Abstract
In the Firmicutes, two-component regulatory systems of the LiaSR type sense and orchestrate the response to various agents that perturb cell envelope functions, in particular lipid II cycle inhibitors. In the current study, we found that the corresponding system in Streptococcus pneumoniae displays similar properties but, in addition, responds to cell envelope stress elicited by murein hydrolases. During competence for genetic transformation, pneumococci attack and lyse noncompetent siblings present in the same environment. This phenomenon, termed fratricide, increases the efficiency of horizontal gene transfer in vitro and is believed to stimulate gene exchange also under natural conditions. Lysis of noncompetent target cells is mediated by the putative murein hydrolase CbpD, the key effector of the fratricide mechanism, and the autolysins LytA and LytC. To avoid succumbing to their own lysins, competent attacker cells must possess a protective mechanism rendering them immune. The most important component of this mechanism is ComM, an integral membrane protein of unknown function that is expressed only in competent cells. Here, we show that a second layer of self-protection is provided by the pneumococcal LiaFSR system, which senses the damage inflicted to the cell wall by CbpD, LytA, and LytC. Two members of the LiaFSR regulon, spr0810 and PcpC (spr0351), were shown to contribute to the LiaFSR-coordinated protection against fratricide-induced self-lysis.
During a transient state termed competence for natural transformation, Streptococcus pneumoniae has the ability to take up naked DNA from the environment and incorporate this DNA into its genome by homologous recombination. A quorum sensing-like system consisting of the secreted competence-stimulating peptide (CSP), a membrane-embedded histidine kinase (ComD), and its cognate cytoplasmic response regular (ComE) controls competence development in this species (15, 16, 37). Upon entering the competent state, pneumococci start transcribing a number of competence-specific genes termed the early and late competence genes (26). A few of these genes are involved in a phenomenon termed fratricide. Naturally competent pneumococci use this mechanism to kill and lyse noncompetent sister cells present in the same environment, presumably to “steal” their DNA (19, 41). The key effector of fratricide in liquid medium is a putative murein hydrolase, CbpD, that together with the autolysins LytA and LytC degrades the cell wall of the noncompetent target cells (7, 12, 22). To avoid succumbing to their own toxins, competent pneumococci protect themselves by producing an immunity protein termed ComM (17). Although the mechanism by which ComM provides protection is not known, we speculated that the murein hydrolases involved in the fratricide mechanism might cause envelope stress in competent pneumococci.
In Gram-positive bacteria, the cell envelope consists of a thick murein cell wall, interspersed with (lipo)teichoic acids and the cytoplasmic membrane. The cell envelope is a crucial structure of the bacterial cell that fulfills a number of important functions. It protects the cell and gives it its shape but also functions as a molecular sieve and communication interface with the environment. Ensuring its integrity is therefore crucial for survival. Accordingly, bacteria have evolved numerous regulatory devices that monitor the integrity of and initiate protective responses in case of damage to the cell envelope. These systems are collectively termed cell envelope stress responses (20).
Most Gram-positive bacteria belonging to the phylum Firmicutes possess homologs of a cell envelope stress sensor system from Bacillus subtilis, designated LiaSR (30). In Lactococcus lactis and Staphylococcus aureus the corresponding two-component regulatory systems are called CesSR and VraSR, respectively (24, 29). In addition to their membrane-spanning histidine kinase sensors (LiaS/VraS/CesS) and their cognate response regulators (LiaR/VraR/CesR), these systems contain a third protein found to be essential for signal sensing. The gene encoding this protein, termed liaF in B. subtilis, is transcribed from the same promoter as the genes encoding the two-component regulatory system. Evidence indicates that LiaF, which is a membrane-anchored protein, acts as a negative modulator of the system through direct interaction with LiaS (21). When LiaS and/or LiaF sense the appropriate stress signal(s), LiaF-mediated inhibition of the LiaSR signal transduction system is relieved, resulting in activation of LiaR-dependent promoters. The exact signal(s) detected by LiaSR, VraSR, and CesSR are not known. In general, they are induced by cell wall-active antibiotics, especially those that interfere with the lipid II cycle (e.g., bacitracin, ramoplanin, vancomycin, and nisin) (20). In addition, direct cell wall damage, for example treatment with lysozyme, may in some cases also activate these systems (45). We therefore decided to investigate whether the pneumococcal two-component system HK03/RR03, which corresponds to LiaSR, VraSR, and CesSR, is activated during competence in response to the activity of CbpD, LytA, and LytC. In what follows, we will refer to the HK03/RR03 system as LiaSR. In accordance with the cell envelope stress-sensing systems from B. subtilis, S. aureus, and L. lactis, the pneumococcal system also contains a third component (spr0342) corresponding to LiaF.
Our results show that CbpD, together with LytA and LytC, activates the pneumococcal LiaFSR system in a comM deletion mutant, providing evidence that both glycan strand cleavage and hydrolysis of the stem peptide-N-acetylmuramyl bond can trigger LiaFSR activation. In addition, we found that the LiaFSR system contributes significantly to protect competent pneumococci against the potentially lethal effects of the fratricide mechanism. The LiaFSR is also induced by cell wall antibiotics that interfere with the lipid II cycle of cell wall biosynthesis. Using a combination of global transcriptional profiling, regulon mining, and comparative genomics, we defined the LiaR regulon in S. pneumoniae. We demonstrate that two regulon members, Spr0810 and PcpC (Spr0351), are important to counteract cellular lysis. Taken together, our data indicate that the LiaFSR system of S. pneumoniae has adapted its function to orchestrate a second, ComM-independent layer of protection against fratricide-induced self-lysis.
MATERIALS AND METHODS
Bacterial growth conditions and storage.
Cultures from which RNA was extracted were grown in Todd-Hewitt broth (THB). All other experiments were carried out in C medium (25). Precultures of the bacteria were grown at 37°C to an optical density at 550 nm (OD550) of 0.3 to 0.35, mixed with 0.5 vol of 50% glycerol, and stored at −80°C.
Construction of mutants and plasmids.
Bacterial strains and plasmids used in this work are listed in Table 1, while the sequences of all primers employed are given in Table S1 in the supplemental material. DNA was introduced into the streptococcal strains by means of natural transformation using 250 ng/ml of CSP to induce the competent state in the DNA recipient. Cultures of S. pneumoniae were grown at 37°C until they reached an OD550 of 0.15 to 0.2. Then, CSP and transforming DNA were added, and the cultures were further incubated for 90 to 120 min at 37°C before plating on selective medium to identify transformants. S. pneumoniae transformants were selected by plating on Todd-Hewitt agar supplemented with chloramphenicol (4.5 μg/ml), kanamycin (400 μg/ml), streptomycin (200 μg/ml), spectinomycin (200 μg/ml), or tetracycline (2.5 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype/relevant featurea | Reference/source |
|---|---|---|
| Strains | ||
| RH4 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) Smr by transformation with CP1200 chromosomal DNA; Eryr Spcr Cmr Smr | 7 |
| RH229 | ΔcomA::ermAM (Eryr) spr1332::luc; Eryr Cmr | This study |
| RH237 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) ΔcomM; Eryr Spcr Cmr Smr | This study |
| RH253 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) ΔcomM liaR::Janus; Eryr Spcr Cmr Kanr | This study |
| RH259 | ΔcomA::ermAM (Eryr) ebg::spc liaR::luc by transformation with pVEL5; Eryr Spcr Smr Cmr | This study |
| RH269 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) ΔcomM, Δspr0810 Eryr Spcr Cmr Kanr | This study |
| RH270 | ΔcomA::ermAM (Eryr) ebg::spc spr0810::luc by transformation with pVEL6; Eryr Spcr Smr Cmr | This study |
| RH271 | ΔcomA::ermAM (Eryr) ebg::spc spr0810::luc ΔcomM::Janus; Eryr Spcr Cmr Kanr | This study |
| RH272 | ΔcomA::ermAM (Eryr) ebg::spc spr0810::luc ΔcomM; Eryr Spcr Cmr Smr | This study |
| RH273 | ΔcomA::ermAM (Eryr) ebg::spc spr0810::luc ΔcomM ΔliaR::Janus; Eryr Spcr Cmr Kanr | This study |
| RH276 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) ΔliaR::Janus; Eryr Spcr Cmr Kanr | This study |
| RH283 | ΔcomA::ermAM (Eryr) ebg::spc spr0810::luc ΔcomM ΔcbpD::Janus; Eryr Spcr Cmr Kanr | This study |
| RH284 | ΔcomA::ermAM (Eryr) ebg::spc spr0810::luc ΔcomM cbpDC75A; Eryr Spcr Cmr Smr | This study |
| RH287 | ΔcomA::ermAM (Eryr) ebg::spc ΔlytC::tet; Eryr Spcr Smr Tetr | This study |
| RH288 | ΔcomA::ermAM (Eryr) ebg::spc ΔlytC::tet; ΔcomM::Janus; Eryr Spcr Tetr Kanr | This study |
| RH289 | ΔcomA::ermAM (Eryr) ebg::spc ΔlytC::tet; ΔcomM; Eryr Spcr Tetr Smr | This study |
| RH290 | ΔcomA::ermAM (Eryr) ebg::spc ΔlytC::tet ΔcomM spr0810::luc; Eryr Spcr Tetr Smr Cmr | This study |
| RH291 | ΔcomA::ermAM (Eryr) ebg::spc ΔlytC::tet ΔcomM spr0810::luc ΔlytA::Janus; Eryr Spcr Tetr Kanr Cmr | This study |
| RH294 | ΔcomA::ermAM (Eryr) ebg::spc spr1332::luc ΔcomM::Janus; Eryr Cmr Kanr | This study |
| TMSP008 | liaR::Spcr | This study |
| TMSP011 | bgaA::pBG0202 | This study |
| TMSP013 | bgaA::pBG0204 | This study |
| TMSP014 | bgaA::pBG0205 | This study |
| TMSP015 | bgaA::pBG0206 | This study |
| SPH-4 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) ΔcomM ΔpcpC::Janus; Eryr Spcr Cmr Kanr | This study |
| SPH-5 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) ΔcomM Δspr0810 Eryr Spcr Cmr Smr | This study |
| SPH-6 | ΔcomA::ermAM (Eryr) ebg::spc hirL::lacZ (Cmr) ΔcomM Δspr0810 ΔpcpC::Janus; Eryr Spcr Cmr Kanr | This study |
| Plasmids | ||
| pR424 | ColE1(pEVP3) derivate, Cmr, carries an ssbB targeting fragment and luc gene; insertion-duplication in S. pneumoniae generates a ssbB::luc (ssbB+) fusion | 4 |
| pPPP2 | ColE1 derivate, Ampr Tetr ′lacZ | 14 |
| pVEL5 | pR424 derivative, Cmr; carries liaR targeting fragment and luc gene; insertion-duplication in S. pneumoniae generates a liaR::luc (rr03+) fusion | This study |
| pVEL6 | pR424 derivative, Cmr; carries spr0810 targeting fragment and luc gene; insertion-duplication in S. pneumoniae generates a spr0810::luc (spr0810+) fusion | This study |
| pR1332 | pR424 derivative, Cmr; carries spr1332 targeting fragment and luc gene; insertion-duplication in S. pneumoniae generates a spr1332::luc (spr1332) fusion | This study |
| pBG0202 | pPPP2 derivative, PliaF(−137-+57)::lacZ | This study |
| pBG0204 | pPPP2 derivative, PliaF(−61-+57)::lacZ | This study |
| pBG0205 | pPPP2 derivative, PliaF(−17-+57)::lacZ | This study |
| pBG0206 | pPPP2 derivative, PliaF(−64-+57)::lacZ | This study |
| pDG1726 | pUC19 derivative with Spcr cassette from pJL74 | 11 |
All Streptococcus pneumoniae strains are R6 derivates. Cmr, chloramphenicol resistance; Eryr, erythromycin resistance; Spcr, spectinomycin resistance; Smr, streptomycin resistance; Kanr, kanamycin resistance, Ampr, ampicillin resistance; Tetr, tetracycline resistance.
RH237, which carries a deletion of the comM locus, was derived from strain RH420 (7), which contains a Janus cassette replacing comM. To construct RH237, the upstream and downstream regions (∼1,000 bp) of comM were amplified with the primer pairs ComMF/ComMRdel and ComMFdel/ComMR, respectively. The fragments were subsequently fused in a PCR using the primers ComMF and ComMR. The resulting DNA fragment was transformed into RH420, resulting in removal of the Janus cassette.
Initially, an allelic replacement mutant of liaR was constructed by long-flanking homology (LFH) PCR, essentially as described previously (31, 46). In brief, a spectinomycin resistance cassette was amplified from plasmid pDG1726 (11). Two primer pairs (designated up-reverse/up-forward and do-reverse/do-forward) were designed to amplify ∼1,000-bp DNA fragments flanking the region to be deleted (liaR) at its 5′ and 3′ ends. The resulting fragments are here called the “liaR-up” and “liaR-do” fragments, respectively. The 3′ end of the liaR-up fragment as well as the 5′ end of the liaR-do fragment extended into the gene to be deleted in a way that all expression signals of genes up- and downstream of the targeted gene remained intact. Extensions of ∼25 nucleotides were added to the 5′ ends of the up-reverse and do-forward primers that were complementary (opposite strand and inverted sequence) to the 5′ and 3′ ends, respectively, of the amplified spectinomycin resistance cassette. Portions (100 ng) of the liaR-up and liaR-do fragments and a 3-fold molar excess of the spectinomycin resistance cassette were used together with the specific up-forward and do-reverse primers at standard concentrations in a second PCR. The PCR product was directly used to transform S. pneumoniae, resulting in strain TMSP008.
RH253 was constructed by transforming RH237 with a Janus cassette targeting liaR. The upstream region of liaR (∼1,000 bp) was amplified with the primers Rr03F and Rr03Rkan and fused together with a Janus cassette in a subsequent PCR using the primers Rr03F and RpsL41R. The downstream region of liaR was amplified using the primers Rr03rpsl and Rr03R and fused together with the upstream region and the Janus cassette in a final PCR employing the primers Rr03F and Rr03R. Both liaR mutants behaved similarly with regard to phenotypes and target gene expression (data not shown).
To construct a spr0810 deletion mutant, 1,000 bp upstream and downstream of spr0810 was amplified using the primer pairs spr0810F/spr0810kanR and spr0810rpslF/spr0810R, respectively, and fused to the 5′ and 3′ends of a Janus cassette. Subsequent transformation of this DNA fragment into RH237 resulted in RH269. Similarly, a pcpC-targeting Janus cassette was amplified using the primer pairs pcpCF/pcpCkanR and pcpCrpslF/pcpCR and transformed into RH237, resulting in SPH-4. In order to construct the strain RH259, carrying a luciferase reporter gene immediately downstream of liaR, the plasmid pR424 (4) was digested with BamHI and HindIII. Next, ∼400 bp of the 3′ region of liaR was amplified with the primers Rr03luc1 and Rr03luc2. The PCR fragment was subsequently cloned into the linearized vector using the In-Fusion Dry-Down PCR cloning kit (Clontech) according to the manufacturer's protocol, giving rise to pVEL5. This plasmid was transformed into RH421 (7), resulting in RH259.
To construct pVEL6 we utilized a method described by Geiser et al. (10). The primers spr0810pR424F and spr0810pR424R, carrying at their 5′ ends sequences homologous to a region immediately upstream of the luc reporter gene of the pR424 plasmid, were used to amplify a 487-bp region starting 184 bp upstream of spr0810 and ending at its stop codon. In order to incorporate the 487-bp sequence upstream of the luc reporter gene in pR424, the PCR product was purified and utilized as a primer (200 ng) for a second PCR with pR424 (100 ng) as a template. To get rid of methylated template plasmid, 20 U DpnI (New England BioLabs) was added to the reaction at the end of the thermal cycling. The new plasmid pVEL6, harboring the gene encoding spr0810 under the control of its own promoter upstream of the luc gene, was incorporated into the genome of the RH421 strain by insertion-duplication mutagenesis, giving rise to the RH270 strain. RH271 was constructed by transforming RH270 with a comM-targeting Janus cassette amplified from RH420 with the primers ComMF and ComMR. The cassette was subsequently removed by transforming RH271 with a fragment targeting the Janus cassette, amplified from RH237 with the same primers, resulting in RH272.
To delete liaR in this background, a Janus cassette targeting liaR was first amplified from RH253 using the primers Rr03F and Rr03R. This PCR product was transformed into RH272, resulting in RH273. Similarly, this Janus cassette was transformed into RH4, resulting in RH276.
In order to replace cbpD with a point-mutated (C75→A) version, cbpD was first deleted in RH272 by transforming the strain with a cbpD-targeting Janus cassette. This cassette was amplified from RH422 (7) using the primers CbpD-1098F and CbpDR. The resulting strain, RH283, was subsequently transformed with a PCR cassette that replaced the Janus insertion with a cbpD allele encoding the C75àA mutation. First, a fragment starting ∼1,100 bp upstream of cbpD and ending just downstream of the active-site cysteine was amplified using the primers CbpD-1098F and CbpDC75AR. The rest of cbpD, including ∼1,000 bp of the cbpD downstream region, was amplified using the primers CbpDC75AF and CbpDR. The two fragments were then fused in a separate PCR using the primers CbpD-1098F and CbpDR. This fragment was transformed into RH283, resulting in RH284.
To construct RH291, RH421 (7) was first transformed with a PCR cassette replacing lytC with a Tetr resistance gene, resulting in RH287. The Tetr cassette was amplified from RH16 (7) using the primers LytCF and LytCR. RH287 was next transformed with the comM-targeting Janus cassette described above, giving rise to RH288. Subsequent removal of the Janus cassette by transformation with the PCR fragment described for RH237 resulted in RH289. This mutant was subsequently transformed with pVEL6 in order to introduce the luciferase reporter gene fusion of spr0810, resulting in RH290. Finally, a lytA-targeting Janus cassette was amplified from RH11 (7) using the primers LytAF and LytAR and transformed into RH290, resulting in RH291.
RH294 was constructed as follows. First, an ∼400-bp internal portion of spr1332 was amplified using the primers spr1332lucF and spr1332lucR. The fragment was cloned into pR424 linearized with BamHI and HindIII using an In-Fusion kit (Clontech) following the protocol supplied by the manufacturer. The resulting vector, pR1332, was transformed into R704 (17), resulting in RH229. This strain was transformed with the comM-targeting Janus cassette described above, giving rise to RH294. To construct SPH-5, the Janus cassette in RH269 was removed from the spr0810 locus. The upstream and downstream regions (∼1,000 bp) of spr0810 were amplified with the primer pairs spr0810F/spr0810delR and spr0810delF/spr0810R, respectively. The fragments were subsequently fused in a PCR using the primers spr0810F and spr0810R. The resulting DNA fragment was transformed into RH269, resulting in removal of the Janus cassette. Subsequently, the pcpC gene was replaced with the Janus cassette described above in order to obtain a spr0810 pcpC comM mutant (SPH-6).
Ectopic integration of PliaF-lacZ fusions were constructed based on the vector pPPP2. This vector carries a tetracycline resistance cassette for selection in S. pneumoniae and integrates into the bgaA locus. Promoter fragments were amplified using the primers TM1044, TM1276, TM1278, TM1279, and TM1339, thereby introducing EcoRI and BamHI sites. Standard cloning techniques were applied (39). The resulting plasmids pBG0202, pBG0204, pBG0205, and pBG0206 were used to transform S. pneumoniae R6 with tetracycline selection (3 μg/ml), resulting in strains TMSP011, TMSP013, TMSP014, and TMSP015.
Luciferase reporter assays.
Mutants harboring the luc gene fused to spr0810 or liaR were grown in 280 μl C medium mixed with 20 μl of a 10 mM solution of d-luciferin from Photinus pyralis in 96-well Corning NBS plates. The cultures were incubated at 37°C in a FluoStar Optima plate-reader (BMG Labtech), and induced to competence by the addition of 250 ng/ml CSP at an OD492 of ∼0.200. Uninduced cultures were run in parallel as controls. Growth and luminescence were measured at 3-min intervals.
β-Galactosidase assays.
Quantification of β-galactosidase release from competence-induced cells and promoter induction assays were performed as described previously (7, 32, 41) based on the protocol of Miller (34).
Preparation of total RNA for DNA microarray analysis and Northern blotting.
TMSP008 was grown anaerobically at 37°C in THB medium to mid-log phase. The culture was split and induced with bacitracin (10 μg/ml final concentration), with one sample remaining as the uninduced control. After 10 min of induction, 30 ml of each sample was mixed with 15 ml cold killing buffer (20 mM Tris-HCl, pH 7.0, 5 mM MgCl2, 20 mM NaN3), harvested by centrifugation, and frozen in liquid nitrogen. For cell disruption, the pellet was resuspended in 200 μl killing buffer, immediately dropped into the Teflon vessel (filled and precooled with liquid nitrogen), and then disrupted with a Mikro-Dismembrator U (Sartorius). The resulting cell powder was resuspended in 3 ml of lysis solution (4 M guanidine-thiocyanate, 0.025 M Na-acetate, pH 5.2, 0.5% N-lauroylsarcosinate), and the RNA was extracted twice by phenol-chloroform/isoamylalcohol (25:24:1) followed by chloroform/isoamylalcohol (24:1) extraction and ethanol precipitation. RNA samples were DNase treated with the RNase-free DNase kit (Qiagen) according to the manufacturer's instructions and purified using RNeasy mini columns (Qiagen).
Probe preparation and Northern blot analysis.
An internal fragment of liaR (∼500-nucleotide length) was amplified by PCR using the primers TM1118 and TM1199 (see Table S1 in the supplemental material). The PCR fragments were purified by using the Qiagen PCR purification kit, and 1 μg of each fragment was labeled with digoxigenin (DIG) by in vitro transcription using the DIG RNA labeling mix (Roche) and the T7 RNA polymerase (Roche) according to manufacturer's protocol.
For Northern blot analysis, 5 μg or 10 μg of total RNA was denatured and loaded on a formaldehyde agarose gel. After electrophoresis, the RNA was transferred to a nylon membrane (Roche) in a downward transfer using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0) as the transfer buffer. The RNA was cross-linked by exposing the damp membrane to UV light. The blot was prehybridized at 68°C for 1 h with prehybridization solution (0.2% sodium dodecyl sulfate [SDS], 0.1% N-lauroylsarcosinate, 5× SSC, 50% formamide, 2% blocking reagent), and labeled probe was added to the hybridization tube. Hybridization was performed overnight at 68°C. On the next day, the membrane was washed twice with low-stringency buffer (2× SSC, 0.1% SDS) at room temperature for 5 min, followed by two high-stringency washes (0.1× SSC, 0.1% SDS) at 68°C for 15 min. The DIG nucleic acid detection kit (Roche) was used for transcript detection. The blot was removed from the hybridization tube and placed in a box with 1× buffer 1 (10× buffer 1 is 1 M maleinic acid, 1.5 M NaCl, 0.3% Tween 20, pH 7.5) for 5 min at room temperature. The membrane was preincubated with buffer 2 (10% 10× buffer 1, 1% blocking reagent) for 30 min, treated with the antibody against DIG conjugated with alkaline phosphatase (AP) (Roche) for 30 min, and washed three times with 1× buffer 1 for 10 min. The blot was wrapped in plastic wrap, treated with the AP substrate CDP-Star (Roche) at a dilution of 1:200, and analyzed using a LumiImager (PeqLab).
The S. pneumoniae R6/TIGR4-Chip.
The R6/TIGR4-Chip is a combined S. pneumoniae R6/TIGR4 oligoarray set, designed by the Department of Microbiology, University of Kaiserslautern, and produced in cooperation with the company OPERON (Huntsville, AL). The oligonucleotides are 70-mer sense oligonucleotides and have a 5′-amino-C6 linker. The oligoarray chip consists of 2,964 oligonucleotides in total: 2,347 oligonucleotides covering genes and unique intergenic regions which are longer than 200 bp of S. pneumoniae R6, 488 oligonucleotides covering genes and unique intergenic regions which are longer than 200 bp of TIGR4, 44 oligonucleotides covering additional sequences (repetitive elements, RNAs [tRNAs, rRNAs, RNaseP, small cytoplasmic RNA, small stable RNA]), and 1 oligonucleotide for the pbp2x variant of the penicillin-resistant isolate S. pneumoniae 2349, a representative of the clone Spain23F-1 (2349.2x). The remaining 84 oligonucleotides represent controls: 12 randomized negative controls (HumV3con_1), 4 eukaryotic control genes, 10 positive controls, and 4 stringency controls (4 oligonucleotides with a 100%, a 90%, an 80%, and a 70% match to the corresponding gene), 32 tracking oligonucleotides (opHsV04NC000001) representing randomly generated 30-mers (tracking oligonucleotides are randomly positioned in the 384-well [4 in each]), and 10 alien controls according to the Stratagene SpotReport alien oligoarray validation system (internal controls for the whole procedure [mRNA-cDNA hybridization]).
There were four types of oligonucleotides: (i) genes that were identical in R6 and TIGR4 (homology >95%); (ii) genes that occurred in only one of the two strains; (iii) variable genes, with one oligonucleotide covering the region conserved in both strains; and (iv) genes which varied over the whole length (on the DNA level approximately 18 to 30%): strain R6 was a reference strain. Altogether, 2,038 oligonucleotides represented R6/TIGR4 genes and 309 oligonucleotides were TIGR4 specific; 328 oligonucleotides represented intergenic regions of R6, and 160 represented those of TIGR4.
Microarray.
Each oligonucleotide was spotted in duplicate onto a Schott Nexterion (Jena, Germany) slide E using the SpotArray TM24 Microarray Spotting System (PerkinElmer, Waltham, MA). Then, 30 μg of purified mRNA was synthesized from random hexamers into labeled cDNA by using the LabelStar array kit (Qiagen). The probes were then resuspended in 50 μl hybridization solution (Nexterion) plus 50 μl formamide and heated at 95°C for 5 min prior to hybridization.
Pretreatment, hybridization, and slide washing.
Hybridizations were performed using a Tecan HS400 hybridization station. Prehybridization was carried out using the following steps: a 30-s wash with 0.5% SDS at 25°C; a 30-s rinse with H2O at 25°C; and a 30-min incubation with prehybridization solution (4× SSC, 0.1% SDS, 0.1 mg/ml bovine serum albumin) at 42°C. The probe was then ready for injection. Hybridization was performed at 40°C with a medium agitation frequency for 16 h. Slide washing consisted of the following steps, all of which were performed at 30°C: 1 min with 2× SSC and 0.1% SDS, 1 min with 1× SSC, and 1 min with 0.1× SSC. The slides were finally dried under nitrogen (2.7 × 105 Pa) for 3 min.
Experimental data and replicates.
For each strain to be analyzed, two independently grown cultures were used. Each mRNA preparation was divided into two parts, and each part was labeled with either Cy3 or Cy5 in order to eliminate any labeling bias during the following analyses. Since the oligonucleotides on the microarray were spotted in duplicate, a final number of four spots per experiment were analyzed. The chips were scanned with a resolution of 10 μm using the ScanArray4000 microarray analysis system. The hybridization spots were analyzed using the EasyScan method in the ScanArrayExpress software, version 4.0. A noise-to-background ratio of three was used as the cutoff. The microarray data were normalized using the LOWESS fit, and the resulting ratios were analyzed using Student's t test, available at the Nano+Bio-Center of the University of Kaiserslautern (http://nbc3.biologie.uni-kl.de/). Only genes that had reproducible changes in the transcript amount of greater than a 2-fold threshold were considered further. Original raw data can be found in the supplemental material or at http://microbial-stress.iab.kit.edu/87.php.
Comparative genomics and regulon predictions.
To analyze the LiaR binding site in sequenced S. pneumoniae genomes, we used the Virtual Footprint algorithm, embedded in the Prodoric database at http://prodoric.tu-bs.de/vfp (36). To calibrate our LiaR-specific position weight matrix (PWM) and determine the parameter settings that enable us to discriminate false-positive hits from true target loci, we first screened the R6 genome. Based on the known positions of LiaR binding sites upstream of their target genes in three species, we restricted our search by applying the following cutoff criterion to filter out false-positive hits: repeats were considered potential LiaR binding sites only when they were located in noncoding regions upstream of genes with a maximum distance of 200 bp. Applying this criterion, we subsequently calibrated the sensitivity parameters, based on the known binding sites in S. pneumoniae R6, to a sensitivity of 0.93 (no threshold) and a core sensitivity of 0.90, with nonoccurrence penalty ON. The search window was limited to DNA regions 150 bp upstream of genes, and both “ignore match orientation” and “remove redundant” options were set ON (see http://prodoric.tu-bs.de/vfp/vfp_help.php#pwm for an explanation of these parameters).
RESULTS
Identification of the LiaR regulon by genome-wide transcriptional profiling.
Bacitracin has been shown to be among the strongest inducers of LiaR-dependent gene expression in B. subtilis, L. lactis, and S. aureus (9, 29, 31). In all three cases, expression of the operon encoding the LiaRS two-component system is positively autoregulated. An initial test verified that the expression of the liaFSR operon of S. pneumoniae is also induced by bacitracin in a LiaR-dependent manner (data not shown). To identify LiaR-dependent genes (i.e., the LiaR regulon) in S. pneumoniae, we performed a DNA microarray analysis to compare the global gene expression profile of the R6 wild-type strain and the isogenic liaR mutant in the presence of sublethal amounts of bacitracin. For this purpose, both strains were grown to mid-logarithmic growth phase and induced with sublethal amounts of bacitracin (final concentration: 10 μg/ml) for 10 min. Cells were harvested and snap-frozen in liquid nitrogen. Subsequently, total RNA was prepared, reverse transcribed, and hybridized to a S. pneumoniae R6/TIGR4 biochip, based on the primer set designed by Eurofins MWG GmbH (Ebersbach, Germany; see Materials and Methods for details). A total of 18 genes, organized in six transcriptional units, were found to be upregulated more than 2-fold in the wild-type strain but not in the corresponding liaR mutant (Table 2). This set of genes includes the monocistronic spr0810 gene, which encodes a protein that belongs to the phage-shock protein C (PspC) superfamily. Another member of this superfamily (Llmg2163) was reported to be strongly upregulated by the corresponding two-component system (CesSR) in L. lactis (29).
TABLE 2.
Identification of the LiaR regulon of S. pneumoniae by DNA microarray analysis
| Locus tag | Gene | (Putative) functiona | Fold changesb |
|---|---|---|---|
| spr0173 | arsC | Arsenate reductase, glutaredoxin family; Spx-like | 2.5 |
| spr0174 | Hypothetical protein | 2.27 | |
| spr0342 | liaF | Conserved membrane protein, putative regulator of LiaSR activity | 2.7 |
| spr0343 | liaS | Histidine kinase, homolog of B. subtilis LiaS | 2.86 |
| spr0344 | liaR | Response regulator, homolog of B. subtilis LiaR | 11.1 |
| spr0345 | alkD′ | DNA alkylation repair enzyme, truncated | 7.7 |
| spr0346 | ′alkD′ | DNA alkylation repair enzyme, truncated | 4.0 |
| spr0347 | ′alkD | DNA alkylation repair enzyme, truncated | 5.56 |
| spr0348 | Hypothetical protein | 2.38 | |
| spr0349 | cpbG′ | Choline binding protein G, truncated | 2.86 |
| spr0350 | ′cpbG | Choline binding protein G, truncated | 2.5 |
| spr0351 | pcpC | Choline binding protein | 2.7 |
| spr0453 | hrcA | Heat-inducible transcription repressor | 2.22 |
| spr0454 | grpE | Heat shock protein (activation of DnaK) | 2.27 |
| spr0810 | Displays low level of homology to PspC from E. coli | 6.0 | |
| spr1080 | Hypothetical protein | 2.22 | |
| spr1183 | ′ABC-NBD | Putative ABC transporter, ATP binding protein; possible multidrug efflux; truncated | 2.04 |
Gene annotations are taken from the MicrobesOnline database (http://www.microbesonline.org/).
Fold changes express the average induction value of four replicates (two biological and two technical replicates) in the wild-type strain relative to an isogenic liaR mutant, both in the presence of bacitracin (final concentration, 10 μg/ml).
Some target genes encode proteins with putative stress-related functions, such as the hrcA-grpE operon, which encodes a heat-shock regulator and a heat shock protein (18). Moreover, the spr0173/0174 operon encodes a putative transcriptional regulator with homology to Spx. Interestingly, Spx was also described as part of the CesSR regulon in L. lactis (45). Finally, spr1183 is a truncated monocistronic gene encoding the nucleotide-binding domain of a putative multidrug-efflux pump.
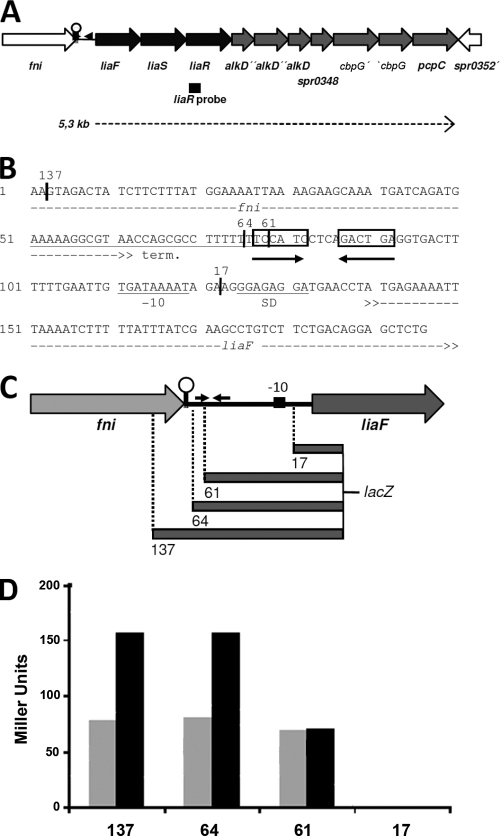
The largest and most strongly induced LiaR-dependent operon consists of 10 annotated genes (spr0342-spr0351; Fig. 1A) and includes the genes encoding the LiaFSR system (spr0343/44), thereby verifying the positive autoregulatory feedback loop observed for the homologous systems in B. subtilis, L. lactis, S. aureus, and Streptococcus mutans (21, 29, 43, 47). Induction of the liaFSR operon and its downstream genes by bacitracin has also recently been described for the S. pneumoniae strain D39 (28). The three truncated genes directly downstream of liaSR encode N-terminal, central, and C-terminal fragments of a potential DNA alkylation repair enzyme, homologous to AlkD. A comparative genomic analysis of this region, using the MicrobesOnline database (1), verified that these authentic frameshift mutations are present in all sequenced strains of S. pneumoniae (data not shown). The last three open reading frames of the operon encode the putative cell wall-anchored choline-binding proteins CbpG and PcpC (spr0351). The latter is a paralogue of CbpF. In strain R6, the cpbG gene is disrupted by an authentic frameshift mutation (spr0349/0350). Comparison with other pneumococcal strains reveals some heterogeneity, with some strains harboring an intact cbpG gene (i.e., CDC1873-00, SP11-BS70, SP14-BS69, ATCC 700669, and G54), while most other strains show various truncations (data not shown). The pcpC gene downstream of cbpG is found only in some pneumococcal strains, i.e., strains R6, D39, CDC1873-00, ATCC 700669, and Hungary19A-6.
FIG. 1.
Expression and promoter deletion analysis of the liaFSR operon. (A) Map of the liaFSR operon. The size of the liaR-specific transcript that was detected by Northern analysis is indicated by the arrow. (B) Sequence of the promoter region upstream of liaF (spr0342). (C) Schematic representation of the fragments cloned for the promoter deletion analysis of the liaF promoter. (D) β-Galactosidase activity of the fragments in the absence (light gray bars) and presence (black bars) of 10 μg/ml bacitracin. The numbers correspond to the fragments illustrated in panel C.
Verification of the LiaR binding site and autoregulation of the lia operon by promoter deletion analysis and Northern blots.
Because of its strong induction and known autoregulation, we next focused our attention on the lia operon (spr0342-0351). First, we analyzed the LiaR-dependent, bacitracin-inducible expression by Northern analysis. Northern analysis with a liaR-specific probe verified that the gene is expressed as part of a single ∼5- to 6-kb transcript, corresponding well with the theoretical size of 5.3 kb (Fig. 1A). Its expression was induced by bacitracin in the wild-type strain but not in an isogenic liaR mutant (data not shown).
Binding sites for LiaR-like response regulators have been identified in all organisms studied in this respect to date, and the potential LiaR/CesR binding sites were predicted bioinformatically (21, 29). To verify the location of the postulated LiaR binding site, we performed a promoter deletion analysis. Promoter fragments upstream of spr0342 were transcriptionally fused to lacZ, using the promoter-probe vector pPPP2 (14) (see Table S1 in the supplemental material for primer sequences and Fig. 1B and C for details). The resulting plasmids (pBG0202-0206) were stably integrated in the chromosome of strain R6, resulting in strains TMSP011 and TMSP013-015 (Table 1). β-Galactosidase assays, performed in the presence and absence of bacitracin, demonstrated that the fragment of strain TMSP015, containing the complete inverted repeat representing the postulated LiaR binding site, was indeed still sufficient to mediate LiaR-dependent induction, whereas strain TMSP013, which lacks most of the 5′ half of the repeat, showed only a constitutive basal promoter activity. In strain TMSP014, where the predicted core promoter is missing, basal promoter activity was lost as well (Fig. 1D). Therefore, we conclude that the previously postulated inverted repeat TcaaTCT—AGAcctA indeed represents the LiaR binding site of S. pneumoniae.
In silico identification of the LiaR binding site upstream of potential target genes.
Using this sequence motif but also the information from previous comparative genomics analyses of LiaR/CesR binding sites (21, 29, 43), we generated a LiaR/CesR consensus motif (Fig. 2). Two groups can be distinguished: the LiaR-like binding site upstream of liaIH-like operons in Bacillus and Listeria species (group 1; Fig. 2A) is a 7-2-7 inverted repeat, while the lactococcal/streptococcal binding motif represents a 6-4-6 motif (group 2; Fig. 2B). The core residues are identical in both groups, but the sequence conservation pattern shows a number of alterations.
FIG. 2.
LiaR binding sites and consensus sequence. (A) LiaR-like binding sites in Bacillus and Listeria species (group 1), as derived from the work of Jordan et al. (21). (B) LiaR/CesR-like binding sites in Lactococcus and Streptococcus species, based on the work of Martinez et al. (29) and this work. LiaR dependently expressed genes, as determined by DNA microarray analysis (Table 2), are highlighted in bold. Core residues (>80% conserved) and highly conserved residues (>60% conserved) are highlighted in black and gray, respectively. WebLogo (5) representations of the position weight matrices derived from these sequences are shown below. The inverted repeat is indicated by the two black arrows.
Subsequently, we screened the promoter regions of potential LiaR target genes identified by our transcriptome analysis for the presence of LiaR binding motifs. This was done both by manual screening and by using the Virtual Footprint algorithm implemented in the Prodoric database (36), as described previously (21). We were able to identify a potential LiaR box in all of the six promoter regions (Fig. 2B). While the motifs upstream of liaF, hrcA, spr0810, and spr1183 were well conserved, the motifs of the two remaining loci (arsC and spr1080) showed some mismatches in core residues of the inverted repeat (Fig. 2B). The resulting motif is very similar for L. lactis, S. mutans, and S. pneumoniae but differs from the signature described for the genera Bacillus and Listeria, as mentioned above (Fig. 2). We therefore combined all identified LiaR boxes shown in Fig. 2B to develop an optimized position weight matrix (PWM) for streptococcal species. This PWM was subsequently used for genome-wide predictions of the LiaR regulon in other strains of S. pneumoniae.
Prediction of the LiaR regulon in pathogenic strains of S. pneumoniae by comparative genomic profiling.
The laboratory strain R6 has undergone extensive degenerative evolution, resulting in the loss of about 10% of the genome compared to pathogenic wild-type isolates (44). Both the number of pseudogenes found in the LiaR regulon and the lack of any antibiotic resistance phenotype linked to the Lia system made us wonder if the integrity of the Lia regulon has also been affected by this domestication. To get an idea of the potential degeneration of the LiaR regulon, we used the PWM of the streptococcal LiaR box described above (Fig. 2B) to screen all completely sequenced strains of S. pneumoniae that are embedded in the Prodoric database. Using the criteria and parameters described in Materials and Methods, we were able to retrieve nine potential motifs, including five of seven potentially LiaR binding sites of S. pneumoniae R6 (that we identified by the transcriptome analysis; see Table 2). The predicted LiaR binding site upstream of the remaining two loci shows significant mismatches in highly conserved residues of the binding site and could therefore not be retrieved (Fig. 2B).
In addition to the already known target loci, only four additional motifs were identified. In three cases (spr1303, spr1320, and pgsA), the potential target genes were located inside operons. For the fourth hit, fucI (spr1964), the motif was located 146 bp upstream of the start codon. Taken together, there is a good overall correlation between PWM-based binding-site predictions and the results from the genome-wide expression profiling (Table 2).
Subsequently, the same PWM and criteria were used to screen the four clinical S. pneumoniae strains implemented in the Prodoric genome database: D39 (NCTC 7466), TIGR4 (ATCC BAA-334), CGSP14, and Hungary19A-6. The results for all strains were consistent with the data retrieved from the scan of the R6 genome. Between seven and nine potential binding motifs were retrieved, including all target loci identified for strain R6 (data not shown). No additional potential LiaR target loci were identified. The data therefore indicate that no potential LiaR regulon members were lost in the course of domestication of the laboratory strain R6.
To further validate the quality of our PWM, we also screened the genome of S. mutans UA159, again using the same criteria and parameters as above. We identified five potential LiaR binding sites, including motifs upstream of the known target loci spr0810 and liaFSR.
Interestingly, the two target genes hrcA and arsC/spx are shared between S. mutans and S. pneumoniae. These results further strengthen our regulon prediction and the quality of the LiaR-specific PWM graphically illustrated in Fig. 2.
LiaSR in S. pneumoniae strain R6 is activated by lipid II-interfering antibiotics but does not confer resistance against them.
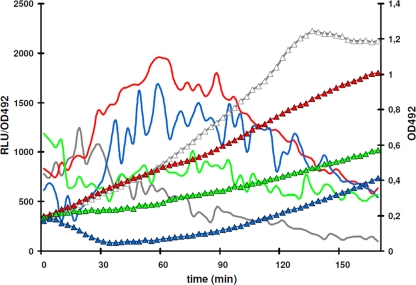
The best-studied inducers of envelope stress-sensing systems of the LiaSR/VraSR/CesSR type are antibiotics interfering with the lipid II cycle, with bacitracin, ramoplanin, vancomycin, and nisin constituting the strongest inducers identified so far (20). We tested various antibiotics inhibiting different steps in the bacterial cell wall synthesis cycle for their ability to stimulate expression of the liaSR genes in S. pneumoniae. This time, we inserted the luciferase reporter gene immediately behind liaR itself, resulting in the strain RH259. Bacitracin, nisin, and tunicamycin were found to induce luc expression in this strain (Fig. 3), whereas a luminescence signal above background level could not be detected with cycloserine (10 to 500 μg/ml), ampicillin (0.0025 to 0.5 μg/ml), or vancomycin (0.1 to 2 μg/ml). The latter finding was unexpected, since vancomycin has previously been reported to upregulate liaSR expression in S. pneumoniae (13). In addition, vancomycin induces the LiaFSR system of B. subtilis (32) and the S. aureus homolog VraSR (23).
FIG. 3.
Induction of the LiaSR system by antibiotics. A luciferase (luc) reporter gene was inserted immediately downstream of liaR (strain RH259). Bacterial growth curves are marked by triangles, whereas expression of liaR::luc transcriptional fusions is shown as unmarked curves. Luciferase activity is presented as relative luminescence units (RLU)/optical density (OD492). Gray curves, untreated cultures; red curves, cultures grown in 5 μg/ml bacitracin; blue curves, cultures grown in 20 μg/ml nisin; green curves, cultures grown in 20 μg/ml tunicamycin. Antibiotics were added at time zero.
For B. subtilis LiaSR, S. aureus VraSR, and L. lactis CesSR, it was shown that the two-component system mediates resistance to at least some of the inducing antibiotics. We therefore examined whether the same is true for S. pneumoniae LiaSR. We performed disk diffusion assays and killing curve experiments for all inducing antibiotics, basically as described previously (32), by comparing the sensitivity of the wild-type strain R6 and the isogenic liaR deletion mutant. No significant differences in the sensitivity toward any of the antibiotics tested were detected (data not shown). Interestingly, a weak increase of bacitracin susceptibility (2-fold) was recently observed for a liaFSR mutant of the ancestor strain D39, relative to the wild-type strain (28). The significance of this difference remains to be investigated.
LiaSR is activated during competence in the absence of the immunity gene comM.
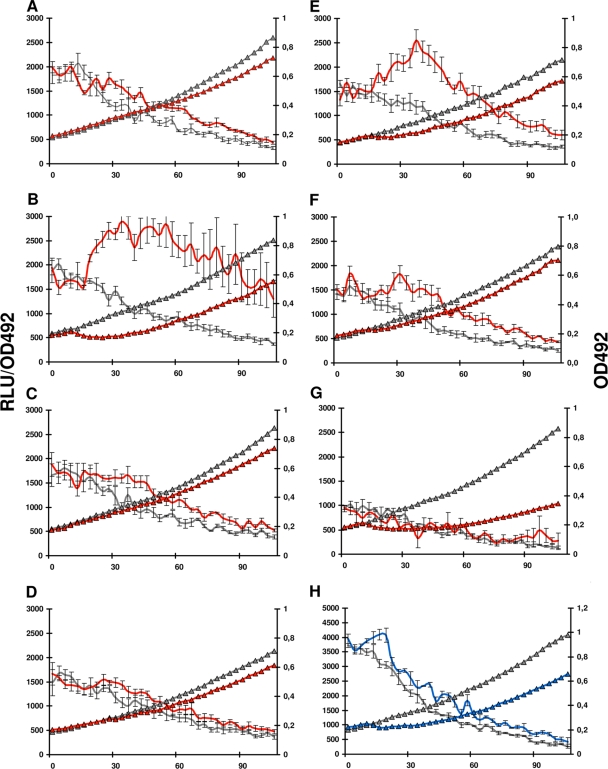
Because of the lack of an antibiotic resistance phenotype linked to LiaSR, we wondered if this system might be important in responding to other cell envelope perturbing conditions. In order to quantify LiaSR activity, the luciferase gene from Photinus pyralis (luc) was inserted immediately downstream of spr0810, one of the most strongly induced genes of the LiaR regulon, as demonstrated by our microarray analysis (Table 2). Expression of Luc in the resulting strain, RH270, was monitored by cultivating the bacteria in a 96-well plate in a Fluostar OPTIMA luminometer at 37°C as described previously (19). When competence was induced by the addition of 250 ng/ml of CSP to the culture (15), a slight increase in luc expression was observed (Fig. 4A). In order to determine whether ComM played a role in protecting against the observed cell envelope stress, comM was deleted using the Janus method (42). Interestingly, induction of competence in strain RH272 (ΔcomM spr0810::luc) resulted in a strong upregulation of spr0810 expression (Fig. 4B). To verify that this upregulation was due to activation of the LiaSR system, liaR was deleted in the RH272 strain, giving rise to RH273. In strain RH273, the overall spr0810 expression was reduced, and the observed increase in luminescence following treatment with CSP was abolished (Fig. 4G). This result verifies that LiaSR is indeed responsible for regulation of spr0810 expression and shows that monitoring spr0810 expression can be used as a proxy for monitoring activation of the LiaSR system.
FIG. 4.
Comparison of liaSR activation in competent and noncompetent pneumococci harboring various mutations in genes involved in the fratricide mechanism. The luc reporter was inserted immediately downstream of spr0810, a gene that is part of the LiaSR regulon. Bacterial growth curves are marked by triangles, whereas expression of spr0810::luc transcriptional fusions is shown as unmarked curves. Luciferase activity is presented as relative luminescence units (RLU)/optical density (OD492). Gray curves represent noncompetent cultures, while colored curves represent CSP-induced competent cultures. CSP was added at time zero. (A) RH270 (spr0810::luc), (B) RH272 (spr0810::luc comM), (C) RH283 (spr0810::luc comM cbpD), (D) RH284 (spr0810::luc comM cbpDC75A), (E) RH290 (spr0810::luc comM lytC), (F) RH291 (spr0810::luc comM lytC lytA), (G) RH273 (spr0810::luc comM liaR), (H) RH294 (spr1332::luc comM). Results presented in panels A to H are the means ± standard errors of results of three to five independent experiments.
To rule out the possibility that the increased luminescence (expressed in relative luminescence units [RLU]/OD492) observed for the comM mutant upon competence induction is an artifact resulting from the transient fratricide-induced drop in OD492, we created a comM mutant carrying the luc gene fused to a aldo-keto reductase (spr1332) that has no role in competence or cell envelope maintenance. Competence induction in this strain resulted in only a brief and very modest increase in RLU/OD492 (Fig. 4H), demonstrating that the strong LiaSR activation observed during competence in cells lacking the ComM immunity protein is real.
Upregulation of LiaSR is attributable to the activity of CbpD, LytA, and LytC.
To decide whether CbpD, the key player and trigger factor of the fratricide mechanism in S. pneumoniae, is responsible for activating LiaSR, the cbpD gene was deleted in strain RH272, giving rise to RH283. When RH283 was induced to competence, expression of the luc reporter gene returned to the level observed for strain RH270, demonstrating that CbpD activity is responsible for activating the LiaSR system in the absence of ComM (Fig. 4C). CbpD contains a proteolytic domain called CHAP (cysteine, histidine-dependent amidohydrolases/peptidases), which due to its strong homology to a number of phage lysins is believed to function as a murein hydrolase. In addition, CbpD contains two SH3b domains and four choline-binding repeats. While it has been firmly established that choline-binding repeats mediate noncovalent binding to cell wall teichoic (TA) and lipoteichoic acids (LTA) in S. pneumoniae (27), the exact function of the SH3b domains remains to be elucidated. The stress response elicited by CbpD is most probably generated by the enzymatic activity of the CHAP domain but could also be caused by the abrupt accumulation of this protein in the periplasm or the cell wall. To discriminate between these possibilities, we exchanged the wild-type cbpD gene with a mutated version in which the active site cysteine (C-75) had been replaced by an alanine residue. Induction of competence in this mutant strain, RH284 (ΔcomM spr0810::luc CbpDC75A), revealed that a functional CHAP domain is essential for the ability of CbpD to activate the LiaSR system (Fig. 4D).
It is well established that LytC is directly or indirectly activated by CbpD during fratricide and that this lysozyme is important for efficient lysis of target cells (7). Activation of the LiaSR system could therefore be attributable to LytC activity in addition to, or instead of, CbpD. We found that deletion of lytC (RH290) led to a change in the kinetics of the stress response. In the absence of LytC, the response reached approximately the same maximum level as in the RH272 strain, but expression of the Luc reporter was delayed and light production decayed more rapidly in the RH290 strain (Fig. 4E). The major autolysin in S. pneumoniae, LytA, has also been shown to be activated by CbpD and to contribute to the fratricide mechanism. It is therefore possible that the remaining stress response detected upon competence induction in strain RH290 was due to the N-acetylmuramoyl-L-alanine amidase (NAM-amidase) activity of LytA and only indirectly depended on the enzymatic activity of CbpD. To further elucidate these matters, the lytA gene was deleted in the RH290 strain, giving rise to RH291 (ΔcomM spr0810::luc ΔlytC ΔlytA). The magnitude of the stress response in this strain was only slightly stronger than in the cbpD deletion mutant (Fig. 4F). Together, our results show that cell wall damage caused by the concerted action of the murein hydrolases CbpD, LytA, and LytC activates the LiaSR system in cells lacking the fratricide immunity protein ComM.
Deletion of liaR significantly increases lysis during competence.
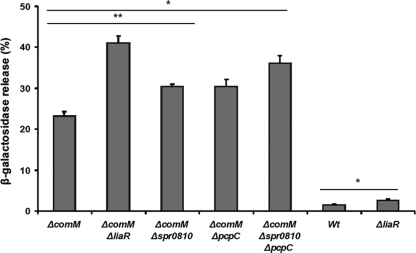
Considering that CbpD is able to elicit envelope stress that activates the LiaSR system, we wondered whether LiaSR in S. pneumoniae plays a part in protecting competent cells against their own lysins. To detect cell lysis caused by the fratricide mechanism, we took advantage of a lacZ reporter gene inserted downstream of the constitutive hirL promoter (41). The percent cell lysis was quantified by measuring β-galactosidase leakage to the supernatant relative to the total content of β-galactosidase in the culture (cells plus supernatant). First, liaR was deleted in the strain RH4 (hirL::lacZ), giving rise to RH276. The RH4 and RH276 strains were then induced to competence, and cell lysis was measured 30 min post CSP induction. Due to the presence of a functional comM gene, the RH4 strain (designated WT in Fig. 5) underwent very limited lysis. Interestingly, the liaR mutant RH276 exhibited a doubling of lysis compared to the level for RH4, but the level of lysis remained low (Fig. 5). This indicates that ComM in itself is sufficient to prevent most cells from lysing, but that some cells succumb to competence-induced stress in the absence of a functional LiaSR system. Next, we deleted liaR in a strain (RH237) lacking ComM. The resulting mutant strain, RH253, turned out to be considerably more prone to lysis during competence than the parental strain. When the RH253 strain was assayed as outlined above, ∼40% of the cell population lysed within 30 min. In contrast, only ∼20% of the RH237 cells underwent lysis when they were assayed by the same procedure (Fig. 5). Together, these results show that without a functional LiaSR system, a doubling in cell lysis takes place in both ComM-proficient and -deficient cells.
FIG. 5.
Competence-induced cell lysis in various pneumococcal deletion mutants quantified by means of β-galactosidase release. All strains contain a hirL::lacZ fusion conferring constitutive production of an intracellular β-galactosidase reporter protein. The level of β-galactosidase activity (in Miller units) present in the cell-free supernatants is given as a percentage of the total activity present in supernatants plus intact cells. Strains used were as follows: RH237 (ΔcomM), RH253 (ΔcomM ΔliaR), SPH-5 (ΔcomM Δspr0810), SPH-4 (ΔcomM ΔpcpC), SPH-6 (ΔcomM Δspr0810 ΔpcpC), RH4 (wild type), and RH276 (ΔliaR). Data represent the averages ± standard errors of results of at least three independent experiments. **, P ≤ 0.01; *, P ≤ 0.05.
We also investigated whether the LiaFSR system plays a role in protecting noncompetent target cells from competent attacker cells during cocultivation. These mixed-culture experiments, containing equal amounts of β-galactosidase-producing target cells and competent attacker cells, were carried out as described previously (41). Under the experimental conditions used, deletion of liaR in noncompetent target cells had no effect on their rate of survival (results not shown). This finding suggests that competent pneumococci experience an additional level or different kind of stress compared to noncompetent cells under attack and that a functional LiaSR system helps alleviate this stress.
Deletion of the spr0810 and pcpC genes.
Based on the microarray analysis, we tried to identify candidate genes mediating the observed LiaR-dependent protection against CbpD, LytA, and LytC. Among the LiaR-dependent gene products upregulated in the microarray screen, we decided to further investigate spr0810 and PcpC. As mentioned previously, spr0810 encodes a protein predicted to belong to the phage shock protein C superfamily. The phage shock protein (Psp) system was first identified in Escherichia coli (3), and has since then been studied extensively in this species and Yersinia enterocolitica (6). The Psp systems of E. coli and Y. enterocolitica have six proteins in common (PspABCD and PspFG) that are also present in many other Gram-negative bacteria. In these bacteria, the Psp system responds to extracytoplasmic stress and appears to play a role in maintaining cytoplasmic membrane integrity and/or the proton motive force (33). All but the spr0810 gene product, which shows a rather low level of homology to E. coli PspC, appear to be absent from the pneumococcal genome. However, since both E. coli PspC and pneumococcal spr0810 are small membrane proteins possessing one centrally located transmembrane helix, they might perform comparable functions. The choline-binding protein PcpC (spr0351) is closely related to CbpF (spr0337), which has been shown to be a negative regulator of LytC activity in S. pneumoniae (35). As shown above, inactivation of the LiaFSR system increases competence-induced cell lysis (Fig. 5). As this effect is most prominent in the absence of ComM, we decided to delete the spr0810 and pcpC genes in a ΔcomM background. We found that deletion of each of these genes led to a significant increase in competence-induced lysis, demonstrating that they contribute to alleviate stress caused by CbpD, LytA, and LytC. Damage caused by the fratricide mechanism was even less tolerated in a spr0810/pcpC double mutant. In this mutant, the level of cell lysis during competence reached ∼36%, compared to about ∼30% for each of the spr0810 and pcpC single mutants (Fig. 5). Taken together, our data clearly demonstrate an important role of LiaR-dependent gene expression as a second layer of defense, in addition to the primary immunity protein ComM, in counteracting fratricide-induced lysis.
DISCUSSION
All LiaFSR-like systems investigated so far are activated by agents or perturbations that affect the integrity of the bacterial cell envelope. The strongest known inducers are lipid II cycle inhibitors such as of bacitracin, nisin, vancomycin, and ramoplanin. Other antibiotics that interfere with cell wall biosynthesis appear to be effective only in some species, suggesting some differences in the specificity of their LiaFSR systems. The eponymous LiaFSR system of B. subtilis is also weakly induced by more unspecific perturbations of the cell envelope, such as alkaline shock and secretion stress (20). For S. aureus VraSR, the antibiotics d-cycloserine and tunicamycin, which inhibit the enzyme (MraY) catalyzing the synthesis of lipid I, and even β-lactams and reduction in the level of pbpB transcription have been found to induce the system (9, 23). Furthermore, suboptimal transcription of murF, an enzyme that adds the d-alanyl-d-alanine dipeptide to the UDP-linked MurNAc-tripeptide, results in increased transcription of the VraSR system in S. aureus (40).
A different kind of cell envelope stress has been reported to induce the CesSR system in L. lactis. Veiga et al. (45) found that treatment of the MG1363 strain of this species with lysozyme increased expression of its ces operon. In contrast, Martínez et al. (29), working with the same strain of L. lactis, found that lysozyme at the concentrations tested was unable to induce expression of a lacZ reporter gene under the control of a CesR-responsive promoter. In the present study, we clearly show that enzymes that degrade peptidoglycan can activate the LiaFSR cell envelope stress-sensing systems of S. pneumoniae. LytC is a lysozyme that cleaves the β-1-4-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine in the bacterial cell wall. Deletion of the gene encoding this lysozyme significantly altered the kinetics of the stress response in S. pneumoniae (Fig. 4). The response took longer to appear and decayed much faster than in the LytC-proficient parental strain used as a control. Further deletion of the gene encoding the NAM-amidase LytA reduced the LiaSR response to a level just above the detection limit of our system. LiaSR activation was totally dependent on the enzymatic activity of the CHAP domain of CbpD and its triggering of the glycan strand-cleaving activity of LytC or the NAM activity of LytA. The CHAP domain of CbpD is highly homologous to corresponding domains found in a number of phage lysins and bacterial murein hydrolases. Characterized members of the CHAP family has been found to act either as endopeptidases that cleave within murein stem peptides or as amidases that cleave the N-acetylmuramyl-l-Ala bond (2, 38). Thus, our findings indicate that both glycan strand-cleaving enzymes and enzymes that cleave amide/peptide bonds can elicit a stress response that activates the LiaSR system in S. pneumoniae.
Very little is known about the natural biological role of LytC. It is relatively highly expressed when pneumococci are grown in liquid culture under laboratory conditions, but mutants lacking a functional lytC gene display no growth defects or other strong phenotypes (8). However, Garcia et al. (8) showed that LytC mediates autolysis in the stationary phase when the pneumococcal cells are grown at 30°C, suggesting that LytC functions as an autolysin that becomes active at temperatures encountered by pneumococci colonizing the nasopharynx (8). Interestingly, it has also been found that LytC is activated by CbpD during fratricide (7). However, it has not been clear whether CbpD activates LytC through direct interaction or by an indirect mechanism. The fact that induction of the competent state in the CbpDC75A mutant did not trigger a LytC-provoked stress response demonstrates that a functional CHAP domain is required to activate LytC. Furthermore, since CbpD is able to trigger a small but significant stress response in the absence of LytC, the natural substrate of the CHAP protease is probably not LytC itself but the cell wall stem peptides. Based on these observations, our data strongly indicate that LytC is activated by cell wall damage inflicted by the enzymatic activity of CbpD.
Microarray analysis identified LiaR-controlled genes that might be involved in the observed LiaR-mediated protection against the murein hydrolases CbpD, LytA, and LytC. Three of the upregulated genes, hrcA, grpE, and spr0810, encode proteins that are involved in responses to various types of stress. HrcA regulates the expression of DnaK and GroEL by repressing the transcription of the operons encoding these molecular chaperones (18). DnaK and GroEL play a crucial role in protein folding, and is important for cell survival during heat shock and other types of environmental stress. The Psp system of Gram-negative bacteria responds to several types of stimuli, all of which have the potential to damage the cell envelope and dissipate the proton motive force. The only possible component of this system that has been identified in S. pneumoniae is the LiaR-controlled spr0810 gene. In Y. enterocolitica the product of the corresponding gene, PspC, functions both as a regulator of psp gene expression and as an effector that acts independently to support growth when the bacterium is subjected to extracytoplasmic stress (33). In contrast to the complex system regulated by HrcA, the Psp-like system in S. pneumoniae appears to consist of a single protein encoded on a monocistronic operon. We therefore decided to investigate whether deletion of the spr0810 gene in a comM mutant would further increase the fraction of cells undergoing CbpD-induced lysis during competence development. We found a modest but significant increase (∼7%), indicating that spr0810 contributes to alleviate cell envelope stress elicited by the fratricide mechanism.
The pcpC gene is located at the 3′-end of the liaFSR operon, which encodes 10 predicted genes. PcpC is a paralog of CbpF, a choline binding protein that is one of the most abundant proteins in the pneumococcal cell wall. It was recently reported by Molina et al. (35) that external addition of 1.2 μM CbpF abolishes LytC-induced autolysis at 30°C in vitro. Since PcpC is highly homologous to CbpF, we speculated that this protein functions as a modulator of murein hydrolase activity as well. We therefore deleted the pcpC gene in a strain lacking ComM and measured the fraction of cells that lysed during competence. Similar to the results obtained with pneumococcal cells deficient in Spr0810, cell lysis increased by about 7% in the pcpC mutant compared to that in the parental strain. Together, our results show that both Spr0810 and PcpC contribute to stress relief during competence.
Fratricide-mediated self-lysis increased from about 23% in cells lacking a functional comM immunity gene (RH237) to 41% in a comM/liaR double mutant (RH253) (Fig. 5). To determine whether the LiaR-controlled gene products Spr0810 and PcpC can fully account for the protection provided by the LiaSR system, we constructed a spr0810/pcpC double mutant designated SPH-6 (Δspr0810 ΔpcpC ΔcomM). The level of self-lysis in a culture of this mutant reached 36%, approximately the sum of the increases measured for each of the spr0810 and pcpC single mutants relative to their parental strain RH237. As the combined effect of deleting spr0810 and pcpC does not fully amount to the observed difference between the RH237 and RH253 strains, it is likely that additional LiaR-controlled gene products are involved in counteracting CbpD-, LytC-, and LytA-induced stress. Possible candidates are HrcA and GrpE, which are involved in regulating the activity of the chaperone DnaK. In a wild-type strain the protective effect of the LiaSR system is mostly masked by ComM, which functions as a specific immunity mechanism against CbpD-induced fratricide. Still, even in a strain with a functional comM gene, fratricide-induced lysis doubles from 1.3% to 2.6% when LiaR is deleted (Fig. 5).
In S. pneumoniae the LiaFSR system does not protect the cells against the lipid II-interacting antibiotics that it responds to. Instead, it provides protection against self-lysis in competent cells. This finding makes it tempting to speculate that the LiaFSR system in S. pneumoniae has diverged from the classical cell envelope stress response to mediate a second layer of resistance against fratricide-induced lysis. It remains to be investigated whether other target genes of the LiaFSR system of S. pneumoniae have additional protective functions against other aspects of cell envelope stress.
Supplementary Material
Acknowledgments
Work in the laboratory of Thorsten Mascher was supported by grants from the Deutsche Forschungsgemeinschaft (DFG grant MA2837/1-3) and the Fonds der Chemischen Industrie (FCI). The KIT Research Group 11-1 received financial support from the Concept for the Future of the Karlsruhe Institute of Technology within the framework of the German Excellence Initiative. Work performed in the laboratory of Leiv Sigve Håvarstein was supported by grants from the Research Council of Norway. Work in the laboratory of Regine Hakenbeck was supported by grants from the BMBF (grant No. 0313801 l), the Stiftung Rheinland-Pfalz für Innovation (15202-38 62 61/580), and the EU (Intafar LSHM-CT-2004-512138).
The technical assistance of Michele Memmer is greatly acknowledged.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alm, E. J., K. H. Huang, M. N. Price, R. P. Koche, K. Keller, I. L. Dubchak, and A. P. Arkin. 2005. The MicrobesOnline web site for comparative genomics. Genome Res. 15:1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 3.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chastanet, A., M. Prudhomme, J.-P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crooks, G. E., G. Hon, J.-M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 7.Eldholm, V., O. Johnsborg, K. Haugen, H. S. Ohnstad, and L. S. Havarstein. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155:2223-2234. [DOI] [PubMed] [Google Scholar]
- 8.Garcia, P., M. P. Gonzalez, E. Garcia, J. L. Garcia, and R. Lopez. 1999. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33:128-138. [DOI] [PubMed] [Google Scholar]
- 9.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiser, M., R. Cèbe, D. Drewello, and R. Schmitz. 2000. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 31:88-92. [DOI] [PubMed] [Google Scholar]
- 11.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 12.Guiral, S., T. J. Mitchell, B. Martin, and J.-P. Claverys. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U. S. A. 102:8710-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas, W., D. Kaushal, J. Sublett, C. Obert, and E. I. Tuomanen. 2005. Vancomycin stress response in a sensitive and a tolerant strain of Streptococcus pneumoniae. J. Bacteriol. 187:8205-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halfmann, A., R. Hakenbeck, and R. Brückner. 2007. A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268:217-224. [DOI] [PubMed] [Google Scholar]
- 15.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 17.Håvarstein, L. S., B. Martin, O. Johnsborg, C. Granadel, and J.-P. Claverys. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59:1297-1307. [DOI] [PubMed] [Google Scholar]
- 18.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 19.Johnsborg, O., V. Eldholm, M. L. Bjørnstad, and L. S. Håvarstein. 2008. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol. Microbiol. 69:245-253. [DOI] [PubMed] [Google Scholar]
- 20.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kausmally, L., O. Johnsborg, M. Lunde, E. Knutsen, and L. S. Havarstein. 2005. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 187:4338-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 25.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39:508-518. [DOI] [PubMed] [Google Scholar]
- 26.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez, R., and E. Garcia. 2004. Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol. Rev. 28:553-580. [DOI] [PubMed] [Google Scholar]
- 28.Majchrzykiewicz, J. A., O. P. Kuipers, and J. J. E. Bijlsma. 2010. Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob. Agents Chemother. 54:440-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez, B., A. L. Zomer, A. Rodríguez, J. Kok, and O. P. Kuipers. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473-486. [DOI] [PubMed] [Google Scholar]
- 30.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 31.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 32.Mascher, T., S. L. Zimmer, T.-A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxson, M. E., and A. J. Darwin. 2006. PspB and PspC of Yersinia enterocolitica are dual function proteins: regulators and effectors of the phage-shock-protein response. Mol. Microbiol. 59:1610-1623. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Molina, R., A. González, M. Stelter, I. Pérez-Dorado, R. Kahn, M. Morales, M. Moscoso, S. Campuzano, N. E. Campillo, S. Mobashery, J. L. García, P. García, and J. A. Hermoso. 2009. Crystal structure of CbpF, a bifunctional choline-binding protein and autolysis regulator from Streptococcus pneumoniae. EMBO Rep. 10:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munch, R., K. Hiller, A. Grote, M. Scheer, J. Klein, M. Schobert, and D. Jahn. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Mol. Microbiol. 21:4187-4189. [DOI] [PubMed] [Google Scholar]
- 37.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 38.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of γ-D,L-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230-234. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Sobral, R. G., A. E. Jones, S. G. Des Etages, T. J. Dougherty, R. M. Peitzsch, T. Gaasterland, A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2007. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189:2376-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinmoen, H., E. Knutsen, and L. S. Havarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. U. S. A. 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suntharalingam, P., M. D. Senadheera, R. W. Mair, C. M. Levesque, and D. G. Cvitkovitch. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 45.Veiga, P., C. Bulbarela-Sampieri, S. Furlan, A. Maisons, M.-P. Chapot-Chartier, M. Erkelenz, P. Mervelet, P. Noirot, D. Frees, O. P. Kuipers, J. Kok, A. Gruss, G. Buist, and S. Kulakauskas. 2007. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282:19342-19354. [DOI] [PubMed] [Google Scholar]
- 46.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 47.Yin, S., R. S. Daum, and S. Boyle-Vavra. 2006. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.