Kuboniwa, T. Nakamura, S. Kawabata, and S. Hamada. 2002. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect. Immun. 70:277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa, I., H. Inaba, T. Yamamura, T. Kato, S. Kawai, T. Ooshima, and A. Amano. 2006. Invasion of epithelial cells and proteolysis of cellular focal adhesion components by distinct types of Porphyromonas gingivalis fimbriae. Infect. Immun. 74:3773-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishikawa, K., and F. Yoshimura. 2001. The response regulator FimR is essential for fimbrial production of the oral anaerobe Porphyromonas gingivalis. Anaerobe 7:255-262. [Google Scholar]
- 31.Nishikawa, K., F. Yoshimura, and M. J. Duncan. 2004. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol. Microbiol. 54:546-560. [DOI] [PubMed] [Google Scholar]
- 32.Nishiyama, S., Y. Murakami, H. Nagata, S. Shizukuishi, I. Kawagishi, and F. Yoshimura. 2007. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology 153:1916-1925. [DOI] [PubMed] [Google Scholar]
- 33.Njoroge, T., R. J. Genco, H. T. Sojar, N. Hamada, and C. A. Genco. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose, R. J., D. Verger, T. Daviter, H. Remaut, E. Paci, G. Waksman, A. E. Ashcroft, and S. E. Radford. 2008. Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc. Natl. Acad. Sci. USA 105:12873-12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoji, M., M. Naito, H. Yukitake, K. Sato, E. Sakai, N. Ohara, and K. Nakayama. 2004. The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol. Microbiol. 52:1513-1525. [DOI] [PubMed] [Google Scholar]
- 36.Skorupski, K., and R. K. Taylor. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003-1009. [DOI] [PubMed] [Google Scholar]
- 37.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 38.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, Y., F. Yoshimura, K. Takahashi, H. Tani, and T. Suzuki. 1988. Detection of fimbriae and fimbrial antigens on the oral anaerobe Bacteroides gingivalis by negative staining and serological methods. J. Gen. Microbiol. 134:2713-2720. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi, Y., M. Davey, H. Yumoto, F. C. Gibson III, and C. A. Genco. 2006. Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell. Microbiol. 8:738-757. [DOI] [PubMed] [Google Scholar]
- 41.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 70:1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umeda, J. E., C. Missailidis, P. L. Longo, D. Anzai, M. Wikström, and M. P. Mayer. 2006. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis. Oral Microbiol. Immunol. 21:415-419. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe-Kato, T., J. Hayashi, Y. Terazawa, C. I. Hoober, K. Nakayama, E. Hibi, N. Kawakami, T. Ikeda, H. Nakamura, T. Noguchi, and F. Yoshimura. 1998. Isolation and characterization of transposon-induced mutants of Porphyromonas gingivalis deficient in fimbriation. Microb. Pathogen. 24:25-35. [DOI] [PubMed] [Google Scholar]
- 44.Wang, M., M. K. Shakhatreh, D. James, S. Liang, S. Nishiyama, F. Yoshimura, D. R. Demuth, and G. Hajishengallis. 2007. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol. 179:2349-2358. [DOI] [PubMed] [Google Scholar]
- 45.Yoshimura, F., K. Takahashi, Y. Nodasaka, and T. Suzuki. 1984. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J. Bacteriol. 160:949-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.