Abstract
Myxococcus xanthus DK1622 contains two paralogous groEL gene loci that possess both different sequences and different organizations within the genome. Deletion of either one of these two genes alone does not affect cell viability. However, deletion of both groEL genes results in cell death unless a complemented groEL1 or groEL2 gene is present. The groEL1 gene was determined to be essential for cell survival under heat shock conditions; a strain with mutant groEL2 caused cells to be more sensitive than the wild-type strain to higher temperatures. Mutants with a single deletion of either groEL1 (MXAN_4895) or groEL2 (MXAN_4467) had a growth curve similar to that of the wild-type strain DK1622 in medium containing hydrolyzed proteins as the substrate. However, when cells were cultured on medium containing either Escherichia coli cells or casein as the substrate, deletion of groEL2, but not groEL1, led to a deficiency in cell predation and macromolecular feeding. Furthermore, groEL1 was found to play an indispensable role in the development and sporulation of cells, but deletion of groEL2 had no visible effects. Our results suggest that, although alternatively required for cell viability, the products of the two groEL genes have divergent functions in the multicellular social life cycle of M. xanthus DK1622.
Myxobacteria are characterized among the prokaryotes by their unique social behavior (4, 30, 36). The social behavior of myxobacterial cells is present in each stage of the life cycle; cells glide on solid surfaces in swarms, feed on macromolecules and other microbial cells in groups, and develop multicellular resting structures called fruiting bodies that contain myxospores when food is exhausted (32). Many genes are required to conduct this complicated social lifestyle. Genome sequencing revealed that myxobacteria have the largest bacterial genome and possess many multicopy genes (8, 31). A major duplication occurs with chaperone genes, which are essential for cell functions by assisting protein folding, assembly, transport, and degradation (21, 26), not only in normal cellular processes but also in response to nonpermissive temperatures (their products thus belong to the heat shock protein family) (5, 23). There are several reports related to the functions of chaperones in the sociality of Myxococcus. For example, in Myxococcus xanthus, there appear to be several duplicated genes encoding the chaperone HSP70 protein (DnaK) (8), of which only one, sglK (MXAN_6671), has been studied. SglK was shown to be essential for social motility and multicellular development in M. xanthus DK1622, but it did not respond to temperature changes or heat shock (35, 37).
GroEL protein, a major type of chaperonin, is ubiquitously distributed in bacteria (11). Bacterial cells usually contain one copy of the groEL gene within the genome. However, approximately 20% of sequenced bacterial genomes were found to have duplicate or multiple copies of the groEL gene (10, 11). The products of duplicated groEL genes have been reported to play divergent roles in various bacteria. For example, in Mycobacterium smegmatis, which possess duplicate groEL genes, the GroEL2 protein was shown to be essential for cell growth. In contrast, inactivation of the groEL1 gene did not affect the normal growth of cells but prevented the formation of mature biofilms and the biosynthesis of mycolic acid (24). There are three copies of the groEL gene in Rhizobium leguminosarum. The cpn60.1 gene (groEL1) has been shown to be indispensable for the growth of this organism, while the other two paralogous genes, cpn60.2 and cpn60.3 (groEL2 and groEL3), can be deleted without affecting cell viability (28). The three GroEL homologues of R. leguminosarum display distinct properties in vitro (7) and preferentially self-assemble rather than form mixed hetero-oligomeric proteins when coexpressed in E. coli cells (10). Furthermore, Sinorhizobium meliloti has five copies of the groEL gene in its genome. However, only the product encoded by groEL1, which is required for symbiosis, is required for viability (2).
In different myxobacteria, there are also two copies of the groEL gene (13), of which little is known with regard to their specific cellular roles, especially in the social life of myxobacterial cells. In this study, the functions of duplicate groEL genes in cell growth, predation feeding, development, and heat shock response were investigated. Our results demonstrate that either copy of the duplicate groEL genes is indispensable for the survival of Myxococcus xanthus DK1622. Furthermore, these studies also suggest that the products of the two groEL genes have played divergent roles in the social life cycle of myxobacterial cells.
MATERIALS AND METHODS
Cultures, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The M. xanthus strains were cultivated in Casitone-based rich-nutrient medium CTT (12) for growth assays and on TPM agar (17) for developmental assays. E. coli strains were routinely grown on Luria-Bertani (LB) agar or in LB broth. Myxococcus strains were incubated at 30°C, and E. coli was grown at 37°C. When required, a final concentration of 40 μg/ml or 20 μg/ml of kanamycin (Km) was added to the solid or liquid media.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| M. xanthus | ||
| DK1622 | Wild-type strain | 15; D. Kaiser, University of Stanford |
| YL0301 | DK1622 ΔMXAN_4895 (groEL1 deletion) | This study |
| YL0302 | DK1622 ΔMXAN_4467 (groEL2 deletion) | This study |
| YL0305 | DK1622::pZC4895 (lacZ fused to groEL1) | This study |
| YL0306 | DK1622::pZC4467 (lacZ fused to groEL2) | This study |
| YL0307 | YL0301::pSWU4895 (groEL1 integrated at attB site) | This study |
| YL0308 | YL0307 ΔMXAN_4467 (groEL2 deletion) | This study |
| YL0309 | YL0302::pSW4467 (groEL2 integrated at attB site) | This study |
| YL0310 | YL0309 ΔMXAN_4895 (groEL1 deletion) | This study |
| E. coli | ||
| DH5α(λpir) | φ80 lacZΔM15 ΔlacU169 recA1 endA1 hsdR17 supE44 thi-1 gyrA relA1 λpir | H. B. Kaplan, University of Texas |
| XL1-Blue MR | Δ(mcrA)183Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| Plasmids | ||
| pBJ113 | Gene replacement vector with KG cassette, Kmr | 14; Z. M. Yang, Virginia Tech |
| pMiniHimar1-lacZ | KmrlacZ | H.B. Kaplan, University of Texas |
| pSWU30 | Site-specific integration vector with Mx8 attP integration site, Tetr | Mignot Tâm, CNRS (Centre national de la recherche scientifique) |
| pSL1180 | Cloning vector, Ampr | Amersham |
| pSL1180-Km | 1.25-kb fragment of kan from pColE1 inserted into SmaI of pSL1180, Ampr Kmr | This study |
| pSW4467 | 2.14-kb fragment of MXAN_4467, with its upstream 495-bp promoter sequence, inserted into XbaI/EcoRI sites of pSWU30, Tetr | This study |
| pSW4895 | 2.93-kb fragment of MXAN_4895, with its upstream 1.28-kb promoter sequence, inserted into XbaI/EcoRI sites of pSWU30, Tetr | This study |
| pZCY11 | 1.56-kb HindIII/EcoRI fragment from pSL1180-Km, blunted with Klenow fragment and ligated with 4.57-kb T4 polymerase-blunted KpnI fragment from pMiniHimar1-lacZ, KmrlacZ | This study |
| pBJ4467 | Upstream and downstream homologous arms of DK1622 MXAN_4467 inserted into SmaI site of pBJ113, Kmr | This study |
| pBJ4895 | Upstream and downstream homologous arms of DK1622 MXAN_4895 inserted into SmaI site of pBJ113, Kmr | This study |
| pZC4467 | 601-bp fragment of DK1622 MXAN_4467, containing a stop codon, inserted into SpeI/KpnI sites of pZCY11, KmrlacZ | This study |
| pZC4895 | 685-bp fragment of DK1622 MXAN_4895, containing a stop codon, inserted into SpeI/KpnI sites of pZCY11, KmrlacZ | This study |
In-frame deletion of groEL genes.
In-frame deletion of MXAN_4467 (groEL2) and MXAN_4895 (groEL1) in M. xanthus was performed using the positive-negative KG cassettes described by Ueki et al. (34). In brief, genomic DNA from DK1622 served as the template for PCR amplification of the upstream and downstream homologous arms using Pfu DNA polymerase (Stratagene). The arms were fused at the BamHI site for both MXAN_4467 and MXAN_4895 to form the internal deletion fragments. The fragments were cloned into SmaI-digested pBJ113 to construct the deletion plasmids pBJ4467 and pBJ4895, which were then transferred by electroporation into M. xanthus DK1622 cells, as previously described (16). Individual Km-resistant colonies were transferred onto CTT agar plates supplemented with 1% galactose (Sigma) for the second round of screening. Deletion mutants were identified by the phenotypes of galactose resistance and kanamycin sensitivity, as well as verified by PCR. The primers for verification are listed in Table 2.
TABLE 2.
PCR primers
| Primer | Sequence (5′-3′)a | Use | Expected size (bp) |
|---|---|---|---|
| 4467-KO-1-for | GGCTGTGGCGGGTGGACAAGGC | Amplification of the upstream homologous arm for deletion of groEL2 | 810 |
| 4467-KO-1-rev | CGCGGATCCGCCGCTCCGGAGGGCTCGGGA | ||
| 4467-KO-2-for | CGCGGATCCCGCAACCTCCTGAATTCACTGGG | Amplification of the downstream homologous arm for deletion of groEL2 | 895 |
| 4467-KO-2-rev | CAGATGAGGCAGTCCGCGGTTC | ||
| 4895-KO-1-for | CGCAGGTGAACAGCTTGTCG | Amplification of the upstream homologous arm for deletion of groEL1 | 979 |
| 4895-KO-1-rev | CGCGGATCCACCGCGTCGGCCAGGATGTT | ||
| 4895-KO-2-for | CGCGGATCCGTCATCGACCCGGCCAAGGT | Amplification of the downstream homologous arm for deletion of groEL1 | 1,072 |
| 4895-KO-2-rev | GTCGCTTGCGGGAAAGGTCT | ||
| 4895-CIs-F | CTAGTCTAGAGTGCGGACCCACGCCTCATA | PCR amplification of groEL1 with its promoter | 2,931 |
| 4895-CIs-R | CGGAATTCGGGGTGAGGGGCGGACTACAT | ||
| 4467-CIs-F | CTAGTCTAGAGAACTCTGCACGATGCTCTCTC | PCR amplification of groEL2 with its promoter | 2,139 |
| 4467-CIs-R | CGGGATTCTCAGTAGTCCATGTCGTCGCCGC | ||
| Km-for | GTGCTGACCCCGGATGAATGTCAG | PCR amplification of kan | 1,251 |
| Km-rev | ATCGAGCCCGGGGTGGGCGAAGAA | ||
| 4467-Rp-for | GGGGTACCCATCCGCACGCAGATTGACTCC | lacZ transcription fusion of groEL2 | 601 |
| 4467-Rp-rev | GGACTAGTTCAGTAGTCCATGTCGTCGCCG | ||
| 4895-Rp-for | GGGGTACCGGACAAGGACAACACCACCATC | lacZ transcription fusion of groEL1 | 685 |
| 4895-Rp-rev | GGACTAGTGGCGGACTACATGCCCATACC | ||
| PV-4895-Rp | GCCGAGGAGAACAAGACCAAG | Reverse primer for verification of proper integration of YL0305 | 2,216 |
| PV-4467-Rp | GTCCGTCACCGAGCCGTCCT | Reverse primer for verification of proper integration of YL0306 | 2,182 |
| PV-Rp-pZCY11 | TTCTTCTGAGCGGGACTCTGGG | Forward primer for verification of proper integration of YL0305 and YL0306 |
Restriction sites that were incorporated into the primers are underlined, and the added bases needed for the restriction enzyme digestion of PCR products are in italics.
Complementary inactivation.
Due to several failed attempts to delete both copies of the groEL gene, complemented deletion strains were constructed (20). The groEL1 gene and its upstream 1,280-bp sequence was PCR amplified, digested with XbaI and EcoRI, whose recognition sites were incorporated into the primers, and ligated with the XbaI-EcoRI-digested site-specific integration plasmid pSWU30 to give pSW4895. After electroporation, pSW4895 was integrated at the Mx8 attB site in the genome of YL0301 (ΔgroEL1) mutant, producing a groEL1 complemented mutant, designated YL0307. The mutant was selected with 10 μg/ml tetracycline and verified by PCR using four primers specific for the 5′ and 3′ ends of the attP and attB regions. Then, the mutant YL0307 was used to delete the groEL2 gene by electroporation of pBJ4467 as described above, producing the final complemented deletion mutant YL0308. Complementary inactivation of groEL2 was performed using a similar procedure.
Construction of the groEL-lacZ reporter genes.
The promoterless lacZ gene (4.57 kb) was obtained from pMiniHimar1-lacZ using KpnI digestion. T4 polymerase was used to blunt the end of the fragment. Separately, the Klenow fragment (MBI Inc.) was used to blunt the ends of a HindIII-EcoRI-digested fragment (1.56 kb) from pSL1180-Km. The fragments were ligated together to produce pZCY11. A 685-bp DNA sequence including the stop codon of groEL1 was amplified from the genome of DK1622 using KpnI- and SpeI-incorporated primers. The PCR fragments and pZCY11 were digested with KpnI and SpeI. Then, the fragments were ligated together, producing pZC4895, in which the lacZ gene was fused to the groEL1 gene. A similar strategy was used to construct the groEL2-lacZ fusion, producing pZC4467. The groEL-lacZ fusion constructions were separately transferred by electroporation into M. xanthus DK1622, and individual kanamycin-resistant clones were selected. The mutants of the groEL-lacZ transcription fusion were verified by PCR amplification (primers are listed in Table 2). The expression levels of lacZ were visualized using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma) as a substrate.
Growth assays.
M. xanthus cells were inoculated in liquid CTT medium to a final concentration of 1 × 107 cells/ml. The cultures were shaken at 30°C and monitored every 4 h by measuring culture turbidity using a Klett-Summerson photoelectric colorimeter equipped with a red filter (Unic Tech, Inc., China). The growth was reported as optical density at 600 nm (OD600) values. Assays were performed in triplicate, and the results are presented with standard deviations.
Predation and feeding assays.
A predation assay was conducted according to a previously published method (1). Briefly, cultures were harvested at mid-log phase and were washed three times in 10 mM MOPS (morpholinepropanesulfonic acid; pH 7.6) buffer. M. xanthus and E. coli cells were concentrated to a final cell density of 5 × 109 cells/ml and 1 × 1011 cells/ml, respectively. Fifty microliters of E. coli cells was pipetted onto the plate and allowed to dry, forming a 1-cm-diameter colony. A 2-μl aliquot of M. xanthus cells was then spotted onto the center of the prey E. coli mat, with an inoculation size of 0.15 cm in diameter. The assay was repeated three times. During a 6-day incubation, the swarming size of M. xanthus cells was measured and monitored every 12 h with a dissection microscope. The swarming size of M. xanthus cells is reported as the predation ability. The liquid feeding assay was carried out by cultivating the mutants and the wild-type strain DK1622 in CT medium, for which casein was used as the only nutrient (29).
Developmental assays.
M. xanthus cells were harvested at mid-log phase and resuspended in TPM buffer (17) at a concentration of 5 × 109 cells/ml. Aliquots of 10 μl were deposited onto TPM agar. The cultures were incubated at 30°C and were detected every 24 h under a dissection microscope to monitor the formation of fruiting bodies. The sporulation was measured on 5-day TPM cultures as previously described (9). The sporulation rate was calculated as the number of colonies divided by the number of inoculated cells. Assays were performed at least three times.
Heat shock assays.
Mid-log-phase cultures of M. xanthus were harvested as described above. After a heat shock of 30 min or 60 min at 42°C, the cells were immediately serially diluted and plated on the CTT agar. After 6 days, the numbers of CFU were calculated, with untreated cells as a control.
Viability staining.
Following heat treatment, the cell viability of the wild-type strain DK1622 and the mutants lacking groEL1 or groEL2 were measured using the LIVE/DEAD BacLight bacterial viability kit (Invitrogen Co.; L7012), as described by Nariya and Inouye (22), with minor modifications. Briefly, the cells collected from cultures were incubated at 42°C for 30 min or 60 min, followed by two washes in TM buffer. The cells were then adjusted to 5 × 108 cells per milliliter, stained according to the manufacturer's protocol, and observed and counted under a fluorescence microscope (Nikon-TE2000s).
Quantitative real-time PCR analysis.
M. xanthus DK1622 cells were collected from a 36-h culture and inoculated in CTT medium to a final concentration of 1 × 107 cells/ml. The culture was harvested every 6 h, and the RNA was extracted immediately using a total RNA extraction kit (Promega) by following the manufacturer's instructions. Genomic DNA contamination was removed by using a DNA-free kit (ABI). The purified RNA extract was reverse transcribed to cDNA and stored in aliquots at −70°C. Quantitative real-time PCR was performed in a Bio-Rad sequence detection system with a total reaction volume of 20 μl, containing 250 nM primers, 10 μl of SYBR green PCR master mix (Bio-Rad Co.), 8.5 μl of RNase-free water (Takara), and 0.5 μl of a 10-fold-diluted cDNA template. The PCR was conducted in the following manner: 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, and 15 s at 72°C. The 16S RNA was used as the normalization signal (3). Calibration curves of groEL1, groEL2, and 16S RNA were generated from 10-fold dilutions of genomic DNA of M. xanthus DK1622. The primer pairs used for each gene were as follows (forward and reverse): for groEL1, 5′-GAGAAGGTGGGCAAGGAAGG-3′ and 5′-TCACGAAGTACGGGGACAGG-3′; for groEL2, 5′-ACAGCCCGATGGACCTCA-3′ and 5′-TGGTCTCATCCCCGTTGG-3′; and for 16S rRNA, 5′-CGGCGTGACAAGTCGGGTGTGAAAG-3′ and 5′-CGTCTCAGCGTCAGTTACCGTCCAG-3′.
Measurement of β-galactosidase activity.
β-Galactosidase-specific activity was assayed as described by Kroos et al. (18), with minor modifications. In brief, the cells were broken with a mini-beadbeater (Biospec) at a speed of 3,000 rpm. Between each agitation, the samples were cooled with ice water. The enzymatic activity of β-galactosidase was determined by using ONPG (o-nitrophenyl-β-galactopyranoside; Sigma) as the substrate and read at 420 nm with a SpectraMax 190 plate reader (Molecular Devices). The amount of proteins in the extracts was determined by a bicinchoninic acid (BCA) protein assay (Pierce), and the specific activity was calculated using the following formula: specific activity = 213 × A420/(sample volume [ml] × protein concentration [mg/ml] × reaction time [min]).
RESULTS
Deletions of groEL genes in Myxococcus xanthus.
The two groEL genes in Myxococcus xanthus DK1622 are MXAN_4467 (groEL2) and MXAN_4895 (groEL1). The levels of similarity of the nucleotide and amino acid sequences of the duplicate groELs are 83% and 79%, respectively. The most significant difference between the two paralogous genes is that groEL1 is arranged with an upstream cochaperone gene, groES, forming a complete bicistronic groESL operon. In contrast, the second groEL gene (groEL2) does not have an accompanying groES gene. The genetic composition and arrangement differences between the two groEL genes suggest they may possess divergent functions in M. xanthus cells.
The groEL1 and groEL2 genes were deleted individually, producing the mutants YL0301 and YL0302. We tried to obtain, in addition to the single mutants, a double mutant with mutations in the two groEL genes by either insertion or deletion mutation of groEL1 in YL0302 or groEL2 in YL0301. However, attempts to produce a double groEL mutant were unsuccessful, suggesting that a double mutant is not viable. In order to confirm that the two groELs could not be inactivated at the same time, complemented double-deletion mutants were constructed by site-specific integration of groEL1 or groEL2 (including the upstream promoter region in each case) at the Mx8 attB site in the genome of YL0301 or YL0302, respectively. Following the complementation, the other groEL gene could be deleted from their native site in the genome.
Heat shock responses of Myxococcus groEL mutants.
Using two-dimensional gel electrophoresis, Otani et al. showed that both of the GroEL proteins in M. xanthus were overexpressed following a heat shock treatment (25). However, their specific roles in the heat shock response remain unclear. Visualized by using the live/dead staining method (22), more than 80% of the YL0301 cells were most likely dead (dyed red) after 30 min of a 42°C heat shock. However, approximately 80% of the YL0302 cells were determined to be alive (dyed green). In comparison, about 90% of the wild-type DK1622 cells survived following the 42°C heat treatment. The survival rates of the groEL mutants were further assayed by calculation of the number of CFU. After a 30-min heat shock treatment at 42°C, relative to the wild-type DK1622 cells, only 2.1% of the YL0302 cells survived, while the YL0301 cells were unviable following the 30-min heat shock. These results suggest that groEL1 is essential for cell survival under heat shock conditions; mutating groEL2 caused cells to be more sensitive than the wild-type strain to higher temperatures.
Growth ability of the groEL mutants.
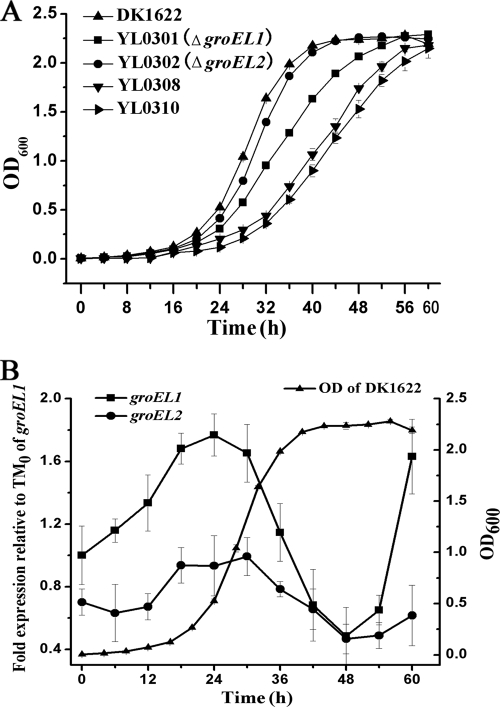
When cultured in CTT medium, both of the groEL mutants YL0301 (ΔgroEL1) and YL0302 (ΔgroEL2) demonstrated a growth curve similar to that of the wild-type strain (Fig. 1A). The generation times during the exponential growth stage were 4.06 ± 0.1, 4.61 ± 0.1, and 4.34 ± 0.15 h for DK1622, YL0301, and YL0302, respectively. The generation times of the mutants showed that deletion of one of the two groELs had minor effects on cell growth. After the exponential growth stage, both of the mutants reached nearly the same cell density as the wild-type strain DK1622. Quantitative PCR showed that expression of the groEL1 gene increased during the first 24-h incubation in CTT medium (Fig. 1B). After 30 h of incubation, the expression of the groEL1 gene was sharply decreased and reached the lowest level at about 48 h of incubation. Then, the expression of groEL1 increased again in the late stationary growth stage. On the other hand, compared to that of groEL1, the expression of groEL2 was relatively stable but lower than that of groEL1 throughout the growth stage. The quantitative PCR results also demonstrated that, after deletion of groEL1 (in YL0301), the expression of the groEL2 gene increased 3.1- and 2.5-fold compared to that in wild-type strain DK1622 at 24 h and 28 h, respectively, during which the expression of groEL2 was relatively stable in the wild-type strain (see Fig. 1B). However, expression of groEL1 in YL0302 was similar to that in the wild-type strain DK1622.
FIG. 1.
(A) Growth curves of the mutants with single deletions of either groEL1 or groEL2 or a double deletion of groEL1 and groEL2 and complementation with groEL1 (YL0308) or groEL2 (YL0310) and of the wild-type strain DK1622. (B) Quantitative PCR analysis of the expression levels of groEL1 and groEL2 of the wild-type strain DK1622 in CTT medium with an overlaid representative growth curve of the wild-type strain DK1622. The values are shown as levels relative to the expression of groEL1 at the onset of inoculation from a 36-h culture; the error bars represent standard deviations from three independent biological replicates. TM0, time zero.
Interestingly, after the introduction of groEL1 or groEL2 (with their respective promoters) into the single groEL deletion mutant, both strains (containing one original groEL1 or groEL2 gene and one transposed groEL2 or groEL1 gene) exhibited growth characteristics similar to those of the wild-type strain in the CTT medium. However, both complemented double-deletion mutants exhibited a significant delay in cell growth (Fig. 1A). The generation time of the groEL1-complemented double-deletion mutant YL0308 was 6.92 ± 0.29 h and that of the groEL2-complemented mutant YL0310 was 7.05 ± 0.23 h in CTT medium, respectively. Several studies have reported reduced expression of genes at the Mx8 attB site (6, 19, 33). The quantitative-PCR results indicated that the levels of expression of the transposed groEL1 and groEL2 genes were approximately 30% to 50% and 75% to 90%, respectively, of that in the wild-type strain DK1622 at 24 h and 28 h in CTT cultures. Thus, both groEL genes are involved in the growth of M. xanthus DK1622.
Predation feeding ability of the groEL mutants.
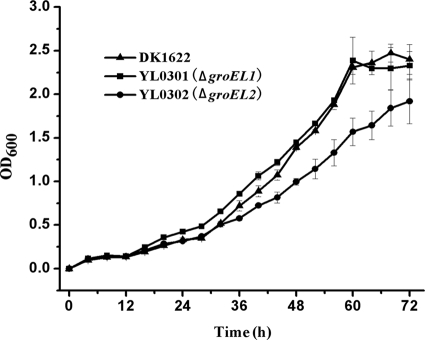
The presence of a single groEL1 and groEL2 gene was able to support the growth of M. xanthus cells and gave a growth curve similar to that of the wild-type strain DK1622, with hydrolyzed proteins as the substrates (CTT medium). However, it was interesting to observe a growth difference among the mutants when casein was used as the substrate (Fig. 2). The growth of both mutants and the wild-type strain was significantly delayed when casein was substituted for hydrolyzed proteins in the CTT growth medium. Unlike with growth in the hydrolyzed-protein-containing medium, YL0301 demonstrated nearly the same growth curve as the wild-type strain DK1622. The generation times of YL0301 and the wild-type strains were 7.63 ± 0.51 h and 7.43 ± 0.32 h, respectively. However, the YL0302 cells showed a growth delay under these culture conditions, exhibiting a generation time of 8.68 ± 0.44 h during the exponential growth stage, and the final cell density of YL0302 at 72 h was lower than those of both the wild-type strain and YL0301.
FIG. 2.
Growth curves in CT medium (29) for mutants with the groEL1 or groEL2 deletion and for the wild-type strain DK1622.
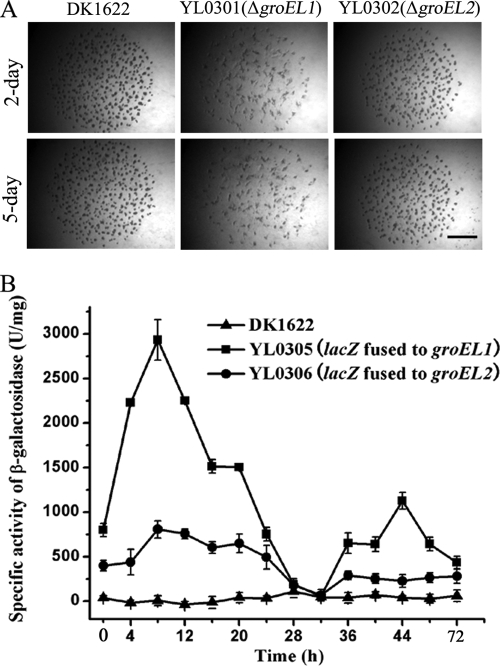
In a natural environment, myxobacteria usually live on solid surfaces by feeding in groups on microbial cells or their debris (27, 32). Based on bioinformatic analysis of the Myxococcus genome sequence, Goldman et al. speculated that the duplication of chaperone proteins may be responsible for the predatory-feeding lifestyle of M. xanthus cells (8). A predation feeding experiment was performed using live E. coli cells on an agar plate (Fig. 3). After the myxobacterial strains were spotted on the E. coli mat containing 1 × 1011 cells/cm2, M. xanthus DK1622 and YL0301 cells formed visible degradation regions as early as 12 h after inoculation. In contrast, the degradation region produced by the YL0302 cells was not visible until 48 h after inoculation. These results suggest that the GroEL2 protein plays an important role in predation feeding on macromolecule substrates and E. coli cells. However, the GroEL1 protein appears not to exhibit the same function. Following exhaustion of E. coli cells, the developmental behaviors of YL0301, YL0302, and the wild-type strain DK1622 were similar to those on TPM medium (see below).
FIG. 3.
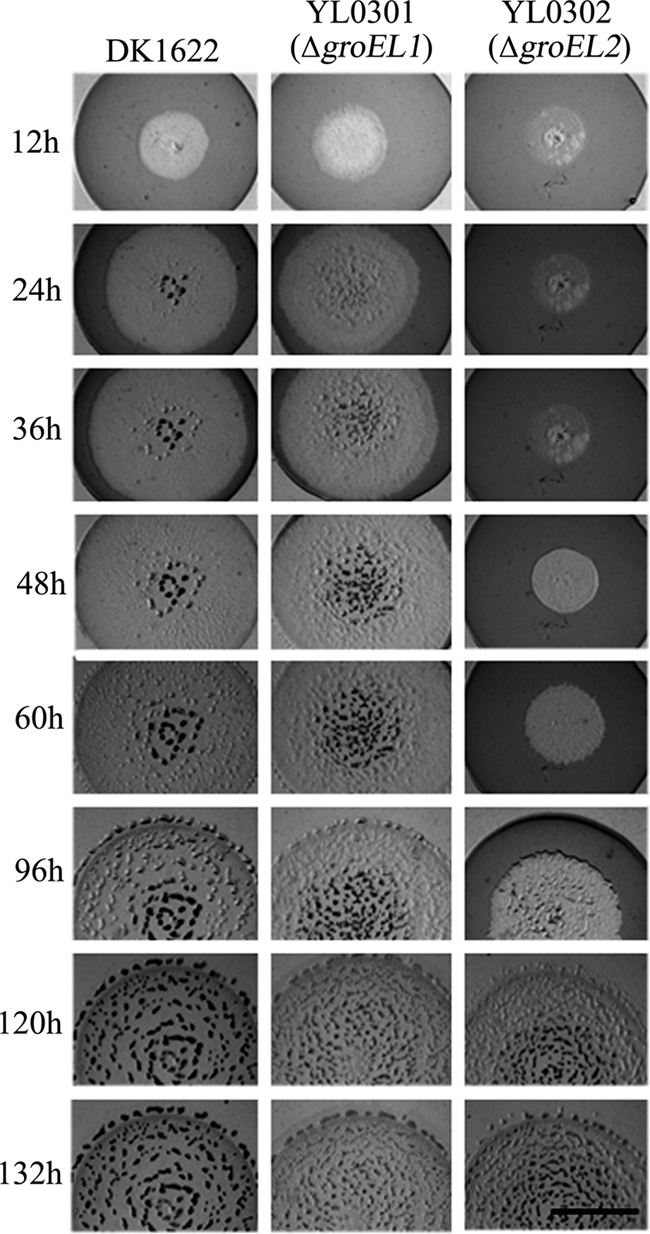
Predation feeding of M. xanthus DK1622, YL0301 (ΔgroEL1), and YL0302 (ΔgroEL2) on an E. coli prey mat. The bar is equal to 5 mm.
Development ability of the groEL mutants.
Upon starvation, myxobacterial cells are able to form multicellular fruiting body structures, in which myxospores develop in the late stage of morphogenesis (32). When cultured on the developmental medium TPM, the groEL1 deletion mutant YL0301 formed irregular aggregates (Fig. 4A). The sporulation ability of the groEL1 mutant was only 3.0% ± 2.1% of that of DK1622 and therefore was also determined to be significantly defective. Conversely, the groEL2 deletion mutant YL0302 exhibited a phenotype similar to that of fruiting body structures of the wild-type strain. The sporulation capacity of the groEL2 mutant was also observed to be similar to that of strain DK1622 (98.3% ± 16.5%). In the case of predation on E. coli cells, after the exhaustion of food, the YL0302 cells were also able to develop normal fruiting body structures, although the developmental process was significantly delayed due to the belated predation behavior (Fig. 3). Upon exhaustion of E. coli cells, YL0301 was also able to form irregular aggregates, but they did not mature. In parallel to the developmental behaviors of the groEL mutants, expression of the lacZ gene transcriptionally fused after the groEL1 gene in the wild-type strain DK1622 (YL0305) was found to be increased in the early stage (Fig. 4B). Peak levels of lacZ product were observed after approximately 8 h of starvation. The enzymatic activity of the groEL1-fused lacZ product sharply decreased during the next 20 h of incubation. Afterwards, expression of the groEL1 gene significantly increased again, forming the second peak. Highly regulated expression of groEL1 during development was correlated with our observations that groEL1 plays important roles in the formation of fruiting bodies and myxospores. On the other hand, assayed by the β-galactosidase activity of the groEL2-lacZ product (YL0306), groEL2 was expressed at much lower levels throughout development, consistent with our observation that groEL2 is not required for the formation of fruiting bodies or myxospores.
FIG. 4.
(A) Development of fruiting bodies of the different groEL mutants and the wild-type strain DK1622 on TPM plates. The bar is equal to 1.5 mm for each panel. (B) β-Galactosidase activity in the wild-type strain DK1622, the fused groEL1-lacZ DK1622 mutant (YL0305), and the fused groEL2-lacZ DK1622 mutant (YL0306) on TPM agar. The datum at each point represents the mean of independent triplicate determinations with standard deviations.
DISCUSSION
Genome sequencing has revealed many duplicated genes within the myxobacterial genome. Many of these duplicated genes have been suggested to have evolved to accommodate the social life cycle of myxobacteria (8). The studies in this paper investigate the duplicate groEL genes of myxobacteria and explore how they may be involved in the social lifestyle of Myxococcus. The GroEL proteins typically participate in growth and in responses to various environmental stresses and many other cellular physiological processes by assisting in the proper folding of many proteins (5). In some bacterial species, duplicate or multiple copies of the groEL gene have evolved, along with divergence of their functions. In most cases, one GroEL is essential, while the others are specialized in their roles to meet the requirements of different bacterial lifestyles (2, 24, 28). In M. xanthus DK1622, there are two groEL gene loci. The sequences of the duplicate groEL genes and their organization in the genome are significantly different, suggesting that they possess divergent cellular roles. The groEL deletion experiments described in this paper have indicated that either one of the duplicate groEL genes could support survival of the cell but that mutation of both groEL genes was lethal for the cell unless one groEL gene was complemented. These results indicate that the two GroEL proteins are partially redundant in function during growth.
Single deletions of groEL genes resulted in almost no observable effects on the growth of Myxococcus cells in CTT medium containing hydrolyzed proteins as the substrates. However, the two groEL genes showed divergent roles for the social life cycle of Myxococcus. GroEL2, but not GroEL1, was found to participate in the predation and feeding processes of the bacteria, while GroEL1, but not GroEL2, was strictly required for the fruiting body development and sporulation of Myxococcus. Previous studies have reported that chaperone HSP70 proteins (DnaK and its homologue) are involved in the control of S motility (35, 37). However, the deletion of a single groEL gene had no apparent effect on S motility (unpublished data). Thus, it appears that different chaperones are involved in different physiological processes in the social life of Myxococcus. Interestingly, whether or not groEL1 and groEL2 functioned, both genes were expressed in the growth and development stages. GroEL1 was almost always expressed at a higher level than GroEL2. However, if groEL1 was deleted, the expression of the groEL2 gene was increased. Thus, there is a balanced expression of the duplicate groEL genes in Myxococcus cells. There are several possibilities for the imbalance of functions and expression levels of the duplicate groEL genes. For instance, our experiments have suggested that deletion of the groEL1 gene led to a higher expression level of the groEL2 gene, possibly as compensation to support the normal growth of cells. However, expression of the groEL1 gene was not affected by deletion of groEL2. It is also possible that other chaperone proteins may be able to partially share the tasks of the GroEL proteins during cell growth, especially when one of the duplicated groEL genes is deleted. Higher expression of GroEL1 might allow GroEL1 to participate in some more physiological processes than GroEL2 during the growth stage. However, in the development stage, no matter whether or not the expression of groEL2 was upregulated by the deletion of groEL1, groEL1 deletion led to severe developmental defects. We thus speculate that the function divergences of the duplicate groELs not only are probably due to the different expression levels but also are the results of their substrate specificity. In order to clearly determine the functions and collaborations of the two groEL genes, further investigations, such as coimmunoprecipitation studies for identification of the proteins that are folded by GroEL1 and GroEL2, are required and are currently being undertaken in our laboratory.
Acknowledgments
The work was financially supported by the National Science Foundation for Distinguished Young Scholars (grant 30825001) and the National Natural Science Foundation (grants 30825001 and 30671192) of China.
We thank Dale Kaiser, Heidi B. Kaplan, Mitchell H. Singer, and Zhaomin Yang for sharing strains and plasmids. We thank anonymous reviewers for helpful comments on the manuscript.
Footnotes
Published ahead of print on 5 February 2010.
REFERENCES
- 1.Berleman, J. E., and J. R. Kirby. 2007. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J. Bacteriol. 189:5675-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittner, A. N., A. Foltz, and V. Oke. 2007. Only one of five groEL genes is required for viability and successful symbiosis in Sinorhizobium meliloti. J. Bacteriol. 189:1884-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bode, H. B., M. W. Ring, G. Schwar, R. M. Kroppenstedt, D. Kaiser, and R. Muller. 2006. 3-Hydroxy-3-methylglutaryl-coenzyme A (CoA) synthase is involved in biosynthesis of isovaleryl-CoA in the myxobacterium Myxococcus xanthus during fruiting body formation. J. Bacteriol. 188:6524-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dworkin, M., and D. Kaiser (ed.). 1993. Myxobacteria II. American Society for Microbiology, Washington, DC.
- 5.Fayet, O., T. Ziegelhoffer, and C. Georgopoulos. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171:1379-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisseha, M., M. Gloudemans, R. E. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George, R., S. M. Kelly, N. C. Price, A. Erbse, M. Fisher, and P. A. Lund. 2004. Three GroEL homologues from Rhizobium leguminosarum have distinct in vitro properties. Biochem. Biophys. Res. Commun. 324:822-828. [DOI] [PubMed] [Google Scholar]
- 8.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorski, L., T. Gronewold, and D. Kaiser. 2000. A sigma(54) activator protein necessary for spore differentiation within the fruiting body of Myxococcus xanthus. J. Bacteriol. 182:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gould, P. S., H. R. Burgar, and P. A. Lund. 2007. Homologous cpn60 genes in Rhizobium leguminosarum are not functionally equivalent. Cell Stress Chaperones 12:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyal, K., R. Qamra, and S. C. Mande. 2006. Multiple gene duplication and rapid evolution in the groEL gene: functional implications. J. Mol. Evol. 63:781-787. [DOI] [PubMed] [Google Scholar]
- 12.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, D. M., L. Zhao, C. Y. Zhang, J. Li, Z. J. Xia, J. Wang, Z. H. Wu, and Y. Z. Li. 2008. Taxonomic analysis of Sorangium strains based on HSP60 and 16S rRNA gene sequences and morphology. Int. J. Syst. Evol. Microbiol. 58:2654-2659. [DOI] [PubMed] [Google Scholar]
- 14.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 17.Kearns, D. B., B. D. Campbell, and L. J. Shimkets. 2000. Myxococcus xanthus fibril appendages are essential for excitation by a phospholipid attractant. Proc. Natl. Acad. Sci. U. S. A. 97:11505-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 19.Li, S. F., and L. J. Shimkets. 1988. Site-specific integration and expression of a developmental promoter in Myxococcus xanthus. J. Bacteriol. 170:5552-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobedanz, S., and L. Sogaard-Andersen. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17:2151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 22.Nariya, H., and M. Inouye. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55-66. [DOI] [PubMed] [Google Scholar]
- 23.Neidhardt, F. C., and R. A. VanBogelen. 1987. Heat shock response, p. 1334-1345. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 24.Ojha, A., M. Anand, A. Bhatt, L. Kremer, W. R. Jacobs, Jr., and G. F. Hatfull. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123:861-873. [DOI] [PubMed] [Google Scholar]
- 25.Otani, M., J. Tabata, T. Ueki, K. Sano, and S. Inouye. 2001. Heat-shock-induced proteins from Myxococcus xanthus. J. Bacteriol. 183:6282-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranson, N. A., H. E. White, and H. R. Saibil. 1998. Chaperonins. Biochem. J. 333:233-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichenbach, H. 1999. The ecology of the myxobacteria. Environ. Microbiol. 1:15-21. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Quinones, F., M. Maguire, E. J. Wallington, P. S. Gould, V. Yerko, J. A. Downie, and P. A. Lund. 2005. Two of the three groEL homologues in Rhizobium leguminosarum are dispensable for normal growth. Arch. Microbiol. 183:253-265. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg, E., K. H. Keller, and M. Dworkin. 1977. Cell density-dependent growth of Myxococcus xanthus on casein. J. Bacteriol. 129:770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg, E. (ed.). 1984. Myxobacteria: development and cell interactions. Springer-Verlag Press, New York, NY.
- 31.Schneiker, S., O. Perlova, O. Kaiser, K. Gerth, A. Alici, M. O. Altmeyer, D. Bartels, T. Bekel, S. Beyer, E. Bode, H. B. Bode, C. J. Bolten, J. V. Choudhuri, S. Doss, Y. A. Elnakady, B. Frank, L. Gaigalat, A. Goesmann, C. Groeger, F. Gross, L. Jelsbak, J. Kalinowski, C. Kegler, T. Knauber, S. Konietzny, M. Kopp, L. Krause, D. Krug, B. Linke, T. Mahmud, R. Martinez-Arias, A. C. McHardy, M. Merai, F. Meyer, S. Mormann, J. Munoz-Dorado, J. Perez, S. Pradella, S. Rachid, G. Raddatz, F. Rosenau, C. Ruckert, F. Sasse, M. Scharfe, S. C. Schuster, G. Suen, A. Treuner-Lange, G. J. Velicer, F. J. Vorholter, K. J. Weissman, R. D. Welch, S. C. Wenzel, D. E. Whitworth, S. Wilhelm, C. Wittmann, H. Blocker, A. Puhler, and R. Muller. 2007. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25:1281-1289. [DOI] [PubMed] [Google Scholar]
- 32.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thony-Meyer, L., and D. Kaiser. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol. 175:7450-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 35.Weimer, R. M., C. Creighton, A. Stassinopoulos, P. Youderian, and P. L. Hartzell. 1998. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J. Bacteriol. 180:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitworth, D. E. (ed.). 2007. Myxobacteria: multicellularity and differentiation. ASM Press, Washington DC.
- 37.Yang, Z., Y. Geng, and W. Shi. 1998. A DnaK homolog in Myxococcus xanthus is involved in social motility and fruiting body formation. J. Bacteriol. 180:218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]