Abstract
Decorin-binding proteins B and A (DbpB and DbpA) are thought to play important roles in Borrelia burgdorferi pathogenesis by serving as adhesins for the extracellular matrix. It has been established that the expression of DbpBA is governed by the Rrp2-RpoN-RpoS regulatory pathway. However, the precise mechanism underlying the control of DbpBA expression has been unclear. In particular, it has been unknown whether RpoS influences DbpBA expression directly or indirectly (through an additional regulatory molecule[s]). Here, employing a wild-type B. burgdorferi strain and a dbpBA-deficient mutant, we analyzed the 5′ genetic elements of the dbpBA operon using deletion analysis, coupled with luciferase reporter assays, quantitative reverse transcription PCR, and immunoblot analyses. A minimal promoter, encompassed within 70 bp upstream of the ATG start codon of dbpBA, was identified and found to be necessary and sufficient to initiate dbpBA transcription. The minimal dbpBA promoter was responsive to environmental stimuli such as temperature, pH, and whole blood. Two in silico-identified inverted repeat elements were not involved in the response of dbpBA expression to in vitro stimulation by environmental factors. The expression of dbpBA from the minimal promoter was abolished when rpoS was inactivated. In addition, the targeted mutagenesis of a C at position −14 within the extended −10 region of dbpBA, which has been postulated to be strategic for EσS binding in Escherichia coli, abolished dbpBA expression in B. burgdorferi. These combined data suggest that the Rrp2-RpoN-RpoS pathway controls dbpBA expression by the direct binding of RpoS to an RpoS-dependent promoter. However, given that there remains a distinct difference between the expression of DbpBA and other genes under the direct control of RpoS (e.g., OspC), our findings do not preclude the existence of another layer of gene regulation that may contribute to the modulation of DbpBA expression via an as-yet unknown mechanism.
Borrelia burgdorferi, the etiological agent of Lyme disease, is maintained in nature through a complex enzootic life cycle, which involves an arthropod vector (Ixodes tick) and a variety of mammalian hosts (10, 50). After B. burgdorferi is transmitted to humans through tick bites, spirochetes can disseminate and spread hematogenously to various target sites, such as heart, joints, and other distant locations, causing a broad spectrum of clinical manifestations, including carditis, arthritis, and neuroborreliosis (42, 49). It has long been presumed that the interactions between surface molecules such as the adhesins of B. burgdorferi and specific molecules in the mammalian hosts are critical for the pathogen to disseminate to and colonize specific niches (14, 16, 17). In this regard, B. burgdorferi expresses several adhesins, including the decorin-binding proteins (Dbp) DbpA and DbpB (22, 23), BBK32 (40), P66 (BB0603) (9, 15), and Bgp (Borrelia-GAG-binding protein BB0588) (37), which are thought to bind to integrin and mammalian extracellular matrix (ECM) components such as fibronectin, decorin, glycosaminoglycans (GAGs), and type I collagen. In addition, B. burgdorferi also expresses OspA, which binds to TROSPA expressed in tick midguts (32, 34, 35).
Among the putative adhesins expressed by the Lyme disease spirochete, DbpB and DbpA are encoded in a bicistronic operon, dbpBA, on linear plasmid lp54 in B. burgdorferi (20, 23, 24). These two surface-exposed proteins were first reported in a study to identify B. burgdorferi molecules that bind decorin, a collagen-binding proteoglycan produced in the connective tissues. Using gel overlay assays, Guo et al. (23) observed two decorin-binding proteins in B. burgdorferi, DbpB and DbpA, of 19 and 20 kDa, respectively. These two proteins are 56% similar, and both proteins contain conserved lysine residues critical for decorin recognition (7, 22, 38). Due to the propensity for the recombinant versions of DbpB and DbpA to bind to decorin and GAGs, DbpB and DbpA continue to be implicated as being important to the colonization and dissemination of B. burgdorferi within mammalian hosts. Furthermore, Brown et al. (8) reported that the ability of B. burgdorferi to disseminate, survive, and cause disease was impaired in decorin-deficient mice. More recently, data from three independent groups have revealed that the deletion of either dbpB, dbpA, or both genes in B. burgdorferi resulted in a dramatic decrease in the infectivity of needle-inoculated mice, suggesting that both DbpA and DbpB proteins contribute to B. burgdorferi's infectivity (3, 46, 47, 54). However, Blevins et al. (3) reported that the deletion mutant lacking dbpBA still was able to infect naïve mice via tick bite (50% [1/2] and 67% [4/6] of mice became infected when challenged with 5 and 10 ticks containing the dbpBA mutant, respectively). These data suggest that although both DbpB and DbpA probably contribute to Borrelia virulence, their precise roles in B. burgdorferi pathogenesis and infectivity still are not fully understood.
Studies have shown that B. burgdorferi alters its expression of DbpBA in response to various environmental stimuli, such as temperature, pH, cell density, and dissolved CO2 and O2, changes that B. burgdorferi ostensibly encounters during its transition between the tick vector and mammalian hosts (28, 44, 45, 52, 56). Moreover, work from our laboratory and others has demonstrated that these signals influence dbpBA expression in B. burgdorferi, likely via the Rrp2-RpoN-RpoS regulatory pathway (5, 6, 11-13, 18, 27, 31, 33, 48, 58). In this pathway, under elevated temperature (e.g., 37°C) or low pH (pH 6.8), the putative response regulator Rrp2, along with the alternative sigma factor RpoN (σN), directly activates the expression of the central alternative sigma factor RpoS (σS) which, in turn, regulates the expression of a number of B. burgdorferi virulence-associated lipoproteins, such as DbpBA, OspC, and the Mlp family. There now are compelling data that the Rrp2-RpoN-RpoS pathway regulates the expression of OspC by the direct interaction of RpoS with the RpoS-dependent ospC promoter (1, 18, 59). However, it has remained unclear how the dbpBA operon is controlled by RpoS. This information gap is of particular interest in view of the fact that dbpA exhibits an expression pattern somewhat different from that of ospC. Although both ospC and dbpA are upregulated by a shift to elevated temperatures, only OspC expression is highly induced by the process of tick feeding (21, 26, 36). DbpA (and presumably DbpB) expression is absent in both flat and fed ticks (25), suggesting that the transient elevation at ambient temperature during tick feeding does not overcome a suppression mechanism involved in tempering dbpBA expression. Furthermore, DbpA likely is persistently expressed during the course of mammalian infection, whereas OspC expression diminishes in late phase or persistent infection in mammalian hosts (29, 30). In addition, relative to gene expression in B. burgdorferi cultivated under certain in vitro growth conditions, the transcription of both rpoS and ospC were upregulated when B. burgdorferi was cultured within intraperitoneal dialysis membrane chambers (DMCs), whereas dbpA transcription was downregulated (13). Nonetheless, the level of DbpA protein, together with the protein levels of RpoS and OspC, was increased when B. burgdorferi was cultured within DMCs (relative to in vitro-cultured Borrelia). This observation suggests that a suppression mechanism or posttranscriptional regulatory mechanism (probably involving signals from the mammalian host) influences dbpBA expression. Finally, Yang et al. (56) showed that ospC and dbpA respond differently to pH changes when B. burgdorferi is cultured in Barbour-Stoenner-Kelly (BSK) medium. These combined observations thus have prompted the hypothesis that, in addition to RpoS, some other regulatory molecule(s) is involved in the modulation of DbpBA expression. As a first step toward assessing the contribution of RpoS-mediated control over dbpBA expression, herein we focused on examining putative cis elements in the 5′ upstream regulatory region(s) of dbpBA, with emphasis on assessing whether dbpBA contains a minimal RpoS-dependent promoter. The use of luciferase reporter constructs (luc fused to the putative dbpBA promoter) within the native Borrelia background facilitated these studies.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All strains and plasmids used in this study are described in Table 1. Barbour-Stoenner-Kelly II (BSK-II) medium or BSK-H medium (Sigma Chemical Co., St. Louis, MO) (39) supplemented with 6% rabbit serum (Pel-Freez Biologicals, Rogers, AR) was used routinely to grow B. burgdorferi. Spirochetes were enumerated using dark-field microscopy. To determine the effects of temperature and pH on gene expression, spirochetes were cultured under various environmental conditions, such as at 23 or 37°C or in BSK medium at pH 7.6 or 6.8, as described previously (33, 56). To determine the influence of blood supplementation on gene expression, various strains were inoculated into BSK-H medium at 1 × 103 spirochetes/ml. The culture was split into two groups when it reached a density of ∼1 × 106 to 5 × 106 cells/ml. To the test group, 6% fresh heparinized rabbit blood (from which the buffy coat was removed) was added, whereas 6% heparin-supplemented BSK-II (∼14 USP/ml), instead of heparinized rabbit blood, was added to the control culture. Cultures were grown at 37°C for 48 h, with periodic mixing to prevent the settling of the red blood cells (RBCs). Cells from both groups were harvested for luciferase assays when the cell density reached late log phase (approximately 5 × 107 bacteria/ml). Immediately prior to harvest, 6% whole blood was added to the control culture. Escherichia coli strain TOP10 (Invitrogen, Carlsbad, CA) was used as the cloning host. When appropriate, kanamycin (Kan) or streptomycin (Strep) was added to BSK medium at a final concentration of 160 or 150 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| B. burgdorferi | ||
| 297 | Infectious, low-passage B. burgdorferi | 27 |
| BbAH206 | 297, rpoS mutant | 27 |
| BbKH500 | 297, dbpBA mutant, Kanr | 3 |
| OY20 | 297 transformed with pOY63, Strr | This study |
| OY16 | 297 transformed with pOY64, Strr | This study |
| OY17 | 297 transformed with pOY66, Strr | This study |
| OY21 | 297 transformed with pOY67, Strr | This study |
| OY22 | 297 transformed with pOY68, Strr | This study |
| OY23 | 297 transformed with pOY69, Strr | This study |
| OY24 | 297 transformed with pOY70, Strr | This study |
| OY25 | 297 transformed with pOY71, Strr | This study |
| OY26 | 297 transformed with pOY72, Strr | This study |
| OY27 | 297 transformed with pJSB165, Strr | This study |
| OY28 | 297 transformed with pJSB175, Strr | This study |
| OY45 | BbKH500 transformed with pOY94, Kanr, Strr | This study |
| OY48 | BbKH500 transformed with pOY98, Kanr, Strr | This study |
| OY49 | 297 transformed with pOY109, Strr | This study |
| OY50 | BbKH500 transformed with pOY107, Kanr, Strr | This study |
| OY51 | BbKH500 transformed with pOY108, Kanr, Strr | This study |
| OY52 | BbAH206 transformed with pOY69, Strr | This study |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pJD7 | B. burgdorferi/E. coli shuttle vector with PflgB-aadA; Spcr, Strr | 4 |
| pJD44 | B. burgdorferi/E. coli shuttle vector with aph[3′]-IIIa; Kanr | 41 |
| pJD48 | pJD44::promoterless lucBb+, Kanr | 4 |
| pJD54 | B. burgdorferi/E. coli shuttle vector with PflgB-aadA; Spcr, Strr | 4, 41 |
| pOY63 | promoterless lucBb+ from pJD48 cloned into pJD54 at BglII and HindIII; Spcr, Strr | This study |
| pOY64 | pOY63::WT PdbpBA (PCR product from primers ZM61 and ZM68); Spcr, Strr | This study |
| pOY66 | pOY63::ΔIR1 PdbpBA (PCR product from primers ZM61 and ZM54); Spcr, Strr | This study |
| pOY67 | pOY63::ΔIR1/2 PdbpBA (PCR product from primers ZM61 and ZM55); Spcr, Strr | This study |
| pOY68 | pOY63::PCR product from primers ZM61 and ZM56; Spcr, Strr | This study |
| pOY69 | pOY63::Min PdbpBA (PCR product from primers ZM61 and ZM57); Spcr, Strr | This study |
| pOY70 | pOY63::Δ−35 PdbpBA (PCR product from primers ZM61 and ZM58); Spcr, Strr | This study |
| pOY71 | pOY63::Δ−35/−10 PdbpBA (PCR product from primers ZM61 and ZM59); Spcr, Strr | This study |
| pOY72 | pOY63::PCR product from primers ZM61 and ZM60; Spcr, Strr | This study |
| pOY94 | pOY63::Min PdbpBA-dbpBA (PCR product from primers ZM102 and ZM104.3); Spcr, Strr | This study |
| pOY98 | pOY63::Δ−35 PdbpBA-dbpBA (PCR product from primers ZM103 and ZM104.4); Spcr, Strr | This study |
| pOY107 | pJD7::WT PdbpBA-dbpBA (PCR product from primers ZM109F and ZM110R); Spcr, Strr | This study |
| pOY108 | pJD7::ΔIR1 PdbpBA-dbpBA (PCR product from primers ZM110F and ZM110R); Spcr, Strr | This study |
| pOY109 | pOY63::Min PdbpBA with −14 C/A (PCR product from primers ZM61 and ZM57.2); Spcr, Strr | This study |
| pJSB165 | pJD7::divergently oriented PospC-Bbluc+; Spcr, Strr | 4 |
| pJSB175 | pJD7::divergently oriented PflaB-Bbluc+; Spcr, Strr | 4 |
Generation of luciferase reporter vectors and DbpBA expression complementation plasmids.
The promoterless luciferase open reading frame (the lucBb+ ORF; referred to as luc hereafter) was excised from pJD48 (4) by digestion with BglII and HindIII and ligated into pJD54 (19, 41, 51) that was digested with the same enzymes; this generated pOY63. The luc gene was codon optimized for the optimal expression of luciferase in B. burgdorferi (4). DNA fragments containing various versions of the proposed dbpBA promoter (PdbpBA) were generated by PCR using Pfx50 DNA polymerase (Invitrogen). The primers for each insert are listed in Table 2. These DNA inserts then were digested using appropriate restriction enzymes (NcoI, BglII, and NdeI) and fused to the luc gene in pOY63 that was linearized by digestion with the same restriction enzymes. In particular, all inserts were amplified using different forward primers and the same reverse primer, ZM61 (CATATGCTTTTCCCGTGGCTTCTTTT). In the sequence complementary to primer ZM61, i.e., AAAAGAAGCCACGGGAAAAGCATATG, the ATG in the NdeI restriction enzyme site (CATATG) serves as the start codon for luc in all of the PdbpBA-luc reporter constructs, whereas AAGAAG serves as a ribosome-binding site (RBS). AAGAAG as an RBS has been employed in E. coli to express proteins efficiently. This strategy places the transcription of luc under the direct control of the cloned PdbpBA. All constructs were verified by restriction digestion and DNA sequence analysis. These constructs then were transformed into B. burgdorferi strain 297, and luciferase activity was assessed to monitor dbpBA transcription.
TABLE 2.
Oligonucleotide primers used in this studya
| Name | 5′-3′ |
|---|---|
| ZM54 | CCATGGCCTTTTAAGCCTGCCAATCC |
| ZM55 | ATTTTAGATCTTTGATTCAATTTGC |
| ZM56 | AGATCTTTGTAATTCCAAACAATGTTACTGC |
| ZM57 | AGATCTATTTTATTTTATTTTTCATAAAGTGGGCTAAA |
| ZM57.2 | TATCCATGGATTTTATTTTATTTTTCATAAAGTGGGATAAAATTTAAATTTAAC |
| ZM58 | AGATCTCATAAAGTGGGCTAAAATTTAAAT |
| ZM59 | AGATCTATGAAAATTGGAAAGCTAAATTCAA |
| ZM60 | GGCAGATCTACATCAACATACTAACTA |
| ZM61 | CATATGCTTTTCCCGTGGCTTCTTTT |
| ZM68 | TATCCATGGTGCTTTCTTCTGCCAGGTC |
| ZM102 | TATCCATGGATTTTATTTTATTTTTCATAAAGTGGGCTAAA |
| ZM103 | TATCCATGGCATAAAGTGGGCTAAAATTTAAAT |
| ZM104.3 | GCCCATATGCTTTGGGTTAATTGCTTTAAC |
| ZM104.4 | TATCATATGTTTAGATTCTAAAGTTTAGATAAAAATTGGTCGGG |
| ZM109F | TAATGGCGCGCCTGCTTTCTTCTGCCAGGTC |
| ZM110F | TAATGGCGCGCCTTTTAAGCCTGCCAATCC |
| ZM110R | TAATGGCGCGCCTTTAGATTCTAAAGTTTAGATAAAAATTGGTCGGG |
| bba25-F | TGGCTATGTTTGACTTAATGCTTGAG |
| bba25-R | GATTCCTCTAAAACACGGGCTTTT |
| bba24-F | GGGTAGTGGGGTATCAGAAAATC |
| bba24-R | GAGCTGTAGTTGGAGGATTCTC |
| flaB-F | ACTCTTAAAGTCCAAGACGCTTGAG |
| flaB-R | TTGGAATGCAGCCTGCAAA |
| aadA-F | TAAGGCTTGATGAAACAACGCGGC |
| aadA-R | CGTCGTGCACAACAATGGTGACTT |
Restriction enzymes sites are underlined. In ZM57.2, the A used to replace the −14 C in the dbpBA promoter is indicated in boldface.
To validate data from luciferase reporter assays, various shuttle vectors harboring different versions of the dbpBA operon were generated. Briefly, various versions of dbpBA promoter were cloned into either pOY63 at the NcoI and NdeI sites or into pJD7 at the AscI site. These vectors were transformed into the dbpBA mutant, BbKH500 (3), and dbpBA expression in these strains was examined using quantitative RT-PCR (qRT-PCR) or immunoblot analyses.
B. burgdorferi transformation.
Plasmid DNA for electroporation was purified using a CompactPrep Plasmid Maxi kit (Qiagen, Valencia, CA). The transformation of B. burgdorferi was carried out as described previously (43, 59), with minor modifications. Briefly, 50 μl (∼2 × 109 spirochetes) of electrocompetent B. burgdorferi suspension was transformed with 30 μg of plasmid DNA. After electroporation, cells were recovered using 25 ml of prewarmed BSK-II medium and incubated overnight at 37°C. Appropriate antibiotics then were added, and cultures were distributed into multiple 96-well tissue culture plates. Transformants were selected based on antibiotics resistance and verified using PCR amplification. To further confirm the presence of the shuttle vector in the transformants, genomic DNA was isolated from Borrelia clones using the Wizard Genomic DNA Purification kit (Promega Corp., Madison, WI) and transformed into E. coli. Plasmid DNA then was isolated from the resulting E. coli clones and verified by PCR, restriction digestion, and sequencing analysis.
Luciferase assays.
Luciferase assays were performed using the Luciferase Assay System (Promega Corp.). Spirochetes were collected by centrifugation at 10,000 × g for 10 min. Cell pellets then were lysed using 100 μl of cell culture lysis buffer containing 25 mM Tris-phosphate (pH 7.8), 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100, 1.25 mg/ml lysozyme, and 2.5 mg/ml bovine serum albumin. Relative luciferase units (RLU) were measured using a Centro LB 960 luminometer (Berthold Technologies, Oak Ridge, TN) as described previously (4). Results are presented as the RLU/1 × 106 spirochetes. At least three independent tests were performed, and the results were analyzed using a paired, two-tailed Student's t test, in which statistical significance was determined when P < 0.05.
Quantitative RT-PCR analysis.
qRT-PCR was employed to examine dbpBA expression. Specific primers (Table 2) were designed by using PRIMEREXPRESS software (Applied Biosystem, Foster City, CA) and validated as described previously (33). Spirochetes were grown in BSK-H medium at 37°C under 5% CO2 and harvested when bacterial growth reached a density of 5 × 107 cells per ml. Total RNA was isolated using TRIzol (Invitrogen) according to the instructions. After genomic DNA was digested using RNase-free DNase I (GenHunter Corporation, Nashville, TN), RNA was further purified using an RNeasy Mini kit (Qiagen). cDNA was generated from 1 μg of RNA using the SuperScript III Platinum two-step qRT-PCR kit according to the manufacturer's protocol (Invitrogen). qPCR (in quadruplicate) using Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) then was performed, and the relative quantification method (ΔΔ threshold cycle [ΔΔCT]) was used to calculate the variation in gene expression between B. burgdorferi strains. The aadA gene (encoding streptomycin-spectinomycin adenylyltransferases) carried by the cloning vectors (19) or the Borrelia flaB gene was used as the endogenous control to normalize all qRT-PCR data.
SDS-PAGE and immunoblot analysis.
SDS-PAGE and immunoblot analysis were performed as previously described (57). Briefly, spirochetes were harvested and washed three times in 0.9% (wt/vol) NaCl. Cell pellets were resuspended in an appropriate volume of SDS sample buffer. A volume of whole-cell lysate equivalent to 4 × 107 bacteria was loaded per lane on a 12.5% acrylamide gel. Resolved proteins either were stained with Coomassie brilliant blue or transferred to nitrocellulose membranes for immunoblot analysis. An anti-DbpB polyclonal antibody, SS65, an anti-DbpA monoclonal antibody, 6B3, and a chicken IgY anti-FlaB antibody were used to detect DbpB, DbpA, and FlaB, respectively (48). Immunoblots were developed colorimetrically using 4-chloro-1-naphthol as the substrate.
RESULTS
In silico analysis of dbpBA promoter.
In a previous study (24), a strong transcriptional initiation site at nucleotide −28 (T) (asterisk in Fig. 1) upstream of the start codon of the dbpB ORF was identified using primer extension analysis. When analyzing the 5′ sequence upstream of the dbpBA operon using BPROM (http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb), a bacterial σ70 promoter recognition program, a typical bacterial σ70 promoter harboring canonical −10/−35 elements, was predicted in the upstream 5′ regulatory region of the dbpBA operon (Fig. 1), which is consistent with the −10/−35 elements previously predicted (13, 24) based on their relative proximity to the transcriptional start. Moreover, the putative dbpBA promoter (PdbpBA), or the upstream 5′ regulatory region of dbpBA operon, does not share significant sequence similarity with the consensus sequence of the σS-dependent promoter of the Borrelia ospC gene (1, 18, 59). Furthermore, possible upstream A/T-rich sequences (UP elements) for σS-specific promoters (53) were not predicted in the 5′ regulatory region of dbpBA. However, given that σS and σ70 promoters are very similar, it is difficult to discern whether PdbpBA is σ70 or σS specific based on sequence information alone (2, 53). In addition, using the Inverted Repeats Finder program (https://tandem.bu.edu/cgi-bin/irdb/irdb.exe?taskid=0), two sets of inverted repeats (IRs) were predicted in the upstream 5′ regulatory region of dbpBA (Fig. 1). Previously, four putative ORFs, including bba26, bba27, bba28, and bba29, were annotated in this region (20). However, given the fact that these four ORFs are extremely short (bba26, 132 bp; bba27, 120 bp; bba28, 126 bp; bba29, 126 bp), they likely do not encode functional proteins in B. burgdorferi. These considerations prompted the hypothesis that the IRs serve as potential binding sites for a putative transcriptional regulator(s) involved in the regulation of dbpBA expression.
FIG. 1.
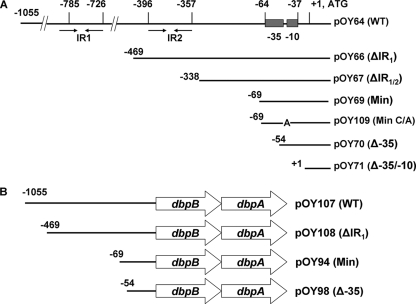
5′ Regulatory sequence of the B. burgdorferi dbpBA upstream region. Pairs of convergent arrows indicate the two putative inverted repeat elements (IR1 and IR2). The numbers in parentheses indicate numbers of nucleotides omitted (dash lines). The transcription start site (marked by the asterisk), the associated −35 and −10 elements (underlined), the ribosome-binding site (RBS) (underlined), and the ATG start codon (in boldface) are shown. Filled arrowheads denote the starting position of the minimal (Min) or the −35 deletion (Δ−35) promoter. The −14 C residue (boxed) within the extended −10 region was targeted for mutagenesis.
Assessing dbpBA expression using luciferase reporter assays.
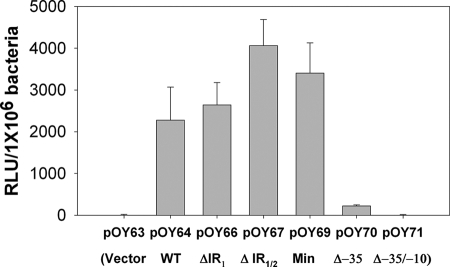
A recently developed luciferase reporter assay (4) was employed to explore how the expression of the dbpBA operon is controlled in B. burgdorferi. Along these lines, initially we created one shuttle vector, pOY64, harboring 1,055 bp of DNA upstream of the dbpB start codon (including the putative PdbpBA) and 161 bp of dbpB ORF DNA (PCR product amplified using primers ZM68 and ZM61) (Fig. 2A). In addition, pOY71 was generated by cloning a 161-bp fragment of dbpB (PCR product amplified using primers ZM59 and ZM61) into the promoterless luc reporter vector pOY63. These constructs then were transformed into the low-passage, virulent, wild-type (WT) B. burgdorferi strain 297. As shown in Fig. 3, substantial luciferase activity was detected in the strain harboring pOY64 (WT PdbpBA, the longest construct) but not from the strains containing the promoterless luc constructs (pOY63 and pOY71). The observation that luciferase activity was not detected from pOY71 indicated that the 161 bp of dbpB ORF DNA cloned into the luc fusion constructs did not contain an occult promoter(s). On the other hand, the expression of luciferase from the PdbpBA-containing vector pOY64 indicated that the 5′ regulatory region of dbpBA cloned into pOY64 contains a functional promoter.
FIG. 2.
Diagram of a series of dbpBA promoter (PdbpBA)-luc reporter constructs (A) and different constructs containing various versions of PdbpBA with the entire dbpBA operon to complement dbpBA expression in trans in the dbpBA deletion strain BbKH500 (B). Nucleotide positions are relative to the ATG start codon, where A is +1. WT, wild-type PdbpBA; ΔIR1, deletion of IR1; ΔIR1/2, deletion of both IR1 and IR2; Min, minimal PdbpBA; Min C/A, minimal PdbpBA with the −14 C mutated to A; Δ−35, deletion of −35 sequence; Δ−35/−10, deletion of both −35 sequence and −10 sequence.
FIG. 3.
Luciferase activity (denoted in RLU/106 bacteria) detected in B. burgdorferi strains transformed with various PdbpBA-luc constructs. Spirochetes were cultured in BSK-H medium at 37°C and harvested at late log phase. Results from three independent experiments are presented as the mean values ± standard errors of the means (SEM). pOY63, cloning vector containing a promoterless luc; pOY64, vector containing WT PdbpBA; pOY66, vector containing ΔIR1 PdbpBA; pOY67, vector containing ΔIR1/2 PdbpBA; pOY69, vector containing minimal (Min) PdbpBA; pOY70, vector containing Δ−35 PdbpBA; and pOY71, vector containing Δ−35/−10 PdbpBA.
Minimal promoter for dbpBA transcription.
As mentioned above, a functional PdbpBA was identified in the 5′ regulatory sequence of dbpBA. To determine the minimal genetic element(s) required for PdbpBA, a series of PdbpBA-luc transcriptional deletion constructs was created (Fig. 2A) and introduced into B. burgdorferi 297. The expression of the luc gene from these constructs was compared to the expression of luc from the promoterless vector (pOY63) and the construct containing the WT PdbpBA (pOY64). As shown in Fig. 3, luciferase activity was readily detected in strains harboring the putative, intact PdbpBA (pOY64, pOY66, pOY67, and pOY69). In particular, pOY69, which contains the sequence from the −35 element of PdbpBA only (the sequence upstream of the −35 element was deleted), still expressed luciferase as efficiently as pOY64. In contrast, the construct pOY70, lacking the putative −35 element, displayed greatly diminished luciferase expression (∼95% decrease compared to that of pOY69). In addition, when both the −35 and −10 elements of PdbpBA were deleted (pOY71), no luciferase activity was detected. These data indicate that pOY69 comprises a minimal sequence encompassed within 70 bp upstream of the ATG start codon of dbpBA, which is essential for PdbpBA to be functional.
To further validate these data, two additional constructs, containing the entire dbpBA operon with either the minimal PdbpBA (same version of PdbpBA as that cloned in pOY69) (pOY94) or PdbpBA lacking the −35 element (Δ−35; the same version of PdbpBA as that cloned into pOY70) (pOY98), were created and introduced into the dbpBA-deficient mutant BbKH500 (3). The expression of DbpB and DbpA then was assessed using qRT-PCR and immunoblotting to determine whether these constructs could complement DbpBA expression in trans. As shown in Fig. 4, when B. burgdorferi was cultured in BSK-H medium at pH 7.6, both DbpB and DbpA were expressed (at both the RNA [Fig. 4A] and protein [Fig. 4B] levels) in strain OY45 (containing the minimal PdbpBA) but not from OY48 (containing the Δ−35 PdbpBA). Similar data were obtained when Borrelia was grown in BSK-H medium at pH 6.8 (data not shown). These data indicate that a minimal −35/−10 dbpBA promoter sequence is necessary and sufficient for dbpBA expression in B. burgdorferi.
FIG. 4.
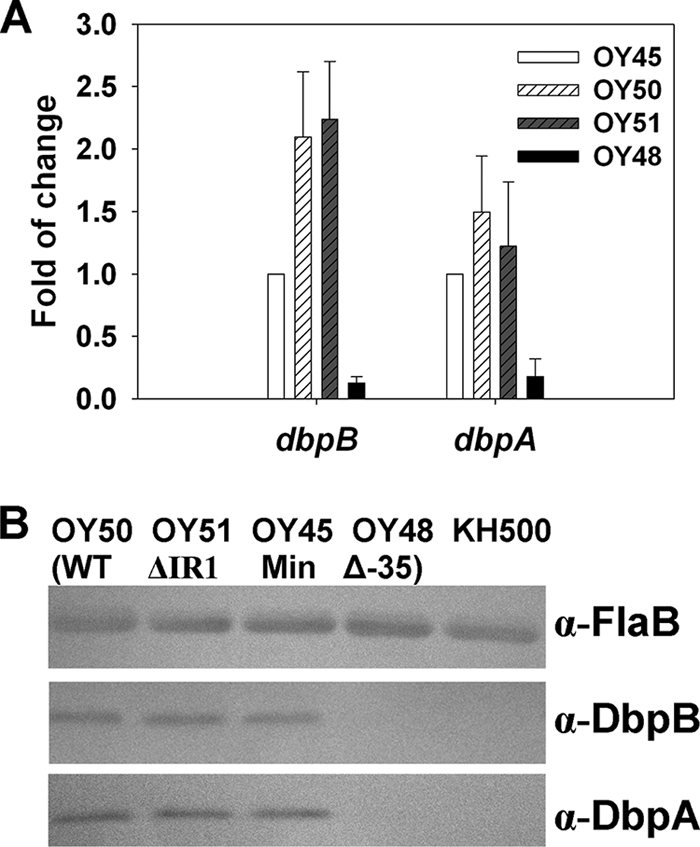
Influence of upstream IRs on dbpBA expression. B. burgdorferi dbpBA deletion mutant BbKH500 transformed with different shuttle vectors were grown in BSK-H medium and harvested at late-log phase. The expression of DbpB and DbpA were assessed using qRT-PCR (A) or immunoblot analysis (B). (A) Results from three tests are presented as the mean fold changes (relative to the gene expression level in OY45) ± SEM. (B) Approximately 4 × 107 spirochetes were loaded onto each lane of an SDS-PAGE gel and transferred to a nitrocellulose membrane. FlaB, DbpB, and DbpA were detected using antibodies described in the Materials and Methods. α, anti; OY45, vector containing the minimal (Min) PdbpBA; OY50, vector containing the WT PdbpBA; OY51, vector containing ΔIR1 PdbpBA; OY48, vector containing Δ−35 PdbpBA.
IR elements in the 5′ dbpBA regulatory sequence.
Two sets of IR elements, putative transcriptional regulator-binding sites, are present in the dbpBA 5′ regulatory sequence. To assess the potential roles of these two IRs in dbpBA regulation, a series of PdbpBA deletions and mutations fused to luc were created (Fig. 2A) and then introduced into strain 297. As shown in Fig. 3, when Borrelia was grown in BSK-H medium at pH 7.6 and harvested at late log phase, comparable luciferase activity was observed from constructs containing WT PdbpBA (pOY64), PdbpBA lacking either IR1 (ΔIR1) (pOY66) or both IR1 and IR2 (ΔIR1/2) (pOY67), or the minimal (Min) PdbpBA (pOY69), although a slightly higher level of luciferase expression was observed when both IRs were deleted. To substantiate these data, we also created constructs harboring various versions of PdbpBA and the entire dbpBA operon (Fig. 2B). We then introduced these constructs into BbKH500 and measured the expression of DbpB and DbpA RNA and protein using qRT-PCR and immunoblotting. As shown in Fig. 4A and B, similar levels of DbpB and DbpA expression were detected in OY50 (containing the WT PdbpBA), OY51 (containing the ΔIR1 PdbpBA), and OY45 (containing the minimal PdbpBA), whereas the expression of DbpB and DbpA was dramatically reduced in OY48 (containing the Δ−35 PdbpBA). This was observed in Borrelia spirochetes grown at either pH 7.6 (Fig. 4) or 6.8 (data not shown). These data suggest that neither IR is involved in the primary regulation of dbpBA, at least not under the conditions tested.
dbpBA expression is influenced by various environmental parameters.
Previous studies have revealed that dbpBA expression is influenced by various environmental factors, such as temperature, pH, cell density, and whole blood (52, 56). As a first attempt to quantitatively determine the influence of environmental factors on dbpBA transcription, and to explore whether these factors impact dbpBA expression through the minimal dbpBA promoter or the IRs located upstream of the 5′ regulatory sequence of dbpBA, luciferase activities were measured in B. burgdorferi harboring different PdbpBA deletion constructs (including WT, ΔIR1, ΔIR1/2, and minimal PdbpBA) under various environmental conditions. As shown in Fig. 5A, luciferase expression from all of these constructs was markedly induced in Borrelia grown at pH 6.8 (compared to that at pH 7.6). Moreover, when fresh whole rabbit blood was added to the culture, luciferase expression was dramatically increased (Fig. 5B). We also examined the effect of temperature on dbpBA expression using these luc constructs. Luciferase expression was barely detected in Borrelia grown at 23°C (data not shown) but was highly expressed in these Borrelia strains cultured at 37°C (Fig. 5), indicating that dbpBA expression is induced by elevated temperature, as previously reported (1, 11-13, 31, 52, 56). These data were further confirmed by probing for DbpB and DbpA in these strains using immunoblotting (data not shown). Of note, luc reporter constructs containing the ospC promoter (PospC) or flaB promoter (PflaB) also were employed in this study as controls. As expected, PospC displayed a response to the stimuli similar to that of PdbpBA, showing induction at elevated temperature, lower pH, or blood addition, whereas PflaB showed no response to these stimuli (Fig. 6).
FIG. 5.
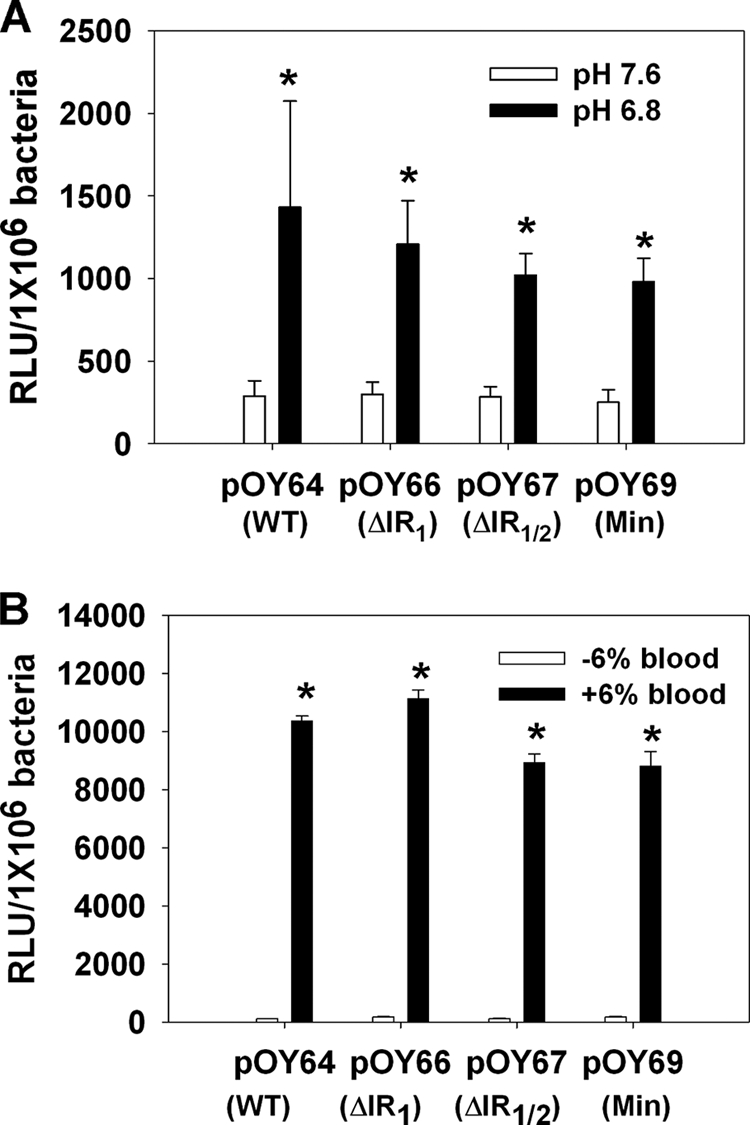
Influence of culture pH (A) and whole blood (B) on luciferase expression from various PdbpBA-luc constructs. Borrelia organisms were cultivated in BSK-H medium and harvested when growth reached a cell density of ∼1 × 106 to 5 × 106 spirochetes/ml. The experiments were replicated thrice, and bars represent the mean measurements ± SEM. The asterisk indicates statistical significance using Student's t test (P < 0.05). pOY64, vector containing WT PdbpBA; pOY66, vector containing ΔIR1 PdbpBA; pOY67, vector containing ΔIR1/2 PdbpBA; and pOY69, vector containing minimal (Min) PdbpBA.
FIG. 6.
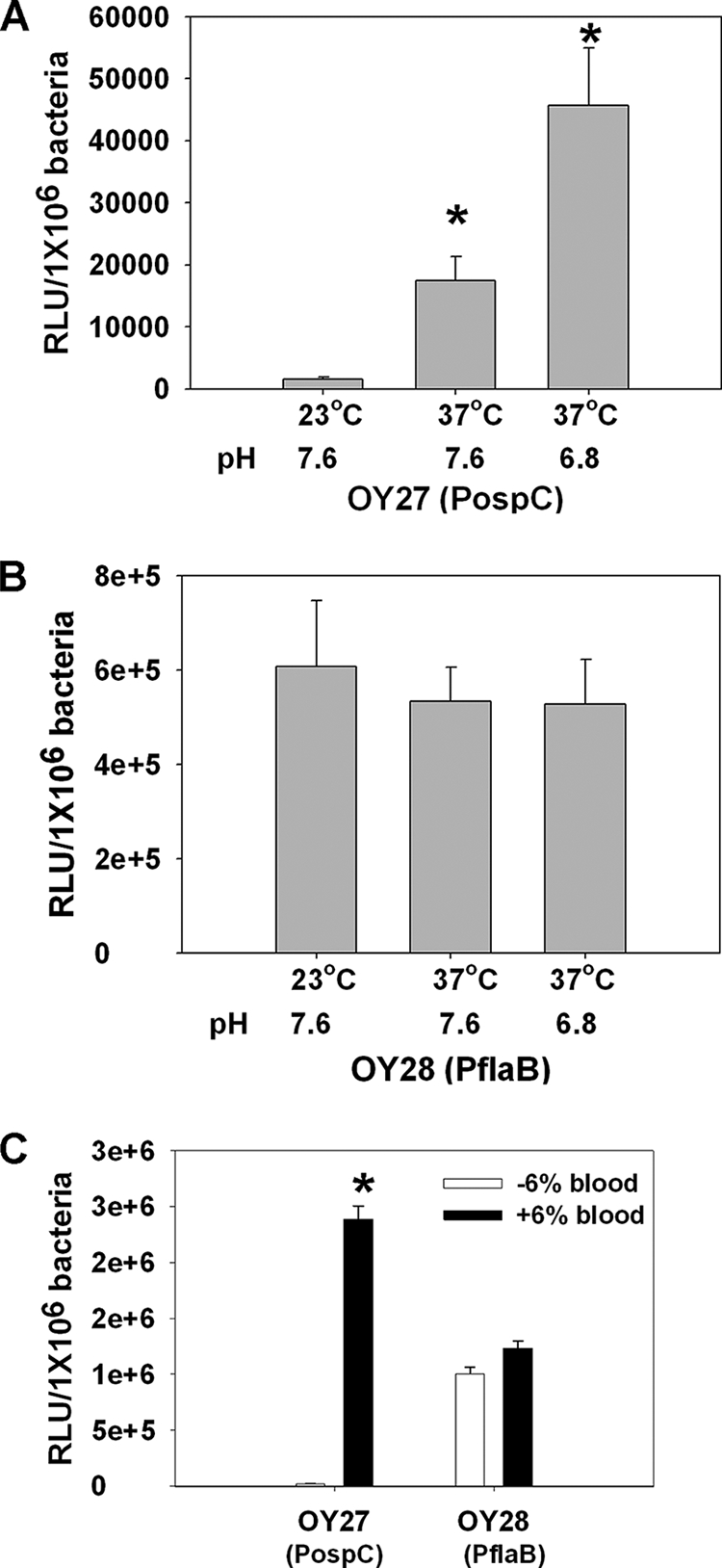
Influence of temperature, pH (A and B), and whole blood (C) on luciferase expression driven by the B. burgdorferi ospC promoter (PospC) or flaB promoter (PflaB). Borrelia organisms were cultivated in BSK-H medium and harvested when growth reached a cell density of ∼1 × 106 to 5 × 106 spirochetes/ml. The experiments were replicated thrice, and bars represent the mean measurements ± SEM. The asterisk indicates statistical significance using Student's t test (P < 0.05).
dbpBA and its minimal promoter are RpoS dependent.
To study how RpoS impacts dbpBA expression, the minimal PdbpBA construct, pOY69, was further introduced into an rpoS-deficient strain, BbAH206 (27). Consistently with previous reports that dbpBA expression is dependent on RpoS in B. burgdorferi (6, 11-13, 18, 27, 31, 33, 58), luciferase expression from pOY69 was abolished in BbAH206 (Fig. 7A). However, given that σS and σ70 are highly related and both σS and σ70 holoenzymes recognize very similar core promoter sequences, it is difficult to distinguish between σS and σ70 promoters based on sequence information alone or using gel shift assays. Nonetheless, in E. coli, studies have shown that a −13 C residue in the extended −10 region is essential for σS activity and is highly conserved in σS-dependent promoters, but not in σ70-dependent promoters (2). Therefore, a C at position −13 introduces σS promoter selectivity and serves as a hallmark of σS-dependent promoters (53). An analogous situation has been verified in B. burgdorferi, in that a −15 C is critical for the activity of the RpoS-dependent ospC promoter (59), although the −15 C also was reported not to be essential for RpoS selectivity (13, 18). In PdbpBA, a −14 C (relative to the transcriptional start of dbpBA [defined as +1]) also is present in the extended −10 region (Fig. 1). Therefore, to garner evidence that RpoS controls dbpBA expression in B. burgdorferi via direct interaction with the minimal PdbpBA, one construct, pOY109, was created by cloning a PCR product amplified using ZM57.2 and ZM61 into pOY63. Thus, pOY109 encompassed a minimal PdbpBA with the −14 C mutated to adenine. Luciferase activity expressed from pOY109 was essentially abolished compared to that of the construct containing the minimal PdbpBA, regardless of whether B. burgdorferi was cultivated at pH 7.6 or 6.8 (Fig. 7B). These data suggest that RpoS governs dbpBA expression via direct interaction with the minimal promoter of dbpBA.
FIG. 7.
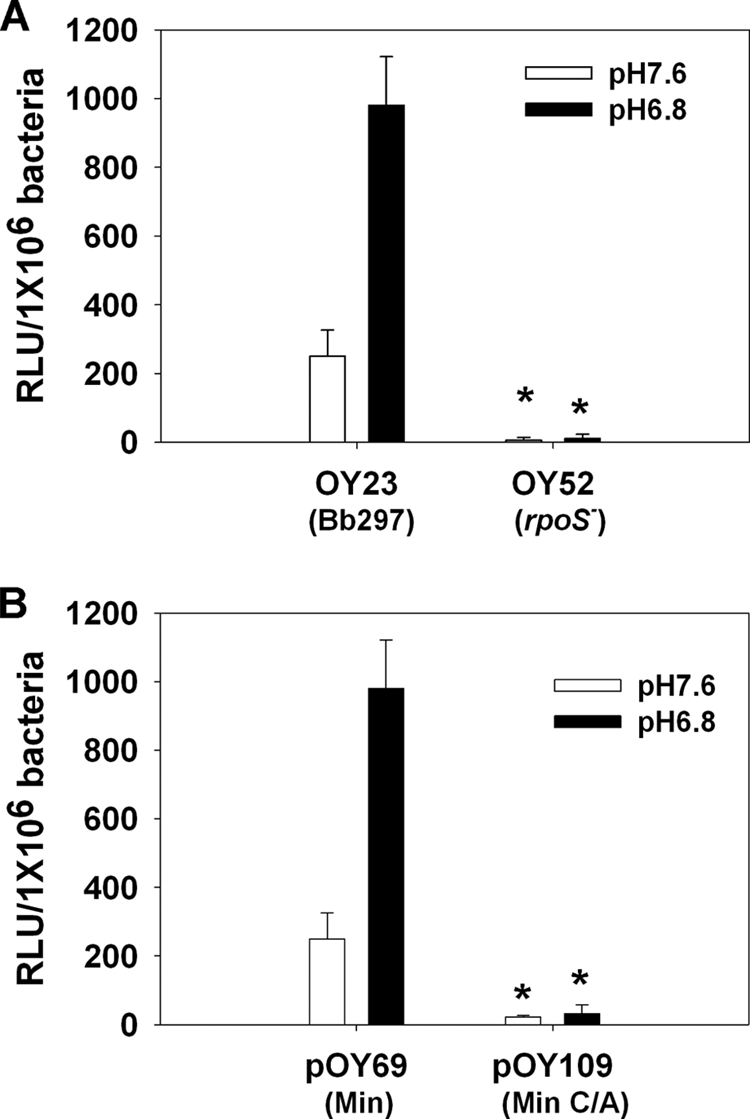
B. burgdorferi PdbpBA is RpoS dependent. (A) Luciferase activity, driven from the minimal PdbpBA, was measured in Borrelia strain Bb297 or an rpoS mutant. (B) Luciferase was expressed from either the minimal PdbpBA (pOY69) or the minimal PdbpBA containing a mutated −14 residue (C replaced by A) (pOY109). Spirochetes were grown in BSK-H medium at a pH of either 7.6 (open bar) or 6.8 (black bar) and harvested at late log phase. Results from three tests are indicated as means ± SEM. The asterisk indicates statistical significance using Student's t test (P < 0.05).
E. coli as a surrogate system for studying B. burgdorferi gene regulation.
Given the limited genetic tools available for Borrelia research, E. coli has been exploited as a surrogate system for the study of B. burgdorferi gene expression. However, there are a number of salient differences between B. burgdorferi and E. coli that warrant consideration. In addition to many obvious cellular differences, B. burgdorferi encodes very few predicted transcriptional regulators, despite a more complex overall genome consisting of a linear chromosome and numerous linear/circular plasmids. Borrelia also is dramatically different from E. coli in metal acquisition and metal homeostasis, systems typically under complex regulatory control. These and many other key differences call into question the suitability of E. coli for assessing B. burgdorferi gene regulation. To further examine the potential utility of E. coli as a surrogate system for assessing B. burgdorferi gene expression and regulation, we examined luciferase expression from the constructs comprising different versions of PdbpBA. As shown in Fig. 8A, pOY70, which harbors a Δ−35 PdbpBA, expressed very low levels of luciferase in Borrelia. However, when pOY70 was introduced into E. coli, it expressed a very high level of luciferase (Fig. 8B). These data indicate that the mutated PdbpBA cloned in pOY70 is capable of promoting gene transcription in E. coli but not in Borrelia, which in turn suggests that E. coli may not be an ideal surrogate system for studying Borrelia gene expression and regulation.
FIG. 8.
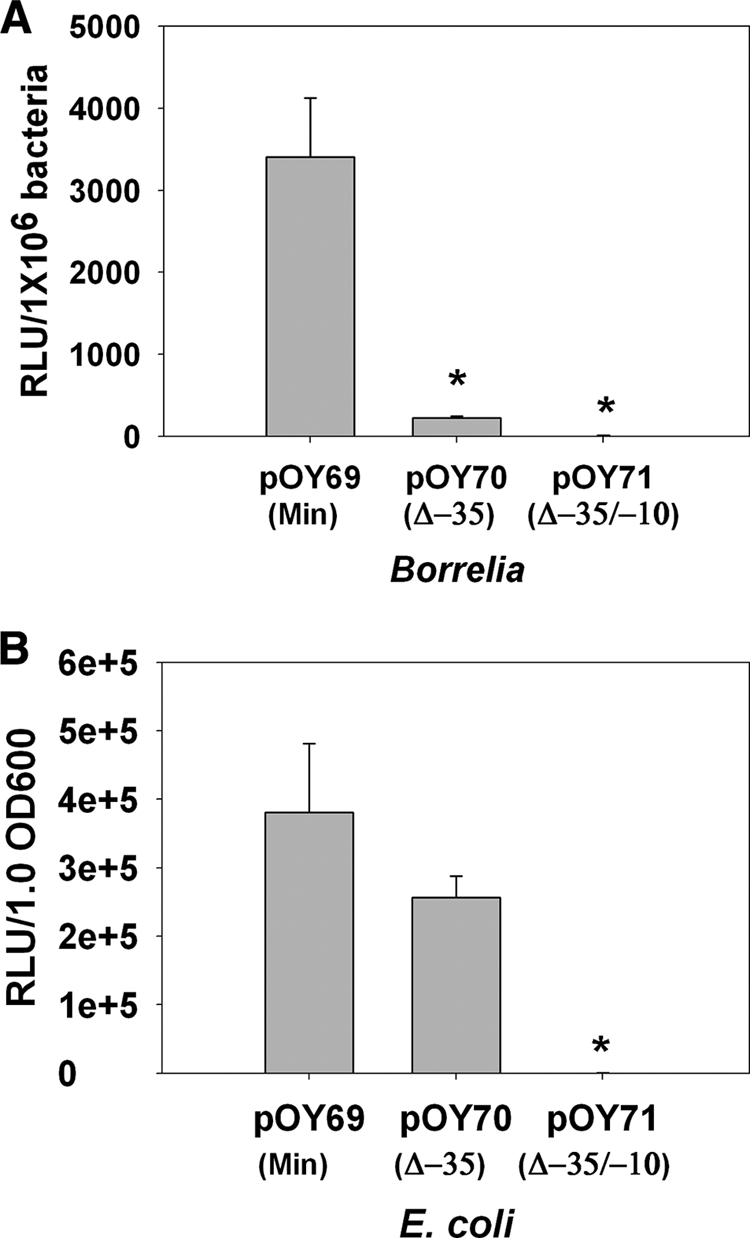
Luciferase expressed from B. burgdorferi (A) or E. coli (B) containing various PdbpBA-luc constructs. Borrelia spirochetes were grown in BSK-H medium and collected at late log phase, whereas E. coli was grown in LB and harvested when growth reached an optical density at 600 nm of ∼0.6. Results from three tests are indicated as means ± SEM. The asterisk indicates statistical significance using Student's t test (P < 0.05).
DISCUSSION
Using a sensitive luciferase reporter assay, we investigated the involvement of the 5′ regulatory sequence of the dbpBA operon in the regulation of dbpBA expression in B. burgdorferi. The luciferase reporter assay employed in our study had several advantages. First, this is a quantitative reporter assay. Moreover, the luc gene used to generate the reporter constructs is codon optimized, thereby rendering optimal luciferase expression in B. burgdorferi (4). In addition, the luciferase reporter assay is sensitive, convenient, and relatively simple. One potential problem for the luciferase reporter assay concerns the copy number of the shuttle vector used to generate the luc reporter constructs. Although it is unclear how many copies of these vectors are maintained in Borrelia, our qRT-PCR data revealed that similar numbers of the aadA gene (encoding streptomycin-spectinomycin adenylyltransferases in the shuttle vectors, including pOY63 and pJD7) were found in strains OY45, OY48, OY50, and OY51 (data not shown), suggesting that similar numbers of copies of plasmid constructs were present among the various strains. We further measured dbpBA expression using qRT-PCR or immunoblotting, which served as additional approaches to corroborate the data obtained from the luciferase reporter assays.
As an initial step for elucidating the molecular mechanism governing dbpBA expression, we created a series of luciferase reporter constructs by fusing various versions of deletion constructs of PdbpBA to the promoterless luc gene. We then examined whether the putative 5′ cis regulatory elements were involved in dbpBA expression. We identified a minimal promoter that is necessary and sufficient to drive dbpBA expression. Moreover, because the expression of dbpBA has been suggested to be influenced by several environmental factors, we also examined the effect of various factors on luciferase expression from these reporter constructs. Consistently with previous studies of dbpA expression (52, 56), our data revealed that luciferase expression driven from the minimal PdbpBA was induced by elevated temperature (37°C) or the supplementation of blood. However, luciferase expression driven from all functional PdbpBA constructs, including WT, IR-deleted, or minimal PdbpBA, was more induced at pH 6.8 than at pH 7.6, suggesting that dbpBA transcription also was induced by lower pH, which is disparate from a previous observation that the expression of DbpA was slightly repressed at pH 6.8 (56). The reason for this discrepancy currently remains unknown. It may emanate from different CO2 levels used in these studies to grow the spirochetes. In the current study, spirochetes were grown at 5% CO2, whereas 1% CO2 was employed to grow Borrelia in the previous study (56). In accord with this possibility, DbpA expression was reported to be influenced by the CO2 level (28). Alternatively, it also might be due to as-yet unknown subtle composition differences among BSK media.
Another goal of this study was to garner further evidence regarding whether RpoS controls dbpBA expression directly or indirectly. Based on in silico information, the dbpBA operon has been proposed to possess a typical −35/−10 σ70 promoter. However, because both σS and σ70 holoenzymes recognize the same core promoter elements, it is not feasible to discern whether the dbpBA promoter is σS or σ70 specific based on sequence information alone. Therefore, to garner direct evidence for how σS (RpoS) influences dbpBA expression in B. burgdorferi, we examined the effect of RpoS on luciferase expression driven from the minimal PdbpBA. Subsequently, we found that (i) the minimal PdbpBA lost its ability to promote luc transcription in an rpoS-deficient mutant, and (ii) the minimal PdbpBA harbors an essential −14 C, similarly to the E. coli σS-dependent promoter and the Borrelia ospC promoter. These compelling data support the notion that RpoS controls dbpBA expression by direct interaction with the RpoS-dependent promoter in the 5′ regulatory region of the dbpBA operon.
Although the data described above that dbpBA expression was induced by elevated temperature, lower pH, or blood, and that RpoS controls dbpBA expression directly (similarly to Borrelia ospC expression), is compelling, there also is abundant evidence that dbpA has an expression pattern that is slightly different from that of ospC (13, 21, 25, 26, 29, 30, 36, 56). In particular, the expression of ospC (and rpoS) (13, 21, 26, 36), but not dbpA (25), has been observed in fed ticks, suggesting that, in addition to RpoS, another regulatory protein(s) (perhaps a repressor) is involved in the fine tuning of dbpBA expression. Relative to this hypothesis, two sets of conserved IRs in the 5′ regulatory sequence upstream of the PdbpBA potentially served as candidate binding sites for transcriptional regulators. However, the deletion of both IRs did not significantly alter the level of luciferase expression from the various reporter constructs in Borrelia (stimulated under various environmental conditions). All four versions of PdbpBA (including WT, IR1 deleted, IR1/2 deleted, and the minimal PdbpBA) displayed comparable (and not significantly different) abilities to promote luc transcription under all tested conditions. Similarly, using immunoblotting or qRT-PCR, the deletion of both IRs (from vectors harboring the entire dbpBA operon) also had no effect on dbpBA expression. These data suggest that the IRs are dispensable for the regulation of dbpBA expression, at least under the in vitro culture conditions tested. In the case of the Borrelia ospC promoter, a deletion of the IRs (ospC operator) located upstream of the ospC promoter also did not affect ospC expression when Borrelia was cultivated in vitro (59). However, the ospC operator assumes functional significance in vivo, wherein its presence is crucial for the repression of in vivo ospC expression in mammalian hosts and thus evasion from specific humoral immunity (55). As such, the ospC operator probably serves as a binding site for an unidentified DNA-binding protein that functions to suppress ospC during in vivo mammalian expression. Given what is now known about the ospC operator (18, 55, 59), it thus remains premature to exclude the possibility that these IRs contribute to the control of dbpBA expression in B. burgdorferi during its natural life cycle. The IRs may serve as binding sites for transcriptional regulator(s) involved in the regulation of dbpBA expression when Borrelia transits between its tick vector and mammalian hosts. Such a regulatory protein, potentially a repressor, may be expressed only when B. burgdorferi colonizes ticks and/or during tick feeding. Alternatively, a dbpBA-specific regulatory protein may be inactive when spirochetes are cultivated in vitro, perhaps first requiring some tick phase-specific cofactor(s) or ligands. Continued efforts are warranted to examine these possibilities by investigating the PdbpBA deletion construct strains in the tick and mammalian host phases of B. burgdorferi's infectious life cycle.
Acknowledgments
We thank Xiaofeng (Frank) Yang and Jon S. Blevins for many helpful discussions.
This work was supported by Public Health Service Grant AI-059062 from the National Institutes of Health.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Alverson, J., S. F. Bundle, C. D. Sohaskey, M. C. Lybecker, and D. S. Samuels. 2003. Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol. Microbiol. 48:1665-1677. [DOI] [PubMed] [Google Scholar]
- 2.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoter sigma(S) dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of sigma(S). Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 3.Blevins, J. S., K. E. Hagman, and M. V. Norgard. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blevins, J. S., A. T. Revel, A. H. Smith, G. N. Bachlani, and M. V. Norgard. 2007. Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl. Environ. Microbiol. 73:1501-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blevins, J. S., H. Xu, M. He, M. V. Norgard, L. Reitzer, and X. F. Yang. 2009. Rrp2, a sigma54-dependent transcriptional activator of Borrelia burgdorferi, activates rpoS in an enhancer-independent manner. J. Bacteriol. 191:2902-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boardman, B. K., M. He, Z. Ouyang, H. Xu, X. Pang, and X. F. Yang. 2008. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect. Immun. 76:3844-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, E. L., B. P. Guo, P. O'Neal, and M. Hook. 1999. Adherence of Borrelia burgdorferi. Identification of critical lysine residues in DbpA required for decorin binding. J. Biol. Chem. 274:26272-26278. [DOI] [PubMed] [Google Scholar]
- 8.Brown, E. L., R. M. Wooten, B. J. Johnson, R. V. Iozzo, A. Smith, M. C. Dolan, B. P. Guo, J. J. Weis, and M. Hook. 2001. Resistance to Lyme disease in decorin-deficient mice. J. Clin. Investig. 107:845-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunikis, J., L. Noppa, and S. Bergstrom. 1995. Molecular analysis of a 66-kDa protein associated with the outer membrane of Lyme disease Borrelia. FEMS Microbiol. Lett. 131:139-145. [DOI] [PubMed] [Google Scholar]
- 10.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease-a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 11.Burtnick, M. N., J. S. Downey, P. J. Brett, J. A. Boylan, J. G. Frye, T. R. Hoover, and F. C. Gherardini. 2007. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol. Microbiol. 65:277-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caimano, M. J., C. H. Eggers, K. R. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 72:6433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caimano, M. J., R. Iyer, C. H. Eggers, C. Gonzalez, E. A. Morton, M. A. Gilbert, I. Schwartz, and J. D. Radolf. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 65:1193-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coburn, J. 2001. Adhesion mechanisms of the Lyme disease spirochete, Borrelia burgdorferi. Curr. Drug Targets Infect. Disord. 1:171-179. [DOI] [PubMed] [Google Scholar]
- 15.Coburn, J., W. Chege, L. Magoun, S. C. Bodary, and J. M. Leong. 1999. Characterization of a candidate Borrelia burgdorferi beta3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926-940. [DOI] [PubMed] [Google Scholar]
- 16.Coburn, J., J. R. Fischer, and J. M. Leong. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 57:1182-1195. [DOI] [PubMed] [Google Scholar]
- 17.de Silva, A. M., K. R. Tyson, and U. Pal. 2009. Molecular characterization of the tick-Borrelia interface. Front. Biosci. 14:3051-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eggers, C. H., M. J. Caimano, and J. D. Radolf. 2004. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 186:7390-7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank, K. L., S. F. Bundle, M. E. Kresge, C. H. Eggers, and D. S. Samuels. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J. Bacteriol. 185:6723-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fuji, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 21.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 101:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, B. P., E. L. Brown, D. W. Dorward, L. C. Rosenberg, and M. Hook. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711-723. [DOI] [PubMed] [Google Scholar]
- 23.Guo, B. P., S. J. Norris, L. C. Rosenberg, and M. Hook. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 63:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagman, K. E., P. Lahdenne, T. G. Popova, S. F. Porcella, D. R. Akins, J. D. Radolf, and M. V. Norgard. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 66:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagman, K. E., X. Yang, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 68:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodzic, E., S. Feng, K. J. Freet, D. L. Borjesson, and S. W. Barthold. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 70:3382-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hübner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Hyde, J. A., J. P. Trzeciakowski, and J. T. Skare. 2007. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J. Bacteriol. 189:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reference deleted.
- 31.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang, F. T., J. Yan, M. L. Mbow, S. L. Sviat, R. D. Gilmore, M. Mamula, and E. Fikrig. 2004. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 72:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lybecker, M. C., and D. S. Samuels. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 64:1075-1089. [DOI] [PubMed] [Google Scholar]
- 34.Neelakanta, G., X. Li, U. Pal, X. Liu, D. S. Beck, K. DePonte, D. Fish, F. S. Kantor, and E. Fikrig. 2007. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathog. 3:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang, Z., J. S. Blevins, and M. V. Norgard. 2008. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology 154:2641-2658. [DOI] [PubMed] [Google Scholar]
- 36.Pal, U., A. M. de Silva, R. R. Montgomery, D. Fish, J. Anguita, J. F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Investig. 106:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pal, U., X. Li, T. Wang, R. R. Montgomery, N. Ramamoorthi, A. M. Desilva, F. Bao, X. Yang, M. Pypaert, D. Pradhan, F. S. Kantor, S. Telford, J. F. Anderson, and E. Fikrig. 2004. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119:457-468. [DOI] [PubMed] [Google Scholar]
- 38.Pal, U., X. Yang, M. Chen, L. K. Bockenstedt, J. F. Anderson, R. A. Flavell, M. V. Norgard, and E. Fikrig. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Investig. 113:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parveen, N., and J. M. Leong. 2000. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:1220-1234. [DOI] [PubMed] [Google Scholar]
- 40.Pikas, D. S., E. L. Brown, S. Gurusiddappa, L. Y. Lee, Y. Xu, and M. Hook. 2003. Decorin-binding sites in the adhesin DbpA from Borrelia burgdorferi: a synthetic peptide approach. J. Biol. Chem. 278:30920-30926. [DOI] [PubMed] [Google Scholar]
- 41.Pollack, R. J., S. R. Telford III, and A. Spielman. 1993. Standardization of medium for culturing Lyme disease spirochetes. J. Clin. Microbiol. 31:1251-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Probert, W. S., and B. J. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003-1015. [DOI] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Revel, A. T., J. S. Blevins, C. Almazan, L. Neil, K. M. Kocan, J. de la Fuente, K. E. Hagman, and M. V. Norgard. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. USA 102:6972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosa, P. A., K. Tilly, and P. E. Stewart. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3:129-143. [DOI] [PubMed] [Google Scholar]
- 46.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seshu, J., J. A. Boylan, F. C. Gherardini, and J. T. Skare. 2004. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect. Immun. 72:1580-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi, Y., Q. Xu, K. McShan, and F. T. Liang. 2008. Both decorin-binding proteins A and B are critical for overall virulence of Borrelia burgdorferi. Infect. Immun. 76:1239-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi, Y., Q. Xu, S. V. Seemanaplli, K. McShan, and F. T. Liang. 2008. Common and unique contributions of decorin-binding proteins A and B to the overall virulence of Borrelia burgdorferi. PLoS One 3:e3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, A. H., J. S. Blevins, G. N. Bachlani, X. F. Yang, and M. V. Norgard. 2007. Evidence that RpoS (sigmaS) in Borrelia burgdorferi is controlled directly by RpoN (sigma54/sigmaN). J. Bacteriol. 189:2139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 113:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 54.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 55.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Typas, A., G. Becker, and R. Hengge. 2007. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol. Microbiol. 63:1296-1306. [DOI] [PubMed] [Google Scholar]
- 57.Weening, E. H., N. Parveen, J. P. Trzeciakowski, J. M. Leong, M. Hook, and J. T. Skare. 2008. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect. Immun. 76:5694-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Xu, Q., K. McShan, and F. T. Liang. 2007. Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol. Microbiol. 64:220-231. [DOI] [PubMed] [Google Scholar]
- 60.Reference deleted.
- 61.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 62.Yang, X., T. G. Popova, K. E. Hagman, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 1999. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect. Immun. 67:6008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, X. F., S. M. Alani, and M. V. Norgard. 2003. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 100:11001-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, X. F., M. C. Lybecker, U. Pal, S. M. Alani, J. Blevins, A. T. Revel, D. S. Samuels, and M. V. Norgard. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J. Bacteriol. 187:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reference deleted.