Abstract
Systematic inactivation of pathways involved in DNA alkylation damage repair demonstrated that inactivation of the ada, ogt, tag, uvrA, and mfd genes is required to detect a Salmonella enterica virulence decrease. Furthermore, the fitness of S. enterica, defective in these genes, is lowered only when the bacterium is orally, but not intraperitoneally, inoculated.
Bacteria are exposed to a wide variety of DNA-injuring agents, including alkylating agents, and their effects have been well studied (12, 17, 18). The repair of alkylated DNA in bacterial cells has been described mainly in Escherichia coli (12, 18), which presents the following two different mechanisms to eliminate alkyl radicals from its DNA: (i) the alkyl-induced expression of genes encoding the necessary repair enzymes (12, 17) and (ii) the constitutive synthesis of such proteins (9).
Many bacterial species have a genetic network, known as the adaptive response, which functions to repair alkyl lesions in the bacterial DNA (18, 19) and is regulated by the Ada protein, which also possesses methyltransferase activity (18). Transfer of a methyl from DNA-methylated phosphates to the Cys-37 residue of Ada (17) triggers a conformational change in this protein that converts it into a positive transcriptional regulator. Once activated, Ada stimulates the expression of its own transcriptional unit, including the alkB gene, which encodes an N1-meA-DNA dioxygenase, and the alkA and aidB genes, which encode a N3-meA-DNA-glycosylase and a flavin-containing DNA binding protein, respectively (9).
Bacteria also possess the following two additional enzymes involved in the repair of alkylated DNA: Ogt (O6-meG-DNA methyltransferase) and Tag (N3-meA-DNA glycosylase) (9). Expression of the ogt and tag genes is constitutive and does not depend on the presence of DNA alkylation (9). Despite the importance of alkylated DNA repair, there is very little information concerning the relevance of this process in pathogenic bacteria.
To determine the implications of DNA alkylation damage repair in Salmonella enterica virulence, mutants defective in the ada gene, or genes under its control (alkA, alkB and aidB), or mutants defective in the ogt and tag genes were constructed using the one-step PCR-based gene replacement method (6). As expected, survival assays showed that each of these single mutants was more sensitive than the wild-type strain to several alkylating agents, including N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), methyl methanesulfonate (MMS), and diethyl sulfate (DES) (data not shown). Furthermore, competition assays between each of these mutants and the wild-type strain, carried out as reported previously (3, 4), demonstrated that the inactivation of neither ada, ogt, nor tag had any effect on the virulence of S. enterica cells, regardless of whether the bacteria were inoculated orally or intraperitoneally (i.p.) in BALB/c mice (data not shown).
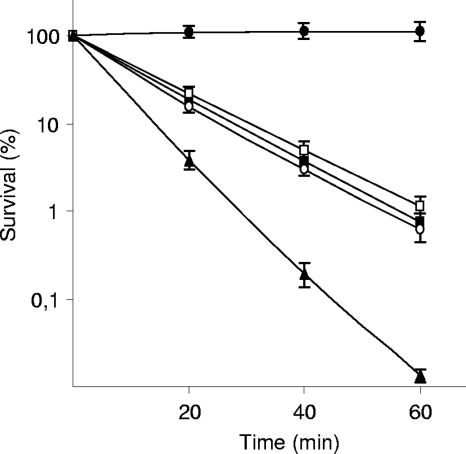
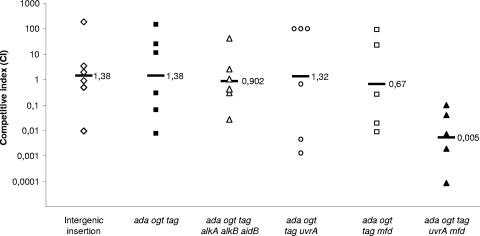
In light of these results, S. enterica strains carrying all possible double combinations of the ada, tag, and ogt mutations were constructed. These double mutants were much more sensitive to MMS, MNNG, or DES than either the wild-type or the single-mutant strains (data not shown). Nevertheless, their virulence was not diminished when inoculated either i.p. or orally, as shown by the competitive index (CI) determined with competition assays (data not shown). An S. enterica ada ogt tag triple mutant was subsequently constructed, and although it was extremely sensitive to the alkylating agents (Fig. 1), its virulence, when inoculated either orally (Fig. 2) or i.p. (data not shown), was not diminished compared with that of the wild-type strain. Likewise, the virulence of an S. enterica ada ogt tag alkA alkB aidB mutant, i.e., defective in all genes specifically involved in DNA alkylation damage repair (9, 17), was not affected (Fig. 2). These results show that the combined inactivation of the ada network and the ogt and tag genes does not decrease the fitness of S. enterica cells during the infective process.
FIG. 1.
Sensitivity of S. enterica wild-type (•), ada tag ogt (▪), ada tag ogt mfd (□), ada tag ogt uvrA (○), and ada tag ogt uvrA mfd (▴) strains to 30 mM DES. Sensitivity is expressed as the survival rate of exponential-phase cells treated with the corresponding alkylating agent. The data are the average results from at least three independent experiments.
FIG. 2.
In vivo competition infection experiments. The S. enterica wild-type strain was mixed with each of the mutant derivatives at a ratio of 1:1 before being used in the oral infection of BALB/c mice. The competitive index (CI) was calculated as the ratio of the mutant to the wild-type strain in the output (bacteria recovered from the host after infection) divided by that ratio in the input (initial inoculum). Each symbol indicates the CI value for one mouse. Bars represent the geometric mean results, with their values written alongside them. A CI of <1 indicates a competitive disadvantage of the mutant compared to that of the wild-type strain. The CI for a wild-type S. enterica derivative strain containing only a chloramphenicol resistance cassette inserted in the chromosomal coordinate 16088, corresponding to an intergenic region and with no effect on virulence (data not shown), was used as the control. Statistical analysis was carried out using a two-tailed t test, which unequivocally demonstrated that the virulence of S. enterica ada ogt tag uvrA mfd was significantly lower than that of the wild type (P = 0.00000096).
The nucleotide excision repair system (associated with the UvrABC excinuclease) is also involved in the repair of alkylated DNA (25). Likewise, the mfd pathway has been shown to participate in the repair of DNA lesions that block bacterial transcription and prevent RNA polymerase elongation, both of which being induced by alkylating agents and by other injurious compounds (16, 20). Accordingly, a mutation in the uvrA gene, the mfd gene, or both was introduced into the S. enterica ada ogt tag mutant. Survival assays showed that only the presence of both mutations (uvrA and mfd) increased the sensitivity of the cells to DES (Fig. 1). Similarly, results obtained from competitive experiments indicated that the presence of either the uvrA or the mfd mutation in the S. enterica ada ogt tag mutant did not lower its CI compared to that of the wild-type strain when inoculated either i.p. (data not shown) or orally (Fig. 2). However, the addition of both mutations (uvrA and mfd) to the triple (ada ogt tag) mutant, generating strain UA1869, decreased the CI by about 200-fold (Fig. 2). However, this decrease in virulence was apparent only when the bacteria were inoculated orally; the fitness of the i.p.-inoculated UA1869 mutant was the same as that of the wild type (data not shown). Moreover, there was no alteration in the virulence of either the S. enterica uvrA or the S. enterica mfd single mutant or in that of the S. enterica uvrA mfd double mutant when the bacteria were administered orally or i.p. (data not shown).
Several in vitro tests were carried out to understand the UA1869 strain fitness reduction. The growth kinetics of this strain under oxic and anoxic conditions were found to be the same as those of the wild-type strain (data not shown). Therefore, the competitive defect of orally inoculated UA1869 cells was not due to any putative DNA-damaging effect of endogenous alkylating agents generated in bacterial cells during anaerobic respiration or metabolically (23). In addition, UA1869 displayed the same sensitivity to pH and bile salts as the wild-type strain (data not shown).
Taken together, these data suggest that the effect of DNA alkylation damage on the S. enterica infective process is not particularly significant, since despite the high sensitivity of ada ogt tag alkA alkB aidB cells to alkylating agents, there is no decrease in their virulence (Fig. 2). However, our results suggest that DNA alkylation damage in S. enterica must occur before the orally ingested bacteria are disseminated in the bloodstream, since the virulence of i.p.-inoculated strain UA1869 was not affected. Indeed, the production of alkylating compounds in the mammalian gut has been reported (15, 24).
Furthermore, it could be argued that spontaneous mutation rates are higher in S. enterica cells harboring several inactivated DNA repair systems, such that their fitness during the infective process is altered. This possibility is unlikely, since it has been widely described that there is no decrease in the virulence of S. enterica hypermutable phenotypes (5, 26).
Although Salmonella lives primarily in the intestinal tracts of animals, dissemination of this pathogen also requires that it be able to survive outside the host. In fact, these bacteria are readily isolated from environmental sources, e.g., water and soil contaminated with Salmonella-containing feces (7), due to the application of manure or sewage sludge as fertilizers on agricultural fields (8, 10, 13, 19, 22). Moreover, the survival of these contaminating pathogens, such as S. enterica, may range from several days to a few months, depending on the environmental conditions (13, 14, 21). Besides its protective role against DNA alkylation damage generated during the intestinal steps of oral infection (Fig. 2), the extensive alkylation repair system of Salmonella may be involved in the long-term selective pressure to increase the survival of these bacteria outside the infected animal and, thus, in enabling them to overcome the potentially massive DNA injuries caused by alkylating agents present in the environment (1, 2, 11).
Acknowledgments
This work was funded by grant BFU2008-01078 from the Ministerio de Ciencia y Innovación (MICINN) de España and by grant 2009SGR1106 from the Generalitat de Catalunya. Gerard Àlvarez and Denis A. Spricigo were the recipients of predoctoral fellowships from the MICINN and CAPES, respectively.
We are deeply indebted to Joan Ruiz for his excellent technical assistance.
Footnotes
Published ahead of print on 5 February 2010.
REFERENCES
- 1.Ayanaba, A., W. Verstraete, and M. Alexander. 1973. Formation of dimethylnitrosamine, a carcinogen and mutagen, in soils treated with nitrogen compounds. Soil Sci. Soc. Am. Proc. 37:565-568. [Google Scholar]
- 2.Ayanaba, A., W. Vestraete, and M. Alexander. 1973. Possible microbial contribution of nitrosamine formation in sewage and soil. J. Natl. Cancer Inst. 50:811-813. [DOI] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 4.Campoy, S., M. Jara, N. Busquets, A. M. Perez De Rozas, I. Badiola, and J. Barbe. 2002. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 70:4721-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campoy, S., A. M. Perez de Rozas, J. Barbe, and I. Badiola. 2000. Virulence and mutation rates of Salmonella typhimurium strains with increased mutagenic strength in a mouse model. FEMS Microbiol. Lett. 187:145-150. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dworking, M., S. Falkow, E. Rosenberg, K. Schleifer, and E. Stackebrandt (ed.). 2006. The prokaryotes. Proteobacteria: gamma subclass, 3rd ed., vol. 6. Springer, New York, NY.
- 8.Franz, E., and A. H. van Bruggen. 2008. Ecology of E. coli O157:H7 and Salmonella enterica in the primary vegetable production chain. Crit. Rev. Microbiol. 34:143-161. [DOI] [PubMed] [Google Scholar]
- 9.Friedberg, E. C. 1995. Out of the shadows and into the light: the emergence of DNA repair. Trends Biochem. Sci. 20:381. [DOI] [PubMed] [Google Scholar]
- 10.Horswell, J., J. Hewitt, J. Prosser, A. Van Schaik, D. Croucher, C. Macdonald, P. Burford, P. Susarla, P. Bickers, and T. Speir. 2010. Mobility and survival of Salmonella Typhimurium and human adenovirus from spiked sewage sludge applied to soil columns. J. Appl. Microbiol. 108:104-114. [DOI] [PubMed] [Google Scholar]
- 11.Khan, S. U., and J. C. Young. 1977. N-nitrosamine formation in soil from the herbicide glyphosate. J. Agric. Food Chem. 25:1430-1432. [DOI] [PubMed] [Google Scholar]
- 12.Kleibl, K. 2002. Molecular mechanisms of adaptive response to alkylating agents in Escherichia coli and some remarks on O(6)-methylguanine DNA-methyltransferase in other organisms. Mutat. Res. 512:67-84. [DOI] [PubMed] [Google Scholar]
- 13.Natvig, E. E., S. C. Ingham, B. H. Ingham, L. R. Cooperband, and T. R. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholson, F. A., S. J. Groves, and B. J. Chambers. 2005. Pathogen survival during livestock manure storage and following land application. Bioresour. Technol. 96:135-143. [DOI] [PubMed] [Google Scholar]
- 15.Povey, A. C., A. F. Badawi, D. P. Cooper, C. N. Hall, K. L. Harrison, P. E. Jackson, N. P. Lees, P. J. O'Connor, and G. P. Margison. 2002. DNA alkylation and repair in the large bowel: animal and human studies. J. Nutr. 132:3518S-3521S. [DOI] [PubMed] [Google Scholar]
- 16.Savery, N. J. 2007. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 15:326-333. [DOI] [PubMed] [Google Scholar]
- 17.Sedgwick, B. 2004. Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 5:148-157. [DOI] [PubMed] [Google Scholar]
- 18.Sedgwick, B., and T. Lindahl. 2002. Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene 21:8886-8894. [DOI] [PubMed] [Google Scholar]
- 19.Sedgwick, B., and P. Vaughan. 1991. Widespread adaptive response against environmental methylating agents in microorganisms. Mutat. Res. 250:211-221. [DOI] [PubMed] [Google Scholar]
- 20.Selby, C. P., and A. Sancar. 1994. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol. Rev. 58:317-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenov, A. V., A. H. van Bruggen, L. van Overbeek, A. J. Termorshuizen, and A. M. Semenov. 2007. Influence of temperature fluctuations on Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in cow manure. FEMS Microbiol. Ecol. 60:419-428. [DOI] [PubMed] [Google Scholar]
- 22.Semenov, A. V., L. van Overbeek, and A. H. van Bruggen. 2009. Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 75:3206-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taverna, P., and B. Sedgwick. 1996. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J. Bacteriol. 178:5105-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tornqvist, M., B. Gustafsson, A. Kautiainen, M. Harms-Ringdahl, F. Granath, and L. Ehrenberg. 1989. Unsaturated lipids and intestinal bacteria as sources of endogenous production of ethene and ethylene oxide. Carcinogenesis 10:39-41. [DOI] [PubMed] [Google Scholar]
- 25.Van Houten, B., and A. Sancar. 1987. Repair of N-methyl-N′-nitro-N-nitrosoguanidine-induced DNA damage by ABC excinuclease. J. Bacteriol. 169:540-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahrt, T. C., N. Buchmeier, and S. Maloy. 1999. Effect of mutS and recD mutations on Salmonella virulence. Infect. Immun. 67:6168-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]