Abstract
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that utilizes a type III secretion system to subvert host innate immunity. Of the 4 known effector proteins injected into eukaryotic cells, ExoS and ExoU are cytotoxic. The cytotoxic phenotype of ExoU depends on the enzymatic activity of the patatin-like phospholipase A2 domain localized to the N-terminal half of the protein. Amino acid residues located within the C-terminal region of ExoU are postulated to be required for trafficking or localization to the plasma membrane of eukaryotic cells. This report describes the characterization of a transposon-based linker insertion library in ExoU. Utilizing an unbiased screening approach and sensitive methods for measuring enzymatic activity, we identified regions of ExoU that are critical for activation of the phospholipase activity by the only known cofactor, SOD1. Insertions at D572 and L618 reduced the rate of substrate cleavage. Enzymatic activity could be restored to almost parental levels when SOD1 concentrations were increased, suggesting that the linker insertion disrupted the interaction between ExoU and SOD1. An enzyme-linked immunosorbent assay (ELISA)-based binding test was developed to measure ExoU-SOD1 binding. These experiments suggest that ExoU activation by SOD1 is hampered by linker insertion. ExoU derivatives harboring minimal phospholipase activity retained biological activity in tissue culture assays. These proteins affected primarily cellular architecture in a manner similar to that of ExoT. Our studies suggest that conformational changes in ExoU are facilitated by SOD1. Importantly, the level of phospholipase activity influences the biological outcome of ExoU intoxication.
Pseudomonas aeruginosa is a Gram-negative bacterium responsible for severe and potentially fatal opportunistic infections. As a contributor to nosocomial infections, P. aeruginosa is a leading cause of hospital-acquired and ventilator-associated pneumonias (40). Furthermore, P. aeruginosa is responsible for ulcerative keratitis and ocular disease found in conjunction with the use of soft contact lenses (2, 10, 54). Infections with this pathogen are of critical concern for individuals admitted with severe burns, due to the bacterium's ability to colonize and persist in damaged tissues (35). Patients suffering from cystic fibrosis often succumb to severe lung infections and inflammation due to colonization with antibi otic-resistant, mucoid strains of P. aeruginosa (3). The expression of multiple efflux pumps and the ability to inactivate and modify antibiotics make P. aeruginosa dangerous and difficult to treat (27). Several investigators are exploring ways, as adjuncts or alternatives to antibiotic treatment, to neutralize virulence factors that contribute to the ability of P. aeruginosa to suppress host innate and adaptive immune responses (17, 21, 22, 52).
Many Gram-negative bacteria, including P. aeruginosa, encode one or more type III secretion systems (T3SS), which are thought to aid in pathogenesis and increase disease severity (19, 32, 39). Four effectors are translocated by the T3SS of P. aeruginosa and include ExoS, ExoT, ExoU, and ExoY (8, 23, 56, 57). The activity of each effector is dependent upon interaction with a cofactor present in eukaryotic but not prokaryotic cells. ExoS and ExoT are bifunctional enzymes that possess both Rho GTPase-activating protein and ADP-ribosyltransferase activities (23, 25, 51). The ADP ribosylation of eukaryotic proteins by ExoS and ExoT requires activation by members of the 14-3-3 family of scaffolding proteins (13). ExoY is an adenylyl cyclase that causes the accumulation of cyclic AMP (cAMP) in intoxicated cells. The eukaryotic cofactor required for ExoY activity has not been identified (57). ExoU, a potent A2 phospholipase responsible for membrane disruption and cellular lysis, requires superoxide dismutase 1 (SOD1) for the detection of enzymatic activity (43, 46).
ExoU is an important virulence factor of P. aeruginosa, as it causes rapid cell death during in vitro infections and is associated with poor clinical outcomes (19, 39, 44). Several studies have used truncation analyses, linker mutagenesis, and site-specific amino acid substitutions to define regions of ExoU important for various functions (7, 36). ExoU is a 74-kDa, hydrophilic, and slightly acidic protein with a pI of 5.9 (8). The first 52 amino acids are required for interaction with the chaperone SpcU and may be important for translocation through the T3SS (7, 9). Enzymatic activity is attributed to the patatin-like phospholipase domain located between residues 107 and 357 (34, 46). Two catalytic residues, S142 and D344, and a sequence encoding an oxyanion hole (112GGAK115) are located within this domain (34, 46). The oxyanion hole is thought to stabilize the negative charge of the intermediate structure during substrate cleavage (5). C-terminal residues of ExoU, specifically the last 137 amino acids, have been implicated in membrane localization after translocation into mammalian cells (37). The domain or region(s) required for the activation of ExoU by SOD1 have not been identified.
In this study, linker-scanning mutagenesis (the insertion of 15 nucleotides randomly throughout the coding sequence) was used to identify regions of exoU that impair activation of phospholipase activity by SOD1. Our data support the model that SOD1 may be facilitating the activation of ExoU by altering the conformational properties of the enzyme. Understanding the molecular mechanisms mediating SOD1 and ExoU interaction may contribute to the design of therapeutics for the treatment of acute P. aeruginosa infections.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Pseudomonas strains were grown at 37°C in Luria-Bertani (LB) broth and plated on Vogel Bonner medium (55). To produce recombinant proteins from linker-containing genes, open reading frames were subcloned from pUCPexoU-LINKER into pET16b and transformed into Escherichia coli BL21(DE3) pLysS. Point mutations in pUCPexoU-LINKER were generated using the Change-IT mutation site-directed mutagenesis kit (USB Corporation). Bacterial strains containing plasmids were grown in the presence of the appropriate antibiotic(s) at a final concentration of 100 μg/ml of ampicillin, 30 μg/ml of chloramphenicol, or 50 μg/ml of kanamycin for E. coli and 400 μg/ml of carbenicillin for P. aeruginosa.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or properties | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 28 |
| E. coli BL21(DE3) pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3) pLysS (Camr) | 50 |
| P. aeruginosa PA103ΔexoUexoT::Tc | Functional secretion apparatus, no effectors | 53 |
| Plasmids | ||
| pUCP19 | Wild type, E. coli-P. aeruginosa shuttle vector, Ampr | 48 |
| pUCPexoT | ADP-ribosyl transferase and Rho-GAP activities | 56 |
| pUCPexoU | Catalytically active phospholipase | 8 |
| pUCPexoU-S142A | Catalytically inactive phospholipase | 46 |
| pUCPL618-S142A | Catalytically inactive phospholipase | This study |
| pUCPL651-S142A | Catalytically inactive phospholipase | This study |
| pet15b | N-terminal His-tag fusion expression vector with thrombin cleavage site, Ampr | Novagen |
| pet16b | N-terminal His-tag fusion expression vector, Ampr | Novagen |
| pet16bpcrV | Pseudomonas V antigen, 294 aa | 47 |
| pet15bexoU | Catalytically active phospholipase, 687 aa | 1 |
| pet16bexoU | Catalytically active phospholipase, 687 aa | 9 |
| pet16bexoU-S142A | Catalytically inactive phospholipase, 687 aa | 46 |
| pet16bexoU-D344A | Catalytically inactive phospholipase, 687 aa | 46 |
| pet16bexoU-P56 | Linker insertion after P56, 692 aa | This study |
| pet16bexoU-L109 | Linker insertion after L109, 692 aa | This study |
| pet16bexoU-A117 | Linker insertion after A117, 692 aa | This study |
| pet16bexoU-P365 | Linker insertion after P365, 692 aa | This study |
| pet16bexoU-A393 | Linker insertion after A393, 692 aa | This study |
| pet16bexoU-D572 | Linker insertion after D572, 692 aa | This study |
| pet16bexoU-D572* | Linker insertion after D572, 692 aa | This study |
| pet16bexoU-L618 | Linker insertion after L618, 692 aa | This study |
ExoU secretion and detection by Western blot analysis.
For ExoU secretion assays, P. aeruginosa strains were grown in deferrated and dialyzed Trypticase soy broth with 10 mM nitrilotriacetic acid (secretion medium) as previously described for ExoS expression and secretion (23). Secretion medium was inoculated with an emulsion of bacteria at a starting optical density at 540 nm (OD540) of 0.2 and cultured at 32°C until the OD540 reached 3 or 4. The culture supernatant was separated from bacteria by centrifugation at 5,000 × g for 10 min at 4°C and concentrated 20-fold by 55% ammonium sulfate precipitation. Proteins were analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) and then by Western blotting. Blots were probed with a monoclonal anti-ExoU antibody, U29F8 (1:20,000), and a secondary mouse anti-immunoglobulin G (IgG) antibody (1:10,000) conjugated to horseradish peroxidase (HRP; Roche). Bound IgG-HRP was detected with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Linker-scanning mutagenesis of exoU.
Nucleotide insertions (encoding five amino acids) were generated by transposon mutagenesis of pUCPexoU using the GPS-LS linker scanning system (New England Biolabs, Inc.). The pUCPexoU transposon mutants were transformed into DH5α, selected for transposon-encoded kanamycin resistance (Kanr), and subsequently screened for plasmid-encoded ampicillin resistance (Ampr). Approximately 498 colonies were picked after transformation; of these, 101 colonies were resistant to both kanamycin and ampicillin. Plasmids were isolated (Montage plasmid miniprep96 kit) and digested with PmeI (Fermentas) to remove the transposon, and the vector ends were ligated together using standard cloning techniques. The ligated plasmid was transformed into E. coli DH5α, and colonies were picked onto LB supplemented with kanamycin (LBkan) and ampicillin (LBamp) to screen for loss of the transposon. Plasmids were isolated from mutants that were Ampr and Kans and transformed (33) into PA103ΔexoUexoT::Tc, abbreviated throughout as PA103ΔUT. Insertions within the exoU open reading frame will be referred to as ExoU-LINKER molecules. Linker sequences and locations were identified by DNA sequencing of each ExoU-LINKER gene using the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). For clarification, the L109 gene encodes an in-frame 5-amino acid linker after the leucine residue at position 109 in wild-type (WT) ExoU.
Recombinant protein expression, purification, and quantitation.
Cultures were grown at 37°C until the OD600 reached 0.5 or 0.6, and isopropyl-β-d-thiogalactopyranoside (0.5 mM) was added to induce protein expression. The temperature was shifted to either 16°C or 30°C, and the cultures were incubated for 2 or 3 h until the OD600 was greater than 1.0. Cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C. The bacterial pellet was washed with binding buffer (50 mM Na2HPO4, 300 mM NaCl, 5 mM imidazole, pH 7.0) in the presence of protease inhibitors (128 μM benzamidine, 84 μM leupeptin, 2.92 μM pepstatin A, and 6.8 μM phosphoramidon) and subjected to centrifugation at 5,000 × g at 4°C for 10 min. Bacterial pellets were suspended in binding buffer in the presence of protease inhibitors, 0.65 μM DNase (Sigma), and 1.5 μM RNase (Sigma). Cells were lysed by probe-tip sonication (Sonifier cell disrupter; Heat Systems, Inc.), and between pulses the samples were cooled on ice for 5 min. The lysate was subjected to centrifugation at 16,000 × g for 15 min at 4°C to remove membrane debris and unlysed cells. The supernatant was then subjected to ultracentrifugation at 100,000 × g for 1 h at 4°C to remove insoluble material. The soluble supernatant was collected, and histidine-tagged proteins were purified by Talon cobalt chromatography per the manufacturer's instructions (Clontech). Eluted fractions were analyzed by SDS-PAGE, pooled, and dialyzed extensively in the following buffer: 50 mM MOPS (morpholinepropanesulfonic acid) (pH 6.3) and 50 mM NaCl. The protein concentration was determined by bicinchoninic acid (BCA) analysis (Thermo Scientific). Proteins were concentrated in Amicon (Millipore) units that retain molecules above 10 kDa. Untagged rExoU was made as previously described and dialyzed against 50 mM MOPS (pH 6.3) and 50 mM NaCl (1). Purified recombinant protein preparations were stored at −80°C.
In vitro fluorescence-based ExoU activity assay.
The fluorescence-based ExoU activity assay utilizes the fluorogenic substrate PED6 (N-((6-(2,4-dinitrophenyl)amino)hexanoyl)-2-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt) (Invitrogen). Cleavage of PED6 at the sn-2 position relieves the intramolecular quenching effect of the dinitrophenyl group, and upon excitation, the BODIPY-FL fluorescent dye releases a photon at 511 nm. Fluorescence intensity was measured at an excitation wavelength of 488 nm and emission wavelength of 511 nm (495-nm cutoff filter, Spectramax M5 microplate reader; Molecular Devices). The assay conditions were modeled after those used by Sato et al. and Benson et al., with modifications (1, 43).
Supernatants from P. aeruginosa cultures grown overnight at 32°C containing secreted ExoU-LINKER molecules were tested for phospholipase activity using the fluorescence-based ExoU activity assay. Briefly, 20 μl of culture supernatant was added to a mixture of 50 mM MOPS (pH 6.3), 50 mM NaCl, 3.125 μM bovine superoxide dismutase 1 (bSOD1; Sigma), 250 mM monosodium glutamate (MSG) (pH 6.3; Sigma), and 30 μM PED6 in a final volume of 50 μl. Background fluorescence was measured under reaction conditions lacking bSOD1. Background relative fluorescence units (RFU) were subtracted from each sample at each time point. Fluorescence intensity was measured every 15 min for 2 h and normalized to the OD540 at the time of harvest. Positive phospholipase activity was judged as at least 150 RFU and possession of an increasing catalytic rate over the duration of the assay.
When activity was measured from recombinant proteins, the reaction conditions included 50 mM MOPS (pH 6.3), 750 mM MSG (pH 6.3), 30 μM PED6, 0.16 μM ExoU/ExoU-LINKER, and 6.3 μM bSOD1. Background fluorescence, the fluorescence intensity recorded from the reaction conditions lacking bSOD1, was subtracted from each sample. For the bSOD1 titration (6.3 to 75.0 μM) analysis, 0.2 μM ExoU/ExoU-LINKER was added to the assay. Fluorescence intensity was recorded every 15 min for 6 h. The data for phospholipase activity and the bSOD1 titration were plotted as the mean RFU ± standard error of 3 independent experiments.
Assessment of cytotoxicity mediated by P. aeruginosa ExoU-LINKER strains.
To determine if the linker insertion in exoU disrupted cytotoxicity of mammalian cells, HeLa cell monolayers were infected with the P. aeruginosa ExoU-LINKER strains. HeLa cells were seeded in a 24-well plate in Dulbecco's modified Eagle medium (DMEM; Invitrogen) with 10% newborn calf serum (Invitrogen) for confluence (1.2 × 105 cells/well) the next day. Fresh streaks from freezer stocks of P. aeruginosa ExoU-LINKER strains were grown overnight at 37°C. Prior to infection, HeLa cell monolayers were rinsed with Hank's buffered salt solution (HBSS) containing calcium and magnesium. The HeLa cell monolayers were infected with a multiplicity of infection (MOI) of 20 and incubated for 4 h at 37°C with 5% CO2. Cells in an uninfected control well were incubated in serum-free DMEM. After infection, the inoculum was removed, and the monolayers were stained with a 0.3% crystal violet solution in 5% isopropanol/ethanol/methanol for 5 min. Stained cells were rinsed three times with water. After the wells were dry, the retained crystal violet was solubilized in 95% ethanol, and the absorbance at 570 nm was measured. The results were interpreted as positive or negative for monolayer (crystal violet) retention. This procedure was also performed to characterize the monolayer retention with human lung epithelial A549 cells, incorporating the following changes. The A549 cells were seeded in a 48-well plate in DMEM with 10% fetal calf serum (Invitrogen) for confluence (2.4 × 105 cells/well) the next day. When the monolayer was intact, cells were not affected by ExoU intoxication and the crystal violet stain was retained (positive retention). Conversely, loss of the cells corresponded to little or no stain retention (negative retention) and indicated that ExoU cytotoxic activity was expressed.
To confirm the cytotoxic/noncytotoxic phenotype of the P. aeruginosa ExoU-LINKER strains, lactate dehydrogenase (LDH) release from A549 cells was measured. A549 cells were seeded in a 24-well plate in DMEM with 10% fetal calf serum for confluence (2.4 × 105 cells/well) the next day. The A549 cell monolayers were infected with an MOI of 20 and incubated for 4 h at 37°C with 5% CO2. Culture medium from infected cells was harvested and subjected to centrifugation at 2,000 × g for 5 min at 4°C to remove cellular debris and bacteria. The supernatant was transferred to a fresh tube, and LDH activity was measured per the manufacturer's instructions (CytoTox96 nonradioactive assay; Promega). These data were compared to those for a detergent-lysed monolayer (100% lysis) and plotted as the mean ± standard error of 3 independent experiments.
Immunofluorescence microscopy.
To determine the biological activity of P. aeruginosa strains expressing ExoU-LINKER molecules possessing minimal phospholipase activity, infected A549 cells were examined by fluorescence microscopy. A549 cells were seeded for confluence (4.8 × 105 cells/well) in a 6-well plate containing one fibronectin (10 μg/ml; Sigma)-coated coverslip per well. Prior to infection, A549 cell monolayers were rinsed once with HBSS containing calcium and magnesium. The A549 cell monolayers were infected with an MOI of 10 for 4 h at 37°C with 5% CO2. Cells in a control well were incubated in serum-free DMEM. After infection, cells were rinsed with PBS and fixed with 3.7% paraformaldehyde for 10 min at room temperature. After fixation, the cells were washed three times with PBS and permeabilized by incubation with 0.5% saponin in PBS buffer (BP) containing 1% BSA for 15 min at room temperature. The A549 cell monolayers were incubated in BP buffer containing the polyclonal anti-LPS antibody T2WB (1:400,000; a kind gift of Gerald Pier) for 30 min at room temperature. A549 cell monolayers were washed three times with BP buffer and incubated with a goat anti-rabbit Alexa488-labeled secondary antibody (1:1,000; Invitrogen) in BP buffer for 30 min at room temperature. The stained cells were rinsed three times with PBS before exposure to Texas Red-X phalloidin in PBS containing 1% BSA per the manufacturer's instructions (Molecular Probes). Cells were rinsed three times with PBS, and the coverslips were mounted on slides with ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). After 24 h, the coverslips were sealed with nail polish and viewed under a 20× or 60× (oil immersion) objective lens using a Nikon Eclipse Ti inverted microscope.
Binding assay for rExoU association with immobilized SOD1.
To determine if the interaction between ExoU and SOD1 was disrupted in different linker-containing derivatives, a plate-binding assay using recombinant proteins was developed. Briefly, bSOD1 (3 μg in 100 μl of 50 mM sodium carbonate buffer [pH 9.6]) was used to coat a 96-well polyvinyl chloride plate (Falcon; BD Bioscience). The immobilization of bSOD1 was performed at room temperature for 2 h. Bovine SOD1 was aspirated, and the wells were washed three times with 10 mM Tris and 0.9% NaCl (pH 8.0) and blocked overnight at 4°C using a solution of 1% gelatin (BD Bioscience) in sodium carbonate buffer (200 μl). Blocking agent was aspirated, and the wells were washed three times with 50 mM MOPS (pH 6.3), 50 mM NaCl (ExoU assay buffer) with 1.3 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfate (CHAPS; Sigma). All subsequent steps for protein binding, detecting with antibodies, or washing were performed in assay buffer containing CHAPS. Reagents were allowed to bind for 2 h at room temperature. Primary antibodies were used at a 1:2,000 dilution. Histidine-tagged ExoU or PcrV protein was detected using monoclonal antibody (MAb) U29F8 or MAb 166 (12), respectively. A secondary mouse anti-IgG antibody conjugated to HRP (1:5,000; Roche) was used to detect the monoclonal antibody binding. Bound IgG-HRP was detected after incubation for 55 min with QuantaBlu substrate solution per the manufacturer's protocol (Pierce). The fluorescence intensity was measured at an excitation wavelength of 325 nm and an emission wavelength of 420 nm. Nonspecific binding was assessed with two types of wells. One set of wells was coated with gelatin, exposed to recombinant ExoU (rExoU), rExoU derivatives, or rPcrV at identical protein concentrations used to assess specific binding, washed, and processed with primary and secondary antisera (nonspecific binding of test protein controls). Other control wells were coated with SOD1 and blocked with gelatin but were not exposed to rExoUs or rPcrV. These wells were processed with primary and secondary antisera and served as a control for nonspecific antibody binding. The cumulative background from both wells was subtracted from values obtained with wells with SOD1, rExoUs, or rPcrV and the appropriate primary and secondary antibodies. Data were plotted as the mean ± standard error of 3 independent experiments.
Statistics and software.
Statistical analysis and graphs were assembled using Prism 5.0 (GraphPad Software, Inc.) and Microsoft Excel. All figures were generated using Adobe Photoshop CS3, and microscopy images were analyzed using NIS-Elements AR 3.0 (Nikon).
RESULTS
ExoU-LINKER library construction.
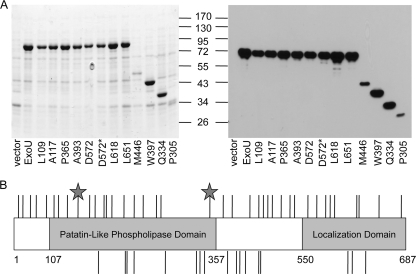
To define regions of ExoU required for cytotoxicity, a previous study used linker-scanning mutagenesis (36). Transposon mutagenesis was performed with a galactose-inducible yeast expression vector containing exoU, insertions within the coding region were resolved, and the resulting plasmids were transformed into a yeast host (36). Cytotoxicity was scored based on growth inhibition in the presence of galactose. As expected, linkers encoding stop codons or insertions disrupting the catalytic residues or oxyanion hole resulted in nontoxic derivatives. Additionally, a C-terminal region (amino acids [aa] 550 to 687) was identified as being required for cytotoxicity, and its functional role appeared to be involved in ExoU localization to the plasma membrane (37). We refined this experimental design to generate linker insertions within a plasmid used for ExoU expression in P. aeruginosa (pUCPexoU). This strategy allowed us to screen for secretion-competent ExoU-LINKER molecules, ensuring that the molecules were capable of passing through the type III needle. We isolated 498 insertions, of which 101 mapped to the exoU open reading frame by restriction endonuclease digestion. The transposons were resolved, and each plasmid was transformed into PA103ΔUT. To screen for full-length molecules, PA103ΔUT strains expressing each insertion were grown under conditions in which type III secretion is induced. Supernatants containing the secreted proteins were analyzed on Coomassie-stained polyacrylamide gels and by Western blot analysis with a primary antibody specific for ExoU to determine if insertion of the linker resulted in an in-frame insertion or a stop codon (Fig. 1A). Of the 101 insertions that mapped within the exoU open reading frame, 69 insertions were in frame and full length, 24 insertions resulted in a stop codon, and 8 insertions resulted in the absence of extracellular ExoU. After removing duplicate insertion molecules, 38 full-length and 17 truncated ExoU-LINKER molecules were identified using this technique. All ExoU-LINKER molecules obtained were analyzed for in vitro phospholipase activity and for cytotoxicity in a HeLa cell infection model. Figure 1B is a summary of the location of each ExoU-LINKER molecule, with full-length ExoU-LINKER molecules above the diagram and truncated molecules below the diagram. D572 and D572* are two ExoU-LINKER molecules with insertions at the same position but with different amino acid sequences encoded by the linker. The two stars in Fig. 1B represent the two catalytic residues of exoU, S142 and D344. Each ExoU-LINKER molecule is identified as the last ExoU-specific amino acid before the linker insertion.
FIG. 1.
Summary of the ExoU-LINKER molecules. (A) Supernatants from strains were screened by SDS-PAGE and Western blot analyses to determine whether the linker insertion encoded a full-length or truncated ExoU derivative. Shown are a Coomassie-stained gel showing the representative full-length and truncated ExoU-LINKER molecules (left) and a Western blot probed with a monoclonal antibody to ExoU (right). pUCPexoU is a control for full-length molecules, and vector is a negative control for ExoU expression. All ExoU-LINKER molecules were competent for secretion from P. aeruginosa under inducing conditions. Each ExoU-LINKER insertion is labeled according to the site of insertion. For example, L109 represents an insertion after amino acid 109 and before amino acid 110. D572* is a second insertion at position 572, with a linker sequence different than that of D572. The sizes of the molecular weight markers (in thousands) are indicated in the middle. (B) The location of each insertion is depicted on a diagram of exoU. The lines above and below the diagram represent the unique full-length and truncated ExoU-LINKER molecules, respectively. The gray stars depict the two catalytic amino acids required for phospholipase activity, S142 and D344.
ExoU phospholipase activity and cytotoxicity screens.
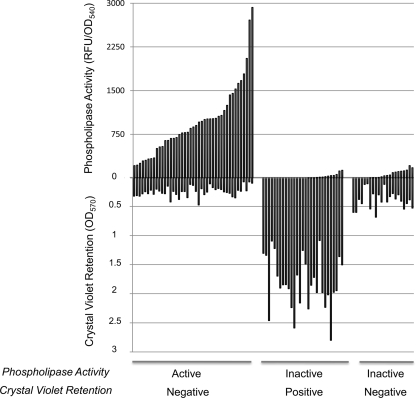
To determine the effect of the linker insertions on ExoU activity, each ExoU-LINKER protein was screened for in vitro phospholipase activity present in bacterial culture supernatants and for cytotoxicity during cellular infections. An optimized activity assay was utilized that measures phospholipase activity by the cleavage of the fluorogenic substrate PED6 (1, 43). To normalize for the phospholipase activity relative to the growth of the culture, RFU values were divided by the OD540 value at the time of harvest. Supernatants of each ExoU-LINKER molecule were scored as active, based on an increase in fluorescence intensity as a function of time, or inactive if no increase in fluorescence intensity was observed within 2 h. An initial increase in fluorescence intensity was observed for a few ExoU-LINKER supernatants, but the rate associated with substrate cleavage did not increase by the 2-h time point. These ExoU-LINKER molecules were considered inactive. A supernatant containing either no ExoU or WT ExoU were the negative or positive control for the assay, respectively. Figure 2 (top) shows the distribution of active or inactive ExoU-LINKER molecules from this high-throughput screen.
FIG. 2.
Compilation of phospholipase and cytotoxic activities for all ExoU-LINKER molecules. Supernatants containing ExoU-LINKER molecules secreted from P. aeruginosa were used to determine phospholipase activity in a fluorescence-based ExoU activity assay. ExoU-LINKER molecules are categorized as either active or inactive based on PED6 cleavage after 2 h (top half). Every ExoU-LINKER strain was tested for cytotoxicity as determined by loss of a crystal violet-stained HeLa cell monolayer. After a 4-h infection, the HeLa cell monolayers were fixed and stained as outlined in Materials and Methods. The stain was solubilized with 95% ethanol, and the absorbance was recorded at 570 nm. Each ExoU-LINKER molecule is categorized as being positive or negative for crystal violet retention (bottom half).
To determine the activity of ExoU-LINKER molecules in the context of a P. aeruginosa infection, confluent HeLa cell monolayers were infected with each P. aeruginosa ExoU-LINKER strain and stained with crystal violet. The retention of crystal violet (positive retention) indicates that the monolayer is not lysed after intoxication with ExoU, while the absence of staining (negative retention) indicates a fully cytotoxic strain. The negative and positive controls (PA103ΔUT vector only and pUCPexoU, respectively) were incorporated into each assay set. ExoU-LINKER strains possessed either a cytotoxic or noncytotoxic phenotype (Fig. 2, bottom).
To understand the results from both high-throughput screenings of the ExoU-LINKER library, data from each screen were first plotted in the order of the lowest to highest values obtained (data not shown). Two trends were observed: ExoU-LINKER strains were either cytotoxic and had phospholipase activity or were noncytotoxic and had no phospholipase activity. However, upon aligning results from both screenings on the same graph, a unique set of ExoU derivatives that were cytotoxic to infected cells (low crystal violet retention) but had no phospholipase activity were identified. These ExoU-LINKER molecules possessing unique phenotypes were investigated further.
The data for each class of molecules identified are summarized in Tables 2 to 4. These tables include information identifying the location of each insertion, the amino acid sequence of the insertion, and the crystal violet retention value of the cytotoxicity assay. For comparison purposes, the crystal violet retention values are reported for the vector control strain and a strain complemented with parental ExoU. The mean phospholipase activity of the extracellular medium for each group is 800 ± 100 RFU for members that were cytotoxic and possessed activity (Table 2), 12 ± 6 RFU for candidates that were noncytotoxic and had no extracellular activity (Table 3), and 46 ± 13 RFU for isolates that were apparently cytotoxic but were below the threshold activity to be considered positive in our screens (Table 4). Insertions affecting the phospholipase activity were found throughout the molecule.
TABLE 2.
Screen summary for cytotoxic ExoU-LINKER molecules with supernatant phospholipase activity
| Designation | Sequence | Membrane retention (OD570) |
|---|---|---|
| Vector | NA | 1.3 |
| WT ExoU | NA | 0.275 |
| S5 | LVFKQ | 0.245 |
| S22 | QVFKQ | 0.262 |
| P56 | EMFKQ | 0.275 |
| Q68 | GCLNK | 0.222 |
| I135 | RCLNI | 0.324 |
| L173 | LFKHL | 0.476 |
| L194 | MFKQL | 0.243 |
| K195 | CLNMK | 0.336 |
| G263 | CLNTG | 0.245 |
| F290 | KQAMF | 0.315 |
| E430 | LFKHE | 0.529 |
| Q498 | LFKHQ | 0.100 |
| G530 | CLNNG | 0.248 |
| R581 | VFKQR | 0.192 |
| V585 | QSLFK | 0.283 |
| S587 | LFKQS | 0.381 |
| Y619 | LFKQY | 0.139 |
| W681 | CLNTW | 0.204 |
NA, not applicable.
TABLE 4.
Screen summary for cytotoxic ExoU-LINKER molecules without supernatant phospholipase activity
| Designation | Sequence | Membrane retention (OD570) |
|---|---|---|
| Vector | NA | 1.3 |
| WT ExoU | NA | 0.275 |
| L104 | TSCLN | 0.332 |
| K161 | CLNIK | 0.373 |
| I254 | CLNTI | 0.301 |
| P365 | LLFKQ | 0.279 |
| A393 | CLNSA | 0.452 |
| N443 | FTCLN | 0.453 |
| A452 | HLLFK | 0.544 |
| L537 | VFKQL | 0.425 |
| W560 | CLNTW | 0.111 |
| D572 | HQCLN | 0.122 |
| D572* | CLNID | 0.123 |
| L618 | CLNTL | 0.302 |
| L651 | MFKQL | 0.426 |
TABLE 3.
Screen summary for noncytotoxic ExoU-LINKER molecules without supernatant phospholipase activity
| Designation | Sequencea | Membrane retention (OD570) |
|---|---|---|
| Vector | NA | 1.3 |
| WT ExoU | NA | 0.275 |
| L109 | MFKHL | 1.306 |
| A117 | AYCLN | 2.461 |
| I190 | V* | 1.719 |
| P222 | LDV* | 1.914 |
| L223 | ERV* | 1.980 |
| D229 | CLNSD | 1.340 |
| V251 | TVV* | 1.223 |
| L272 | SAV* | 1.846 |
| N300 | V* | 1.854 |
| P305 | DLV* | 1.975 |
| Q334 | AGV* | 1.086 |
| A384 | PDV* | 1.044 |
| W397 | VVV* | 1.697 |
| V399 | V* | 1.902 |
| L412 | EGV* | 2.588 |
| T445 | IV* | 2.261 |
| M446 | V* | 2.799 |
| L458 | QVFKH | 1.504 |
| G463 | EHCLN | 1.488 |
| A606 | V* | 2.234 |
| N608 | V* | 2.238 |
| G622 | GV* | 1.678 |
* denotes a stop codon. NA, not applicable.
Phospholipase activity of recombinant ExoU-LINKER molecules.
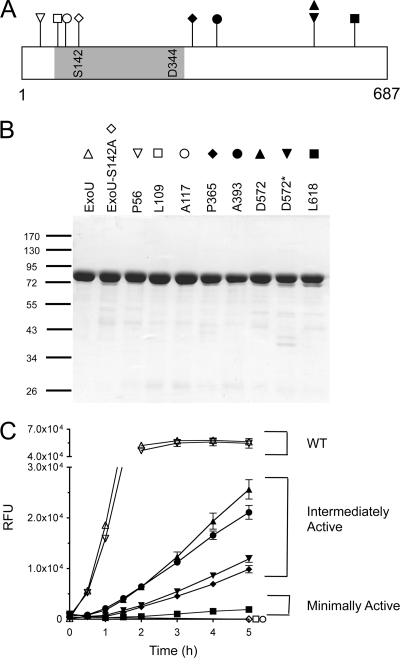
Although standardized by the OD540 value at the time of harvest, supernatants are a crude source of enzyme for quantifying activity. Different amounts of ExoU-LINKER molecules could be present in the culture medium due to differences in production rate or protein degradation. We addressed this issue by expressing and purifying recombinant, histidine-tagged ExoU-LINKER molecules. The locations of the ExoU-LINKER molecules chosen for recombinant protein analysis are shown in Fig. 3A. These derivatives were selected based on the apparent uncoupling of phospholipase activity and cytotoxicity as measured by crystal violet retention (P365, A393, D572, D572*, and L618) and represent a subset of the total number of strains with this phenotype. P56 was chosen as a linker control molecule that displayed parental extracellular phospholipase and cytotoxicity activities in the screen. Two insertions mapping adjacent to the oxyanion hole were selected (L109 and A117) to verify that phospholipase activity was eliminated (36). Histidine-tagged parental ExoU and ExoU-S142A were purified as positive and negative controls, respectively. All recombinant proteins were purified using affinity chromatography and analyzed by SDS-PAGE and Coomassie staining (Fig. 3B). Recombinant L651 appeared to be labile when expressed from E. coli and was excluded from in vitro phospholipase activity analyses (data not shown).
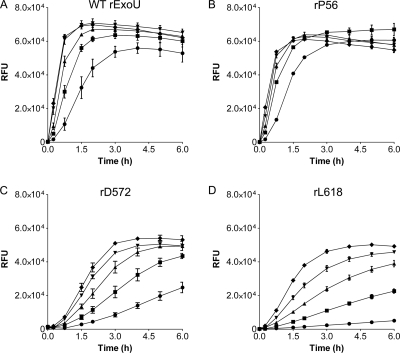
FIG. 3.
Purified ExoU-LINKER proteins and phospholipase activity. (A) Location of ExoU-LINKER molecules chosen for analysis using recombinant proteins. The symbol above each location site in panel A corresponds to the symbol used for each ExoU derivative in panels B and C. The gray region represents the patatin-like phospholipase A2 domain of ExoU. The catalytic sites are marked within the gray region of the diagram. The ExoU-LINKER molecules chosen for analysis include P56 (▿), L109 (□), A117 (○), S142A (⋄), P365 (♦), A393 (•), D572 (▴), D572* (▾), and L618 (▪). Not depicted in the diagram is WT rExoU (▵). Two insertions were obtained at D572, but with different sequences. (B) SDS-PAGE analysis of ExoU-LINKER recombinant proteins used in the fluorescence-based ExoU activity assay. Each well of the Coomassie-stained gel contains approximately 1.5 μg of recombinant protein. The sizes of the molecular weight markers (in thousands) are indicated on the left. (C) Phospholipase activity of recombinant proteins was measured in the fluorescence-based ExoU activity assay. The assay reaction included 0.16 μM ExoU/ExoU-LINKER, 6.3 μM bSOD1, 50 mM MOPS (pH 6.3), 50 mM NaCl, 750 mM MSG (pH 6.3), and 30 μM PED6. RFU were measured every 15 min for 2 h and every h thereafter for a total of 5 h. Recombinant L109 and rA117 possessed the same activity curve as a noncatalytic molecule, S142A. Each point represents the average of 3 independent experiments, with error bars for standard error.
WT rExoU and rP56 displayed a rapid increase in fluorescence intensity and resulted in a substrate cleavage maximum of approximately 50,000 RFU by 2 h (Fig. 3C). The ExoU-LINKER proteins rP365, rA393, rD572, and rD572* exhibited phospholipase activity as the fluorescence intensity increased over time but at a rate significantly lower than that of WT rExoU. Recombinant L618 exhibited a significantly impaired, but measurable, increase in fluorescence intensity over the 5-h time period. The negative controls for the assay, rExoU-S142A and two linker molecules located close to the oxyanion hole, rL109 and rA117, displayed no phospholipase activity after 5 h. The position of the linkers and the phospholipase activity allowed grouping of the recombinant ExoU-LINKER proteins into two broad categories, enzymes with either intermediate activity or minimal activity. Proteins belonging to the first category display phospholipase activity at a level less than that of WT rExoU and include P365, A393, D572, and D572*. The second category includes L618 and possibly L651 (based on activity assays of P. aeruginosa culture supernatants), which possess a minimal increase in fluorescence intensity as a function of time. Together, these data suggest that of the insertions analyzed, those mapping within the C terminus impair the phospholipase activity of ExoU.
Crystal violet retention and LDH release in A549 cells.
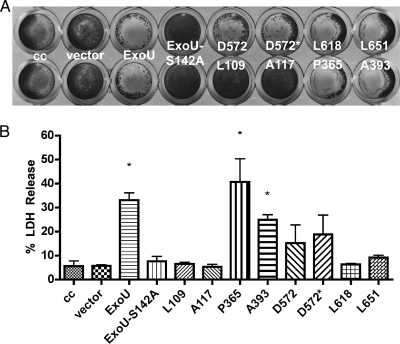
Results from the fluorescence-based ExoU activity assays suggested that the linker insertions in ExoU resulted in a collection of molecules with variable but detectable enzyme activity. Cytotoxicity assays using crystal violet staining, however, indicated that these molecules were still relatively potent. Based on these observations, we postulated that either a small amount of phospholipase activity was sufficient for cytotoxicity or that ExoU may express a second activity that is masked by membrane disruption. Alternatively, HeLa cells may be particularly susceptible to either the phospholipase or the postulated second activity. To determine if differences in the assay outcomes were due to different cell lines being tested, confluent A549 cell monolayers were infected with each ExoU-LINKER expression strain and stained with crystal violet (Fig. 4A). Infection with strains containing the vector control ExoU-S142A, L109, and A117 were no different than uninfected-cell controls. Infections with strains expressing ExoU or ExoU-LINKER molecules P365, A393, D573, D572*, L618, and L651 resulted in a loss of the monolayer and minimal crystal violet retention. These results were identical to those observed when HeLa cells were infected, suggesting that both cell lines are susceptible to the action(s) of ExoU.
FIG. 4.
ExoU-mediated cytotoxicity analysis in A549 cells. (A) Crystal violet retention was determined for selected ExoU-LINKER strains in A549 cells. A549 cell monolayers were infected with an MOI of 20 for 4 h and stained with crystal violet. The positive controls for crystal violet membrane retention include an uninfected cell control (cc), an empty vector control, and catalytically inactive ExoU-S142A. The negative control for crystal violet membrane retention is WT ExoU. (B) LDH release from A549 cells. The negative controls for LDH release include cell control (cc), vector, and catalytically inactive ExoU-S142A. A549 monolayers were infected with the same PA103ΔUT strains, and LDH release was compared to a maximum lysis control to calculate a percent LDH release for each sample. The data are plotted as mean ± standard error of 3 independent experiments. The asterisks represent values statistically significant compared to ExoU-S142A (Student's t test, P < 0.004).
To analyze cytotoxicity quantitatively, we infected A549 monolayers with the same strain set and measured LDH release (Fig. 4B). Infections with strains expressing ExoU, P365, or A393 resulted in LDH release and were clearly cytotoxic relative to negative controls (uninfected cells, vector control, and ExoU-S142A). LDH release from cells infected with strains expressing D572 and D572* trended toward being cytotoxic, but the values were variable between experiments, indicating that other factors may be dictating the susceptibility of cells to the intermediate phospholipase activity of these variants. Cells infected with L109, A117, L618, and L651 released LDH at levels statistically identical to ExoU-S142A and were judged as noncytotoxic by this assay (Student's t test, P < 0.004). While the results of the LDH assay and crystal violet staining are consistent for L109 and A117, they are diametrically opposed for L618 and L651. These data suggest that the minimal phospholipase activities exhibited by these linker insertion molecules may result in changes in cell association with the tissue culture plate substrate and extracellular matrix rather than membrane permeability and cell death.
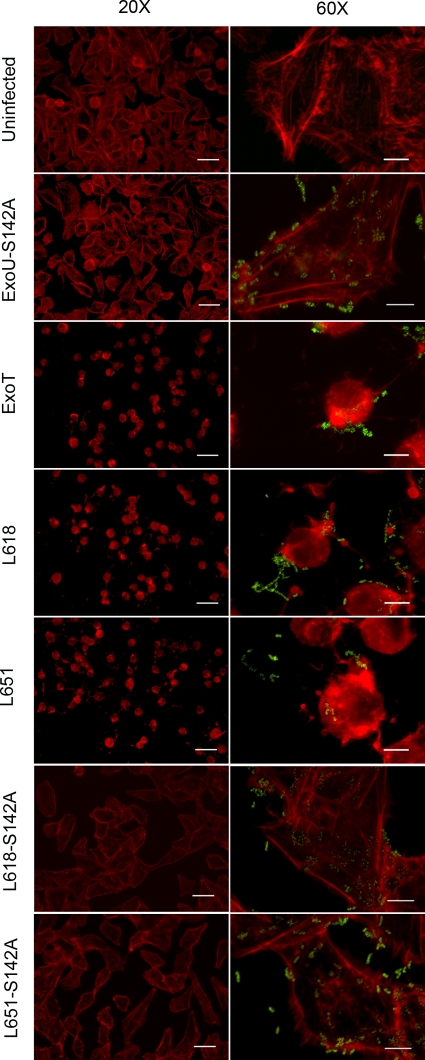
Minimal phospholipase activity affects the actin cytoskeleton.
To determine the biological effect of minimal phospholipase activity, A549 cells were infected with the PA103ΔUT ExoU-LINKER L618 and L651 mutant strains. These two strains have minimal phospholipase activity in vitro and result in loss of HeLa and A549 cell monolayers in the crystal violet retention assay but do not release LDH. A549 cell monolayers on coverslips were infected, and the actin cytoskeleton morphology was examined by phalloidin staining (Fig. 5). In addition to an uninfected control, A549 cell monolayers were also infected with PA103ΔUT ExoU-S142A or the same host strain that is complemented with an ExoT expression construct. The actin cytoskeleton does not change shape after intoxication with a catalytically inactive phospholipase (ExoU-S142A). However, infection with an ExoT-expressing strain resulted in distinct changes to the cytoskeleton, as previously reported (15). Infection with the minimally active ExoU-LINKER molecules L618 and L651 also shows a rearrangement of the actin cytoskeleton similar to that of ExoT. To ensure that phospholipase activity was responsible for the cell-rounding phenotype, we introduced the S142A mutation into molecules L618 and L651 and repeated the in vitro infections. Introducing the catalytic mutation completely abrogated the effects on the actin cytoskeleton, confirming that minimal amounts of phospholipase activity are generating the observed cell rounding. In addition to crystal violet staining, these data confirm that both ExoU-LINKER molecules are translocated into A549 cells. Minimal phospholipase activity appears to have either specific or nonspecific effects on the cellular cytoskeleton that result in rounding and detachment.
FIG. 5.
Minimal activity of ExoU-LINKER molecules affect the actin cytoskeleton of infected cells. A549 cell monolayers were infected with PA103ΔUT strains at an MOI of 10 for 4 h. After infection, samples were permeabilized and Texas Red-X phalloidin was used to visualize the actin cytoskeleton. Polyclonal anti-LPS antibody, followed by a secondary goat anti-rabbit Alexa488-labeled antibody, was used to detect P. aeruginosa. Slides were examined at ×20 and ×60 magnification of an objective lens to observe phalloidin staining of the actin cytoskeleton (left, ×20), in conjunction with the presence of the bacteria (right, ×60). The scale bars for ×20 and ×60 magnification are 50 and 10 μm, respectively. A549 cells infected with PA103ΔUT ExoT served as a positive control for translocation and cell rounding.
Activation of ExoU-LINKER molecules with bSOD1.
The C terminus of ExoU has previously been postulated to be important for the interaction with the cellular activator SOD1 (45). To determine if the activation of each molecule could be restored to a wild-type level with additional cofactor present, bSOD1 (6.3 to 75 μM) was titrated into reaction mixtures containing purified rExoU, rP56, rD572, or rL618. Recombinant ExoU and rP56 exhibited dose-dependent increases in substrate cleavage, which reached a maximum fluorescence intensity of approximately 60,000 RFU (Fig. 6A and B). A dose-dependent increase in phospholipase activity was also observed for rD572 and rL618, but the level of substrate cleavage saturation was lower than that of WT rExoU or rP56 (Fig. 6C and D). Table 5 lists the enzymatic rates of rExoU, rP56, rD572, and rL618 upon activation by bSOD1. The rates were statistically different when rExoU or rP56 was compared to rD572 or rL618 (Student's t test, P < 0.0394). This further validates the assignment of D572 and L618 to intermediate and minimally active categories, respectively. These results suggest that either the association with or activation by SOD1 is affected by the linker insertion within ExoU.
FIG. 6.
SOD1 titration with ExoU-LINKER molecules. Phospholipase activity was measured in the presence of increasing concentrations of bSOD1. Progression curves for rExoU (A), rP56 (B), rD572 (C), and rL618 (D) show a dose-dependent increase in phospholipase activity over 6 h. The concentrations of bSOD1 used include 6.3 (•), 18.8 (▪), 37.5 (▴), 56.3(▾), and 75.0 (⧫) μM. A blank lacking bSOD1 was measured for each recombinant protein to subtract background cleavage of PED6. Each point represents the average of 3 independent experiments, with error bars for standard error.
TABLE 5.
Enzymatic rates in response to bSOD1 concentrationa
| μM bSOD1 | rExoU | P56 | D572 | L618 |
|---|---|---|---|---|
| 6.3 | 24,950 ± 2,654 | 32,992 ± 617 | 4,978 ± 559 | 839 ± 41 |
| 18.8 | 40,287 ± 1,339 | 41,418 ± 826 | 7,863 ± 386 | 3,948 ± 222 |
| 37.5 | 65,740 ± 3,914 | 59,393 ± 1,710 | 12,124 ± 735 | 6,977 ± 275 |
| 56.3 | 81,178 ± 3,378 | 71,781 ± 1,018 | 15,884 ± 373 | 12,797 ± 684 |
| 75.0 | 82,780 ± 1,686 | 91,102 ± 143 | 17,792 ± 517 | 19,871 ± 456 |
Values (RFU/h) are reported as mean ± standard error.
SOD1-ExoU association.
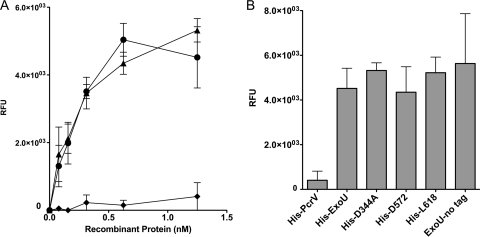
To measure the association between bSOD1 and rExoU parental or derivative molecules, we developed an ELISA-based test. The highest specific binding was achieved when bSOD1 was immobilized to the solid phase. The inclusion of CHAPS, a zwitterionic detergent, in the wash and binding buffers increased the sensitivity of the assay. Finally, a highly sensitive horseradish peroxidase substrate was incorporated to detect potentially minor populations of interactants. Assay buffers, time, and incubation temperatures mimicked the in vitro conditions used to detect phospholipase activity. Since bSOD1 binds copper and zinc, assay conditions with and without 1 mM EDTA were tested to suppress the possible nonspecific binding of tagged molecules to the metals associated with bSOD1. The inclusion of EDTA in binding, wash, and assay buffers made no difference (data not shown). Various histidine-tagged proteins, including WT rExoU, rPcrV, rD344A, rD572, and rL618 and an untagged version of rExoU were tested.
The nonspecific binding control protein rPcrV associated with bSOD1 to a limited extent but was significantly different than WT rExoU and rD344A (P < 0.0143; Fig. 7A). All rExoU derivatives bound to immobilized bSOD1 similarly and were significantly different than rPcrV (P < 0.0313; Fig. 7B). The binding of untagged rExoU to bSOD1 was similar to that of WT rExoU but was more variable from experiment to experiment. These results indicate that the linker insertions within ExoU likely disrupt the ability of the molecule to be activated, perhaps by preventing ExoU from assuming an optimal conformation.
FIG. 7.
The ExoU-SOD1 interaction is maintained for C-terminal linker insertion molecules. An ELISA was developed to study ExoU-SOD1 binding. Recombinant protein binding, wash conditions, and controls for nonspecific interactions are described in Materials and Methods. (A) Control histidine-tagged proteins tested include WT rExoU (•), rD344A (▴), and rPcrV (♦). (B) Binding of 1.25 nM recombinant protein to bSOD1, with cumulative background control wells subtracted. The data are plotted as the average of 3 independent experiments, with error bars for standard error.
DISCUSSION
The long-term goal of our studies is to identify the mechanism of ExoU activation. Our strategy was to map regions important for cytotoxicity and enzyme activation. Previous biochemical analyses identified three regions encompassing an oxyanion hole and the two catalytic residues located within the patatin-like phospholipase domain (34, 46). Linker insertion mutagenesis of exoU indicated that in addition to these important regions, cytotoxic activity also required an uncharacterized region within the patatin-like phospholipase domain and a membrane recognition signal (36, 37). Although the techniques used in this study to generate linker insertion molecules in ExoU were identical to those of Rabin et al., our expression, screening, and catalytic analyses differed (36). Our initial screening for insertions was unbiased, in the sense that only clones with linkers within the exoU open reading frame were selected for study; expression or function was not assessed at this stage. Additionally, functional screens were performed with P. aeruginosa to eliminate molecules whose gross structure was disrupted to the extent that they could not be secreted by the P. aeruginosa T3SS. This approach allowed us to assay for activation by a eukaryotic cofactor after injection occurred in a biologically relevant manner. Finally, a highly sensitive and defined enzymatic assay was used that allowed characterization of marginally active molecules. Although no single screen can identify all the residues or regions required for the biologic activities of ExoU, the combined data from both studies represents a fairly saturated coverage of the molecule and reveals novel functional information.
The optimized enzymatic assay allowed characterization of ExoU molecules that differed in activity. To date, only fully enzymatic and catalytically inactive ExoU proteins have been examined for phospholipase activity (43, 46). However, upon linker insertion, two additional classes of enzymes were identified that had intermediate and minimal phospholipase activity. Possession of some enzymatic activity argues against a general disruption of ExoU structure and indicates that the linker affects only a localized region of the molecule. The modulating properties of the identified residues in conjunction with SOD1 suggest that these regions may be involved in conformational changes that are perhaps required for ExoU to recognize its phospholipid substrates. This could be achieved by an interaction with SOD1 that removes an inhibitory domain or an interaction that opens a catalytic pocket. The linker insertions analyzed did not have a significant effect on interactions with immobilized SOD1 in an ELISA-based test, suggesting that SOD1 binding was not disrupted.
The impairment of the activation of ExoU by SOD1 is further supported by the SOD1 titration experiment. Wild-type levels of phospholipase activity were not achieved even when physiological concentrations of SOD1 were added to the reaction mixture. Under the conditions of our assay, we observed saturating kinetics for WT ExoU in the presence of 56.3 and 75 μM bSOD1. These concentrations appear high, although biologically, they are similar to those present in mammalian cells (4, 38). The high concentrations of SOD1 required for activation may be related to the aggregation state of SOD1 or suggest that only a subpopulation of molecules is competent to interact and activate ExoU (1). Conversely, the intracellular aggregation state of ExoU may be important, and SOD1 may be functioning to oligomerize or disrupt oligomers of ExoU.
The ability to target the innate immune system prevents elimination of invading bacteria (29). Specifically, neutrophils and macrophages are targeted by type III secretion systems (6, 20, 29). As a result, bacterial replication can proceed beyond control of this first line of defense. During acute P. aeruginosa infections, bacterial burden is not the only issue worth consideration. P. aeruginosa strains possessing or not possessing a T3SS colonize the lung to the same extent; however, expression of T3SS effectors facilitates systemic dissemination (49). Neutralization of innate immunity as well as systemic dissemination appears to be promoted by the activities of the P. aeruginosa effectors secreted by the T3SS. Each enzyme utilizes eukaryotic cofactors to potentiate their activity. ExoS, ExoT, and ExoY have profound effects on the cellular cytoskeleton and prevent phagocytosis (16, 53). ExoS is also cytotoxic and likely contributes to the tissue destruction that leads to systemic spread (11). Our analysis demonstrates that ExoU affects the cellular cytoskeleton as well. These effects are detectable only during in vitro infections in which strains express molecules with compromised activity. The effects do not appear to be related to another functional domain, as infection with strains expressing noncatalytic ExoU site-specific mutants (the S142A and D344A mutants) do not alter cellular architecture. Importantly, catalytic mutations introduced into the linker molecules abrogate the cell rounding activity, verifying that phospholipase activity itself is critical. The diminished activity of the mutants may provide a valuable clue to the primary activity of ExoU in a biological context. One might imagine that, in tissue, fully motile bacteria would be capable of injecting and moving to another location. Thus, the effects on the cytoskeleton by a low dose (single administration or small number of administrations) of ExoU may mimic in vivo events and suggest that ExoU's primary role may be to breach epithelial barriers rather than destroy them.
The phospholipase A2 activity of ExoU has profound implications for lipid metabolism. The release of lysophospholipids and arachidonic acid from a membrane substrate results in the activation of several enzymes, including lipoxygenase and cyclooxygenase. Lipoxygenase and cyclooxygenase catalyze the conversion of arachidonic acid to leukotrienes and prostaglandins, respectively, which aid in generating an inflammatory response (26). ExoU and the T3SS have both been shown to be important for the release of arachidonic acid and the induction of cyclooxygenase 2 after infection with P. aeruginosa (41, 42). Furthermore, arachidonic acid has been shown to prime and activate NADPH oxidase, an enzyme that produces large amounts of superoxide radicals (18). SOD is responsible for catalyzing the conversion of the superoxide radical to molecular oxygen and hydrogen peroxide to prevent any further oxidative damage to surrounding tissues (4). Thus, SOD1, which is generally in high concentrations within undamaged cells, is also in high concentration in inflammatory fluids in which P. aeruginosa replicates rapidly (24). Considering the activity of ExoU during tissue culture infections, it is conceivable that the inflammatory response coupled with necrosis could release a fair amount of extracellular SOD1 that might activate ExoU secreted by rapidly replicating bacteria. An extracellular phospholipase activity could contribute to tissue destruction and the systemic spread of P. aeruginosa. Additionally, SOD3 is abundant in the lungs of humans and mice and may also participate in activating extracellular ExoU (30, 31). Based on our analysis, the ExoU derivatives with reduced activity may be fully capable of affecting single cells but may lose extracellular properties due to the diminished activity. The analysis of these strains in vivo will be a worthwhile experiment.
Our data suggest that a domain that interacts with SOD1 has yet to be mapped. T3SS effectors are assumed to be unfolded during injection since the needle cannot accommodate folded molecules (14). We have considered the hypothesis that ExoU begins to fold within the cell in response to contact with SOD1. Folding may be a potential rate-limiting step in achieving an active conformation. In this model, SOD1 functions as a scaffold or protein chaperone for ExoU, perhaps explaining the high concentration of cofactor required for activation. Alternatively, SOD1 may alter the conformation of partially folded ExoU to remove an inhibitory domain. Finally, regions of the C terminus of ExoU may be involved in catalysis by positioning important residues within the N-terminal patatin domain. Clearly, future studies would benefit from structural analyses and a determination of the role of SOD1 or other molecules in ExoU activation.
Acknowledgments
We thank Eric Danielson for help in generating the ExoU-LINKER library, Monika Casey for her expert technical assistance, Hiromi Sato for her microscopy expertise, and Owen Griffith for his help in interpreting and designing kinetic experiments. We are grateful to the Human and Molecular Genetics Center at the Medical College of Wisconsin for use of their high-throughput sequencing facility.
This work was supported by funds from the National Institute of Health (A149577 to D.W.F.), the Center for Biopreparedness and Infectious Disease, and the Advancing a Healthier Wisconsin Foundation at the Medical College of Wisconsin.
Footnotes
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Benson, M. A., K. M. Schmalzer, and D. W. Frank. 2010. A sensitive fluorescence-based assay for the detection of ExoU-mediated PLA(2) activity. Clin. Chim. Acta 411:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bharathi, M. J., R. Ramakrishnan, R. Meenakshi, C. S. Kumar, S. Padmavathy, and S. Mittal. 2007. Ulcerative keratitis associated with contact lens wear. Indian J. Ophthalmol. 55:64-67. [DOI] [PubMed] [Google Scholar]
- 3.Bjarnsholt, T., P. O. Jensen, M. J. Fiandaca, J. Pedersen, C. R. Hansen, C. B. Andersen, T. Pressler, M. Givskov, and N. Hoiby. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547-558. [DOI] [PubMed] [Google Scholar]
- 4.Chang, L. Y., J. W. Slot, H. J. Geuze, and J. D. Crapo. 1988. Molecular immunocytochemistry of the CuZn superoxide dismutase in rat hepatocytes. J. Cell Biol. 107:2169-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dessen, A., J. Tang, H. Schmidt, M. Stahl, J. D. Clark, J. Seehra, and W. S. Somers. 1999. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 97:349-360. [DOI] [PubMed] [Google Scholar]
- 6.Diaz, M. H., C. M. Shaver, J. D. King, S. Musunuri, J. A. Kazzaz, and A. R. Hauser. 2008. Pseudomonas aeruginosa induces localized immunosuppression during pneumonia. Infect. Immun. 76:4414-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finck-Barbançon, V., and D. W. Frank. 2001. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J. Bacteriol. 183:4330-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finck-Barbançon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. Fleiszig, C. Wu, L. Mende-Mueller, and D. W. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 9.Finck-Barbançon, V., T. L. Yahr, and D. W. Frank. 1998. Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J. Bacteriol. 180:6224-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleiszig, S. M., N. Efron, and G. B. Pier. 1992. Extended contact lens wear enhances Pseudomonas aeruginosa adherence to human corneal epithelium. Investig. Ophthalmol. Vis. Sci. 33:2908-2916. [PubMed] [Google Scholar]
- 11.Fleiszig, S. M., J. P. Wiener-Kronish, H. Miyazaki, V. Vallas, K. E. Mostov, D. Kanada, T. Sawa, T. S. Yen, and D. W. Frank. 1997. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 65:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank, D. W., A. Vallis, J. P. Wiener-Kronish, A. Roy-Burman, E. G. Spack, B. P. Mullaney, M. Megdoud, J. D. Marks, R. Fritz, and T. Sawa. 2002. Generation and characterization of a protective monoclonal antibody to Pseudomonas aeruginosa PcrV. J. Infect. Dis. 186:64-73. [DOI] [PubMed] [Google Scholar]
- 13.Fu, H., J. Coburn, and R. J. Collier. 1993. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. U. S. A. 90:2320-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567-573. [DOI] [PubMed] [Google Scholar]
- 15.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. Hauser, and J. N. Engel. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrity-Ryan, L., S. Shafikhani, P. Balachandran, L. Nguyen, J. Oza, T. Jakobsen, J. Sargent, X. Fang, S. Cordwell, M. A. Matthay, and J. N. Engel. 2004. The ADP ribosyltransferase domain of Pseudomonas aeruginosa ExoT contributes to its biological activities. Infect. Immun. 72:546-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillis, R. J., K. G. White, K. H. Choi, V. E. Wagner, H. P. Schweizer, and B. H. Iglewski. 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 49:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartfield, P. J., and J. M. Robinson. 1998. Arachidonic acid activates NADPH oxidase by a direct, calmodulin-regulated mechanism. Prostaglandins Other Lipid Mediat. 56:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Hauser, A. R., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 20.Hauser, A. R., and J. N. Engel. 1999. Pseudomonas aeruginosa induces type-III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect. Immun. 67:5530-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huigens, R. W., J. J. Richards, G. Parise, T. E. Ballard, W. Zeng, R. Deora, and C. Melander. 2007. Inhibition of Pseudomonas aeruginosa biofilm formation with Bromoageliferin analogues. J. Am. Chem. Soc. 129:6966-6967. [DOI] [PubMed] [Google Scholar]
- 23.Iglewski, B. H., J. Sadoff, M. J. Bjorn, and E. S. Maxwell. 1978. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc. Natl. Acad. Sci. U. S. A. 75:3211-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinnula, V. L., and J. D. Crapo. 2003. Superoxide dismutases in the lung and human lung diseases. Am. J. Respir. Crit. Care Med. 167:1600-1619. [DOI] [PubMed] [Google Scholar]
- 25.Krall, R., J. Sun, K. J. Pederson, and J. T. Barbieri. 2002. In vivo rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuehl, F. A. J., and R. W. Egan. 1980. Prostaglandins, arachidonic acid, and inflammation. Science 210:978-984. [DOI] [PubMed] [Google Scholar]
- 27.Lambert, P. A. 2002. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J. R Soc. Med. 95(Suppl. 41):22-26. [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Marketon, M. M., R. W. DePaolo, K. L. DeBord, B. Jabri, and O. Schneewind. 2005. Plague bacteria target immune cells during infection. Science 309:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marklund, S. L. 1984. Extracellular superoxide dismutase in human tissues and human cell lines. J. Clin. Investig. 74:1398-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marklund, S. L. 1984. Extracellular superoxide dismutase and other superoxide dismutase isoenzymes in tissues from nine mammalian species. Biochem. J. 222:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller, C. A., P. Broz, and G. R. Cornelis. 2008. The type III secretion system tip complex and translocon. Mol. Microbiol. 68:1085-1095. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, R. H., G. DeBusscher, and W. R. McCombie. 1982. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J. Bacteriol. 150:60-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips, R. M., D. A. Six, E. A. Dennis, and P. Ghosh. 2003. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 278:41326-41332. [DOI] [PubMed] [Google Scholar]
- 35.Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, J. Pirson, M. Struelens, L. Duinslaeger, P. Cornelis, M. Zizi, and A. Vanderkelen. 2003. Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone. J. Clin. Microbiol. 41:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabin, S. D., and A. R. Hauser. 2005. Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 73:573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabin, S. D., J. L. Veesenmeyer, K. T. Bieging, and A. R. Hauser. 2006. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect. Immun. 74:2552-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rae, T. D., P. J. Schmidt, R. A. Pufahl, V. C. Culotta, and T. V. O'Halloran. 1999. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284:805-808. [DOI] [PubMed] [Google Scholar]
- 39.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 40.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadikot, R. T., H. Zeng, A. C. Azim, M. Joo, S. K. Dey, R. M. Breyer, R. S. Peebles, T. S. Blackwell, and J. W. Christman. 2007. Bacterial clearance of Pseudomonas aeruginosa is enhanced by the inhibition of COX-2. Eur. J. Immunol. 37:1001-1009. [DOI] [PubMed] [Google Scholar]
- 42.Saliba, A. M., M. C. de Assis, R. Nishi, B. Raymond, A. Marques Ede, U. G. Lopes, L. Touqui, and M. C. Plotkowski. 2006. Implications of oxidative stress in the cytotoxicity of Pseudomonas aeruginosa ExoU. Microbes Infect. 8:450-459. [DOI] [PubMed] [Google Scholar]
- 43.Sato, H., J. B. Feix, and D. W. Frank. 2006. Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry 45:10368-10375. [DOI] [PubMed] [Google Scholar]
- 44.Sato, H., and D. W. Frank. 2004. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 53:1279-1290. [DOI] [PubMed] [Google Scholar]
- 45.Sato, H., and D. W. Frank. 2007. Type III secretory proteins in Pseudomonas aeruginosa, p. 3-22. In Brogden, K. A., F. C. Minion, N. Cornick, T. Stanton, Q. Zhang, L. K. Nolan, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC.
- 46.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawa, T., T. L. Yahr, M. Ohara, K. Kurahashi, M. A. Gropper, J. P. Wiener-Kronish, and D. W. Frank. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 5:392-398. [DOI] [PubMed] [Google Scholar]
- 48.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 49.Shaver, C. M., and A. R. Hauser. 2006. Interactions between effector proteins of the Pseudomonas aeruginosa type III secretion system do not significantly affect several measures of disease severity in mammals. Microbiology 152:143-152. [DOI] [PubMed] [Google Scholar]
- 50.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 51.Sun, J., and J. T. Barbieri. 2003. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J. Biol. Chem. 278:32794-32800. [DOI] [PubMed] [Google Scholar]
- 52.Tateda, K., R. Comte, J. C. Pechere, T. Kohler, K. Yamaguchi, and C. Van Delden. 2001. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vallis, A. J., V. Finck-Barbancon, T. L. Yahr, and D. W. Frank. 1999. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect. Immun. 67:2040-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verhelst, D., C. Koppen, J. Van Looveren, A. Meheus, and M. J. Tassignon. 2006. Contact lens-related corneal ulcers requiring hospitalization: a 7-year retrospective study in Belgium. Acta Ophthalmol. Scand. 84:522-526. [DOI] [PubMed] [Google Scholar]
- 55.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 56.Yahr, T. L., J. T. Barbieri, and D. W. Frank. 1996. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J. Bacteriol. 178:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. U. S. A. 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]