Abstract
Campylobacter jejuni is a leading cause of gastroenteritis in humans and a commensal bacterium of the intestinal tracts of many wild and agriculturally significant animals. We identified and characterized a locus, which we annotated as rdxAB, encoding two nitroreductases. RdxA was found to be responsible for sensitivity to metronidazole (Mtz), a common therapeutic agent for another epsilonproteobacterium, Helicobacter pylori. Multiple, independently derived mutations in rdxA but not rdxB resulted in resistance to Mtz (Mtzr), suggesting that, unlike the case in H. pylori, Mtzr might not be a polygenic trait. Similarly, Mtzr C. jejuni was isolated after both in vitro and in vivo growth in the absence of selection that contained frameshift, point, insertion, or deletion mutations within rdxA, possibly revealing genetic variability of this trait in C. jejuni due to spontaneous DNA replication errors occurring during normal growth of the bacterium. Similar to previous findings with H. pylori RdxA, biochemical analysis of C. jejuni RdxA showed strong oxidase activity, with reduction of Mtz occurring only under anaerobic conditions. RdxB showed similar characteristics but at levels lower than those for RdxA. Genetic analysis confirmed that rdxA and rdxB are cotranscribed and induced during in vivo growth in the chick intestinal tract, but an absence of these genes did not strongly impair C. jejuni for commensal colonization. Further studies indicate that rdxA is a convenient locus for complementation of mutants in cis. Our work contributes to the growing knowledge of determinants contributing to susceptibility to Mtz (Mtzs) and supports previous observations of the fundamental differences in the activities of nitroreductases from epsilonproteobacteria.
Nitroreductases form a large family of enzymes whose physiological roles have been implicated or proposed to function in diverse processes, such as the generation of nitrogen sources for metabolism, degradation of potentially toxic nitro compounds, vitamin and bioluminescence production, redox balancing, and oxidative stress responses (20, 31, 32, 35, 41, 43, 58). These enzymes can been subdivided into two main categories based on characteristics of their reductive processes, including the mechanism of electron transfer and sensitivity to oxygen. Type I (O2-insensitive) nitroreductases catalyze a sequential two- or three-step reduction of the nitro group on heterocyclic compounds via paired-electron transfer to produce either hydroxylamine or amino derivatives. Type II (O2-sensitive) nitroreductases catalyze a single-electron reduction of heterocyclic nitro compounds that is reversible in the presence of oxygen (40). Nitroreductases are common in bacteria, with a given bacterial species often containing multiple paralogs that presumably reduce different substrates. The genes encoding nitroreductases have received intense study due to their unusual nature in degrading or transforming xenobiotic chemicals. Consequently, these enzymes have become attractive candidates for bioremediation processes, and some are utilized in cancer chemotherapies (27, 49). However, the nitroreductases are also a puzzling class of enzymes, because the natural substrates for most remain unknown and these proteins likely did not evolve to exclusively manipulate xenobiotic compounds.
Metronidazole (Mtz) has been used in multidrug therapy for Helicobacter pylori infections due to production of factors that convert this 5-nitroimidazole product to a toxic form (1, 45, 53). Therapeutic failure with Mtz has been predominantly associated with mutations occurring in one of two genes of H. pylori encoding the nitroreductases RdxA and FrxA. A previous biochemical study characterized the Mtz reductase activity of RdxA (37). Even though RdxA was capable of reducing other nitro compounds under aerobic conditions, the enzyme was unable to reduce Mtz. However, under anaerobic conditions, RdxA was shown to catalyze the reduction of Mtz, and its specific activity for this reaction was 60-fold greater than that of the NfsB nitroreductase of Escherichia coli under similar conditions. In addition, this work revealed that RdxA exhibited a potent NADPH oxidase activity not appreciated in other nitroreductases. Not only did this study demonstrate a direct reduction of Mtz by a nitroreductase, but results from this work implied that RdxA of H. pylori possessed novel biochemical properties relative to other nitroreductases.
Like H. pylori, Campylobacter jejuni is a Gram-negative bacterium belonging to the epsilonproteobacteria class. C. jejuni is a common commensal bacterium of the intestinal tracts of wild and agriculturally significant animals, especially poultry. In contrast, C. jejuni causes acute diarrhea in humans, ranging from a mild enteritis to a bloody diarrheal syndrome, and is one of the most prevalent causes of food-borne gastritis (4, 5, 34, 38). Additionally, postinfectious sequelae can develop in a small percentage of patients following a C. jejuni infection. One major complication is Guillan-Barré syndrome, a temporary and partial paralysis of the peripheral nervous system (21).
Many individuals with C. jejuni enteritis resolve the infection without therapeutic treatment. If antibiotics are administered, fluoroquinolones or macrolides, such as ciprofloxacin or erythromycin, are common drugs of choice, with therapeutic use of Mtz for C. jejuni infections being unconventional. However, Mtz-resistant (Mtzr) C. jejuni isolates have been recovered from humans and animals. In agriculture, 19 to 92% of C. jejuni isolates from avian species (including chickens and turkeys) and 6 to 20% of isolates from lambs, sheep, and cows were Mtzr (13, 47). One study also demonstrated that 62% of C. jejuni clinical isolates from humans were Mtzr (47). These data are curious, since these C. jejuni isolates would have likely developed Mtzr during infections in the absence of selection.
Because susceptibility to Mtz (Mtzs) was also found in isolates associated with each host in studies described above and since C. jejuni is closely related to H. pylori, we hypothesized that C. jejuni may produce a nitroreductase to reduce Mtz to its toxic form, leading to Mtzs. In this report, we identify and characterize the gene required for Mtzs in C. jejuni. Mutations in this gene, encoding a putative nitroreductase, but not in a downstream paralog were linked to the development of Mtzr, indicating that Mtzr in C. jejuni appears to be linked to mutation of only one nitroreductase. Supporting our findings, we provide evidence that a proportion of Mtzr isolates of C. jejuni are due to spontaneous errors during DNA replication in the absence of Mtz exposure, resulting in a variety of mutations. Biochemical analysis of these C. jejuni nitroreductases demonstrated that these proteins had potent NADPH oxidase activity and could reduce Mtz under anaerobic conditions. These results, along with previous biochemical analysis of H. pylori RdxA (37), demonstrate that the nitroreductases of epsilonproteobacteria have unique characteristics in comparison to other bacterial counterparts.
MATERIALS AND METHODS
Strains and culture conditions.
C. jejuni strain 81-176 is a clinical isolate that is able to colonize the intestinal tract of chicks and cause disease in human volunteers (3, 18, 28). DRH212 is a streptomycin-resistant (Smr) derivative of 81-176 with an rpsLSm allele (17). C. jejuni strains were routinely grown from frozen stocks on Mueller-Hinton (MH) agar containing trimethoprim (TMP) under microaerobic conditions (10% CO2, 5% O2, 85% N2) at 37°C for 48 h, followed by reinoculation onto MH agar containing TMP for growth for 16 h. Antibiotics for C. jejuni strains were used at the following concentrations: TMP, 10 μg ml−1; cefoperazone, 30 μg ml−1; chloramphenicol, 15 μg ml−1; and Mtz, 0, 0.2, 0.5, 1, 2.5, 5, 10, 25, 50, 100, and 200 μg ml−1. All C. jejuni strains were stored in 85% MH broth-15% glycerol at −80°C. Escherichia coli DH5α, XL-1 Blue, and BL21(DE3) were grown in Luria Bertani (LB) agar or broth with antibiotics when needed at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 15 μg ml−1; Mtz, 10 μg ml−1. All E. coli strains were stored in 80% LB broth-20% glycerol at −80°C.
Identification of Mtzr transposon mutants.
A C. jejuni 81-176 transposon library constructed with the darkhelmet transposon, a mariner-based transposon, was grown as described above (16). Bacteria were resuspended and diluted in MH broth to an optical density at 600 nm (OD600) of 0.4. The culture was diluted 1:5,000, and 100 μl was spread on two MH agar plates containing either 5, 10, 25, or 50 μg ml−1 Mtz. Serial dilutions were also spread on MH agar containing TMP to determine the bacterial density of the culture. Plates were incubated for 4 days at 37°C under microaerobic conditions. At least one Mtzr colony was obtained per plate, with a total of nine Mtzr isolates recovered. The location of the transposon in each mutant was determined as previously described (16).
Construction of rdxA and rdxB mutants in C. jejuni 81-176.
Insertional inactivation of rdxA or rdxB was accomplished by first amplifying 1.6-kb fragments containing rdxA or rdxB with approximately 500 nucleotides of flanking DNA sequence from C. jejuni 81-176 genomic DNA by PCR with primers containing 5′ BamHI sites (9). After the DNA fragments were cloned into BamHI-digested pUC19 to create pDAR473 (containing rdxA) or pDRH2879 (containing rdxB), PCR-mediated mutagenesis was used to create an internal EcoRV restriction site in each gene. pDAR475 contains an EcoRV site in rdxA by an A222G mutation, and pDAR476 contains an EcoRV site within rdxB by a G293C mutation. These plasmids were used to insert a SmaI-digested cat-rpsL cassette into the EcoRV site of rdxA (creating pDAR505) or rdxB (creating pDAR609) (17). pDAR505 or pDAR609 was electroporated into C. jejuni 81-176 Smr (DRH212) to create DAR521 (81-176 Smr rdxA::cat-rpsL) or DAR562 (81-176 Smr rdxB::cat-rpsL), respectively.
Analysis of colonization capacity of C. jejuni mutants.
One-day-old White Leghorn strain Δ chicks were orally infected with wild-type or mutant C. jejuni 81-176 strains as previously described (2, 18). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 days at 37.8°C with appropriate humidity and rotation in a Sportsman II model 1502 incubator (Georgia Quail Farms Manufacturing Company). Approximately 12 to 36 h after hatching, chicks were orally infected with 100 μl of phosphate-buffered saline (PBS) containing approximately 102 or 104 CFU of wild-type or mutant C. jejuni strains. To prepare strains for infection, bacteria were grown from frozen stocks as described above. Bacteria were resuspended from plates and diluted to the appropriate inoculum in PBS. Dilutions of the inocula were plated to determine the number of bacteria in the inoculation dose. At day 7 postinfection, chicks were sacrificed and the cecal contents were recovered. After suspension in PBS, serial dilutions were spread on MH agar containing TMP and cefoperazone to determine the number of CFU per gram of cecal contents. Statistical analysis of results from colonization experiments was performed by the Mann-Whitney U test.
Isolation of Mtzr mutants after in vitro and in vivo growth.
Seven Mtzs C. jejuni 81-176 isolates were identified, and low-passage frozen stocks of each were prepared. To determine the rate of generation of Mtzr during in vitro growth, each strain was grown from frozen stocks as described above. After suspension of each culture to an OD600 of 1.0, serial dilutions were spread on MH agar containing TMP (to determine total CFU per ml) or 10 μg ml−1 Mtz to recover Mtzr isolates. After growth for 5 days at 37°C under microaerobic conditions, the number of bacteria recovered on MH agar with TMP or Mtz was determined. Twelve random Mtzr isolates from each culture were saved, and the rdxAB operon was PCR amplified from each and sequenced.
Isolation of Mtzr mutants after commensal colonization of the chick ceca was performed by orally gavaging six 1-day-old White Leghorn strain Δ chicks with 100 μl PBS containing approximately 104 CFU of Mtzs C. jejuni 81-176. An aliquot of the inoculum was spread on MH agar containing 10 μg ml−1 Mtz to confirm spontaneous Mtzr C. jejuni were not present in the inoculum. Chicks were given food and water ad libitum and were not administered Mtz during the course of infection. At day 7 postinfection, chicks were sacrificed and the bacteria from the ceca were recovered. Serial dilutions were plated on MH agar containing TMP and cefoperazone (to determine the total C. jejuni load in the cecum of each chick) or MH agar containing TMP, cefoperazone, and 10 μg ml−1 Mtz (to determine the number of Mtzr C. jejuni CFU in the cecum of each chick). Sixty-four Mtzr isolates were recovered. The rdxAB operon was PCR amplified from each isolate and sequenced.
Determination of MIC of Mtz for C. jejuni strains.
MICs of Mtz for various C. jejuni strains were determined by the method of Jeong et al. (23). Briefly, C. jejuni strains were grown from frozen stocks under microaerobic conditions for 48 h at 37°C. Strains were reinoculated on MH agar and grown for another 16 h. Bacteria were suspended and diluted to an OD600 of 1.0. Tenfold serial dilutions were made, and 10 μl of each dilution was spotted onto MH agar containing Mtz at the following concentrations: 0, 0.2, 0.5, 1, 2.5, 5, 10, 25, and 50 μg ml−1. After growth for 3 days at 37°C under microaerobic conditions, the MIC was determined by examining the lowest Mtz concentration resulting in a 10-fold decrease in CFU compared to growth on MH agar without Mtz.
Mtz-induced killing and mutation assays.
Wild-type Mtzs C. jejuni 81-176 was grown on three sets of MH agar plates from frozen stocks for 48 h at 37°C under microaerobic conditions. The strains were then restreaked on MH agar and grown for another 16 h. Bacteria were suspended and diluted to an OD600 of 1.0 in 24 ml of MH broth to give three independent cultures. The cultures were split into two volumes, with one volume receiving 10 μg ml−1 Mtz. The cultures were incubated under microaerobic conditions at 37°C for 6 h. At time zero, 2, 4, and 6 h after addition of Mtz, the number of viable bacteria in cultures with or without Mtz were determined by plating 10-fold serial dilutions on MH agar containing TMP. In addition, the number of Mtzr bacteria at each time point was determined by plating dilutions on MH agar containing 10 μg ml−1 Mtz. Plates were incubated for up to 5 days at 37°C under microaerobic conditions before determining the number of viable CFU per ml of culture.
The ability of Mtz to induce mutations to generate Smr was analyzed in strain 81-176 rpsLSm cjj0275::cat-rpsL (LKB635). To construct this strain, 81176_cjj0275, a gene that has been proposed to be involved in flagellar motility (17), was first PCR amplified from the C. jejuni 81-176 chromosome with primers containing 5′ BamHI restriction sites, resulting in a 2.9-kb fragment containing the gene. The amplified DNA was then cloned into BamHI-digested pUC19 to create pDRH1962. PCR-mediated mutagenesis was then performed to create an MscI site within the coding sequence of cjj0275, resulting in pDRH1975. After digestion with MscI, a SmaI-digested cat-rpsL cassette was then ligated into the plasmid to interrupt cjj0275 to generate pDRH2024 (17). This plasmid was then used to electroporate DRH212 (81-176 rpsLSm), resulting in LKB635.
Methods similar to the experiment described above to test for Mtz-induced mutations and killing with wild-type C. jejuni 81-176 were performed with LKB635 to determine the extent of killing by addition of Mtz. In addition, the number of viable Smr bacteria was determined by plating 10-fold serial dilutions of cultures with or without Mtz at time zero, 2, 4, or 6 h after exposure to Mtz on MH agar containing 2 mg ml−1 Sm.
Expression and purification of RdxA and RdxB.
The coding sequences of rdxA and rdxB from codon 2 to the stop codon were PCR amplified from C. jejuni 81-176 genomic DNA with primers containing in-frame 5′ BamHI restriction sites and cloned into pGEX-4T-2, creating fusion proteins of glutathione S-transferase (GST) to the N terminus of RdxA (pDAR538) or RdxB (pDAR558). Plasmids were transformed into BL21(DE3) for protein expression and purification. For purification of GST, BL21(DE3)/pGEX-4T-2 was used. For induction of proteins, 500 ml LB with ampicillin was inoculated with a 1:40 dilution of overnight culture and grown at 37°C to an OD600 of 0.5. After induction with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 100 μM for 3 h, bacteria were recovered, washed once in cold PBS, and then resuspended in 30 ml PBS. After lysis by passage four times through an EmusliFlex-C5 cell disrupter (Avesin) at 15,000 to 20,000 lb/in2, GST-tagged proteins were purified from the soluble fraction with glutathione Sepharose 4B by gravity flow according to the manufacturer's instructions (GE Healthcare). For purification of native RdxA and RdxB, GST-tagged proteins bound to the Sepharose slurry were resuspended in 2 ml 1× PBS and transferred to a microcentrifuge tube to which 50 U of thrombin (GE Healthcare) was added. Cleavage of the GST tag occurred by overnight incubation at 37°C. Thrombin was removed from purified RdxA and RdxB using benzamidine Sepharose 6B according to the manufacturer's instructions (GE Healthcare). Protein concentrations for purified GST-RdxA, RdxA, GST-RdxB, RdxB, and GST were determined by Bradford assay (Bio-Rad), and purity was examined by SDS-PAGE. Similar to other flavin proteins, purified RdxA and RdxB and their fusions were yellow in color (43).
Antiserum generation and immunoblotting analysis.
The coding sequence of rdxB from codon 2 to the penultimate codon was PCR amplified from C. jejuni 81-176 genomic DNA with primers containing in-frame 5′ and 3′ BamHI restriction sites. The DNA fragment was inserted into BamHI-digested pQE30 (Qiagen) to create pDAR520 and transformed into XL1-Blue. For protein induction, 500 ml LB with ampicillin was inoculated with a 1:40 dilution of overnight growth, and the culture was grown at 37°C to an OD600 of 0.5. The culture was induced with 1 mM IPTG for 4 h, and the bacterial pellet was recovered. Bacteria were lysed as described above, and His6-RdxB was purified from the soluble fraction using nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) according to the manufacturer's instructions. GST-RdxA was purified using glutathione Sepharose 4B according to the manufacturer's instructions. GST-RdxA and His6-RdxB were used to immunize mice by standard procedures for antiserum generation by a commercial vendor (Cocalico Biologicals).
Immunoblotting analyses were performed by growing C. jejuni strains from frozen stocks as described above. Bacteria were suspended from plates and diluted to an OD600 of 0.8. Whole-cell lysates (WCLs) were prepared by pelleting 1 ml of bacteria, washing once with PBS, and then resuspending the bacteria in 50 μl of SDS-PAGE loading buffer. For total membrane preparations, 5 ml of culture prepared as described above was pelleted and washed once in 10 mM HEPES, pH 8.0. Bacteria were sonicated, and total membranes were recovered as the insoluble material as previously described (2). Ten or 20 μl of WCL preparations were separated by SDS-PAGE for detection of RdxA and RdxB, respectively, and proteins were then transferred to an Immobolin-P membrane. Membranes were incubated with a 1:1,000 dilution of M100 anti-RdxA antiserum or a 1:500 dilution of M95 anti-RdxB antiserum for 2 h, washed, and then incubated with a 1:12,500 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse antiserum (Bio-Rad) for 1 h. For detection of FlhB in total membranes, the membrane pellet recovered from 5 ml of culture for each strain was resuspended in 50 μl of SDS-PAGE loading buffer. After SDS-PAGE, proteins were transferred to a membrane and incubated overnight at 4°C with 1:1,000 dilution of Rab476 anti-FlhB antiserum (25), followed by incubation for 4 h at room temperature with a 1:7,500 dilution of HRP-conjugated goat antirabbit antiserum (Bio-Rad).
Biochemical analysis of RdxA and RdxB.
Aerobic reductase or oxidase activities of proteins for various substrates were analyzed as described by Sisson et al. (45). Briefly, the initial rates of activity of purified proteins were determined spectrophotometrically at 25°C in a 1-ml reaction volume containing 50 mM Tris-HCl (pH 7.5), 0.1 mM NADPH, and 0.1 mM substrate. Reactions were initiated by the addition of 10 μg of enzyme, and absorbance readings were measured every 15 s for 5 min by a Beckman DU 530 spectrophotometer. Anaerobic measurement of Mtz reduction was also performed according to the method of Olekhnovich et al. (37). Briefly, reaction mixtures (50 mM Tris [pH 7.5], 0.1 mM NADPH, 0.1 mM substrate, 25 mM glucose, and 6 U ml−1 of both glucose oxidase and catalase) were allowed to undergo anaerobic generation for 5 min. Purified proteins (10 μg) were added to initiate reactions, and absorbance readings were examined as described previously (37). Blank cuvettes did not contain Mtz. The reduction of substrates was measured at the indicated wavelengths, and specific activity for each was calculated from the initial rate, extinction coefficient, and protein concentration: nitrofurazone at 400 nm (E = 12.6 mM−1 cm−1), nitrofurantoin at 420 nm (E = 12 mM−1 cm−1), and furazolidone at 400 nm (E = 18.8 mM−1 cm−1). The oxidation of NADPH was measured at 340 nm (E = 6.22 mM−1 cm−1). Endogenous NADPH oxidase activity for each substrate tested was also determined and was used to correct the specific activities calculated for those substrates under aerobic conditions (37). Assays were performed in duplicate from two independent batches of purified protein.
Analysis of flavin cofactor binding to the GST-RdxA and GST-RdxB fusion proteins was determined by the method of Zenno et al. (57). Purified GST, GST-RdxA, and GST-RdxB were boiled for 20 min and then centrifuged for 20 min at 10,000 × g. Supernatants were analyzed by thin-layer chromatography (TLC) using cellulose polyethyleneimine (PEI) sheets and a solvent system of 0.4 M K2HPO4 and 0.7 M boric acid. Control preparations of flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), and riboflavin were run alongside supernatants. Spots were visualized by UV.
Transcriptional analysis of the rdxAB operon.
Reverse transcriptase PCR (RT-PCR) was performed to analyze cotranscription of rdxA and rdxB. DRH212 (81-176 Smr) was grown from freezer stocks as described above. RNA was isolated using an RNAeasy minikit with the RNAprotect bacterial reagent (Qiagen). After DNase treatment to remove genomic DNA, RNA was reverse transcribed into cDNA in the presence and absence of Superscript II reverse transcriptase (Invitrogen). The products of the cDNA synthesis reactions were used in PCRs to amplify regions internal to rdxA or rdxB or spanning rdxA and rdxB. Products from the cDNA synthesis reactions in the absence of RT served as a negative control to verify that no amplification originated from possibly contaminating genomic DNA. Genomic DNA was used as a positive control in PCR to verify amplification and size of generated fragments.
Primer extension reactions were carried out to determine the transcriptional start site of rdxA. RNA was prepared as described above and used in a RT reaction with the primer rdxA PE (5′-TCCACTTTCTAAGATAAAAT-3′). The same primer was used in a sequencing reaction with pDAR473. Products from the sequencing reaction were run alongside the product of the RT reaction on a 6% sequencing gel to align the cDNA product to the sequencing ladder. The gel was dried and exposed to a Phosphorimager cassette and visualized using the ImageQuant software program.
For real-time RT-PCR analysis, three 1-day-old chicks were orally gavaged with 104 CFU of DRH212 (81-176 Smr). At day 7 postinfection, chicks were sacrificed and the cecal contents were recovered. RNA from the cecal contents containing C. jejuni was isolated using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA from in vitro-grown bacteria was isolated from DRH212 as described above. RNA was DNase treated (GenHunter) and diluted to a final concentration of 50 ng μl−1. Real-time RT-PCR analysis was performed with a 25-μl volume in 1× Sybr green PCR master mix (Applied Biosystems), 0.2 μM forward and reverse primers, and 2.5 μg RNA. RT-positive samples also contained 0.1 μl of Multiscribe reverse transcriptase (Applied Biosystems). Reactions were performed on a 7500 real-time PCR system (Applied Biosystems) under the following conditions: 42°C for 30 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Results were normalized using gyrA or 16S rRNA as a control, and analysis was performed using the ΔΔCT method. The level of gene expression from in vitro-grown samples was calibrated to 1.
Complementation of C. jejuni mutants in cis.
A 1.2-kb fragment containing flhB with its native promoter was PCR amplified from genomic DNA of C. jejuni 81-176 with primers to create 5′ and 3′ SmaI restriction sites. After digestion with SmaI, the PCR product was cloned into the EcoRV site in rdxA in pDAR475 to create pDRH3031. Similarly, a SmaI-digested cat cassette encoding chloramphenicol acetyltransferase from pRY109 was inserted into the EcoRV site in rdxA in pDAR475 to create pDRH3033 (55). Plasmids were electroporated into SNJ471 (81-176 Smr ΔflhB [25]) or DRH1827 (81-176 Smr ΔastA ΔflhB flgDE2::nemo [19]). Transformants were recovered on MH agar containing 10 μg ml−1 Mtz. Mtzr colonies appeared after 4 to 5 days of growth at 37°C under microaerobic conditions. For identification of transformants with rdxA::cat, 80 to 200 colonies were patched on MH agar containing 10 μg ml−1 chloramphenicol. One or two Mtzr colonies were identified that were also resistant to chloramphenicol, and colony PCR was performed to confirm the rdxA::cat mutation.
DRH3107 (81-176 Smr ΔflhB rdxA::flhB) was recovered by stabbing Mtzr transformants after electroporation of SNJ471 with pDRH3031 in MH motility medium containing 0.4% agar. After 24 h of incubation under microaerobic conditions at 37°C, one motile transformant was identified. An agar plug containing the motile bacteria was vortexed in 1 ml MH broth, and dilutions were plated on MH agar containing 10 μg ml−1 Mtz. Colonies were then verified by colony PCR to confirm the rdxA::flhB mutation.
DRH3057 (81-176 Smr ΔastA ΔflhB flgDE2::nemo rdxA::flhB) was recovered by electroporating DRH1827 with pDRH3031 and then plating transformants on MH agar containing 10 μg ml−1 Mtz and 35 μg ml−1 5-bromo-4-chloro-3-indolyl sulfate, the chromogenic substrate for AstA. After incubation for 5 days at 37°C under microaerobic conditions, one transformant displayed a blue-colony phenotype, indicating complementation of expression of the flgDE2-astA transcriptional fusion. Colony PCR was performed to confirm the rdxA::flhB mutation.
Motility assays and arylsulfatase assays to monitor expression of flagellar genes.
Motility phenotypes of C. jejuni wild-type, mutant, and complemented strains were assessed as previously described (26). Strains were grown from frozen stocks as described above, suspended in MH broth to an OD600 of 0.8, and stabbed into semisolid MH motility agar by using a sterilized inoculating needle. The plates were incubated for 30 h at 37°C under microaerobic conditions and then visualized for motility. Wild-type, mutant, and complemented strains containing flgDE2::nemo, a transcriptional fusion of astA to the flgDE2 operon, were grown from frozen stocks as described above. Arylsulfatase production from the transcriptional fusions in these strains was measured by a previously published method (14, 19, 56).
RESULTS
Mutation of rdxA is responsible for Mtzr.
For ease in communicating our findings, we propose annotation of the C. jejuni 81-176 gene cjj81176_1083 as rdxA and cjj81176_1084 as rdxB, which were not annotated in the genome sequence of this strain (9). Our proposed annotation counters that of another C. jejuni strain (NCTC11168), for which cj1064 (cjj81176_1083 in the 81-176 genome) was annotated as a gene encoding a nonfunctional protein due to a frameshift mutation and cj1066 (cjj81176_1084 in the 81-176 genome) was annotated as rdxA (9, 39). As described in detail below, we found that the function of Cjj81176_1083 rather than Cjj81176_1084 was similar to that of RdxA of H. pylori in regard to being the determinant for Mtzs, and our proposed annotation better reflects these findings. Because the coding sequence of cjj81176_1084 has homology to type I nitroreductases and the gene is immediately downstream and in an apparent operon with cjj81176_1083 (see below, Fig. 1A) (9), we propose to annotate cjj81176_1084 as rdxB.
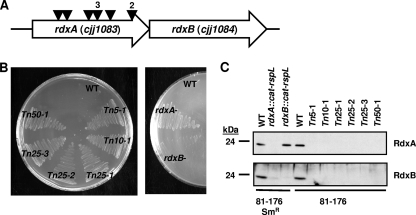
FIG. 1.
Mtzs and Mtzr phenotypes and production of RdxA and RdxB in C. jejuni transposon and site-directed mutants. (A) Identification of Mtzr darkhelmet transposon mutants of C. jejuni 81-176. Triangles represent locations of the darkhelmet transposons within rdxA. Numbers above triangles indicate the numbers of mutants identified with transposon insertions at identical locations, suggesting that these mutants were likely siblings. (B) Mtzs and Mtzr phenotypes of wild-type C. jejuni 81-176 (WT), isogenic darkhelmet transposon mutant strains (left), and wild-type 81-176 Smr (WT) and isogenic site-directed mutant strains (right) after 48 h of growth on MH agar containing 10 μg ml−1 Mtz. (C) Immunoblot analysis of RdxA and RdxB production in whole-cell lysates of wild-type C. jejuni 81-176 or 81-176 Smr strains and derived transposon or site-directed mutants. For panels B and C, strains include wild-type DRH212 (81-176 Smr; WT producing RdxA and RdxB), DAR521 (81-176 Smr rdxA::cat-rpsL, which produces neither RdxA nor RdxB), DAR562 (81-176 Smr rdxB::cat-rpsL, which produces only RdxA), and wild-type 81-176 (WT).
In preliminary studies, C. jejuni strain 81-176 was unable to grow on MH agar containing more than 5 μg ml−1 Mtz, suggesting that this strain may contain a gene encoding a protein that converts Mtz to a toxic form, thus resulting in Mtzs. We used a transposon mutant library of C. jejuni strain 81-176 made with the darkhelmet transposon, a mariner-based transposon, to identify mutants with transposon insertions interrupting such a gene to result in Mtzr (16). Nine Mtzr transposon mutants were recovered on MH agar containing Mtz ranging from 5 to 50 μg ml−1. All mutants had a darkhelmet insertion in rdxA (cjj81176_1083), with six independent insertions identified in the gene (Fig. 1A). rdxA is organized on the C. jejuni genome upstream of rdxB (cjj81176_1084) with the stop codon of rdxA overlapping the start codon of rdxB, suggesting an operonic construction and cotranscription. Both rdxA and rdxB encode putative nitroreductases, which are similar in size (201 to 206 amino acids) and amino acid sequence (34% identity and 60% similarity across the entire sequences) (Fig. 2A). RdxA and RdxB show a general level of similarity to a variety of type I nitroreductases, with conservation of key residues and putative structures for biochemical activity (as reviewed in reference 43).
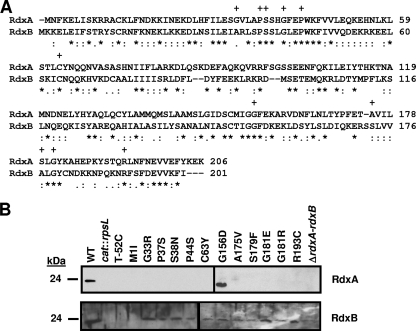
FIG. 2.
Comparison of RdxA and RdxB amino acid sequences and production of RdxA and RdxB in in vitro- and in vivo-isolated Mtzr C. jejuni 81-176 mutants. (A) ClustalV alignment of the amino acid sequences of RdxA and RdxB from C. jejuni 81-176. Asterisks (*) indicate identical residues, semicolons (:) indicate highly conserved residues, and periods (.) indicate weakly conserved residues. Plus signs (+) above residues of RdxA indicate residues changed by point mutations in Mtzr C. jejuni isolates after in vitro or in vivo growth. (B) Immunoblot analysis of RdxA and RdxB production in whole-cell lysates of Mtzr C. jejuni 81-176 isolates with point mutations in rdxA isolated after in vitro or in vivo growth. The wild-type strain is 81-176 Smr (DRH212) (WT), and the cat::rpsL mutants used in the immunoblots are DAR521 (81-176 Smr rdxA::cat-rpsL) and DAR562 (81-176 Smr rdxB::cat-rpsL), respectively. Most Mtzr isolates are identified by the type of change occurring in the protein sequence after spontaneous mutation. T-52C refers to an Mtzr isolate with a nucleotide change occurring 52 bases upstream of the rdxA start codon. ΔrdxA-rdxB refers to an Mtzr isolate with a deletion of DNA encompassing the 3′ end of rdxA through the 5′ end of rdxB.
All 81-176 darkhelment transposon mutants and a reconstructed 81-176 Smr rdxA::cat-rspL mutant promoted growth on MH containing 10 μg ml−1 Mtz, unlike the wild-type strain 81-176 or 81-176 Smr (DRH212) (Fig. 1B). However, production of both RdxA and RdxB was eliminated by interruption of rdxA with these transposons or the cat::rpsL cassette (Fig. 1C), hindering confirmation that rdxA is the determinant responsible for Mtzs and that mutation solely of rdxA contributed to Mtzr in these mutants. We next investigated if interruption of rdxB contributed to Mtzr. Disruption of rdxB did not affect production of RdxA and did not change the Mtzs phenotype of the parental C. jejuni wild-type strain (Fig. 1B and 1C).
As a more quantitative and sensitive method to verify that mutation solely of rdxA contributed to Mtzr, we determined the MIC of Mtz for growth of wild-type and mutant C. jejuni strains. The MIC of Mtz for wild-type C. jejuni 81-176, 81-176 Smr, and 81-176 Smr rdxB::cat-rpsL, which produces RdxA but not RdxB (Fig. 1C), was 2.5 μg ml−1, demonstrating that a lack of RdxB did not result in an increase in Mtzr. In contrast, a spontaneous Mtzr mutant isolated during in vitro growth (see below) with an rdxA(A175V) mutation that lacked production of RdxA but produced RdxB displayed a MIC of Mtz of 50 μg ml−1 (Fig. 2B and data not shown). In addition, all nine 81-176 darkhelmet transposon mutants that failed to produce RdxA and RdxB displayed a MIC of Mtz of 50 μg ml−1. These combined results suggest that Mtzr appeared to be due to mutation of rdxA. Furthermore, mutation of rdxB did not contribute to an increase in the MIC of Mtz in the presence or absence of RdxA at least as analyzed in this strain, 81-176.
Mtzr can be generated in a stepwise process through accumulated mutations in various alleles of H. pylori, with each mutation allowing for growth in the presence of increasing levels of Mtz (1, 23, 24). We attempted to recover additional mutants in the rdxA mutant background that allowed for growth on higher concentrations of Mtz. We used the rdxA(A175V) mutant described herein, which failed to produce RdxA but produced wild-type RdxB and grew in the presence of 50 μg ml−1 Mtz. Several attempts were made to isolate colonies of the rdxA(A175V) mutant on media containing 100 and 200 μg ml−1 Mtz, but we were unable to obtain any colonies after incubation of bacteria for several days. These findings suggest that rdxA is the major determinant influencing growth in the presence or absence of Mtz, and other mutations that may allow growth at higher concentrations of Mtz could not be identified by this approach or are lethal to the bacterium.
Mtzr is generated during in vitro or in vivo growth due to diverse mutations within rdxA.
Mtzr occurs in H. pylori during in vitro growth and in vivo growth in humans due to mutations mainly within rdxA (22, 24, 30, 48). We examined if Mtzr isolates could be recovered after in vitro or in vivo growth of C. jejuni in the absence of selection. After growth on MH agar without Mtz, Mtzr C. jejuni from seven independent cultures was isolated at a rate of 1.06 × 10−6 ± 0.32 × 10−6. Sequencing of rdxA from 12 randomly selected isolates from each culture revealed that at least 87% of the isolates were derived independently from each other, with the remainder possibly representing siblings.
We next determined if Mtzr could be generated during in vivo growth of C. jejuni in chicks, a natural host for the bacterium to promote a harmless, commensal colonization. Six 1-day-old chicks were orally gavaged with wild-type Mtzs C. jejuni 81-176, and C. jejuni bacteria from the ceca were recovered at day 7 postinfection on selective agar with or without 10 μg ml−1 Mtz. Mtzr C. jejuni was isolated from the cecum of each chick, with all isolates from five of the six chicks arising from independent mutations (see Table S1 in the supplemental material). Compared to the number of C. jejuni bacteria that colonized the chick ceca, the best approximation for the rate of Mtzr was determined to be 8.62 × 10−6 ± 0.34 × 10−6.
Mtzr in both the in vitro- and in vivo-isolated mutants was due to many different types of mutations, including point, frameshift, insertion, and deletion mutations, occurring mostly within rdxA (see Tables S2 and S3 in the supplemental material). Approximately 44 to 50% of the in vivo- or in vitro-grown isolates, respectively, had lost or gained one or two nucleotides within the putative promoter region of rdxA or within the coding sequence causing frameshifts. Seventeen to 35% had transition or transversion mutations, mostly occurring in rdxA, resulting in either premature stop codons or amino acid changes that truncated, inactivated, or destabilized the protein (Fig. 2B). One transition mutation in the putative promoter of rdxA changed T-52 relative to the start codon, which prevented production of not only RdxA but also RdxB (Fig. 2B), suggesting that the two genes likely share a promoter and are cotranscribed. Two transition mutations resulted in coding sequences creating the RdxA(M1I) and RdxA(A175V) proteins, which were not detected in the mutants (Fig. 2B). RdxA(A175V) was likely an unstable protein, whereas RdxA(M1I) was may not have been translated due to alteration of the start codon. Curiously, RdxB was not detected in the rdxA(M1I) mutant, suggesting that the mutation may have caused instability of a putative bicistronic mRNA for rdxAB or lack of translation of the entire transcript. An additional missense mutation caused a mutant to produce RdxA(G156D), which was produced at wild-type levels but likely was enzymatically inactive (Fig. 2B).
The remaining 18 to 32% of Mtzr isolates had insertion or deletion of DNA segments of at least 7 and up to 902 nucleotides in length within rdxA or the rdxAB operon to cause frameshifts in the rdxA coding sequence (see Tables S2 and S3 in the supplemental material). Mutants with DNA insertions in rdxA resulted from a direct duplication of a DNA segment to create a two-unit heteropolymeric nucleotide repeat of 7 to 179 nucleotides in length. For the mutants that had lost 22 to 902 bp of DNA within rdxA or rdxAB, the event leading to deletion of the DNA usually occurred at an apparently random site within the gene or locus. However, the DNA that was lost in three of the mutants was flanked by identical DNA repeats 8 to 12 nucleotides in length. These DNA repeats may have influenced a recombination event leading to excision of the intervening DNA. Only one in vivo-grown Mtzr isolate did not possess a mutation within rdxAB, and the mutation leading to Mtzr remains unidentified.
Of note, a small collection of C. jejuni strains in our laboratory was analyzed for Mtzs. We found that those strains that were Mtzs appeared to contain an intact rdxA allele with only one or two amino acid changes, suggesting that a functional RdxA protein was produced. Strains that were Mtzr all had multiple point mutations and frameshift mutations in rdxA, corroborating data of our studies with C. jejuni 81-176 that disruption of rdxA contributes to Mtzr (data not shown).
Due to the reported mutagenic nature of reduced Mtz (46), Mtzr isolates we recovered after in vitro and in vivo growth could represent two populations: (i) those that spontaneously mutated rdxA by DNA replication errors prior to Mtz exposure to result in Mtzr or (ii) those that are Mtz induced once plated on agar containing 10 μg ml−1 Mtz. To help distinguish the origins of these two possible populations, we exposed in vitro-grown cultures of Mtzs wild-type C. jejuni 81-176 to 10 μg ml−1 Mtz and compared the number of Mtzr isolates generated over time to those without Mtz. We detected a 225-fold decrease in viability of 81-176 over a 6-h period with exposure to Mtz (Fig. 3A). Examination of the Mtzr population after exposure to Mtz for 2 h indicated that 31% of this population was Mtzr prior to any Mtz exposure (Fig. 3A; compare time 2 h with time zero h) and suggested that approximately one-third of the total Mtzr isolates were generated by spontaneous mutation through DNA replication errors in the absence of Mtz. Furthermore, we did not detect any increase in the number of Mtzr isolates with exposure to Mtz over time (Fig. 3A).
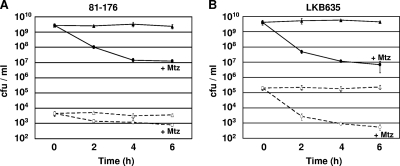
FIG. 3.
Analysis of Mtz-induced killing and mutation over time. (A and B) Mtzs wild-type C. jejuni 81-176 or 81-176 Smr cjj0275::cat-rpsL (LKB635) was grown and then not exposed (triangles) or exposed (circles) to 10 μg ml−1 Mtz over the course of the assay. At each time point, bacteria were plated on MH agar containing TMP (closed symbols, solid lines or 10 μg ml−1 Mtz (open symbols, dashed lines)) (A) or MH agar containing TMP (closed symbols, solid lines) or 2 mg ml−1 Sm (open symbols, dashed lines) (B). The average number of viable bacteria at each time point is reported as the number of CFU per ml. Each assay was performed in triplicate, and the standard deviation is indicated by bars.
We next examined if Mtz exposure caused nonspecific mutation at a second locus to result in an increase in the number of antibiotic-resistant mutants. To this end, we used strain LKB635, a C. jejuni 81-176 derivative that has a wild-type rpsL allele and a recessive streptomycin-resistant (Smr) rpsL allele (rpsLSm). Analogous to mutations in rdxA that result in Mtzr, any type of disruption to the wild-type rpsL allele leading to aberrant protein production will result in Smr. We incubated this strain in the presence and absence of Mtz and then monitored viability and generation of Smr over time. Similar to the results with exposure of 81-176 to Mtz, we detected a 600-fold reduction in viability over 6 h (Fig. 3B). In addition, we did not detect an increase in Smr isolates over time with Mtz exposure (Fig. 3B). Taken together, these results suggest that many of the Mtzr isolates recovered after in vitro or in vivo growth were due to spontaneous DNA replication errors rather than strictly being induced by the mutagenic potential of Mtz.
RdxA and RdxB reduce nitro compounds.
In vitro biochemical analysis of RdxA was performed to provide evidence that the protein reduces Mtz, which would corroborate our findings that production of RdxA in C. jejuni is responsible for Mtzs. Additional nitro compounds, such as those that have been shown to be modified by RdxA and FrxA of H. pylori, were also used to more fully characterize C. jejuni RdxA as a potential nitroreductase (45). Since RdxB is also a putative nitroreductase, we analyzed the reductase activity of this protein as well.
We first cloned rdxA and rdxB into an expression vector in E. coli BL21(DE3) to create N-terminal GST fusion proteins. After induction and purification, both GST-RdxA and GST-RdxB protein preparations, but not GST alone, were yellow in color, suggesting that the flavin cofactor was bound to the fusion proteins. The flavin cofactor was determined to be FMN by protein denaturation followed by TLC (data not shown). Further analysis in E. coli BL21(DE3), which is normally Mtzr, on medium containing 10 μg ml−1 Mtz showed that expression via induction of GST-RdxA or GST-RdxB rendered E. coli more Mtzs, indicating that our recombinant proteins were active.
A potential problem of using GST fusion proteins for biochemical characterization of enzymes is that the GST component could artificially contribute to the activity under investigation. To ensure that the nitroreductase component of these fusions was largely responsible for enzymatic activity, the GST component from each fusion was removed by thrombin-mediated cleavage. The recovered nitroreductases were almost identical to the native proteins in C. jejuni with just the initial methionine replaced with a glycine and serine residue.
RdxA, RdxB, and the respective uncleaved GST fusion proteins were then examined for their ability to oxidize NADPH using oxygen as an electron acceptor or reduce Mtz, nitrofurazone, furazolidone, and nitrofurantoin using NADPH as an electron donor. As shown in Table 1, RdxA, RdxB, and the respective GST fusion proteins oxidized NADPH. Because the activities of RdxA and RdxB were similar with the respective GST fusion proteins and GST alone did not promote oxidation of NADPH, we conclude that these activities were specific to the nitroreductases. Furthermore, RdxA demonstrated a higher specific activity of approximately 2- to 8-fold over that of RdxB. This potent oxidase activity is in line with what has been observed with RdxA of H. pylori (37).
TABLE 1.
Reductase and oxidase activities of RdxA and RdxB and respective GST fusions for various substrates
| Activity type | Substrate (acceptor) | Sp act (μmol/min/mg of protein) ± SEa |
||||
|---|---|---|---|---|---|---|
| GST-RdxA | RdxAb | GST-RdxB | RdxBb | GST | ||
| Oxidase | NADPH (O2) | 2.4 ± 0.50 | 2.1 ± 0.01 | 0.3 ± 0.04 | 1.25 ± 0.01 | <0.01 |
| Reductase | Mtz, anaerobic | 0.18 ± 0.07 | 0.19 ± 0.02 | 0.03 ± 0.01 | 0.06 ± 0.01 | <0.01 |
Specific activities were calculated as the means of results from two assays from two independent purifications of the protein ± standard errors.
GST-RdxA or GST-RdxB was treated with thrombin to cleave the fusion proteins and release the free nitroreductase protein. Thrombin was removed from the nitroreductases by benzamidine Sepharose 6B.
Assays to ascertain the reductase activities of these enzymes were initially performed under aerobic conditions. These experiments revealed that RdxA and RdxB were able to reduce all nitro compounds (data not shown). However, when we accounted for the specific endogenous NADPH oxidase activity present for each nitro compound, we could not demonstrate any significant reductase activity for any of the compounds tested, including Mtz, under aerobic conditions. As reported by Olekhanovich et al. (37), reduction of Mtz by RdxA of H. pylori occurred only under strict anaerobiosis, because trace amounts of molecular oxygen inhibited reduction. Considering these findings, we tested RdxA and RdxB for Mtz reduction under anaerobic conditions. We observed Mtz reduction under these conditions that was specific for the nitroreductases and not due to GST. Again, RdxA and GST-RdxA demonstrated 3- to 6-fold higher activity than RdxB and GST-RdxB, possibly corroborating our genetic analysis that RdxA influences Mtzs and Mtzr in C. jejuni. Furthermore, these data support previous conclusions of the different enzymatic characteristics of nitroreductases of epsilonproteobacteria (as demonstrated by H. pylori RdxA [37]) compared to those of other bacteria.
rdxAB is a cotranscribed operon induced during in vivo growth.
The genomic organization of rdxA and rdxB with the coding sequences of the genes overlapping suggests a cotranscribed operon. Inactivation of rdxA with transposons or antibiotic resistance-encoding cassettes and spontaneous mutation of the rdxA promoter region all resulted in the absence of RdxB, also suggesting cotranscription of these genes (Fig. 1C and 2B). By using reverse transcriptase PCR (RT-PCR), we detected an mRNA transcribing both rdxA and rdxB, verifying that these genes are cotranscribed (Fig. 4A). Primer extension analysis revealed that the start site of transcription for rdxA (and perhaps rdxAB) was located at T-28 relative to the start codon of rdxA (Fig. 4B). A putative −10 binding site for σ70 was located upstream of this site, suggesting T-28 as the most likely start site of transcription.
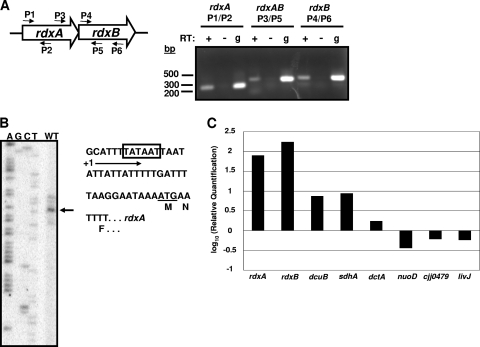
FIG. 4.
Analysis of expression of the rdxAB operon during in vitro and in vivo growth. (A) Examination of cotranscription of the rdxAB operon. Left, diagram of the rdxAB locus and locations of primers used for reverse transcriptase PCR (RT-PCR) analysis. Right, DNA fragments generated after RT-PCR analysis. Three sets of PCRs were performed, each using the indicated primer pair. Each PCR set included a reaction mixture containing cDNA generated from C. jejuni 81-176 RNA after in vitro growth (RT +), a reaction of the same RNA incubated in the absence of RT (RT −) to verify the absence of DNA in the RNA preparation, and a positive-control reaction using genomic DNA (g) as a template for amplification of a DNA product with the primers used. (B) Primer extension analysis to identify the transcriptional start site for rdxAB. Left, the arrow indicates the most prominent start site of transcription next to the sequencing ladder generated with the same primer used for the primer extension reaction. Right, partial sequence of the promoter region for rdxAB. The box indicates the proposed −10 binding site for σ70, +1 indicates the start site of transcription, and the start codon is underlined. The amino acids of the protein are indicated below each codon. (C) Semiquantitative real-time RT-PCR analysis of the level of expression of rdxA and rdxB during in vivo and in vitro growth. Expression analysis was also performed on other genes shown and described in the text as controls for detecting specific induction of in vivo expression. Values indicated are the log10-fold increase or decrease in expression relative to expression of in vitro-grown samples.
Preliminary real-time RT-PCR analysis suggested that the rdxAB operon was not highly expressed during in vitro growth of C. jejuni (data not shown). We considered if this operon may be preferentially expressed in vivo during commensal colonization of the ceca of chicks. One-day-old chicks were infected with C. jejuni 81-176 Smr (DRH212), and the chicks were sacrificed at day 7 postinfection. Cecal contents were isolated, and RNA was extracted. In comparison to in vitro-grown DRH212, expression of rdxA and rdxB were approximately 75- to 175-fold higher in vivo, respectively (Fig. 4C). We also analyzed expression of other genes to determine if enhanced expression of rdxA and rdxB was specific for in vivo growth. A previous study examining expression of genes of C. jejuni strain 11168 after a 12-h infection of chicks compared to results for in vitro-grown cultures found no differential expression of dctA and nuoD and a 19- to 32-fold-increased expression of dcuB and sdhA, respectively, in vivo (54). We found similar trends in expression of these genes in our analysis (Fig. 4C). Additionally, we included analyses of expression of two additional genes, livJ and cjj0479, identified in a previous study as ones required for optimal colonization of chicks (18). Both of these genes did not show evidence of increased expression in vivo (Fig. 4C). Thus, augmented expression of rdxA and rdxB was specific for in vivo growth, although the precise mechanism behind this increased expression remains unknown at this time.
Considering these data, we speculated that RdxA or RdxB may be necessary for wild-type levels of colonization of the chick ceca. One-day-old chicks were infected with approximately 100 or 104 CFU of wild-type C. jejuni 81-176 or 81-176 mutants lacking RdxA, RdxB, or both. For these experiments, we used 81-176 rdxB::cat-rpsL as a mutant lacking production of RdxB but unaffected for production of RdxA (data not shown). Due to the inability to generate an insertionally inactivated rdxA mutant that did not affect production of RdxB, we took advantage of one of the Mtzr isolates recovered after in vitro growth, rdxA(A175V). This mutant did not produce RdxA but was unaffected for production of RdxB (Fig. 2B). For an rdxAB double mutant, we took advantage of another Mtzr isolate recovered after in vitro growth, which lacked a portion of DNA from nucleotide 601 of rdxA through nucleotide 620 of rdxB (see Table S3 in the supplemental material). This strain produced neither RdxA nor RdxB (Fig. 2B).
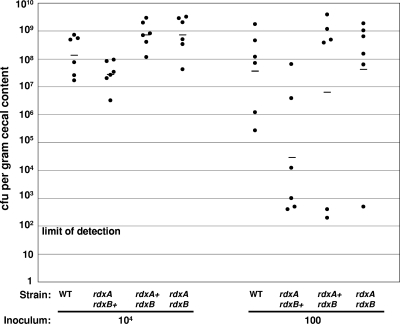
At 7 days postinfection, the levels of wild-type and mutant C. jejuni strains in the ceca were determined. In chicks infected with approximately 104 CFU of C. jejuni strains, only the rdxA mutant showed an average colonization defect of approximately 5-fold, which was not statistically significant (Fig. 5). When the inoculum was reduced to approximately 100 CFU, the rdxA mutant was present at a lower level in four out of six chicks than the wild-type strain in respective infected birds. These levels resulted in an average colonization defect of approximately 1,200-fold compared to the wild-type strain, which was marginally significant (P < 0.05, two-tailed Mann-Whitney U test). In no instances did we detect significant colonization defects of the rdxB mutant or the rdxAB double mutant, even though RdxA was absent in the latter strain. These results suggest that the rdxA mutant may have a dose-dependent colonization defect; however, this defect is not recapitulated in a mutant lacking both rdxA and rdxB.
FIG. 5.
Commensal colonization capacity of wild-type C. jejuni 81-176 and 81-176 mutant strains. One-day-old chicks were orally gavaged with 3.5 × 103 to 1.16 × 104 CFU (to approximate an inoculum of 104 CFU) or 11 to 85 CFU (to approximate an inoculum of 100 CFU) of wild-type C. jejuni 81-176 or 81-176 derivatives lacking production of RdxA, RdxB, or both RdxA and RdxB. At seven days postinfection, the number of C. jejuni CFU in the cecal contents of each chick was determined. Each dot represents the load of C. jejuni in the cecum of an individual chick. The geometric mean of the bacterial loads from each set of chicks is denoted by the horizontal bar. The status of rdxA and rdxB is indicated for each strain analyzed. Statistical analysis was performed using the two-tailed Mann-Whitney U test (P < 0.05).
rdxA as a site for cis complementation of mutants in C. jejuni.
The rdxA locus of H. pylori has been used as a site on the chromosome for insertion of genes to express in cis for complementation of mutants (6, 33). The advantage of this method is that Mtzr can be used as a marker to identify transformants containing rdxA interrupted with the gene for cis-chromosomal complementation. We expected that a similar complementation and selection strategy could be used to complement C. jejuni mutants, which would be an additional tool beneficial for the C. jejuni field.
To test rdxA for accepting genes for the purpose of complementation in cis, we inserted DNA containing the putative promoter and coding sequence of flhB, which encodes a component of the flagellar export apparatus, into rdxA (17, 19, 25). A ΔflhB mutant of C. jejuni lacks flagellar biosynthesis and motility, due in part to the lack of expression of σ54-dependent flagellar genes encoding many components of the flagellar basal body and hook (17, 19, 25). rdxA of two different C. jejuni flhB mutants, SNJ471 (81-176 Smr ΔflhB) and DRH1827 (81-176 Smr ΔastA ΔflhB flgDE2::nemo), was replaced with rdxA::flhB to test the ability of this construct to complement the mutants for restoration of motility and expression of the σ54-dependent flgDE2 operon, encoding two flagellar-hook-associated proteins. As a control, we also transformed these mutants with rdxA interrupted by cat, encoding chloramphenicol acetyltransferase (55). Approximately 1 in 100 Mtzr colonies contained rdxA interrupted with the complementing wild-type flhB allele. As shown in Fig. 6A, wild-type and complemented strains containing rdxA::flhB but not rdxA::cat produced equal amounts of FlhB in the total membrane fraction of the strains.
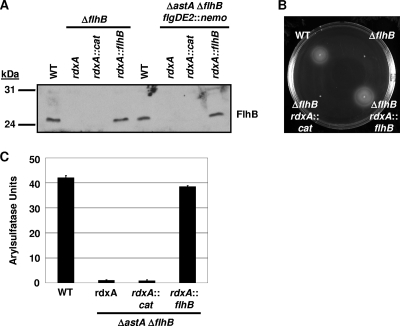
FIG. 6.
rdxA as a locus for genomic complementation of mutants in C. jejuni in cis. (A) Immunoblot analysis of FlhB production in wild-type, mutant, and complemented strains of C. jejuni. Total membranes from wild-type C. jejuni strains and respective mutant or complemented derivatives were analyzed for production of FlhB. Strains (from left to right) include DRH212 (wild-type 81-176 Smr), SNJ471 (81-176 Smr ΔflhB), DRH3067 (81-176 Smr ΔflhB rdxA::cat), DRH3107 (81-176 Smr ΔflhB rdxA::flhB), DRH533 (wild-type 81-176 Smr ΔastA flgDE2::nemo), DRH1827 (81-176 Smr ΔastA ΔflhB flgDE2::nemo), DRH3056 (81-176 Smr ΔastA ΔflhB flgDE2::nemo rdxA::cat), and DRH3057 (81-176 Smr ΔastA ΔflhB flgDE2::nemo rdxA::flhB). (B) Flagellar motility phenotypes of wild-type C. jejuni 81-176 Smr (DRH212) derivatives. Motility was assessed 30 h after inoculation in MH motility agar and incubation at 37°C under microaerobic conditions. Strains include DRH212, SNJ471, DRH3067, and DRH3107. (C) Arylsulfatase assays for analysis of expression of flgDE2::nemo, encoding an flgDE2-astA transcriptional fusion in C. jejuni 81-176 Smr ΔastA flgDE2::nemo derivatives. The results are from a typical assay with each strain tested in triplicate. The values reported for each strain are the average arylsulfatase activity ± standard deviation. Strains include DRH533, DRH1827, DRH3056, and DRH3057.
We then tested the strains for restoration of motility and σ54-dependent flagellar gene expression. We observed that the ΔflhB mutant containing rdxA::flhB was restored for motility to wild-type levels but an isogenic mutant with rdxA::cat was not (Fig. 6B). Similarly, expression of the σ54-dependent flagellar operon flgDE2 as analyzed by a flgDE2-astA transcriptional fusion (encoded by flgDE2::nemo) was restored to wild-type levels in the ΔflhB mutant with rdxA::flhB but not in the mutant containing rdxA::cat (Fig. 6C). These results indicated the functionality of using rdxA as a site for chromosomal insertion of genes for cis complementation of C. jejuni mutants.
DISCUSSION
Despite the almost ubiquitous presence of nitroreductases in diverse bacterial species, the biological function and the natural substrate of most nitroreductases are unknown. These enzymes, however, have been shown to have useful applications in industry and human medicine by reducing certain xenobiotic compounds. In this study, we found the RdxA and RdxB nitroreductases of C. jejuni can reduce Mtz, a prodrug that is used in therapeutic treatment of the closely related bacterium H. pylori and certain parasitic infections (53). Production of wild-type RdxA but not RdxB was associated with Mtzs, presumably due to in vivo conversion of Mtz to a toxic form that is lethal to C. jejuni. Even though we demonstrated the ability of RdxA to reduce Mtz in vitro, Mtz is a xenobiotic compound and likely not the natural substrate for RdxA. Thus, we suspect that RdxA and also RdxB function in other biological processes in C. jejuni whose pathways and natural substrates have yet to be elucidated.
In H. pylori, Mtzr is typically due to a loss of RdxA or production of catalytically inactive RdxA (11). Mutation of a gene encoding a second nitroreductase, FrxA, has been shown to contribute to Mtzr in some strains (22-24, 30, 48). RdxA and RdxB of C. jejuni are paralogs of RdxA and FrxA of H. pylori but are as similar to each other as they are to other bacterial nitroreductases. Contrary to findings of some studies in H. pylori, we found that mutation of only rdxA contributed to Mtzr in C. jejuni strain 81-176. Transposon mutagenesis of C. jejuni identified six independent insertions in rdxA, but none in rdxB, leading to Mtzr. Furthermore, all spontaneous Mtzr isolates generated during in vitro or in vivo growth had mutations in rdxA or rdxAB, but none had only a mutation in rdxB. An exception was one Mtzr isolate for which the location of the mutation could not be identified. These data indicate that elimination of wild-type RdxA resulted in Mtzr.
To confirm that mutation of rdxB did not contribute to Mtzr in C. jejuni 81-176, we constructed a mutant that produced wild-type RdxA but lacked RdxB. The MIC of Mtz for this rdxB mutant was equal to that of the Mtzs parental wild-type strain (approximately 2.5 μg ml−1). Additionally, mutations that eliminated RdxA and RdxB did not increase the MIC value relative to that for a Mtzr strain lacking only RdxA (approximately 50 μg ml−1). In vitro biochemical analysis revealed reduction of Mtz by both proteins, suggesting that RdxB could contribute to Mtzs. However, the specific activity of RdxB for Mtz was 3- to 6-fold lower than that of RdxA, which may not be physiologically significant in C. jejuni 81-176 to promote conversion of Mtz to a toxic form for Mtzs in the absence of RdxA. It is possible that in other C. jejuni strains, RdxB may influence Mtzr, and this possibility is currently under examination. Although we could not show a role for RdxB in Mtzr in strain 81-176, we believe that the major determinant influencing growth of this strain in the presence or absence of Mtz is rdxA.
In H. pylori, it has been shown that Mtzr is largely due to mutations within rdxA, and additional mutations in frxA and other genes can cause a stepwise increase in the MIC of Mtz for the mutant derivatives (1, 23, 24). In our hands, a C. jejuni 81-176 derivative lacking production of wild-type RdxA had a MIC of Mtz of 50 μg ml−1. We attempted to isolate mutants in this background that were able to grow on higher concentrations of Mtz, hopefully identifying other genes potentially mutated that would contribute further to Mtzr, similar to what has been observed in H. pylori. However, we were never able to isolate a mutant that could grow on Mtz above 50 μg ml−1. The lack of isolation of such a mutant suggests one of the following possibilities: (i) mutation of a gene that would contribute to growth on higher levels of Mtz, like those identified in H. pylori, may be lethal when generated in C. jejuni; (ii) Mtz at concentrations higher than 50 μg ml−1 may inhibit growth of C. jejuni by a mechanism independent of reduction by a bacterial nitroreductase; or (iii) an exhaustive screen, more intensive than the one used in the current study, may be required to find rare, spontaneous mutants in the rdxA mutant background that allow for growth on concentrations of Mtz higher than 50 μg ml−1. These findings possibly suggest that the mechanisms of Mtzr show some deviations from the multiple ones in H. pylori that allow for growth in the presence of this prodrug.
Olekhnovich et al. have recently characterized the biochemical activity of H. pylori RdxA for reduction of Mtz (37). Significant findings from this work included the finding that RdxA of H. pylori demonstrated a 60-fold higher activity than the NfsB nitroreductase of E. coli under strict anaerobic conditions. Additionally, RdxA was found to possess a strong NADPH oxidase activity. These data imply that the H. pylori RdxA nitroreductase and possibly other nitroreductases from epsilonproteobacteria may have unique biochemical and functional properties. As such, we found that the C. jejuni nitroreductases could also reduce Mtz in vitro only under anaerobic conditions and demonstrated a robust NADPH oxidase activity. Future work could reveal interesting findings by examining nitroreductases of other epsilonproteobacteria to discern if these enzymes universally share characteristics that differentiate them from those of other bacteria.
Another possibly interesting technique we discovered that may be of interest to the nitroreductase field is the use of GST fusions to purify and analyze the enzymology of nitroreductases. In the case of RdxB, we could easily purify a GST fusion protein from E. coli that contained the flavin cofactor and demonstrated enzymatic activity. Removal of GST by a thrombin-mediated cleavage enhanced enzymatic activity. For RdxA, the GST fusion protein appeared to contain a specific activity comparable to that of native RdxA. Thus, the use of GST fusion proteins may be a convenient method for purification of soluble, stable, and active nitroreductases.
In our analyses, we found a specific increase in expression of rdxA and rdxB during in vivo growth in the chick ceca, implying that these genes may be involved in colonization processes. Therefore, we assessed the ability of mutants lacking RdxA, RdxB, or both RdxA and RdxB to promote commensal colonization of the chick ceca. An rdxB mutant did not demonstrate an appreciable colonization defect. An Mtzr mutant lacking RdxA did not show a significant colonization defect when administered at an inoculum of 104 CFU. At an inoculum of 100 CFU, we did observe a lower colonization rate for four of six chicks compared to that for chicks infected with wild-type bacteria. Confusing these data is the observation that the possible colonization defect due to a lack of RdxA was not maintained in a mutant lacking both RdxA and RdxB, which colonized at wild-type levels. Currently we cannot provide an explanation for this discrepancy. A limited contribution of RdxA to commensalism may not be surprising considering the prevalence of Mtzr C. jejuni isolated from both wild and agriculturally significant birds (13, 47), presumably due to a loss of RdxA. Thus, the loss of rdxA most likely does not significantly hinder the ability of the bacterium to colonize avian hosts in nature. The possibility remains that RdxA or RdxB could be important for infection of humans to promote disease or in maintaining viability in the environmental reservoirs for transmission to humans and animals. Testing of these hypotheses will require development of better in vitro and in vivo model systems.
During analysis of the rdxAB locus, we frequently identified Mtzr isolates after in vitro or in vivo growth of C. jejuni in the absence of selection. Examination of such mutants revealed that point and frameshift mutations, in addition to insertion and deletion of DNA fragments up to approximately 0.9 kb in length, mainly within rdxA, were responsible for generation of Mtzr. These results possibly suggest that these mutations are generated randomly in C. jejuni during normal growth. However, since Mtz is a mutagen (46), these mutations could have been induced by exposure to Mtz rather than having been generated spontaneously through DNA replication errors. Over a series of experiments, we determined that a significant proportion of the population of C. jejuni (at least 31%) developed Mtzr due to spontaneous mutation and that Mtz did not contribute to increased Mtzr in wild-type C. jejuni or Smr in an appropriate C. jejuni mutant over time. While these experiments are not perfect in their execution, they provide evidence that C. jejuni does undergo spontaneous mutation of rdxA resulting in Mtzr.
Some of the spontaneous mutations that resulted in disruption of rdxA to give the Mtzr phenotype are likely caused by the absence in C. jejuni of a complete methyl-directed mismatch repair (MMR) system consisting of MutL, MutH, and MutS, usually present in other bacteria (9, 10, 39). MMR normally corrects base pair mismatches and insertions or deletions of small pieces of DNA, usually under four nucleotides in length (29). Instead, C. jejuni produces only MutS, but the protein is a member of the MutS2 family, of which a homolog in H. pylori has been shown to prevent oxidative damage to DNA (8, 50). Loss of MMR would presumably contribute to some mutations, such as point and small frameshift mutations, within rdxA, causing Mtzr. The lack of MMR may also contribute to a high degree of phase and antigenic variation of important virulence and colonization factors, such as lipooligosaccharide (LOS) glycosylation, capsule production, and flagellar motility (7, 12, 36, 42, 44, 51, 52). Recently we identified flgS and flgR, encoding the sensor kinase and response regulator of a two-component system required for expression of σ54-dependent flagellar genes, as two genes undergoing phase variation (15, 16). However, analysis of spontaneous revertants of flgS and flgR “OFF” variants restored for flagellar motility revealed that not all mutations in the “ON” revertants correctly repaired the original lesion in flgS or flgR. Instead, second-site suppressor mutations and intragenic recombinations, including insertion of repeated DNA sequences and deletions of DNA, were detected in flgS and flgR to create functional mutant proteins that restored flagellar gene expression and motility (15, 16). These mutations that occurred in flgS are similar to the ones contributing to alterations in rdxA, conferring Mtzr (16). Since the revertant flgS alleles were generated in the absence of Mtz, these findings are indirect, corroborative evidence that genetic variability occurs often and randomly to affect production of many phenotypes of C. jejuni, including flagellar motility and Mtzr.
Through the contribution of mutation of rdxA leading to Mtzr, we were able to complement mutants of C. jejuni by a very similar approach that has been useful in H. pylori research (6, 33). By interrupting rdxA with flhB in flhB mutants of C. jejuni, we showed the functionality of cis complementation of motility mutants by recovery of Mtzr transformants. We expect this type of complementation to be useful in complementing a wide range of mutants in the bacterium. Thus, we have added another genetic tool to the C. jejuni field that is likely to assist the molecular analysis of C. jejuni.
In this study, analysis of the rdxAB locus has revealed various interesting aspects of the biology of C. jejuni, ranging from DNA mutation, antibiotic resistance, and biochemistry of metabolic enzymes. Furthermore, we provide supportive evidence that the biochemical mechanisms of Rdx proteins of C. jejuni and H. pylori demonstrate characteristic anaerobic reduction of Mtz, as well as potent oxidation of NADPH, which may be common to those of epsilonproteobacteria and are intrinsically different from those of the nitroreductases of enteric bacteria. In addition, continued exploration of C. jejuni RdxA or RdxB in other potential infection models or assays may reveal a natural substrate for these nitroreductases and a biological function in infection, environmental survival, or transmission.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 AI065539 and by National Research Initiative grants 2006-35201-17382 and 2009-35201-05039 from the USDA Cooperative State Research, Education, and Extension Service Food Safety Program.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 2.Bingham-Ramos, L. K., and D. R. Hendrixson. 2008. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 76:1105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 States, United States, 2005. MMWR Morb. Mortal. Wkly. Rep. 55:392-395. [PubMed] [Google Scholar]
- 5.Coker, A. O., R. D. Isokpehi, B. N. Thomas, K. O. Amisu, and C. L. Obi. 2002. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croxen, M. A., G. Sisson, R. Melano, and P. S. Hoffman. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188:2656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 8.Eisen, J. A. 1998. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 26:4291-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaasbeek, E. J., F. J. van der Wal, J. P. van Putten, P. de Boer, L. van der Graaf-van Bloois, A. G. de Boer, B. J. Vermaning, and J. A. Wagenaar. 2009. Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J. Bacteriol. 191:3785-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin, A., D. Kersulyte, G. Sisson, S. J. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 12.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hariharan, H., S. Sharma, A. Chikweto, V. Matthew, and C. DeAllie. 2009. Antimicrobial drug resistance as determined by the E-test in Campylobacter jejuni, C. coli, and C. lari isolates from the ceca of broiler and layer chickens in Grenada. Comp. Immunol. Microbiol. Infect. Dis. 32:21-28. [DOI] [PubMed] [Google Scholar]
- 14.Henderson, M. J., and F. H. Milazzo. 1979. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. J. Bacteriol. 139:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646-1659. [DOI] [PubMed] [Google Scholar]
- 16.Hendrixson, D. R. 2008. Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol. Microbiol. 70:519-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 18.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 19.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 20.Iwaki, H., T. Muraki, S. Ishihara, Y. Hasegawa, K. N. Rankin, T. Sulea, J. Boyd, and P. C. Lau. 2007. Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoate mutase and a chemoreceptor involved in 2-nitrobenzoate chemotaxis. J. Bacteriol. 189:3502-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs, B. C., A. Van Belkum, and H. P. Endtz. 2008. Guillain-Barre syndrome and Campylobacter infection, p. 245-262. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 22.Jenks, P. J., R. L. Ferrero, and A. Labigne. 1999. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J. Antimicrob. Chemother. 43:753-758. [DOI] [PubMed] [Google Scholar]
- 23.Jeong, J. Y., A. K. Mukhopadhyay, J. K. Akada, D. Dailidiene, P. S. Hoffman, and D. E. Berg. 2001. Roles of FrxA and RdxA nitroreductases of Helicobacter pylori in susceptibility and resistance to metronidazole. J. Bacteriol. 183:5155-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong, J. Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatino, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Wong, S. K. Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H. K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joslin, S. N., and D. R. Hendrixson. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J. Bacteriol. 191:2656-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joslin, S. N., and D. R. Hendrixson. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J. Bacteriol. 190:2422-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knox, R. J., F. Friedlos, P. J. Biggs, W. D. Flitter, M. Gaskell, P. Goddard, L. Davies, and M. Jarman. 1993. Identification, synthesis and properties of 5-(aziridin-1-yl)-2-nitro-4-nitrosobenzamide, a novel DNA crosslinking agent derived from CB1954. Biochem. Pharmacol. 46:797-803. [DOI] [PubMed] [Google Scholar]
- 28.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 29.Kunkel, T. A., and D. A. Erie. 2005. DNA mismatch repair. Annu. Rev. Biochem. 74:681-710. [DOI] [PubMed] [Google Scholar]
- 30.Kwon, D. H., J. A. Pena, M. S. Osato, J. G. Fox, D. Y. Graham, and J. Versalovic. 2000. Frameshift mutations in rdxA and metronidazole resistance in North American Helicobacter pylori isolates. J. Antimicrob. Chemother. 46:793-796. [DOI] [PubMed] [Google Scholar]
- 31.Lei, B., M. Liu, S. Huang, and S. C. Tu. 1994. Vibrio harveyi NADPH-flavin oxidoreductase: cloning, sequencing and overexpression of the gene and purification and characterization of the cloned enzyme. J. Bacteriol. 176:3552-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liochev, S. I., A. Hausladen, and I. Fridovich. 1999. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:3537-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGee, D. J., M. L. Langford, E. L. Watson, J. E. Carter, Y. T. Chen, and K. M. Ottemann. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molbak, K., and A. Havelaar. 2008. Burden of illness of Campylobacteriosis and sequelae, p. 151-162. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 35.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 36.Nuijten, P. J., A. J. van den Berg, I. Formentini, B. A. van der Zeijst, and A. A. Jacobs. 2000. DNA rearrangements in the flagellin locus of an flaA mutant of Campylobacter jejuni during colonization of chicken ceca. Infect. Immun. 68:7137-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olekhnovich, I. N., A. Goodwin, and P. S. Hoffman. 2009. Characterization of the NAD(P)H oxidase and metronidazole reductase activities of the RdxA nitroreductase of Helicobacter pylori. FEBS J. 276:3354-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olson, C. K., S. Ethelberg, W. van Pelt, and R. V. Tauxe. 2008. Epidemiology of Campylobacter jejuni infections in industrialized nations, p. 163-189. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 39.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 40.Peterson, F. J., R. P. Mason, J. Hovsepian, and J. L. Holtzman. 1979. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J. Biol. Chem. 254:4009-4014. [PubMed]
- 41.Pollich, M., and G. Klug. 1995. Identification and sequence analysis of genes involved in late steps in cobalamin (vitamin B12) synthesis in Rhodobacter capsulatus. J. Bacteriol. 177:4481-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ridley, A. M., M. J. Toszeghy, S. A. Cawthraw, T. M. Wassenaar, and D. G. Newell. 2008. Genetic instability is associated with changes in the colonization potential of Campylobacter jejuni in the avian intestine. J. Appl. Microbiol. 105:95-104. [DOI] [PubMed] [Google Scholar]
- 43.Roldan, M. D., E. Perez-Reinado, F. Castillo, and C. Moreno-Vivian. 2008. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol. Rev. 32:474-500. [DOI] [PubMed] [Google Scholar]
- 44.Scott, A. E., A. R. Timms, P. L. Connerton, C. Loc Carrillo, K. Adzfa Radzum, and I. F. Connerton. 2007. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sisson, G., A. Goodwin, A. Raudonikiene, N. J. Hughes, A. K. Mukhopadhyay, D. E. Berg, and P. S. Hoffman. 2002. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sisson, G., J. Y. Jeong, A. Goodwin, L. Bryden, N. Rossler, S. Lim-Morrison, A. Raudonikiene, D. E. Berg, and P. S. Hoffman. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA(+) (nitroreductase) gene. J. Bacteriol. 182:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanley, K. N., and K. Jones. 1998. High frequency of metronidazole resistance among strains of Campylobacter jejuni isolated from birds. Lett. Appl. Microbiol. 27:247-250. [DOI] [PubMed] [Google Scholar]
- 48.Tankovic, J., D. Lamarque, J. C. Delchier, C. J. Soussy, A. Labigne, and P. J. Jenks. 2000. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob. Agents Chemother. 44:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmis, K. N., and D. H. Pieper. 1999. Bacteria designed for bioremediation. Trends Biotechnol. 17:200-204. [DOI] [PubMed] [Google Scholar]
- 50.Wang, G., P. Alamuri, M. Z. Humayun, D. E. Taylor, and R. J. Maier. 2005. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol. Microbiol. 58:166-176. [DOI] [PubMed] [Google Scholar]
- 51.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141:95-101. [DOI] [PubMed] [Google Scholar]
- 52.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolle, K., and P. Malfertheiner. 2007. Treatment of Helicobacter pylori. Best Pract. Res. Clin. Gastroenterol. 21:315-324. [DOI] [PubMed]
- 54.Woodall, C. A., M. A. Jones, P. A. Barrow, J. Hinds, G. L. Marsden, D. J. Kelly, N. Dorrell, B. W. Wren, and D. J. Maskell. 2005. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73:5278-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 56.Yao, R., and P. Guerry. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 178:3335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zenno, S., H. Koike, M. Tanokura, and K. Saigo. 1996. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J. Biochem. 120:736-744. [DOI] [PubMed] [Google Scholar]
- 58.Zenno, S., K. Saigo, H. Kanoh, and S. Inouye. 1994. Identification of the gene encoding the major NAD(P)H-flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J. Bacteriol. 176:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.