Abstract
The Streptococcus mutans hdrRM operon encodes a novel two-gene regulatory system induced by high cell density. Previous studies identified hdrM as the only known negative regulator of competence development in S. mutans. In the present study, we demonstrated that the HdrRM system bypasses the prototypical competence gene regulators ComC and ComDE in the transcriptional regulation of the competence-specific sigma factor comX and the late competence genes. Similarly, the HdrRM system can abrogate the requirement for ComE to produce the bacteriocin mutacin IV. To further probe the regulatory mechanism of hdrRM, we created an hdrR overexpression strain and showed that it could reproduce each of the hdrM competence and mutacin phenotypes, indicating that HdrM acts as a negative regulator of HdrR activity. Using a mutacin IV-luciferase reporter, we also demonstrated that the hdrRM system utilizes the same promoter elements recognized by ComE and thus appears to comprise a novel regulatory pathway parallel to ComCDE.
Streptococcus mutans is a gram-positive oral commensal species often associated with the development of dental caries (tooth decay) (2, 3, 22, 25, 30, 32, 36). Similar to numerous other species of Streptococcus, S. mutans is naturally competent and thus actively internalizes exogenous DNA from the environment, which can lead to genetic transformation (6). Furthermore, the regulatory machinery of the competence system has been shown to affect a variety of virulence factors in S. mutans such as acid tolerance, biofilm formation, and bacteriocin (mutacin) production (11, 13, 14, 35).
Among the streptococci, Streptococcus pneumoniae has the most thoroughly characterized competence system, and consequently, its regulatory scheme has become the prototypical model for competence gene regulation among other naturally competent Streptococcus species. Competence initiation begins with the product of the gene comC, which is a secreted cell signaling peptide referred to as the competence-stimulating peptide (CSP) (5, 14, 17, 26). When the optimal cell density, and thus CSP concentration, has been achieved, CSP is sensed by a two-component regulatory system composed of the ComD sensor kinase and the ComE response regulator (5, 8, 26). Upon binding CSP, ComD phosphorylates ComE, which subsequently stimulates its transcription factor activity, resulting in the expression of a competence-specific sigma factor, comX (12, 14, 17). Finally, ComX is ultimately responsible for transcribing the late competence genes, which comprise all of the machinery required for exogenous DNA internalization and recombination (12, 18, 20). In S. mutans, evidence suggests that the competence cascade seems to function quite similarly, except that the comC and comED genes are not absolutely essential for transformability (1). However, they are essential for the expression of numerous bacteriocins (7, 9, 11, 34), which supports the suggestion that bacteriocin production and competence development are functionally linked in S. mutans (10).
Recently, we reported the identification of a previously uncharacterized two-gene operon (hdrRM) that encodes a predicted transcription regulator (HdrR) and membrane protein (HdrM). The operon was found to be induced by conditions of extremely high cell density, while hdrM was shown to be a potent negative regulator of genetic competence (21). The connection to competence development was of particular interest, as hdrM is currently the only reported negative regulator of competence in S. mutans. Surprisingly, this and other phenotypes were only observable with a mutation in hdrM; an in-frame deletion of hdrR and an hdrRM double deletion both produced wild-type behavior. This suggested that hdrR is likely part of the pathway regulated by hdrM, since the hdrM phenotype was only observable with hdrR present. In the present study, we further investigated the competence phenotype in order to examine the mechanism of regulation by the HdrRM system. We show that in S. mutans, the HdrRM system appears to regulate competence and mutacin IV production similarly to ComDE, but both systems apparently function in response to distinct stimuli. Furthermore, HdrM appears to function as a negative regulator of HdrR, which functions as a positive regulator of competence and mutacin gene expression. These results suggest that the mechanisms governing competence development in S. mutans are potentially much more elaborate than suggested by the S. pneumoniae model.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. S. mutans UA140 and its derivatives were grown in brain heart infusion broth (BHI; Difco) and on BHI agar plates. When testing for mutacin IV production, cells were grown on Todd-Hewitt (TH; Difco) agar plates. For the selection of antibiotic-resistant colonies, BHI plates were supplemented with 800 μg ml−1 kanamycin (Sigma), 15 μg ml−1 erythromycin (MP Biomedicals), 15 μg ml−1 tetracycline (Sigma), or 1,000 μg ml−1 spectinomycin (Sigma). S. mutans strains were grown anaerobically (85% N2, 10% CO2, 5% H2) at 37°C. Escherichia coli cells were grown in Luria-Bertani (LB; Difco) medium with aeration at 37°C. E. coli strains carrying plasmids were grown in LB medium containing 100 μg ml−1 spectinomycin or 100 μg ml−1 kanamycin.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning strain | |
| UA140 | Wild-type S. mutans | 28 |
| NY101 | Wild-type S. sanguinis | 33 |
| TO-m | UA140::pHdrM-i hdrM Spr | This work |
| TO-c | UA140ΔcomC Kmr | This work |
| TO-e | UA140::pComD-KO comE Emr | 14 |
| TO-x | UA140::pComX-KO comX Emr | 14 |
| TO-yad | UA140ΔcomYA-D Emr | 20 |
| TO-mc | UA140::pHdrM-i ΔcomC Sper Kmr | This work |
| TO-me | UA140::pHdrM-i/pComD-KO Sper Emr | This work |
| TO-mx | UA140::pHdrM-i/pComX-KO Sper Emr | This work |
| TO-myad | UA140::pHdrM-i ΔcomYA-D Sper Kanr | This work |
| TO-Roe | UA140/pHdrRoe Spr | This work |
| TO-Roe− | UA140/pHdrRoe− Spr | This work |
| TO-RMoe | UA140/pHdrRMoe Spr | This work |
| TO-eRoe | UA140::pComD-KO/pHdrRoe Emr Spr | This work |
| TO-nLuc | UA140::pNlmA-luc2 Spr | This work |
| TO-nLucRoe | UA140::pNlmA-luc2/pHdrRoe Spr Kmr | This work |
| TO-nLucRMoe | UA140::pNlmA-luc2/pHdrRMoe Spr Kmr | This work |
| TO-nLucE | UA140::pNlmA-luc2/pComD-KO Spr Kmr Emr | This work |
| TO-nLucERoe | UA140::pNlmA-luc2/pComD-KO/pHdrRoe Spr Kmr Emr | This work |
| TO-nLucERMoe | UA140::pNlmA-luc2/pComD-KO/pHdrRMoe Spr Kmr Emr | This work |
| TO-nLucDR | UA140::pNlmA-DR-luc Spr Kmr | This work |
| TO-nLucDRRoe | UA140::pNlmA-DR-luc/pHdrRoe Spr Kmr | This work |
| TO-nLucDRRMoe | UA140::pNlmA-DR-luc/pHdrRMoe Spr Kmr | This work |
| Plasmids | ||
| pFW5 | Suicide vector; Spr | 27 |
| pDL278 | E. coli-Streptococcus shuttle vector; Spr | 4 |
| pBS-Kan | pBluescript with aphAIII in MCS;b Apr Kmr | G. Niu, unpublished data |
| pHdrM-i | pFW5 + internal fragment of hdrM; Spr | This work |
| pHdrRoe | pDL278::φ(ldhp-hdrR); Spr | This work |
| pHdrRoe- | pDL278::φ(ldhp[no RBS]-hdrR); Spr | This work |
| pHdrRMoe | pDL278::φ(ldhp-hdrR)/φ(ldhp-hdrM); Spr | This work |
| pNlmA-luc | pFW5::φ(nlmAp-luc); Spr | This work |
| pNlmA-luc2 | pFW5::φ(nlmAp-luc); Spr Kmr | This work |
| pNlmA-DR-luc | pFW5::φ(nlmAp[ΔDR]-luc); Spr Kmr | This work |
| pComC-del | pFW5 + allelic replacement of comC; Spr | This work |
Apr, ampicillin resistance; Emr, erythromycin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance.
MCS, multiple cloning site.
Construction of hdrM mutants.
The primers used in this study are described in Table 2. An internal DNA fragment of hdrM was amplified by PCR using the primer pair 1690 F and 1690 R. The PCR fragment was then digested with XhoI and HindIII and cloned into the vector pFW5 digested with the same enzymes. The resulting construct was confirmed by restriction analysis and PCR before integration into the chromosome of UA140 via single-crossover homologous recombination.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) | Purpose |
|---|---|---|
| 1690 F | CCCCTCGAGTTTTGGTACGGTCTGCTTGG | hdrM mutation |
| 1690 R | CCCAAGCTTTGTTTAAAATAACGCCGATAATGA | hdrM mutation |
| comC up F | GGCCTGCAGCTGTACCCATTCAGAAATCTC | comC mutation |
| comC up R | GCCCCATGGAAGTCATTTTTTAATGATAGTGTTTTTTTC | comC mutation |
| comC dn F | GGGAAGCTTTGGGAAAATAAGATAGGCTAACAT | comC mutation |
| comC dn R | CCCCTCGAGCCTCTTTCTCAGTGTGTTC | comC mutation |
| hdrR RT F | TCAGCAAATTGGCAAGAGTCA | hdrR qPCRa |
| hdrR RT R | GGAGAAAGACTTATTGATGGCAGAA | hdrR qPCR |
| comC RT F | GACTGATGAATTAGAGATTATCATTGG | comC qPCR |
| comC RT R | TTTCCCAAAGCTTGTGTAAAACT | comC qPCR |
| comE RT F | GAGTTCTCCCACCGCATTGA | comE qPCR |
| comE RT R | ACCATTCTTCTGGCTGTTTTCC | comE qPCR |
| comX RT F | TTATTTCGTGATAGTTTGCTTGCAT | comX qPCR |
| comX RT R | CAAGCGCTCAAACAGCTCTTG | comX qPCR |
| comYA RT F | CTTTTTTCTGGACGTCACGATTT | comYA qPCR |
| comYA RT R | TCGCCCCTTGATTTCATTTAAA | comYA qPCR |
| ldhp F | GCCGGATCCCCGAGCAACAATAACACTC | hdrR/M overexpression |
| ldhp R | CGCACTAGTAACATCTCCTTATAATTTATTAAGTATATATTCTAT | hdrR/M overexpression |
| ldh-RBS R | CCCACTAGTTATAATTTATTAAGTATATATTCTATACATTTTCATTCTAAC | hdrR overexpression |
| ldhp F2 | GCCAAGCTTCCGAGCAACAATAACACTC | HdrM overexpression |
| hdrR oe F | GCCACTAGTATGGAGACAAGATACATTTTTGATG | hdrR overexpression |
| hdrR oe R | CCCAAGCTTTCATAGTAAACTCCTTTTTTTCATAAGT | hdrR overexpression |
| hdrM oe F | CCCACTAGTATGAAAAAAAATTATTTTTGGTACGGT | hdrM overexpression |
| hdrM oe R | CCCAAGCTTTTAATATTGAATGTTTAGAGATCCCATAG | hdrM overexpression |
| nlmA DR F | AAAATAAATTGTTATACTAAAGATGTTGGTTG | nlmA reporter |
| nlmA DR R | TATTTTGTCTTAAACGGTCATTTTTGA | nlmA reporter |
qPCR, quantitative PCR.
Construction of the comC mutant.
Two fragments corresponding to approximately 1 kb upstream and downstream of comC were generated by PCR with Accuprime Pfx high-fidelity polymerase (Invitrogen) and the primers comC up F, comC up R, comC dn F, and comC dn R. The resulting fragments were cloned sequentially into the vector pFW5. The upstream fragment was cut with PstI and NcoI, and the downstream fragment was cut with HindIII and XhoI. Both fragments were ligated to identical sites in pFW5, which occur on either side of a spectinomycin resistance cassette. After confirmation of the resulting plasmid by restriction analysis and PCR, the linearized plasmid was transformed to S. mutans UA140.
Construction of the comE, comX, and comY mutants.
The comE, comX, and comY mutants were previously constructed (14, 20). These mutations were moved into UA140 and its derivatives by transforming genomic DNA.
Construction of overexpression strains.
To generate the hdrR overexpression strain, the entire predicted hdrR open reading frame (ORF) and the highly expressed constitutive lactate dehydrogenase (ldh) promoter were generated by PCR using Accuprime Pfx and the primers hdrR OE F and hdrR OE R (hdrR ORF), as well as ldh-p F and ldh-p R (ldh promoter). Next, the ldh amplicon was digested with BamHI and SpeI and the hdrR amplicon was digested with SpeI and HindIII. Both fragments were ligated together with the shuttle vector pDL278 digested with BamHI and HindIII. To create the ribosome binding site (RBS)-lacking hdrR overexpression strain, a similar strategy was used, except that the ldh promoter fragment was amplified using the primers ldh-p F and ldh-RBS R. The overexpression plasmids were confirmed by DNA sequencing to confirm sequence integrity and the expected fusion of the hdrR ORF. Confirmed plasmids were then transformed to S. mutans UA140. To generate the hdrRM overexpression plasmid, we first amplified the ldh promoter with ldh-p F2 and ldh-p R and the hdrM ORF with hdrM OE F and hdrM OE R. Next, both amplicons were digested with HindIII and SpeI and ligated together. The ligation mixture was amplified by PCR using Accuprime Pfx and subsequently digested with HindIII. The digested fragment was then ligated to the hdrR overexpression plasmid pldh-hdrR digested with HindIII. The hdrRM overexpression plasmid was confirmed by DNA sequencing to confirm sequence integrity and the in-frame fusion of the hdrM ORF.
Construction of nlmA-luciferase reporter gene fusions.
The nlmA-luc plasmid pFW5::φ(nlmAp-luc) was constructed previously (10). In order to use this construct with our hdrR and hdrRM overexpression constructs, it was necessary to first add a kanamycin resistance marker to the reporter plasmid. The kanamycin resistance cassette was excised from the vector pGNaa3 (24) with PvuII and ligated to a SmaI site in pNlmA-luc to create the plasmid pNlmA-luc2. In order to construct an nlmA-luc reporter plasmid containing a deletion of a single ComE binding site direct repeat, pNlmA-luc2 was used as a template for inverse PCR using the primers nlmA DR F and nlmA DR R, both of which had previously been phosphorylated with T4 polynucleotide kinase (NEB). The PCR mixture was then treated with DpnI to remove the template, and the reaction product was ligated and transformed into E. coli. The plasmids were confirmed by DNA sequencing to confirm sequence integrity and the expected deletion of the direct repeat. The plasmids were transformed into UA140 and integrated into the nlmA promoter region via single-crossover recombination. These strains were used as recipients for the hdrR and hdrRM overexpression constructs.
Luciferase assays.
Luciferase assays were performed using previously described methods (16, 19). Briefly, 25 μl 1 mM d-luciferin (Sigma) suspended in 100 mM (pH 6) citrate buffer was added to 100 μl of cell culture. Luciferase activity was measured using a TD 20/20 luminometer (Turner Biosystems). Overnight cultures of reporter strains were diluted 1:20 and grown to an optical density at 600 nm (OD600) of 0.7 to 0.8 before measurement of luciferase activity.
Transformation assays.
Determination of transformation efficiency was performed using a previously described methodology (14, 20). UA140 and its derivatives were diluted 1:30 from overnight cultures and grown to an OD600 of 0.2 to 0.3 in BHI plus 0.4% (wt/vol) bovine serum albumin (BSA) before the addition of DNA. Transforming genomic DNA containing a tetracycline marker was added at a final concentration of 10 μg ml−1 for each reaction, and the cultures were subsequently allowed to grow for an additional 2 h. Reaction mixtures assaying the effect of CSP also included 1 μg ml−1 CSP with the transforming DNA. After the incubation period, cultures were briefly sonicated to disperse cell chains and plated on tetracycline-containing BHI agar plates, as well as on nonselective BHI plates. Successful transformation was scored based on acquired tetracycline resistance following transformation, and the total number of viable cells was determined by counting the colonies growing on nonselective plates. Transformation efficiency was determined by calculating the ratio of transformants to the total number of viable cells.
RNA extraction and transcriptional analysis.
Overnight cultures of UA140 and its derivatives were diluted 1:30 in 300 ml BHI plus 0.4% (wt/vol) BSA and collected at three separate OD600 values, 0.2 to 0.3, 0.5 to 0.6, and 0.8 to 0.9. Cells were harvested by centrifugation at 4°C and then stored at −80°C. RNA was extracted from cell pellets using a previously described methodology (24). Total RNA (300 ng) was used for cDNA synthesis using Stratascript reverse transcriptase (Stratagene) according to the manufacturer's protocol. For real-time reverse transcription (RT)-PCR, primers were designed using Primer Express 3.0 software (ABI), the reaction mixtures were prepared using Applied Biosystems SYBR green PCR mastermix, and an Applied Biosystems 7300 was used for detection. Relative changes in gene expression were calculated using the ΔΔCT method described previously (24). Total cDNA abundance between samples was normalized using primers specific to the 16S rRNA gene.
Plate assay for mutacin production.
To assay for mutacin IV production, wild-type UA140 and its derivatives were first grown overnight in liquid cultures under standard anaerobic conditions. A 5-μl volume of each overnight culture was spotted onto TH agar plates and incubated anaerobically at 37°C overnight. The following day, the plates were overlaid with a soft-agar suspension of the indicator strain (Streptococcus sanguinis strain NY101) and incubated anaerobically for an additional 16 h. Zones of inhibition were indicative of mutacin IV production.
RESULTS
Epistatic analysis of the competence cascade in the hdrM background.
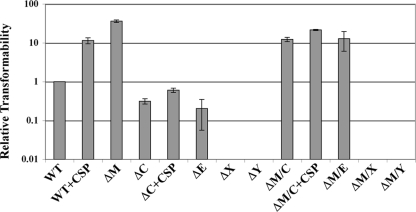
In our previous studies, we determined that a mutation of hdrM caused several phenotypes, including greatly increased genetic competence (21). This phenotype was highly unusual, as the only other reported comparable competence phenotype came from an uncharacterized mutation created by chemical mutagenesis (29). Thus, we were interested in determining the connection between hdrM and competence. Since the major components of the competence cascade were already well established, we reasoned that the competence phenotype of the hdrM mutant must require the function of the comC, comED, comX, and/or comY genes. Since the comY genes are late competence genes known to be absolutely essential for DNA uptake (20), this operon was assumed to be the minimal gene set required for the hdrM competence phenotype. Thus, we created double mutations of hdrM with each of these components of the competence cascade and assayed transformation efficiency. For comparison, we also measured transformation in the presence of exogenously added synthetic CSP. As shown in Fig. 1, CSP addition increased the transformation efficiency of wild-type UA140 >10-fold, which was considerably less than the >35-fold higher transformation efficiency of the hdrM mutant. As expected, both the comC and comED mutants had noticeably lower rates of transformation, while no transformants were detected in the comX and comY backgrounds. In contrast, in the hdrM background, both the comC and comED mutants were highly transformable and, in fact, exhibited transformation efficiencies >10-fold higher than the wild-type level (Fig. 1). As expected, CSP was able to increase the transformation efficiency of the comC (CSP gene) mutant. Interestingly, CSP could also further increase the elevated transformation efficiency of the hdrM comC double mutant. Thus, the classical competence pathway and the hdrM mutant pathway appeared to have additive effects upon competence, which suggested they are likely parallel pathways. However, hdrM was not able to rescue the severe competence defects of the comX and comY mutants, which indicated that the hdrM phenotype likely functioned through comX, since it is upstream of comY.
FIG. 1.
Transformation efficiencies of various hdrM mutants. Transformation efficiency was measured as described in Materials and Methods, and all values were normalized to the wild-type UA140 value (3.14 × 10−6). The strains are identified as follows: WT (wild-type UA140), WT+CSP (UA140 with CSP), ΔM (hdrM mutant), ΔC (comC mutant), ΔC+CSP (comC mutant with CSP), ΔE (comE mutant), ΔX (comX mutant), ΔY (comY mutant), ΔM/C (hdrM comC double mutant), ΔM/C+CSP (hdrM comC double mutant with CSP), ΔM/E (hdrM comE double mutant), ΔM/X (hdrM comX double mutant), and ΔM/Y (hdrM comY double mutant). The results presented here are the average of three independent experiments. Each experiment measured three independent clones for each sample.
HdrM controls competence via transcriptional regulation of comX.
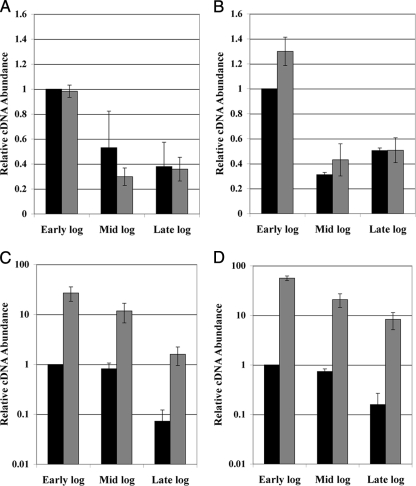
The results described above suggested that the hdrM mutation affected competence downstream of the ComCDE system, possibly through transcriptional regulation of comX or comY. To test this, we measured the transcription of the competence genes in the hdrM background to determine whether the competence phenotype could be attributed to altered transcription. Moreover, we had previously determined that comY operon expression correlated with transformation efficiency (20), so there was reason to suspect that, at a minimum, the comY operon would exhibit altered transcription. Given that competence development is transient during early log phase (15), we further reasoned that it was possible that competence gene transcription was not necessarily higher than the wild-type level but actually lasted longer. Therefore, in order to reconcile these two possible transcriptional effects, we measured the transcription of each competence gene at three different growth phases to determine whether the hdrM mutation simply induced greater competence gene expression or created an alteration in the temporal pattern of gene expression. Samples were taken at the early (OD600, 0.2 to 0.3), mid (OD600, 0.5 to 0.6), and late (OD600, 0.8 to 0.9) log phases. As shown in Fig. 2A and B, comC gene expression and comE gene expression were both quite similar between the wild type and the hdrM mutant throughout the different growth phases, although during the early log phase we had observed a slight but reproducible increase in comE gene expression in the hdrM mutant. In contrast, we observed a dramatic increase in comX and comY gene expression in the hdrM background. At the peak expression of competence in the early log phase, comX expression was >35-fold higher in the hdrM background (Fig. 2C), whereas comY was >50-fold increased (Fig. 2D). Expression of both the comX and comY genes remained higher than the wild-type level at all three time points, although the effect was consistently larger for comY. Interestingly, despite the tremendous increase in comX and comY gene expression in the hdrM mutant, these genes seemingly retained a wild-type pattern of gene expression (Fig. 2C and D). These results suggested that in the hdrM background, the transcription of comX and comY is increased over that in the wild type but the temporal pattern of gene expression is largely unaffected.
FIG. 2.
Transcription of competence genes in the hdrM background. Wild-type UA140 and the hdrM mutant were tested for the expression levels of (A) comC, (B) comE, (C) comX, and (D) comY during the early, mid, and late log phases. Data are presented relative to the transcript abundance of the wild-type early log phase sample, which was arbitrarily assigned a value of 1. Black bars represent wild-type UA140, whereas the hdrM mutant is represented by dark gray bars. These results are the average of three independent experiments. Each experiment measured three independent clones for each sample. All experiments measuring transcription via real-time RT-PCR were normalized by using the 16S rRNA gene as a housekeeping control.
The hdrM mutation increases mutacin IV production independently of ComE.
In S. mutans, multiple bacteriocins (mutacins) have been demonstrated to be under the control of the competence regulon (7, 9, 11, 31, 34). In addition, the comE gene has been recently shown to be an essential regulator for a variety of mutacins and uncharacterized mutacin-like genes (11). Given the apparent link between mutacin production and competence, we were curious as to whether a mutation of hdrM could rescue the mutacin IV deficiency seen in the comE background. As shown in Fig. 3, we performed a deferred antagonism assay to measure the growth inhibition of the mutacin IV-sensitive strain S. sanguinis NY101. As expected, no evidence of mutacin IV production was seen in the comE mutant. In contrast, both the hdrM comE double mutant and the hdrM single mutant produced more mutacin IV than the wild type, suggesting a regulatory mechanism similar to the competence phenotype. The hdrM comE nlmA triple mutant was also tested to ensure that the mutacin phenotype could be attributed to mutacin IV rather than another induced bacteriocin. In addition, we performed the same assay utilizing a comC mutant and obtained identical results (data not shown). Thus, mutacin IV, like genetic competence, is likely to have an additional hdrM-regulated pathway governing its expression that is independent of ComCDE.
FIG. 3.
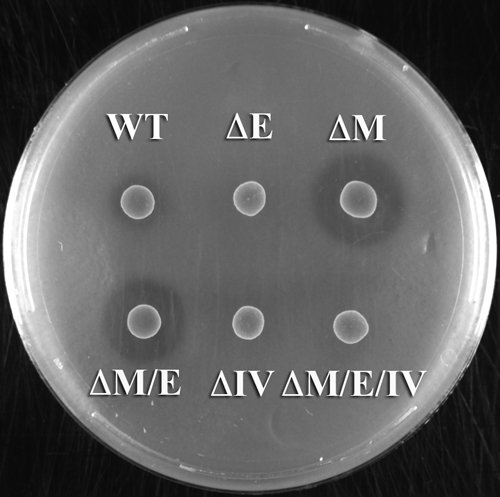
Analysis of mutacin IV (nlmA) production in the hdrM background. The production of mutacin IV was tested by the deferred antagonism assay as described in Materials and Methods. The development of a growth inhibition halo is indicative of the presence of mutacin IV inhibiting the sensitive strain S. sanguinis NY101. S. mutans strains are identified as follows: WT (wild-type UA140), ΔE (comE mutant), ΔM (hdrM mutant), ΔM/E (hdrM comE double mutant), ΔIV (nlmA mutant), and ΔM/E/IV (hdrM comE nlmA triple mutant). This experiment was performed three times with similar results.
Overexpression of hdrR reproduces hdrM phenotypes.
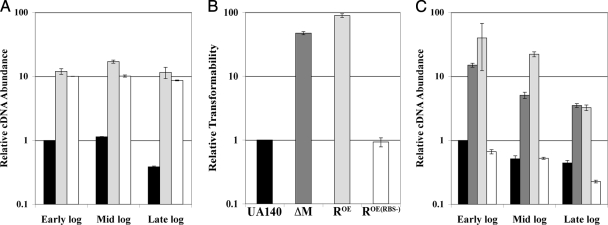
As mentioned previously, genetic evidence suggested that hdrR was involved in the same pathway as hdrM; however, a deletion of hdrR yielded none of the phenotypes seen in the hdrM mutant (21). From these results, we hypothesized that hdrM may act as a negative regulator of hdrR and thus overexpressing hdrR might generate the hdrM phenotype. To this end, we created an HdrR overexpression construct by creating a translational fusion to the highly expressed constitutive lactate dehydrogenase (ldh) gene. As a negative control, we also created another construct containing the ldh promoter but lacking an RBS (see Materials and Methods). Both constructs were then transformed into the wild-type UA140 strain. We first tested the expression of hdrR from our overexpression strains and confirmed that both constructs transcribed hdrR constitutively during growth (Fig. 4A). Next, we tested the transformation efficiency of these strains against the wild type and the hdrM mutant. Surprisingly, we found that the translational fusion hdrR overexpression strain not only reproduced the increased-competence phenotype of the hdrM mutant but also had almost twofold higher competency than the hdrM mutant strain (Fig. 4B). As expected, the RBS-negative hdrR overexpression strained was virtually indistinguishable from the wild type. Since we had previously determined that comX was the likely mediator of the hdrM competence phenotype, we also tested comX gene expression in the hdrR overexpression backgrounds. As shown in Fig. 4C, in the RBS-positive strain, the transcription of comX was greatly increased throughout the growth period and its expression was generally higher than in the hdrM mutant, whereas the RBS-negative strain never showed comX expression above wild-type levels. This result mirrored the transformation results. As a final independent confirmation of these results, we tested mutacin IV production in the hdrR overexpression strains. Similar to what was found in the hdrM mutant, the translational fusion hdrR overexpression strain produced more mutacin IV than the wild type and was able to rescue the mutacin IV deficiency of a comE mutant (Fig. 5). In contrast, no rescue was observed in the RBS-negative overexpression strain. These results suggested that HdrR is likely a positive regulator of both competence and mutacin IV gene expression.
FIG. 4.
Analysis of the hdrR overexpression strain. (A) The expression of hdrR was measured by real-time RT-PCR during the early, mid, and late log phases, and results are presented relative to the early log phase transcript abundance of the wild type. Wild-type UA140 is represented by black bars, the translational fusion hdrR overexpression strain is represented by light gray bars, and the RBS-negative hdrR overexpression strain is represented by solid white bars. (B) Comparison of the transformation efficiencies of wild-type UA140 (black), the hdrM mutant (dark gray), the translational fusion hdrR overexpression strain (light gray), and the RBS-negative hdrR overexpression strain (white). Values are presented relative to the wild type, which was arbitrarily assigned a value of 1. (C) Comparison of the expression of comX among wild-type UA140 (black), the hdrM mutant (dark gray), the translational fusion hdrR overexpression strain (light gray), and the RBS-negative hdrR overexpression strain (white). Samples were measured by real-time RT-PCR during the early, mid, and late log phases, and results are presented relative to the early log phase transcript abundance of the wild type. The data in panels A to C are the average of three independent experiments. Each experiment measured three independent clones for each sample.
FIG. 5.
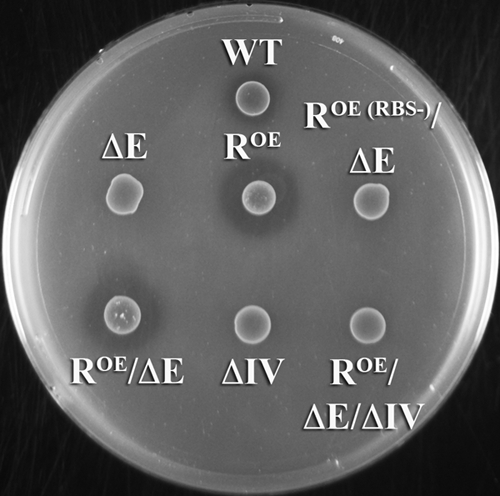
Analysis of mutacin IV production in the hdrR overexpression strain. The production of mutacin IV was detected with the deferred antagonism assay using S. sanguinis NY101. S. mutans strains are identified as follows: WT (wild-type UA140), ΔE (comE mutant), ROE (translational fusion hdrR overexpression strain), ROE(RBS−)/ΔE (RBS-negative hdrR overexpression strain in the comE background), ROE/ΔE (translational fusion hdrR overexpression strain in the comE background), ΔIV (nlmA mutant), and ROE/ΔE/ΔIV (translational fusion hdrR overexpression strain in the comE nlmA background). This experiment was performed three times with similar results.
HdrR activity is regulated by HdrM and functions similarly to ComE.
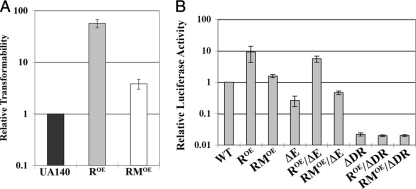
From our analysis of hdrR and hdrM, it seemed likely that both genes function together to regulate competence development and bacteriocin production. Also, from our hdrR overexpression data, we speculated that hdrM somehow serves to antagonize the function of hdrR. In order to test this further, we created another overexpression construct containing separate ldh translational fusions to both hdrR and hdrM. Next, we assayed the transformation efficiency of this strain and determined that the addition of hdrM to the overexpression construct reduced the transformation efficiency to nearly wild-type levels (Fig. 6A). Thus, overexpression of hdrM together with hdrR was indeed able to counteract the competence phenotype associated with hdrR overexpression.
FIG. 6.
Investigation of the effect of hdrRM operon overexpression upon competence and mutacin IV production. (A) Comparison of transformation efficiencies among wild-type UA140 (black), the hdrR overexpression strain (gray), and the hdrRM overexpression strain (white). (B) Luciferase activities of the nlmA-luc reporter strain in the following backgrounds: WT (wild-type UA140), ROE (hdrR overexpression strain), RMOE (hdrRM overexpression strain), ΔE (comE mutant), ROE/ΔE (hdrR overexpression strain in the comE background), RMOE/ΔE (hdrRM overexpression strain in the comE background), ΔDR [UA140 in nlmA(Δdirect repeat)-luc], ROE/ΔDR [hdrR overexpression strain in nlmA(Δdirect repeat)-luc], and RMOE/ΔDR [hdrRM overexpression strain in nlmA(Δdirect repeat)-luc]. The competence and luciferase assays were normalized to the wild-type UA140 values. All results are the average of three independent experiments. Each experiment measured three independent clones for each sample.
Next, we decided to take a similar approach to test whether this was true for mutacin IV regulation as well. For this purpose, we created luciferase reporter constructs for the mutacin IV structural gene nlmA. As shown in Fig. 6B, the hdrR overexpression strain increased nlmA-luc reporter activity, which agreed well with the mutacin IV phenotype of this strain. However, overexpressing hdrR and hdrM together reduced the reporter activity to nearly wild-type levels. The same result was repeatable in the comE background. Since the ComE binding site for competence-regulated bacteriocins has been previously determined (9, 11), we also decided to test whether the mutacin IV phenotype of the hdrR overexpression strain functioned through the ComE recognition sequence. We hypothesized that this may be the case since hdrR appeared to regulate the same genes as comE. Additionally, both hdrR and comE belong to the LytTR family of transcription regulators and it has been proposed that members of this family recognize a conserved consensus pattern of imperfect direct repeats separated by 12- to 13-bp spacers (23). This is also the same motif recognized by ComE in competence and mutacin gene promoters of S. mutans (9). When the hdrR overexpression mutant was tested in an nlmA-luc reporter containing a deletion of just one of these two direct repeats, we observed a severe reduction in nlmA-luc reporter activity (Fig. 6B). This suggested that HdrR or possibly a downstream regulator recognizes the same nlmA promoter elements as ComE. Since a similar LytTR-type recognition sequence does not exist in the S. mutans comX promoter, it is unknown whether potential ComE or HdrR binding sites exist within the comX promoter.
DISCUSSION
In this study, we investigated the mechanism by which the novel HdrRM system regulates competence and mutacin production in S. mutans. Through a combination of epistatic analysis and gene expression studies, we determined that the hdrM mutation affects competence through the transcription of comX and is independent of the comC and comED genes. The hdrM mutation also affected the production of mutacin IV, which has been previously demonstrated to require ComE (10, 11, 21). Similar to competence, the hdrM mutation was able to increase mutacin IV production, even in the comE background. In addition, each of these phenotypes was reproducible when HdrR was overproduced in a wild-type background. This provides an explanation for our previous observation that an hdrR deletion does not result in the same phenotypes as an hdrM mutation (21). Furthermore, the phenotypes of the hdrR overexpression strain could be reverted by overexpressing hdrM together with hdrR. This suggests that hdrM is probably a negative regulator of hdrR activity. Finally, using the mutacin reporter nlmA-luc, we demonstrated that HdrR likely recognizes the same direct repeats in the mutacin IV promoter as ComE but does not function through ComE.
Based upon these results, we propose a model in which the S. mutans competence regulon has at least two independent upstream regulatory systems, the ComCDE system and the HdrRM system (Fig. 7). The two pathways appear to be parallel, as the competence and mutacin IV phenotypes were fully independent of comC and comED in the hdrM mutant or hdrR overexpression background. Yet, the two systems strongly overlap in their ability to regulate the competence regulon. Both pathways converge on comX in order to regulate competence, and they both regulate mutacin IV through conserved direct repeats in the nlmA promoter. However, based upon our genetic and transcriptional data, it appears that the two systems utilize different mechanisms to achieve similar results. Unlike the ComDE two-component system, in the HdrRM system, the transcription regulator (HdrR) does not require the membrane protein (HdrM) for functionality. Rather, we propose that HdrM serves to antagonize the function of HdrR (Fig. 7). Thus, in the hdrM background, the ability to antagonize HdrR would be lost, which could account for the increased expression of comX and nlmA. This model is also consistent with our previous observation that a double mutation of hdrR and hdrM can suppress the hdrM phenotype (21). Our present data suggest that HdrR is the mediator of this phenotype. We have yet to determine whether the antagonism of HdrR occurs directly between HdrM and HdrR or through some other mechanism, but these studies are under way.
FIG. 7.
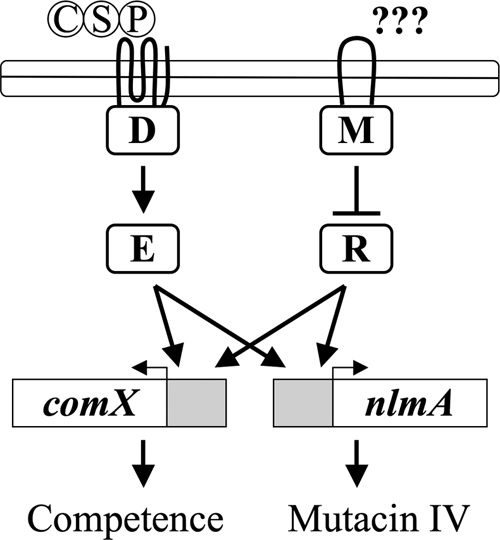
Model of the hdrRM-regulated competence pathway. HdrM (M) responds to unknown environmental signals which regulate the antagonism of HdrR (R) transcription factor activity. When the environmental conditions are such that HdrM is unable to antagonize HdrR, this will allow HdrR to activate the transcription of comX for natural competence and nlmA for mutacin IV production. This pathway functions in parallel with ComD (D) and ComE (E), which respond to the presence of CSP.
An obvious question arising from this study is why S. mutans might require multiple parallel regulatory systems for competence and mutacin production. Part of the answer may be the natural life cycle of S. mutans in the oral cavity. For example, current data suggest that the ComCDE system is the primary competence regulatory system in planktonic cultures of S. mutans. However, as a primarily biofilm-dwelling organism, S. mutans also routinely proliferates in an environment that contains a cell density 2 to 3 orders of magnitude greater than that of a planktonic culture. Under these vastly different conditions, an entirely separate set of stimuli may activate another regulatory system, such as HdrRM, to stimulate competence. In fact, both mutacin IV and the hdrRM operon have been previously shown to be inducible by conditions of extremely high cell density, similar to that obtained in a biofilm or plate colony (10, 21). Furthermore, we speculate that the standard transformation assay probably does not contain the proper stimulus required to counteract the antagonistic effect of hdrM, which would explain why a deletion of hdrR produces a wild-type competence phenotype. Under these conditions, the negative regulatory function of hdrM might predominate, which would then be the functional equivalent of an hdrR mutation (i.e., no HdrR transcription factor activity). However, if under these same conditions HdrR were forced to be active through a mutation of hdrM or through constitutive overexpression, then it would induce competence together with the ComCDE system. Presumably, the additive effect of both pathways working simultaneously would result in greatly increased competence. These results imply that the competence system can be regulated by a variety of stimuli sensed by distinct regulatory systems. Considering the substantial expenditure of energy and genetic information devoted to controlling the competence regulon, as well as producing the competence machinery and competence-regulated bacteriocins, it seems that competence must play a significant role in the survival of the species in the oral cavity.
From the data presented in the present study, two inferences can be made regarding the functionality of the hdrRM system. First, it seems that the antagonist function of HdrM is likely to be sensitive to the concentration of HdrR. Even though overexpressing hdrM together with hdrR strongly inhibited the competence and nlmA gene expression phenotypes seen in the hdrR overexpression strain, the phenotypes were never fully restored to wild-type levels (Fig. 6A and B). In the hdrRM overexpression construct, both genes were expressed as separate translational fusions to the same ldh promoter sequence. Thus, HdrR and HdrM protein levels should be nearly equivalent. Further support for this notion may be contained within the hdrR gene itself. Upstream of the annotated hdrR start codon, there is no Shine-Dalgarno-type RBS, nor is there a potential alternative translation start site with an RBS sequence. In fact, we examined five other strains of S. mutans and found the same result (data not shown). In contrast, the hdrM ORF is preceded by an obvious RBS sequence. Given the apparent lack of an RBS in hdrR, it is possible that fewer HdrR molecules are translated relative to HdrM. Future studies will determine if translational efficiency is indeed a component required for the proper regulation of the system. Second, we speculate that HdrR functions similarly to ComE, albeit under different growth conditions (Fig. 7). Conspicuously, both HdrR and ComE belong to the LytTR family of transcription regulators, which recognize a conserved but flexible consensus sequence (23). Consistent with this prediction, both ComE and HdrR regulate competence and mutacin production and require the same consensus LytTR family DNA binding motif in the nlmA promoter. Interestingly, we identified five other putative LytTR family transcription regulators within the genome of S. mutans in addition to hdrR and comE. An alignment of each of these putative proteins with HdrR suggests that the homology between four of these proteins (SMU.294, SMU.433, SMU.576c, and SMU.1070c) and HdrR is even greater than that between HdrR and ComE. Therefore, it will be interesting to determine whether any of these proteins are similarly able to influence the transcription of the competence regulon. These genes may well comprise additional uncharacterized competence regulatory systems. It would also be interesting to determine whether S. mutans is unique in its ability to control competence through multiple regulatory systems or other species such as S. pneumoniae contain similar parallel competence regulatory pathways yet to be discovered.
Acknowledgments
This work was supported by an NCRR COBRE P20-RR018741-05 grant and an NIDCR DE018725 grant to J.M. and an NIDCR DE014757 grant to F.Q.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74:1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barsamian-Wunsch, P., J. H. Park, M. R. Watson, N. Tinanoff, and G. E. Minah. 2004. Microbiological screening for cariogenic bacteria in children 9 to 36 months of age. Pediatr. Dent. 26:231-239. [PubMed] [Google Scholar]
- 3.Bowden, G. H. 1990. Microbiology of root surface caries in humans. J. Dent. Res. 69:1205-1210. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y. Y., and D. J. LeBlanc. 1992. Genetic analysis of scrA and scrB from Streptococcus sobrinus 6715. Infect. Immun. 60:3739-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Q., E. A. Campbell, A. M. Naughton, S. Johnson, and H. R. Masure. 1997. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23:683-692. [DOI] [PubMed] [Google Scholar]
- 6.Cvitkovitch, D. G. 2001. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:217-243. [DOI] [PubMed] [Google Scholar]
- 7.Hale, J. D., B. Balakrishnan, and J. R. Tagg. 2004. Genetic basis for mutacin N and of its relationship to mutacin I. Indian J. Med. Res. 119(Suppl.):247-251. [PubMed] [Google Scholar]
- 8.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 9.Kreth, J., D. C. Hung, J. Merritt, J. Perry, L. Zhu, S. D. Goodman, D. G. Cvitkovitch, W. Shi, and F. Qi. 2007. The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 153:1799-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57:392-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kreth, J., J. Merritt, L. Zhu, W. Shi, and F. Qi. 2006. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 265:11-17. [DOI] [PubMed] [Google Scholar]
- 12.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindler, L. E., and F. L. Macrina. 1986. Characterization of genetic transformation in Streptococcus mutans by using a novel high-efficiency plasmid marker rescue system. J. Bacteriol. 166:658-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loimaranta, V., J. Tenovuo, L. Koivisto, and M. Karp. 1998. Generation of bioluminescent Streptococcus mutans and its usage in rapid analysis of the efficacy of antimicrobial compounds. Antimicrob. Agents Chemother. 42:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lunsford, R. D., and J. London. 1996. Natural genetic transformation in Streptococcus gordonii: comX imparts spontaneous competence on strain Wicky. J. Bacteriol. 178:5831-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt, J., J. Kreth, F. Qi, R. Sullivan, and W. Shi. 2005. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J. Microbiol. Methods 61:161-170. [DOI] [PubMed] [Google Scholar]
- 20.Merritt, J., F. Qi, and W. Shi. 2005. A unique nine-gene comY operon in Streptococcus mutans. Microbiology 151:157-166. [DOI] [PubMed] [Google Scholar]
- 21.Merritt, J., L. Zheng, W. Shi, and F. Qi. 2007. Genetic characterization of the hdrRM operon: a novel high-cell-density-responsive regulator in Streptococcus mutans. Microbiology 153:2765-2773. [DOI] [PubMed] [Google Scholar]
- 22.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu, G., T. Okinaga, L. Zhu, J. Banas, F. Qi, and J. Merritt. 2008. Characterization of irvR, a novel regulator of the irvA-dependent pathway required for genetic competence and dextran-dependent aggregation in Streptococcus mutans. J. Bacteriol. 190:7268-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobre dos Santos, M., L. Melo dos Santos, S. B. Francisco, and J. A. Cury. 2002. Relationship among dental plaque composition, daily sugar exposure and caries in the primary dentition. Caries Res. 36:347-352. [DOI] [PubMed] [Google Scholar]
- 26.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 27.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 28.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao, L., T. J. MacAlister, and J. M. Tanzer. 1993. Transformation efficiency of EMS-induced mutants of Streptococcus mutans of altered cell shape. J. Dent. Res. 72:1032-1039. [DOI] [PubMed] [Google Scholar]
- 30.Thenisch, N. L., L. M. Bachmann, T. Imfeld, T. Leisebach Minder, and J. Steurer. 2006. Are mutans streptococci detected in preschool children a reliable predictive factor for dental caries risk? A systematic review. Caries Res. 40:366-374. [DOI] [PubMed] [Google Scholar]
- 31.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Houte, J. 1993. Microbiological predictors of caries risk. Adv. Dent. Res. 7:87-96. [DOI] [PubMed] [Google Scholar]
- 33.Weerkamp, A., L. Bongaerts-Larik, and G. D. Vogels. 1977. Bacteriocins as factors in the in vitro interaction between oral streptococci in plaque. Infect. Immun. 16:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yonezawa, H., and H. K. Kuramitsu. 2005. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob. Agents Chemother. 49:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zambon, J. J., and S. A. Kasprzak. 1995. The microbiology and histopathology of human root caries. Am. J. Dent. 8:323-328. [PubMed] [Google Scholar]