Abstract
For construction of the bacterial flagellum, many of the flagellar proteins are exported into the central channel of the flagellar structure by the flagellar type III protein export apparatus. FlhA and FlhB, which are integral membrane proteins of the export apparatus, form a docking platform for the soluble components of the export apparatus, FliH, FliI, and FliJ. The C-terminal cytoplasmic domain of FlhA (FlhAC) is required for protein export, but it is not clear how it works. Here, we analyzed a temperature-sensitive Salmonella enterica mutant, the flhA(G368C) mutant, which has a mutation in the sequence encoding FlhAC. The G368C mutation did not eliminate the interactions with FliH, FliI, FliJ, and the C-terminal cytoplasmic domain of FlhB, suggesting that the mutation blocks the export process after the FliH-FliI-FliJ-export substrate complex binds to the FlhA-FlhB platform. Limited proteolysis showed that FlhAC consists of at least three subdomains, a flexible linker, FlhACN, and FlhACC, and that FlhACN becomes sensitive to proteolysis by the G368C mutation. Intragenic suppressor mutations were identified in these subdomains and restored flagellar protein export to a considerable degree. However, none of these suppressor mutations suppressed the protease sensitivity. We suggest that FlhAC not only forms part of the docking platform for the FliH-FliI-FliJ-export substrate complex but also is directly involved in the translocation of the export substrate into the central channel of the growing flagellar structure.
The bacterial flagellum, which is responsible for motility, is a supramolecular complex of about 30 different proteins, and it consists of at least three substructures: the basal body, the hook, and the filament. Flagellar assembly begins with the basal body, followed by the hook and finally the filament. Many of the flagellar component proteins are translocated into the central channel of the growing flagellar structure and then to the distal end of the structure for self-assembly by the flagellar type III protein export apparatus (11, 16, 22). This export apparatus consists of six integral membrane proteins, FlhA, FlhB, FliO, FliP, FliQ, and FliR, and three soluble proteins, FliH, FliI, and FliJ (18, 21). These protein components show significant sequence and functional similarities to those of the type III secretion systems of pathogenic bacteria, which directly inject virulence factors into their host cells (11, 16).
FliI is an ATPase (4) and forms an FliH2-FliI complex with its regulator, FliH, in the cytoplasm (20). FliI self-assembles into a homo-hexamer and hence exhibits full ATPase activity (1, 8, 17). FliH and FliI, together with FliJ and the export substrate, bind to the export core complex, which is composed of the six integral membrane proteins, to recruit export substrates from the cytoplasm to the core complex (14) and facilitate the initial entry of export substrates into the export gate (23). FliJ not only prevents premature aggregation of export substrates in the cytoplasm (13) but also plays an important role in the escort mechanism for cycling export chaperones during flagellar assembly (3). The export core complex is believed to be located in the central pore of the basal body MS ring (11, 16, 22). In fact, it has been found that FlhA, FliP, and FliR are associated with the MS ring (5, 9). The FliR-FlhB fusion protein is partially functional, suggesting that FliR and FlhB interact with each other within the MS ring (29). The export core complex utilizes a proton motive force across the cytoplasmic membrane as the energy source to drive the successive unfolding of export substrates and their translocation into the central channel of the growing flagellum (23, 27). Here we refer to the export core complex as the “export gate,” as we have previously (8, 16, 23, 24).
FlhA is a 692-amino-acid protein consisting of two regions: a hydrophobic N-terminal transmembrane region with eight predicted α-helical transmembrane spans (FlhATM) and a hydrophilic C-terminal cytoplasmic region (FlhAC) (12, 15). FlhATM is responsible for the association with the MS ring (9). FlhAC interacts with FliH, FliI, FliJ, and the C-terminal cytoplasmic domain of FlhB (6, 12, 21, 24) and plays a role in the initial export process with these proteins (28). It has been shown that the V404M mutation in FlhAC increases not only the probability of FliI binding to the export gate in the absence of FliH (14) but also the efficiency of substrate translocation through the export gate in the absence of FliH and FliI (23). Recently, it has been shown that FlhAC is also required for substrate recognition (7). These observations suggest that an interaction between FlhAC and FliI is coupled with substrate entry, although it is not clear how.
In order to understand the mechanism of substrate entry into the export gate, we characterized a temperature-sensitive Salmonella enterica mutant, the flhA(G368C) mutant, whose mutation blocks the flagellar protein export process at 42°C (28). We show here that this mutation severely inhibits translocation of flagellar proteins through the export gate after the FliH-FliI-FliJ complex binds to the FlhA-FlhB platform of the gate and that the impaired ability of the flhA(G368C) mutant to export flagellar proteins is restored almost to wild-type levels by intragenic second-site mutations that may alter the interactions between subdomains of FlhAC for possible rearrangement for the export function.
MATERIALS AND METHODS
Bacterial strains, plasmids, transductional crosses, DNA manipulations, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. P22-mediated transduction was carried out with p22HTint as described previously (30). An fliH null mutation (ΔfliH) was introduced into SJW2228 using the λ Red homologous recombination system (2). DNA manipulations, site-directed mutagenesis, and DNA sequencing were carried out as described previously (28). L broth (LB) and soft tryptone agar plates were prepared as described previously (18, 21). Ampicillin and chloramphenicol were added to LB to final concentrations of 100 and 30 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Escherichia coli BL21(DE3)/pLysS | Overproduction of proteins | Novagen |
| Salmonella strains | ||
| SJW1103 | Wild type for motility and chemotaxis | 31 |
| SJW1364 | flhA | 10 |
| SJW1368 | Δ(cheW-flhD); master operon mutant | 25 |
| SJW2228 | flhA(G368C) | 28 |
| SJW2228iH | flhA(G368C) ΔfliH | This study |
| MMA2228-1 | flhA(G368C/L359F) | This study |
| MMA2228-3 | flhA(G368C/G364R) | This study |
| MMA2228-7 | flhA(G368C/R370S) | This study |
| MMA2228-9 | flhA(G368C/P550S) | This study |
| MMHI001 | ΔfliH ΔfliI | 17 |
| MMHI0117 | ΔfliH ΔfliI flhB(P28T) | 23 |
| MMHI0117-28 | ΔfliH ΔfliI flhB(P28T) flhA(G368C) | This study |
| Plasmids | ||
| pGEX-6p-1 | Expression vector | GE Healthcare |
| pTrc99A | Expression vector | GE Healthcare |
| pTrc99AFF4 | Expression vector | 26 |
| pGKK1702 | pGEX-6p-1/GST-FliI | 8 |
| pMM102 | pTrc99A/His-FlhAC | 21 |
| pMM102(G368C) | pTrc99A/His-FlhAC(G368C) | This study |
| pMM104 | pET19b/His-FlhAC | 21 |
| pMM104(G368C) | pET19b/His-FlhAC(G368C) | This study |
| pMM104(G368C/L359F) | pET19b/His-FlhAC(G368C) | This study |
| pMM104(G368C/G364R) | pET19b/His-FlhAC(G368C/G364R) | This study |
| pMM104(G368C/R370S) | pET19b/His-FlhAC(G368C/R370S) | This study |
| pMM104(G368C/P550S) | pET19b/His-FlhAC(G368C/P550S) | This study |
| pMM130 | pTrc99AFF4/FlhA | 9 |
| pMM130(L359F) | pTrc99AFF4/FlhA(L359F) | This study |
| pMM130(G364R) | pTrc99AFF4/FlhA(G364R) | This study |
| pMM130(R370S) | pTrc99AFF4/FlhA(R370S) | This study |
| pMM130(P550S) | pTrc99AFF4/FlhA(P550S) | This study |
| pMM306 | pTrc99A/His-FliH | 21 |
| pMM310 | pET19b/His-FliH | 21 |
| pMM1702 | pTrc99A/His-FliI | 21 |
| pMM309iI | pTrc99AFF4/FliH+His-FliI | 20 |
| pMMJ1001 | pGEX-6p-1/GST-FliJ | This study |
| pMMHB1001 | pGEX-6p-1/GST-FlhBC | 24 |
| pTIH001 | pGEX-6p-1/GST-FliH | T. Ibuki, unpublished data |
Motility assays on soft agar plates.
Fresh colonies were inoculated onto soft tryptone agar plates and incubated at 30°C or 42°C.
Preparation of whole-cell and culture supernatant fractions and immunoblotting.
Salmonella cells were grown at 30°C or 42°C with shaking until the optical density at 600 nm (OD600) was ca. 1.2 to 1.4. Cellular and culture supernatant fractions were prepared as described previously (18). Cell pellets were resuspended in SDS loading buffer normalized using the cell density to obtain the same number of cells. The proteins in the culture supernatants were precipitated with 10% trichloroacetic acid, suspended in a Tris-SDS loading buffer, and heated at 95°C for 5 min. After SDS-PAGE, immunoblotting with polyclonal anti-FlhAC and anti-FlgD antibodies was carried out as described previously (18). Detection was performed with an enhanced chemoluminescence (ECL) immunoblot detection kit (GE Healthcare).
Purification of GST, GST-FlhBC, GST-FliH, GST-FliI, and GST-FliJ.
Soluble fractions prepared from SJW1368(ΔcheW-ΔflhD) expressing glutathione S-transferase (GST), GST-FlhBC, GST-FliH, GST-FliI, and GST-FliJ were loaded onto a glutathione Sepharose 4B column (GE Healthcare). After washing with phosphate-buffered saline (PBS) (containing [per liter] 8 g of NaCl, 0.2 g of KCl, 3.63 g of Na2HPO4·12H2O, and 0.24 g of KH2PO4; pH 7.4), proteins were eluted with 50 mM Tris-HCl (pH 8.0), 10 mM reduced glutathione. Fractions containing GST, GST-FlhBC, GST-FliH, GST-FliI, or GST-FliJ were pooled and dialyzed overnight against PBS at 4°C with three changes of PBS.
Pull-down assay.
Soluble fractions prepared from cultures of SJW1103 (wild type) carrying pMM102 or pMM102(G368C) were incubated at 42°C and loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) agarose column (Qiagen) whose bed volume was 1 ml. After the column was washed with 20 ml of a binding buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl) containing 10 mM imidazole and then with 5 ml of binding buffer containing 60 mM imidazole at 42°C, proteins were eluted with binding buffer containing imidazole at 42°C; during elution the imidazole concentration was increased stepwise to 100, 200, 300, and 600 mM. Eluted fractions were analyzed by both Coomassie brilliant blue (CBB) staining and immunoblotting with polyclonal anti-FliI, anti-FliH, and anti-FliJ antibodies.
Purified His-FlhAC and His-FlhAC(G368C) were incubated with purified GST, GST-FlhBC, GST-FliH, GST-FliI, or GST-FliJ at 42°C for 1 h and loaded onto a glutathione Sepharose 4B column (bed volume, 1 ml) preincubated at 42°C. After the column was washed with 20 ml PBS at 42°C, proteins were eluted with 50 mM Tris-HCl (pH 8.0), 10 mM reduced glutathione at 42°C. Eluted fractions were analyzed by both CBB staining and immunoblotting with polyclonal anti-FlhAC antibody.
Purification of His-FlhAC and mutant variants of this construct and limited proteolysis.
His-FlhAC and mutant variants were purified by Ni-NTA affinity chromatography as described previously (21) and dialyzed overnight against 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT).
His-FlhAC and the mutant variants (0.5 mg/ml) were incubated with the Glu-C endoproteinase (Roche Diagnostics) at a protein-to-protease ratio of 300:1 (wt/wt) in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM DTT at 42°C. After incubation was started at 42°C, aliquots were collected at 0, 5, 15, 30, 60, 120, and 180 min, and trichloroacetic acid was added to a final concentration of 10%. The molecular masses of proteolytic cleavage products were determined by using a mass spectrometer (PerSeptive Biosystems) as described previously (28).
RESULTS
Dominance properties of FlhAC(G368C).
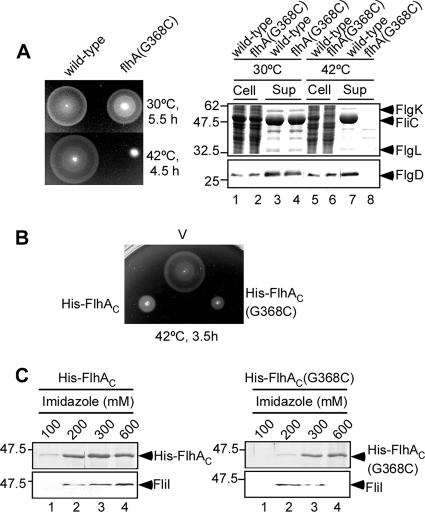
The temperature-sensitive flhA(G368C) mutation substantially reduced motility (Fig. 1A, left panel) and flagellar protein export (Fig. 1A, right panel) at 42°C, in agreement with a previous report (28). To investigate how this mutation affects the flagellar protein export process, we analyzed the effect of the G368C mutation on the interaction of FlhAC with the soluble components of the export apparatus. As FlhAC exerts a strong inhibitory effect on wild-type motility by titrating away other essential proteins involved in flagellar protein export (21, 28), we first analyzed the motility of wild-type cells carrying a plasmid that overexpresses His-FlhAC(G368C) on soft tryptone agar plates containing 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 42°C (Fig. 1B). The motility of wild-type cells overexpressing His-FlhAC(G368C) was much poorer than that of the vector control and even poorer (although slightly) than that of the wild type overexpressing His-FlhAC. Immunoblotting with polyclonal anti-FlhAC antibody revealed that the cellular level of His-FlhAC(G368C) was the wild-type level (data not shown). These results suggest that His-FlhAC(G368C) is able to interact with and sequester the soluble components of the export apparatus and/or export substrates.
FIG. 1.
Characterization of the temperature-sensitive flhA(G368C) mutant. (A) (Left panel) Swarming motility of SJW1103 (wild type) and SJW2228 [flhA(G368C)] on soft agar plates at 30°C and 42°C. (Right panel) Secretion assays. Secretion of FliC, FlgK, and FlgL was analyzed by Coomassie brilliant blue (CBB) staining (upper panel). Secretion of FlgD was measured by immunoblotting with a polyclonal anti-FlgD antibody (lower panel). Cell, whole-cell proteins; Sup, culture supernatant fractions. The positions of molecular mass markers (in kDa) are indicated on the left. (B) Dominant negative effect on wild-type swarming motility of SJW1103 transformed with pTrc99A (V), pMM102 (His-FlhAC) or pMM102(G368C) [His-FlhAC(G368C)] on soft agar plates containing 0.1 mM IPTG at 42°C. (C) Pull-down assay. The soluble fractions prepared from SJW1103 overproducing His-FlhAC (left panel) or His-FlhAC(G368C) (right panel) were loaded onto an Ni-NTA agarose column. After washing, proteins were eluted with a buffer containing 100 mM, 200 mM, 300 mM, and 600 mM imidazole. Pull-down assays were done at 42°C. Eluted His-FlhAC was analyzed by CBB staining, while eluted FliI was detected by immunoblotting with a polyclonal anti-FliI antibody.
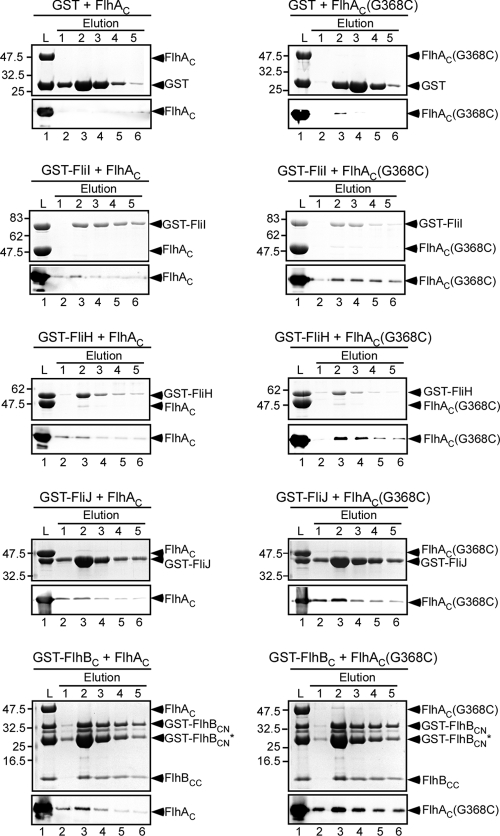
To identify which components are sequestered by overproduction of His-FlhAC or His-FlhAC(G368C), we carried out pull-down assays by using Ni-NTA affinity chromatography at 42°C (Fig. 1C). FliI copurified with both His-FlhAC and His-FlhAC(G368C). No FliI was seen in the elution fractions of the vector control or a fliI null mutant overexpressing His-FlhAC or His-FlhAC(G368C) (data not shown). In agreement with this, pull-down assays performed by using GST affinity chromatography showed that a slightly larger amount of His-FlhAC(G368C) copurified with GST-FliI (Fig. 2, second row). These results indicate that the interaction between FliI and FlhAC is not eliminated by the G368C mutation. However, neither FliH nor FliJ copurified (data not shown). Since it has been shown that FlhAC binds to these proteins (6, 12, 21, 24), it was possible that FliH and FliJ were dissociated by stringent washes during purification.
FIG. 2.
Interactions of FlhAC with FliH, FliI, FliJ, and FlhBC. Purified His-FlhAC (FlhAC) (left panels) or His-FlhAC(G368C) [FlhAC(G368C)] (right panels) was mixed with purified GST (first row), GST-FliI (second row), GST-FliH (third row), GST-FliJ (fourth row), or GST-FlhBC (fifth row) and incubated at 42°C for 1 h. Then the mixtures (lanes L) were loaded onto a GST column preincubated at 42°C. After washing with PBS at 42°C, proteins were eluted with 50 mM Tris-HCl (pH 8.0), 10 mM reduced glutathione. The eluted proteins were analyzed by both CBB staining (upper panels) and immunoblotting with polyclonal anti-FlhAC antibody (lower panels). Although GST-FlhBC is cleaved between Asp-269 and Pro-270, GST-FlhBCN and FlhBCC associate with each other even after cleavage (19). The arrowheads indicate the positions of GST, GST-FliI, GST-FliH, GST-FliJ, GST-FlhBCN, FlhBCC, His-FlhAC, and His-FlhAC(G368C). The arrowhead labeled GST-FlhBCN* indicates a degraded product just under the main band.
Bypass effects of FliI on motility of the flhA(G368C) ΔfliH double mutant.
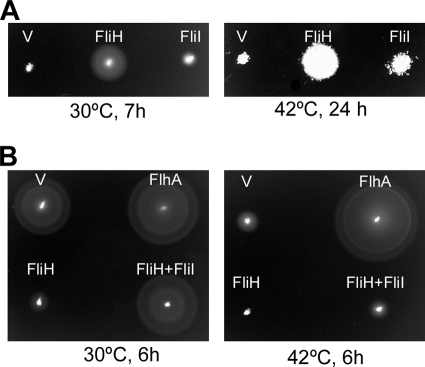
Since the FliH null mutation is suppressed by overproduction of FliI or by mutations in FlhA and FlhB, FliH is thought to be involved in the effective docking of FliI with the export substrate with the FlhA-FlhB platform of the export gate (14). Therefore, we investigated whether the G368C mutation allows FliI overproduction to bypass the FliH defect at 42°C. An flhA(G368C) ΔfliH double mutant was transformed with a plasmid encoding His-FliI, and then the motility of the resulting transformants was analyzed at 30°C and 42°C (Fig. 3A). Overproduction of FliI improved the motility of the double mutant to some extent, suggesting that the G368C mutation does not interfere with the association between FliI and the platform at 42°C.
FIG. 3.
In vivo interaction of FlhA(G368C) with FliH and FliI. (A) Bypass effect of FliI on the motility of SJW2228iH [flhA(G368C) ΔfliH]. The swarming motility of the double mutant transformed with pTrc99A (V), pMM310 (FliH), and pMM1702 (FliI) was determined on soft agar plates at 30°C and 42°C. (B) Multicopy effect of FliH on the motility of the flhA(G368C) mutant. A swarming motility assay was performed with the flhA(G368C) mutant carrying pTrc99AFF4 (V), pMM130 (FlhA), pMM306 (FliH), or pMM309iI (FliH+FliI) at 30°C and 42°C.
Interaction between FlhAC and FliH.
It has been shown that the extreme N-terminal region of FliH is required not only for the interaction with the C-ring protein FliN but also for effective association with the export gate (24). Therefore, we tested if the G368C mutation interferes with the association between FliH and the export gate. Since the presence of free FliH eliminates the export process when FliI is missing, probably due to the binding of FliH to the export gate that inhibits substrate entry into the gate (21, 23, 24), we examined the multicopy effect of FliH on the motility of the flhA(G368C) mutant on soft agar plates at 30°C and 42°C (Fig. 3B). FliH overproduction strongly inhibited the motility of the flhA mutant, and this inhibition of motility was substantially relieved by co-overproduction of FliI (Fig. 3B) but not by co-overproduction of FliN alone or by co-overproduction of FliN plus FliM (data not shown). These results suggest that FliH can bind to the export gate even in the presence of the G368C mutation at 42°C.
Pull-down assays performed by using GST affinity chromatography have shown that there is an interaction between FlhAC and FliH (24). Therefore, we also carried out pull-down assays using GST affinity chromatography at 42°C (Fig. 2). We used GST as a negative control. His-FlhAC copurified with GST-FliH but not with GST (Fig. 2, third and first rows, respectively). A very small amount of His-FlhAC(G368C) was detected in the elution fractions derived from GST alone (Fig. 2, row 1), presumably due to its nonspecific binding to the column. However, the amount of His-FlhAC(G368C) that coeluted with GST-FliH was 20-fold larger than the amount that coeluted with GST when the amounts were normalized using the amounts of GST and GST-FliH. These results indicate that the G368C mutation does not decrease the FlhAC-FliH interaction at 42°C.
Interaction of FlhAC with FliJ and the C-terminal cytoplasmic domain of FlhB.
FlhAC also interacts with FliJ and the C-terminal cytoplasmic domain of FlhB (FlhBC) (6, 12, 21). To investigate whether the G368C mutation interferes with these interactions, we carried out pull-down assays using GST affinity chromatography at 42°C (Fig. 2). Interactions of FlhAC with GST-FliJ and GST-FlhBC were clearly detected even in the presence of the G368C mutation (Fig. 2, fourth and fifth rows, respectively). Interestingly, the amount of His-FlhAC(G368C) that coeluted with GST-FliJ or GST-FlhB was much larger than the amount of His-FlhAC that coeluted (Fig. 2, fourth and fifth rows, respectively), indicating that the mutation somehow increases the binding affinity of FlhAC for both FliJ and FlhBC.
Effect of the G368C mutation on substrate translocation through the export gate.
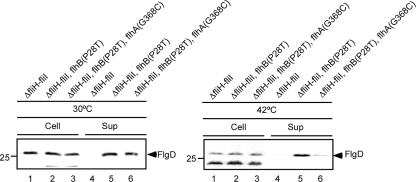
We found that the G368C mutation in FlhAC does not interfere with the interactions with FliH, FliI, FliJ, and FlhBC even at 42°C (Fig. 1 and 2), raising the possibility that this mutation directly blocks translocation of an export substrate through the export gate. Since the P28T mutation of FlhB greatly increases the export efficiency in the absence of FliH and FliI, probably by increasing the probability of entry of flagellar proteins into the export gate (23), we analyzed the levels of secretion of the hook capping protein FlgD by a ΔfliH ΔfliI flhB(P28T) flhA(G368C) mutant (Fig. 4). At 30°C, the level of FlgD secretion by this mutant was similar to the level of FlgD secretion by the ΔfliH ΔfliI flhB(P28T) mutant (Fig. 4, left panel). In contrast, the G368C mutation reduced the levels of secretion considerably at 42°C (Fig. 4, right panel). These results suggest that this mutation significantly reduced the efficiency of protein translocation into the central channel of the growing flagellar structure.
FIG. 4.
Assay of secretion of FlgD: immunoblotting with polyclonal anti-FlgD antibody of whole cells and culture supernatants of MMHI001 (ΔfliH ΔfliI), MMHI0117 [ΔfliH ΔfliI flhB(P28T)], and MMHI0117-28 [ΔfliH ΔfliI flhB(P28T) flhA(G368C)].
Isolation of pseudorevertants from a culture of the flhA(G368C) mutant.
To understand how the G368C mutation interferes with flagellar protein export at 42°C, pseudorevertants were isolated from an flhA(G368C) mutant overnight culture by streaking the culture on soft agar plates, incubating the plates at 42°C for 2 days, and looking for motility halos emerging from the streaks. A total of 16 motile colonies were purified from such halos. The motility of these pseudorevertants was considerably better than that of the parent strain at 42°C, although it was not as good as that of the wild-type strain (Fig. 5A). In agreement with this, the levels of flagellar proteins secreted by these pseudorevertants were recovered to a significant degree (data not shown).
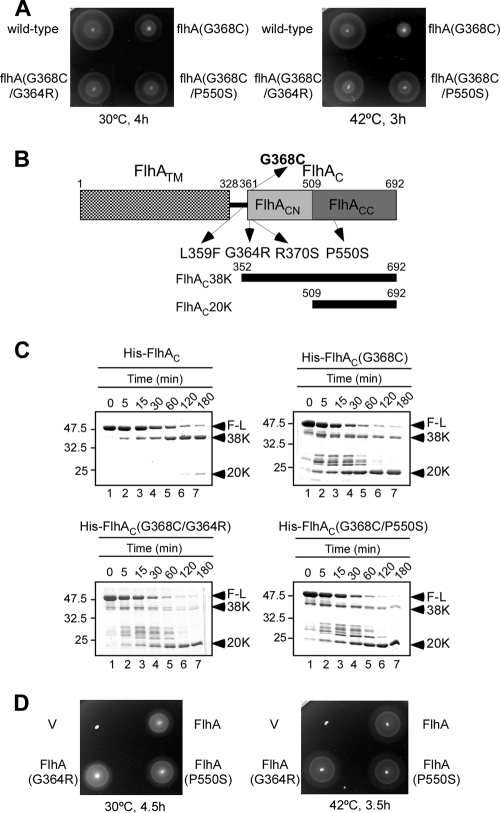
FIG. 5.
Characterization of pseudorevertants isolated from the flhA(G368C) mutant. (A) Swarming motility assay performed with the wild type, the flhA(G368C) mutant, and flhA(G368C) mutant pseudorevertants, including MMA2228-3 [flhA(G368C/G364R)] and MMA2228-9 [flhA(G368C/P550S)], at 30°C and 42°C. (B) Locations of the G368C mutation and intragenic suppressor mutations in stable fragments of FlhA produced by limited proteolysis. FlhA consists of an N-terminal transmembrane region (FlhATM) and a C-terminal cytoplasmic domain (FlhAC) (15). The present study established that FlhAC can be divided into three subdomains: the linker (black bar), FlhACN, and FlhACC. The sites of mutations in FlhA of a temperature-sensitive flhA(G368C) mutant and pseudorevertants of this mutant are indicated by arrows. The N and C termini of FlhA are labeled 1 and 692, respectively. The regions corresponding to FlhAC38K and FlhAC20K are also indicated. (C) Limited proteolysis of His-FlhAC, FlhAC(G368C), and two suppressor variants of FlhAC(G368C), His-FlhAC(G368C/G364R) and His-FlhAC(G368C/P550S), by the Glu-C endoproteinase at 42°C. The time course of digestion was monitored by SDS-PAGE and CBB staining. The arrowheads indicate the positions of full-length FlhAC (F-L) and its 38-kDa (38K) and 20-kDa (20K) cleavage products. (D) Swarming motility assay performed with the mutants with only second-site mutations. SJW1364 (flhA) was transformed with pTrc99A (V), pMM130 (wild type) (FlhA), pMM130(G364R) [FlhA(G364R)], or pMM130(P550S) [FlhA(P550S)] and incubated on soft agar plates at 30°C and 42°C.
P22-mediated transduction experiments showed that all of the suppressor mutations were cotransduced with the parental G368C mutation, indicating that they were located near the first-site mutation (data not shown). Therefore, we sequenced the flhBAE operon of the pseudorevertants, and the mutations were all missense mutations in FlhA; these mutations included L359F (isolated nine times), G364R (isolated three times), R370S (isolated twice), and P550S (isolated twice) (Fig. 5B). The L359F mutation is in the flexible linker region formed by residues 328 to 361, while the other three mutations are in the relatively stable FlhAC38K fragment (28).
Limited proteolysis by endoproteinase Glu-C.
To test if the G368C mutation causes a conformational change in FlhAC at 42°C, we purified His-FlhAC(G368C), carried out limited proteolysis at 42°C with an endoproteinase, Glu-C, which cleaves the C-terminal side of Glu residues, and determined the molecular masses of the fragments by matrix-assisted laser desorption ionization-time of flight mass spectrometry. The time course of degradation, as monitored by SDS-PAGE, is shown in Fig. 5C. In agreement with a previous report (28), most of wild-type FlhAC was degraded into a stable 38-kDa fragment (FlhAC38K) (Fig. 5C, top left panel), which consisted of residues 352 to 692 (Fig. 5B). A very small amount of a 20-kDa fragment (FlhAC20K) was also seen (Fig. 5C, top left panel). In contrast, the FlhAC38K fragment derived from the G368C mutant variant was very unstable and quickly degraded into the FlhAC20K fragment through several intermediate fragments (Fig. 5C, top right panel), indicating that the G368C mutation reduced the structural stability of FlhAC considerably. Based on the molecular mass (20,236 Da), FlhAC20K was identified as a fragment consisting of amino acid residues 509 to 692 with a deduced molecular mass of 20,252 Da (Fig. 5B).
To examine if the structural instability of the FlhAC38K fragment caused by the G368C mutation is reversed by the second-site mutations, we purified four intragenic suppressor mutant variants of His-FlhAC and carried out limited proteolysis at 42°C. As shown by the data for the G368C/G364R and G368C/P550S mutants (Fig. 5C, bottom left and right panels, respectively), the FlhAC38K fragments were still highly susceptible to the protease compared with the wild type and were as unstable as the His-FlhAC(G368C) fragment. These results indicate that the second-site mutations do not restore the structural stability of FlhAC38K, suggesting that the loss-of-function phenotype of the flhA(G368C) mutant at a restrictive temperature (42°C) is not an unavoidable consequence of the structural instability caused by the G368C mutation.
Motility of the second-site flhA mutants.
To test the effect of each second-site mutation by itself on motility, we carried out site-directed mutagenesis and analyzed the motility of the second-site flhA mutants on soft tryptone agar plates. The motility of all of the mutants was normal at both 30°C and 42°C (Fig. 5D), as shown by the data for the flhA(G364R) and flhA(P550S) mutants, indicating that the second-site mutations alone displayed no phenotype. These results suggest that the mutated residues are not important for flagellar protein export.
DISCUSSION
The G368C mutation blocks substrate entry into the export gate.
The flagellar type III protein export process requires the following three steps: delivery of export substrates to the export gate located in the central pore of the basal body MS ring, insertion of the natively disordered N-terminal segment into the presumably narrow pore of the export gate, and successive unfolding and translocation of the compactly folded domains of export substrates by the export gate (16). The FliH2-FliI complex, possibly together with FliJ, delivers export substrates to the export gate, which is composed of six integral membrane proteins, including FlhA and FlhB (14). The interaction between the FliH-FliI complex and the FlhA-FlhB platform allows the unfolded N-terminal segment of an export substrate to be efficiently inserted into the export gate (23).
FlhAC is directly involved in the substrate delivery process along with FliH, FliI, FliJ, and FlhBC (6, 21, 24, 28). It has also been shown that the V404M mutation in FlhAC increases the export efficiency in the absence of the FliH and FliI, suggesting that FlhAC is involved in the process of substrate insertion into the export gate (23). However, it was not clear how FlhAC functions in this process. Here, we characterized the flhA(G368C) mutant; in this mutant the secretion activity of flagellar proteins was considerably reduced at a restrictive temperature (42°C) but not at a permissive temperature (30°C) (Fig. 1A). FlhAC(G368C) was still able to interact with FliH, FliI, FliJ, and FlhBC (Fig. 1 and 2). Introduction of the flhA(G368C) mutation into the ΔfliH ΔfliI flhB(P28T) mutant strain, which can efficiently export flagellar proteins even in the absence of FliH and FliI (23), resulted in a significant decrease in the level of secretion of flagellar proteins at 42°C but not at 30°C (Fig. 4), indicating that the G368C mutation directly inhibits the translocation of export substrates into the central channel of the growing flagellar structure. This suggests that FlhAC not only forms part of the docking platform of the export gate for efficient substrate delivery but also is directly involved in the translocation process. Since the P28T mutation of FlhB allows efficient entry of flagellar proteins and translocation of these proteins into the export gate even in the absence of FliH and FliI (23), the G368C mutation seems to block the export process after docking of the FliH-FliI-FliJ-export substrate complex to the FlhA-FlhB platform of the export gate.
The G368C mutation causes partial disorder in the FlhAC structure.
Limited proteolysis of wild-type FlhAC gave rise to a relatively stable FlhAC38K fragment, which consists of amino acid residues 352 to 692 of FlhA (Fig. 5B and 5C). FlhAC38K consists of at least two subdomains, FlhACN and FlhACC, which consist of residues 352 to 508 and residues 509 to 692, respectively (Fig. 5B). In contrast to wild-type FlhAC38K, FlhAC38K(G368C) was susceptible to the protease, and it was easily degraded into a more stable FlhAC20K fragment identified as FlhACC (Fig. 5C, top right panel), indicating that the G368C mutation induces structural disorder or partial unfolding in FlhACN. Thus, FlhACN appears to be unstable due to the G368C mutation. This observation is consistent with the crystal structure of FlhAC that shows a relatively independent domain arrangement of FlhACN and FlhACC (28a).
Gly-368 is buried in the FlhACN domain in the crystal structure. However, since FlhAC(G368C) tends to form a dimer in the absence of a reducing reagent (data not shown), Cys-368 seems to be surface exposed a significant fraction of the time. This is consistent with the observation obtained by limited proteolysis that the G368 mutation destabilizes FlhACN to expose cleavage sites in FlhACN to solvent.
The second-site mutations do not affect the protease sensitivity of FlhACN.
Suppressor analysis of the G368C mutant identified one suppressor mutation in the flexible linker region, two suppressor mutations in FlhACN, and one suppressor mutation in FlhACC. The improved motility of the suppressor mutants on soft agar plates was due to restoration of flagellar protein export almost to wild-type levels (Fig. 5A). The level of motility of the mutant with each second-site mutation was essentially the same as the wild-type level (Fig. 5D). Thus, there is no phenotype that is associated with the suppressor mutations alone, implying that the residues of the second-site mutations are not directly involved in flagellar protein export. Limited proteolysis revealed that the suppressor mutant variants of FlhAC are partially disordered at 42°C, similar to FlhAC(G368C) (Fig. 5C), indicating that the structural instability of FlhACN caused by the first-site mutation is not reversed by the second-site mutations. Therefore, the structural instability does not appear to be directly responsible for the loss-of-function phenotype of the flhA(G368C) mutant, although this is rather unexpected for a temperature-sensitive mutation.
The L359F and P550S mutations, which are located in the flexible linker and FlhACC, respectively, suppress the G368C mutation located in FlhACN at 42°C (Fig. 5), even though they are located in different domains and are relatively far from one another in the crystal structure of FlhAC (Saijo-Hamano et al., submitted). This suggests that the second-site mutations compensate for the first-site mutation by altering the interactions among the linker, FlhACN, and FlhACC. As discussed above, the G368C mutation inhibits the export process after docking of the FliH-FliI-FliJ-export substrate complex with the FlhAC-FlhBC platform of the export gate. Therefore, we propose that the interactions of the FliH-FliI-FliJ-export substrate complex with the FlhAC domain may need to induce a relative conformational arrangement of the three subdomains of FlhAC to allow export substrates to be translocated into the central channel of the growing flagellar structure. Since this process occurs on the cytoplasmic side of the export gate, it is likely that the conformational rearrangement of FlhAC is responsible for substrate entry into the gate.
Acknowledgments
We acknowledge T. Ibuki for his kind gift of pTIH001 and M. Kinoshita for technical assistance.
This work was supported in part by Grants-in-Aid for Scientific Research 16087207 and 21227006 to K.N., by grants from the Targeted Proteins Research Program to T.M. K.N., and K.I. from the Ministry of Education, Science and Culture of Japan, and by U.S. Public Health Service grant AI12202 to Robert M. Macnab, who was a professor at Yale University and died suddenly on 7 September 2003.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Claret, L., C. R. Susannah, M. Higgins, and C. Hughes. 2003. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol. Microbiol. 48:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datsenko, K. A., and Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans, L. D., G. P. Stafford, S. Ahmed, G. M. Fraser, and C. Hughes. 2006. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc. Natl. Acad. Sci. U. S. A. 103:17474-17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan, F., and R. M. Macnab. 1996. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271:31981-31988. [DOI] [PubMed] [Google Scholar]
- 5.Fan, F., K. Ohnishi, N. R. Francis, and R. M. Macnab. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26:1035-1046. [DOI] [PubMed] [Google Scholar]
- 6.Fraser, G. M., B. González-Pedrajo, J. R. H. Tame, and R. M. Macnab. 2003. Interactions of FliJ with the Salmonella type III flagellar export apparatus. J. Bacteriol. 185:5546-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano, T., S. Mizuno, S.-I. Aizawa, and K. T. Hughes. 2009. Mutations in Flk, FlgG, FlhA, and FlhE that affect the flagellar type III secretion specificity switch in Salmonella enterica. J. Bacteriol. 191:3938-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazetani, K.-I., T. Minamino, T. Miyata, T. Kato, and K. Namba. 2009. ATP-induced FliI hexamerization facilitates bacterial flagellar protein export. Biochem. Biophys. Res. Commun. 388:323-327. [DOI] [PubMed] [Google Scholar]
- 9.Kihara, M., T. Minamino, S. Yamaguchi, and R. M. Macnab. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutsukake, K., and T. Iino. 1985. Refined genetic analysis of the region II che mutants in Salmonella typhimurium. Mol. Gen. Genet. 199:406-409. [DOI] [PubMed] [Google Scholar]
- 11.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 12.McMurry, J. L., J. S. Van Arnam, M. Kihara, and R. M. Macnab. 2004. Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery. J. Bacteriol. 186:7586-7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minamino, T., R. Chu, S. Yamaguchi, and R. M. Macnab. 2000. Role of FliJ in flagellar protein export in Salmonella. J. Bacteriol. 182:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minamino, T., B. González-Pedrajo, M. Kihara, K. Namba, and R. M. Macnab. 2003. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J. Bacteriol. 185:3983-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamino, T., T. Iino, and K. Kutsukake. 1994. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J. Bacteriol. 176:7630-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minamino, T., K. Imada, and K. Namba. 2008. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 4:1105-1115. [DOI] [PubMed] [Google Scholar]
- 17.Minamino, T., K.-I. Kazetani, A. Tahara, H. Suzuki, Y. Furukawa, M. Kihara, and K. Namba. 2006. Oligomerization of the bacterial flagellar ATPase FliI is controlled by its extreme N-terminal region. J. Mol. Biol. 360:510-519. [DOI] [PubMed] [Google Scholar]
- 18.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minamino, T., and R. M. Macnab. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182:4906-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minamino, T., and R. M. Macnab. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37:1494-1503. [DOI] [PubMed] [Google Scholar]
- 21.Minamino, T., and R. M. Macnab. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35:1052-1064. [DOI] [PubMed] [Google Scholar]
- 22.Minamino, T., and K. Namba. 2004. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 7:5-17. [DOI] [PubMed] [Google Scholar]
- 23.Minamino, T., and K. Namba. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485-488. [DOI] [PubMed] [Google Scholar]
- 24.Minamino, T., S. D. J. Yoshimura, Y. V. Morimoto, B. González-Pedrajo, N. Kami-ike, and K. Namba. 2009. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol. Microbiol. 74:1471-1483. [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi, K., Y. Ohto, S.-I. Aizawa, R. M. Macnab, and T. Iino. 1994. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 176:2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnishi, K., F. Fan, G. J. Schoenhals, M. Kihara, and R. M. Macnab. 1997. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J. Bacteriol. 179:6092-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul, K., M. Erhardt, T. Hirano, D. F. Blair, and K. T. Hughes. 2008. Energy source of flagellar type III secretion. Nature 451:489-492. [DOI] [PubMed] [Google Scholar]
- 28.Saijo-Hamano, Y., T. Minamino, R. M. Macnab, and K. Namba. 2004. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J. Mol. Biol. 343:457-466. [DOI] [PubMed] [Google Scholar]
- 28a.Saijo-Hamano, Y., K. Imada, T. Minamino, M. Kihara, M. Shimada, A. Kitao, and K. Namba. Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol. Microbiol., in press. [DOI] [PubMed]
- 29.Van Arnam, J. S., J. L. McMurry, M. Kihara, and R. M. Macnab. 2004. Analysis of an engineered Salmonella flagellar fusion protein, FliR-FlhB. J. Bacteriol. 186:2495-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi, S., S.-I. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi, S., H. Fujita, K. Sugata, T. Taira, and T. Iino. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130:255-265. [DOI] [PubMed] [Google Scholar]