Abstract
Cystic fibrosis transmembrane conductance regulator (CFTR) inhibitory factor (Cif) is a virulence factor secreted by Pseudomonas aeruginosa that reduces the quantity of CFTR in the apical membrane of human airway epithelial cells. Initial sequence analysis suggested that Cif is an epoxide hydrolase (EH), but its sequence violates two strictly conserved EH motifs and also is compatible with other α/β hydrolase family members with diverse substrate specificities. To investigate the mechanistic basis of Cif activity, we have determined its structure at 1.8-Å resolution by X-ray crystallography. The catalytic triad consists of residues Asp129, His297, and Glu153, which are conserved across the family of EHs. At other positions, sequence deviations from canonical EH active-site motifs are stereochemically conservative. Furthermore, detailed enzymatic analysis confirms that Cif catalyzes the hydrolysis of epoxide compounds, with specific activity against both epibromohydrin and cis-stilbene oxide, but with a relatively narrow range of substrate selectivity. Although closely related to two other classes of α/β hydrolase in both sequence and structure, Cif does not exhibit activity as either a haloacetate dehalogenase or a haloalkane dehalogenase. A reassessment of the structural and functional consequences of the H269A mutation suggests that Cif's effect on host-cell CFTR expression requires the hydrolysis of an extended endogenous epoxide substrate.
Pseudomonas aeruginosa is a Gram-negative bacterium that acts as an opportunistic pathogen. In colonizing the urinary tract, eye, and lung, as well as the surfaces of implanted medical devices, it forms antibiotic-resistant biofilms (12). In nosocomial infections, such as ventilator-associated pneumonia, P. aeruginosa is the second most common bacterial agent, and it represents the leading cause of death due to hospital-acquired infection (1). Among patients with compromised pulmonary function, P. aeruginosa frequently establishes persistent lung infections, exacerbating outcomes in chronic obstructive pulmonary disease (42) and cystic fibrosis (30). Overall, nearly 80% of patients with cystic fibrosis have a chronic P. aeruginosa infection in the lung by age 18 (22). Preventing infection by limiting exposure to the pathogen is difficult due to its ubiquitous distribution in the environment (47). On the other hand, the treatment of chronic lung infections is similarly challenging due to the formation of antibiotic-resistant biofilms. As a result, there currently is no effective treatment to eradicate a chronic P. aeruginosa infection from the lung once established (15, 54).
P. aeruginosa secretes a multitude of virulence factors that assist the bacterium during the initial process of airway colonization and biofilm formation (32), in some instances acting directly on host cells. In particular, it was shown recently in a coculture model that the presence of P. aeruginosa causes a decrease in the quantity of cystic fibrosis transmembrane conductance regulator (CFTR) at the apical membrane of human airway epithelial cells (49). CFTR is the ion channel responsible for chloride secretion into the airway surface liquid (ASL) in the lung. The removal of CFTR from the cell surface leads to reduced chloride efflux, ASL dehydration, and decreased mucociliary clearance, thus facilitating the establishment of a bridgehead for bacterial infection.
The downregulation of plasma membrane CFTR is mediated by a single secreted protein, the CFTR inhibitory factor (Cif), which is encoded at the PA14_26090 or cif locus (37) and is delivered into the host cell by outer membrane vesicles (5). The CFTR inhibitory effect also can be replicated by the application of purified, recombinant Cif protein directly to the apical surface of human airway epithelial cells. Within an hour after treatment with Cif, the levels of CFTR in the apical membrane are significantly reduced (37). While the mechanism of Cif action is incompletely understood, Cif has been shown to inhibit the recycling of CFTR to the apical membrane following endocytic uptake (49). Additional work has shown that Cif treatment causes a similar effect on some ABC transporters while having no effect on others (56). The mechanism by which Cif is able to generate this selectivity currently is unknown.
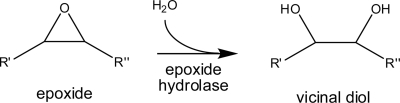
Based on sequence comparisons, Cif was predicted to belong to the α/β hydrolase family (37), which contains several different classes of enzymes with closely related sequences. Specifically, Cif showed the greatest degree of sequence similarity to the class of epoxide hydrolases (EHs), which catalyze the conversion of epoxide moieties to vicinal diols (Fig. 1). The EHs are conserved between bacteria and mammals and are used to detoxify products of oxidative metabolism as well as xenobiotic compounds. In mammals, they metabolize potent chemical signal mediators (9). As a family, they also are of potential biocatalytic interest (46). EHs have not previously been reported as bacterial virulence factors, but Pseudomonas is adept at exploiting a wide variety of biochemical strategies to subvert host cell functions.
FIG. 1.
Epoxide hydrolase activity. The EH class of enzymes is responsible for the catalytic addition of a water molecule to an epoxide ring, creating a vicinal diol.
Preliminary mutagenesis experiments targeting Cif suggested a link between EH activity and host cell effects, but these studies relied on low-identity sequence alignments and activity assays performed with an artificial EH substrate (37) that also is susceptible to esterase activity (18). Sequence alignment of Cif with known EHs revealed substitutions in several conserved EH motifs thought to be required for the formation of the enzyme active site. Furthermore, sequence relationships suggest that several haloacetate dehalogenases (HADs) previously had been misclassified as EHs, all of which cluster in the EH subgroup that includes Cif (52). As a basis for a detailed structure-function analysis of Cif, we have determined its crystal structure and assayed its activity against a variety of candidate substrates for EHs and related α/β hydrolases both for wild-type protein and for a mutation that abrogates Cif's host cell activity.
MATERIALS AND METHODS
Purification of Cif protein.
Wild-type Cif (Cif-WT) and Cif-H269A proteins possessing a C-terminal hexahistidine tag were encoded on plasmids pDPM73 and pDPM77 (37), respectively, under the control of a pBAD arabinose-inducible promoter. Cif-WT and Cif-H269A proteins were expressed in Escherichia coli and purified using immobilized metal affinity chromatography as previously described (3). Residue numbering refers to the complete sequence, including the N-terminal secretion signal. Inductively coupled plasma mass spectrometry of purified Cif was performed at the Dartmouth Trace Element Analysis Core.
Selenomethionine-labeled Cif-WT was expressed from cells grown in selenomethionine M9 minimal medium (0.6% Na2HPO4, 0.3% KH2PO4, 0.1% NH4Cl, 0.05% NaCl, 3 × 10−4% CaCl2, 1.2 × 10−2% MgSO4, 5 × 10−5% thiamine, 1.5 × 10−2% ampicillin, 0.2% [vol/vol] glycerol, 1 × 10−2% l-lysine, 1 × 10−2 % l-phenylalanine, 1 × 10−2% l-threonine, 5 × 10−3% l-isoleucine, 5 × 10−3% l-leucine, 5 × 10−3% l-valine, 5 × 10−3% l-selenomethionine, 0.2% l-arabinose) for 140 h. Labeled Cif-WT was purified as previously described (3). Selenomethionine incorporation was verified by matrix-assisted laser desorption-ionization time-of-flight spectrometry on an Applied Biosystems Voyager DE Pro System following desalting using Omix C18 pipette tips (Varian) and target preparation using sinapinic acid matrix (Sigma).
Crystallization of Cif.
Cif protein crystals were obtained by vapor diffusion against 400 μl of reservoir solution in a 4-μl hanging drop (5 mg/ml protein was mixed in a 1:1 ratio with reservoir solution) at 291 K (3). For Cif-WT, the reservoir solution consisted of 14% (wt/vol) polyethylene glycol (PEG) 8000, 125 mM CaCl2, 100 mM Na acetate (pH 5). For selenomethionine Cif-WT, the reservoir solution consisted of 20% (wt/vol) PEG 8000, 150 mM CaCl2, and 100 mM Na acetate (pH 5). For Cif-H269A, the reservoir solution consisted of 20% (wt/vol) PEG 8000, 550 mM CaCl2, 100 mM Na acetate (pH 5).
Prior to data collection, crystals were soaked in a cryoprotectant consisting of the reservoir solution supplemented with 20% (wt/vol) glycerol and then were flash cooled in the nitrogen stream of an Oxford Cryostream 700 operating at 100 K.
Data collection, processing, structure refinement, and analysis.
Oscillation data for all crystals were collected at the X6A beamline of the National Synchrotron Light Source at Brookhaven National Laboratory. For selenomethionine Cif-WT, anomalous diffraction data were collected at the selenium absorption peak; E = 12.667 keV and λ = 0.9787 Å. Diffraction data also were collected at λ = 0.9781 Å and 0.9464 to 1.8-Å resolution for Cif-WT and λ = 0.9464 to 1.5-Å resolution for Cif-H269A. All data sets were indexed, integrated, and scaled with the XDS package (31), and the Rfree test set was selected in thin shells for Cif-WT and at random for Cif-H269A using SFTOOLS from the CCP4 program suite (11). Phase determination by single-wavelength anomalous diffraction (SAD), automated building of a preliminary structure (see Fig. S1 in the supplemental material), automated molecular replacement searches against native data, and iterative rounds of structure refinement were carried out using Phenix 1.3 (3). Manual adjustment of the model using WinCoot (21) was carried out between rounds of automated refinement. Noncrystallographic symmetry restraints were employed during initial rounds of refinement and were released during the final rounds as refinement statistics converged. The selenomethionine Cif-WT structure was used as a molecular replacement search object for the initial phasing of the Cif-H269A data. To avoid potential phase bias from the WT structure, composite iterative-build OMIT maps (50) were used to determine the initial Cif-H269A model. Structure refinement proceeded as it did for the WT structure. Root mean square deviation (rmsd) calculations (using Cα atoms) and structural alignments were carried out using DaliLite v.3 (28). Images of the models were prepared using PyMOL (17). Pathways to the active site were calculated using Caver (45), and the enclosed volume was determined using Pocket-Finder (26). Coordinates and structure factors have been deposited in the Protein Data Bank (PDB; entries 3KD2 and 3KDA for Cif-WT and Cif-H269A, respectively).
Hydrodynamic analysis.
Velocity sedimentation ultracentrifugation was carried out with 8.4 μM Cif-WT and Cif-H269A (yielding an A280 of 0.4 per cm) in 100 mM NaCl, 20 mM sodium phosphate (pH 7.4) as previously described (13). Protein partial specific volume, buffer viscosity, and buffer density were determined using the program SEDNTERP (34), and data were analyzed using SEDFIT87 (14).
Size-exclusion chromatography (SEC) was carried out with a Superdex 200 HR 10/30 column (GE Healthcare) calibrated with aldolase, ovalbumin, chymotrypsinogen A, and RNase A. The SEC buffer consisted of 100 mM NaCl and 20 mM sodium phosphate (pH 7.4).
Enzymology.
Fluoroacetate dehalogenase activity was assayed by measuring the production of glycolic acid using a protocol adapted from Wolff (53) and Eegriwe (20). Briefly, 20 μM enzyme was incubated with 10 mM fluoroacetate and 100 mM glycine (pH 9) for 30 min at 30°C in a final volume of 200 μl. Two ml of 0.1% 2,7-dihydroxynaphthalene in concentrated H2SO4 then was added, and the solution was incubated at 100°C for 20 min. After the solution was cooled on ice, its optical density was measured at 540 nm. A standard curve was generated with a dilution series of glycolic acid and used to determine the relationship between substrate hydrolysis and colorimetric readout. Similar techniques were used to monitor the dehalogenation of chloro-, bromo-, and iodoacetate.
Haloacetate dehalogenase activity was determined at pH 8.2 using a phenol red reporter assay according to Holloway et al. (27) with 2.3 μM Cif-WT and 10 mM 1,2-dichloroethane as the substrate.
The assay for epibromohydrin (EBH) hydrolysis was adapted from Cedrone et al. (7). Briefly, 20 μM Cif was incubated with 10 mM epibromohydrin for 30 min at 37°C in a total volume of 100 μl in 100 mM NaCl, 20 mM sodium phosphate (pH 7.4). The reaction was stopped by the addition of 50 μl of 5 mM NaIO4 in 90% acetonitrile and incubated for 30 min at room temperature. Fifty μl of 6 mM epinephrine-HCl then was added, and the samples were spun for 5 min at 13,000 rpm in a table-top centrifuge to remove protein precipitate. One hundred μl of supernatant from each sample then was added to a 96-well plate, and the optical density was measured at 490 nm. A standard curve was generated using 3-bromopropane-1,2-diol.
4-Nitrophenyl-2S,3S-epoxy-3 phenylpropyl carbonate (S-NEPC) hydrolysis was measured based on a protocol described by Dietze et al. (18). Briefly, samples were incubated for 1 h at 37°C in a 200-μl volume containing 100 μg protein, 500 mM NaCl, 20 mM Tris (pH 8.5), 100 μM S-NEPC, and 1% (vol/vol) dimethyl sulfoxide. The assay was carried out in a 96-well plate, and the optical density was measured at 405 nm in 1-min intervals. A standard curve was generated using 4-nitrophenol.
The assays for hydrolysis of trans-stilbene oxide (TSO), cis-stilbene oxide (CSO), trans-1,3-diphenylpropene oxide (TDPPO), and juvenile hormone (JH3) were carried out using radiolabeled substrates according to Gill et al. (23), with the enzyme reaction carried out at 37°C for 1 h in a 100-μl volume containing 1 μM Cif, 25 mM sodium phosphate (pH 7.4), 50 μM radiolabeled epoxide substrate, and 1% (vol/vol) dimethyl sulfoxide. Unhydrolyzed epoxides were removed from the reaction by organic extraction, and residual radioactivity was determined.
Biochemical determination of apical membrane CFTR.
The determination of apical membrane CFTR abundance was performed using CFBE4lo− cells stably transduced with WT-CFTR (CFBE-WT) (4) and domain-selective cell surface biotinylation using EZ-LinkTM Sulfo-NHS-LC-Biotin (Pierce), as described previously in detail (37, 41, 48).
RESULTS AND DISCUSSION
Cif has an α/β hydrolase fold.
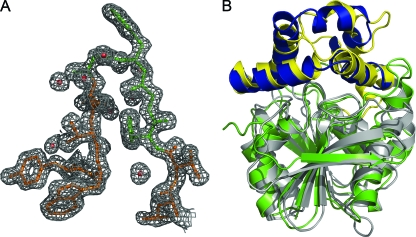
Initial structure determination was performed using 2.0-Å resolution SAD data (35) from a crystal of selenomethionine-labeled Cif (Table 1; also see Fig. S1 in the supplemental material). To determine the structure of the native protein, the preliminary selenomethionine structure was used as a search model for molecular replacement calculations using 1.8-Å resolution diffraction data from a crystal of unlabeled Cif-WT (Table 1). As expected based on Matthews volume calculations, the crystals exhibit noncrystallographic symmetry, with four copies of the protein in the asymmetric unit, corresponding to a solvent content of 46%. Following structure refinement, in all four chains electron density is visible beginning at Ala25, the first residue of the mature, secreted protein. Electron density is not observed for the C-terminal hexahistidine tag or the preceding two residues, and these residues are omitted from the model. The final model of Cif-WT contains 1,172 residues in four chains and shows excellent agreement with the diffraction data (Table 1) and the final refined electron density map of the unlabeled protein (Fig. 2A).
TABLE 1.
Data collection and refinement statistics
| Measurement | Cif-WT | Cif-H269A | Cif-WT SeMet |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Unit cell dimensions | |||
| a, b, c (Å) | 168.2, 83.9, 89.0 | 168.0, 83.6, 88.9 | 168.3, 83.6, 89.0 |
| α, β, γ (°) | 90, 100.5, 90 | 90, 100.5, 90 | 90, 100.5, 90 |
| Resolutiona (Å) | 29.55-1.80 (1.87-1.80) | 48.98-1.50 (1.53-1.50) | 19.74-2.00 (2.15-2.00) |
| Rsymb (%) | 7.4 (27.6) | 6.3 (31.4) | 13.6 (37.5) |
| I/σI | 19.5 (5.2) | 22.6 (5.3) | 10.6 (4.4) |
| Completeness (%) | 99.8 (99.9) | 99.7 (99.8) | 97.9 (97.5) |
| Phase determination | |||
| SigAnoc | 1.10 (0.86) | ||
| Figure of merit (Phaser) | 0.41 | ||
| Figure of merit (Resolve) | 0.71 | ||
| Molecular replacement | |||
| Rotation function search | |||
| Peak no. | 2 | 1 | |
| Log-likelihood gain | 3,560 | 4,470 | |
| Z score | 75.9 | 24.6 | |
| Translation function search | |||
| Peak no. | 1 | 1 | |
| Log-likelihood gain | 6,793 | 7,647 | |
| Z score | 63.9 | 72.9 | |
| Overall log-likelihood gain | 11,262 | 12,556 | |
| Refinement | |||
| Total no. of reflections | 112,388 | 192,673 | |
| Reflections in the test set | 5,733 | 9,636 | |
| Rworkd/Rfreee | 16.7/18.2 | 16.9/18.3 | |
| No. of atoms | |||
| Protein | 9,436 | 9,561 | |
| Solvent | 881 | 1,111 | |
| Ramachandran plotf (%) | 90.4/9.2/0.4/0 | 91.0/8.6/0.4/0 | |
| Bav (Å2) | |||
| Protein | 14.4 | 13.4 | |
| Solvent | 25.0 | 24.5 | |
| Bond length rmsd (Å) | 0.004 | 0.005 | |
| Bond angle rmsd (°) | 0.825 | 1.066 |
Values in parentheses are for data in the highest-resolution shell.
Rsym = ΣhΣi |I(h) − Ii(h)|/ΣhΣi Ii(h), where Ii(h) and I(h) values are the i-th and mean measurements of the intensity of reflection h, respectively.
SigAno = <(|F(+) − F(−)|/σΔ)>.
Rwork = Σh |Fobs(h) − Fcalc(h)|/Σh Fobs(h), hɛ {working set}.
Rfree = Σh |Fobs(h) − Fcalc(h)|/Σh Fobs(h), hɛ {test set}.
Core/allowed/generously allowed/disallowed.
FIG. 2.
Crystal structure of Cif. (A) 2Fo-Fc map of the final refined electron density of Cif-WT, contoured to 1σ; the HGFG motif (residues 61 to 64) is colored green. A density mask with a 1.8-Å radius has been applied to prevent the display of electron density from neighboring residues. (B) Ribbon diagram depicting the superposition of ArEH and Cif-WT by DaliLite v.3 (28). The Cif core domain is colored in gray, and the cap domain is in yellow; ArEH is colored with a green core and a blue cap domain.
In addition to the protein and water moieties, the electron density map contains five prominent difference peaks not accounted for by the model. The shape, electrostatic environment, and coordination geometry of the peaks are suggestive of elemental ion-binding sites for two cations and three anions. However, inductively coupled plasma mass spectrometry of purified Cif failed to reveal significant levels of any candidate cation constitutively bound to the protein. It therefore is most likely that the sites are occupied by weakly bound calcium and chloride ions derived from the crystallization buffer. However, given the tentative nature of this assignment, we have not included ions in the model.
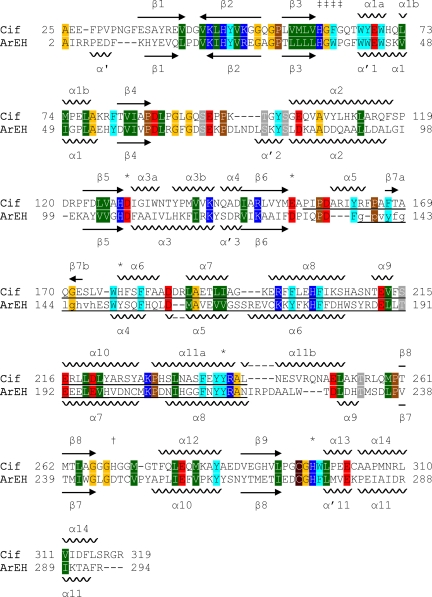
Overall, the Cif structure consists of two domains, one that is largely α helical and one that has a mixed α/β structure. Initial sequence alignments suggested that Cif is a member of the α/β hydrolase family (37). To test this hypothesis at the structural level, we performed a distance matrix alignment (28) of Cif with proteins of known structure. The closest structures all are α/β hydrolases and include the EH from Agrobacterium radiobacter AD1 (ArEH; PDB entry 1EHY) (43). Despite sharing only 21.7% sequence identity, these two structures align with a Z score of 32.4 and a Cα rmsd of 1.9 Å (Fig. 2B and 3), which is consistent with a close agreement between their tertiary structures. Like ArEH, Cif can be divided into two domains: a core and a cap (Fig. 2B). The core domain of Cif exhibits the archetypal fold of the α/β hydrolase family (6, 44): a β-sheet containing seven parallel strands and one antiparallel strand is sandwiched between α helices and spans residues 25 to 154 and 243 to 319. The cap domain consists of residues 155 to 242 and forms a lid-like structure attached to one end of the α/β hydrolase core domain. The amino-terminal boundary between the domains is located in a region of undefined electron density in ArEH, corresponding to residues that are missing in the model. In contrast, this region is well defined in the structure of Cif and contains helix α5 (Fig. 3), which we have assigned to the cap domain due to its surface localization and proximity to the dimer interface.
FIG. 3.
Sequence and structural similarity to ArEH. The sequences of Cif and ArEH were aligned, and the secondary structure was determined using DaliLite v.3 (28). Residues 138 to 148, which are shown in lowercase letters, are absent from the crystal structure of ArEH (PDB ID 1EHY) and were aligned using ClustalW (51). Arrows indicate β strands, and the relative orientation within a sheet is indicated by the direction of the arrow. The cap domain of each protein is underlined. The secondary structure elements of ArEH are named according to Nardini et al. (43). Amino acids are colored as follows: small side chains are orange (Gly and Ala), Pro is brown, Cys is maroon, polar side chains are gray (Ser, Thr, Asn, and Gln), acidic side chains are red (Asp and Glu), basic side chains are blue (His, Arg, and Lys), nonpolar side chains are green (Ile, Leu, Met, and Val), and aromatic side chains are cyan (Phe, Tyr, and Trp). Symbols: ‡, HGFG motif; *, active-site residues, including His177; †, residue His269 of Cif.
Like ArEH, Cif forms homodimers in the crystal lattice via the cap domain. There are small but measurable differences between the two Cif molecules of the homodimer, occurring mainly in the cap domain at the dimer interface. The two core domains align with a 0.2 Å rmsd, while the cap domains align with a 0.4 Å rmsd, giving an overall rmsd of 0.3 Å. These differences likely are due to the distinct environments created by lattice packing within the crystal. The two dimers in the asymmetric unit are related by noncrystallographic symmetry and align with a 0.6 Å rmsd. The dimerization interface encompasses an average surface area of 1,365 Å2 on each monomer. Dimerization is a feature commonly seen in the crystal structures of bacterial α/β hydrolases (29, 43, 57).
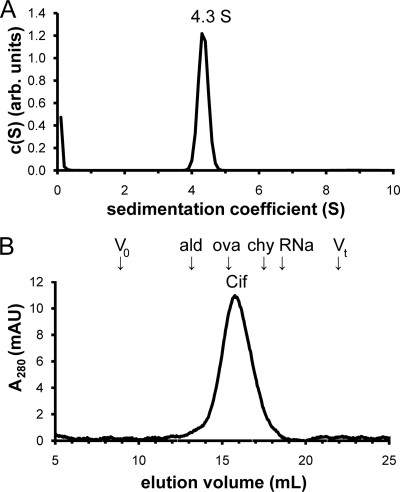
To test whether Cif forms a homodimer in solution, we performed hydrodynamic analyses. First, we performed velocity sedimentation experiments. Cif-WT sedimented as a single species at 4.3S ± 0.1S (Fig. 4A) . We next performed analytical SEC (Fig. 4B). Cif-WT eluted in a single peak. When calibrated relative to standard proteins, the elution volume of Cif corresponds to a Stokes' radius (Rs) of 3.2 nm and, together with the sedimentation coefficient, yields a shape-independent relative molecular mass (Mr) estimate of 60 kDa. Since the calculated Mr of a Cif monomer is 34 kDa, these data support the hypothesis that Cif-WT forms a homodimer in solution.
FIG. 4.
Cif forms a homodimer in solution. (A) The sedimentation coefficient concentration distribution c(S) was determined for Cif using velocity sedimentation analysis. The single peak is shown at 4.3S. (B) SEC of Cif-WT reveals a single peak at Ve = 15.76 ml. Arrows indicate the elution volumes of standard proteins used to calibrate the Rs of Cif. Void volume (V0), 8.90 ml; aldolase (ald), 13.04 ml; ovalbumin (ova), 15.72 ml; chymotrypsinogen A (chy), 17.54 ml; RNase A (RNa), 18.54 ml; and total volume (Vt), 21.93 ml.
Cif is an epoxide hydrolase.
Within the α/β hydrolase family, there are three enzyme classes that utilize an aspartic acid as the nucleophile in the catalytic mechanism (10, 52). These are the EHs, the HADs, and the haloalkane dehalogenases (HLDs). While there are many known physiologically relevant epoxides, most notably in the arachidonic acid pathway (39), EHs traditionally have been characterized by the use of small, artificial substrates. This is due primarily to compound availability and the existence of standardized assays (40). Cif previously has been shown to catalyze epibromohydrin (EBH) by an indirect assay (see Fig. S2 in the supplemental material) (36). After the enzymatic reaction in which Cif is allowed to hydrolyze EBH to form 3-bromo-1,2-propanediol, the protein is denatured with acetonitrile to stop the reaction and the oxidizing agent sodium periodate is added. The periodate does not react with the epoxide. However, it oxidizes the vicinal diol, cleaving it to produce two aldehyde compounds, and is consumed in the process. Residual periodate is reduced by the addition of epinephrine, forming the colored compound adrenochrome, which is detected via A490 measurements (see Fig. S2 in the supplemental material). We measured a specific activity of EBH hydrolysis of 0.25 U/mg (1 U = 1 μmol substrate hydrolyzed per minute) for Cif-WT. Since the structural evidence was consistent with the presence of potentially modulatory divalent cations, we performed the EBH hydrolysis assay in the presence of a panel of divalent cations to test their influence on Cif enzymatic activity. No significant effect was observed (see Fig. S3 in the supplemental material).
Given the central role of an oxidizing agent in the indirect assay for EBH epoxide hydrolysis and Cif's homology to HLDs, it was formally possible that Cif was catalyzing the debromination of EBH, and that the debromination product glycidol was producing the signal detected by the adrenochrome assay. To evaluate this possibility, we added glycidol directly to the adrenochrome assay. It produced no discernible signal (see Fig. S4 in the supplemental material), confirming that the specific activity detected reflects EBH hydrolysis. This clearly establishes Cif as a bona fide EH.
However, EBH is not a standard substrate used in the characterization of mammalian EHs. Microsomal or soluble EHs are demarcated by their cellular localization and can be further differentiated based on their preference to catalyze the hydrolysis of cis- or trans-stilbene oxide, respectively (25). To characterize the enzymatic activity of Cif further, we tested its action against these compounds. While Cif did not have measurable hydrolytic activity against TSO, it did catalyze the hydrolysis of CSO with a specific activity of 0.003 ± 0.001 U/mg (Fig. 5A). While this specific activity is low, there is a robust difference at the level of raw signal between reactions with and without Cif (P = 0.00016; n = 3). Thus, based on comparison to a limited number of mammalian species, Cif appears to be an EH with a substrate selectivity for cis- versus trans-stilbene oxide that is more similar to microsomal than soluble EH (mEH and sEH, respectively).
FIG. 5.
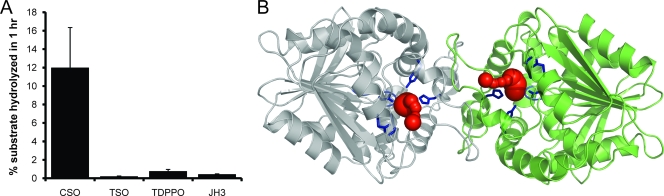
Substrate selectivity of Cif. (A) Buffer-subtracted hydrolysis of 50 μM radiolabeled canonical epoxide hydrolase substrates by 1 μM Cif-WT. Cif exhibited significant hydrolytic activity only for CSO. (B) Ribbon diagram of the Cif homodimer, as seen down the 2-fold axis. The side chains of active-site residues Asp129, Glu153, His177, Try239, and His297 and the HGFG motif ribbon are shown in blue. The calculated tunnel from the solvent to Asp129 for each monomer is shown in red.
To characterize Cif's substrate selectivity further, we assayed enzyme activity with two additional epoxide compounds, TDPPO and JH3, both classically used as EH substrates. Typically, an mEH will have lower but significant activity against TDPPO. JH3 is a substrate for a subclass of mEHs known as the juvenile hormone epoxide hydrolases (16). We were not able to detect hydrolytic activity against either TDPPO or JH3 for Cif. Interestingly, mouse sEH had roughly 100-fold greater activity for CSO than Cif, even though it is not considered an ideal substrate. Considering Cif's poor activity against CSO, the lack of measurable activity against TSO, TDPPO, or JH3, and strong enzyme activity against EBH, we conclude that Cif is an EH with novel substrate selectivity.
One possible explanation for Cif's apparent reduction in specific activity and narrow substrate selectivity involves the access pathway to the active site. Both human and mouse sEH have an open tunnel through the protein with the active site in the middle (9). This configuration has two openings to the solvent from the active site, allowing the substrate and product to enter and exit from different sites. In contrast, the putative substrate access pathway of Cif to the active site begins at the edge of the dimer interface and has only one outlet to the solvent (Fig. 5B), forming a dead-end cavity with an enclosed volume of 347 Å3. As a consequence, the product of the enzymatic reaction is predicted to exit from the active-site pocket before fresh substrate can enter, potentially limiting the substrate turnover rate. The substrate specificity of an enzyme is determined largely by the residues lining the active site and pathway to the active site. It has been demonstrated for other bacterial α/β hydrolases that the mutation of surface residues at the active-site tunnel entrance can alter substrate selectivity (8). By completely blocking access from one side of the active site, Cif may be restricting the majority of traditional epoxide substrates from being hydrolyzed. It also is possible that a conformational change in the cap domain regulates access to the active site.
Cif exhibits novel stereochemical features for an epoxide hydrolase.
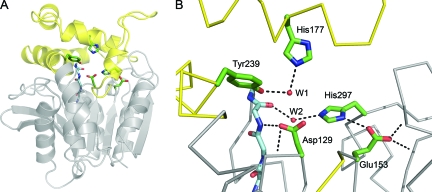
The structural alignment of Cif with ArEH highlights residues conserved between Cif and the EH family (Fig. 3). In agreement with conserved EH sequences (38), Cif has a catalytic triad consisting of residues Asp129, His297, and Glu153 (Fig. 6). Thus, for the Cif protein, it appears that Asp129 serves as the nucleophile responsible for the initial attack on a carbon of the substrate epoxide ring, generating a covalently linked enzyme-substrate complex (2). His297, activated by hydrogen bonding to Glu153, positions and activates an adjacent water molecule (W2 in Fig. 6B) to hydrolyze the enzyme-substrate ester linkage, releasing the diol product.
FIG. 6.
Cif active site. (A) Ribbon diagram of Cif-WT. The cap domain, consisting of residues 155 to 242, is colored yellow, and the core is in gray. The side chains of active-site residues are modeled as sticks, and the main chain of the HGFG motif is shown with the carbons colored light blue. (B) A detailed view of the active site. Hydrogen bonds are shown as dotted lines and the main chain as a Cα trace. The carboxylate of Glu153 participates in three hydrogen bonds. One is accepted from His297 and two from the protein backbone via the amide nitrogens of Gly266 and Met272. Asp129 is positioned by hydrogen bonds donated from the amide nitrogens of neighboring residue Ile130 and of Phe63 of the HGFG motif. W1 is coordinated by hydrogen bonds to Tyr239 and His177. W2 donates hydrogen bonds to the carbonyl oxygen of Phe63 and the imidazole of His297.
Cif diverges from canonical EH sequence motifs at key accessory residues in the active site surrounding the catalytic triad. Conventionally, EHs have two tyrosine residues positioned above the nucleophile to assist in the initial attack step (38), and a recent phylogenetic analysis showed that all experimentally confirmed EHs utilize two tyrosines during epoxide ring opening (52). These generate an oxyanion hole for initial attack and facilitate the opening of the epoxide ring (55). Sequence alignment reveals that Cif has only one of these Tyr residues. While Tyr239 is positioned in a conserved location, the structural alignment reveals that His177 replaces the second Tyr in the active site (Fig. 3). The His side chain at this position is capable of donating a hydrogen bond to stabilize the oxyanion hole. Together with Tyr239, His177 coordinates a water molecule (W1 in Fig. 6B) that occupies the likely position of a substrate epoxide oxygen based on its proximity to Asp129. Thus, it appears that the His substitution, although unprecedented among EHs, is compatible with the observed EH activity of Cif.
The second divergence from consensus motifs is in the standard HGxP motif (33), where x is any residue and the P is a cis-Pro. Cif has a Gly replacing the cis-Pro, creating a novel HGFG sequence at residues 61 to 64 (Fig. 3). The primary role of the HGxP motif is to position the backbone nitrogen of the variable residue x to act as a hydrogen bond donor for the nucleophile. In Cif, Phe63 occupies the corresponding position, and the amide nitrogen is oriented to hydrogen bond with Asp129 (Fig. 6B). The key role of the cis-Pro at the fourth position of this motif is to generate a sharp turn in the protein backbone. Gly64 has enough flexibility that it can accommodate a similar turn (Fig. 2A), preserving the essential stereochemical characteristics of the active site. As a result, despite sequence divergence from canonical EH motifs, the overall structure of the Cif active site is consistent with EH function and, thus, with the enzymology described above. Corresponding noncanonical sequences also are observed in proteins of related bacterial species and therefore may be characteristic for distinct subsets of the epoxide hydrolase family (see Fig. S5 in the supplemental material).
Assessing alternative Cif dehalogenase activity.
While confirming the predicted similarity between ArEH and Cif, our distance matrix alignment also revealed that the most closely related known structure was actually that of the fluoroacetate dehalogenase from Burkholderia sp. strain FA1 (FAc-DEX FA1), with a Z score of 35.7 and an rmsd of 1.9 Å. FAc-DEX FA1 is an α/β hydrolase that defluorinates fluoroacetate to form glycolic acid (29), thus detoxifying this compound, which can form a potent aconitase inhibitor if ingested (24). As a family, the HADs are closely related in sequence to the EHs and also utilize an aspartic acid side chain for initial nucleophilic attack (52). Like ArEH, FAc-DEX FA1 has a catalytic triad that aligns with that of Cif. Moreover, the noncanonical His177 residue of Cif is shared with FAc-DEX FA1. However, HADs are characterized by an arginine-rich motif (DRXXRXXXR, where D is the nucleophile) (52) that is not found in Cif. Cif also differs from the FAc-DEX FA1 sequence at four positions that are specific to the FAc-DEX reaction mechanism (Arg105, Arg108, Trp150, and Tyr212) (29).
Despite these motif mismatches, we experimentally investigated the possibility that Cif exhibits HAD activity. We tested Cif for the ability to dehalogenate fluoroacetate, chloroacetate, bromoacetate, and iodoacetate. In all cases, we found the signal from the Cif-containing sample to be indiscernible from the negative protein control across pH 5 to 9, from 1 to 10 mM substrate, or in the presence or absence of 1 mM MgSO4, which stimulates the enzymatic activity of the FAc-DEX from Pseudomonas fluorescens DSM 8341 (19 and data not shown). Using the substrate fluoroacetate, we determined the specific activity of Cif to be 0.0003 ± 0.0001 U/mg, while FAc-DEX FA1 was determined to have a specific activity (at the assay endpoint, not at Vmax) of 0.22 ± 0.04 U/mg (see Fig. S6 in the supplemental material). There was no statistically significant difference between samples incubated with and without Cif at the level of raw assay signal (P = 0.27; n = 3). In contrast, the FAc-DEX FA1 sample gave a statistically significant signal (P = 0.0015; n = 3). We therefore conclude that Cif does not exhibit HAD activity under the conditions tested, which is consistent with the absence of the active-site motifs identified by the FAc-DEX FA1 structure (29).
The third group of α/β hydrolases utilizing Asp as the nucleophile is the HLD class. Although the HLDs share the Cif catalytic triad, they also differ at key positions. The variable residue of the HGxP motif is an aromatic residue in the EHs, and Cif has a Phe at this position. In contrast, HLDs have a polar or charged residue at this position (10). Furthermore, Cif utilizes Tyr and His residues when opening the epoxide ring of a substrate, while HLDs utilize an Asn or Trp in the dehalogenation reaction (10, 52). Nevertheless, we attempted to measure HLD activity for Cif using the substrate 1,2-dichloroethane. As was seen with assays for HAD activity, the signal was indistinguishable from that of the non-Cif protein control (data not shown). Thus, given the peripheral sequence mismatches and the lack of detectable HLD activity, the possibility that Cif is an HLD was not investigated further.
The structure of Cif-WT reveals the location of residue His269.
Based on initial sequence alignment, His269 had been predicted as the catalytic triad His (37). In apparent agreement with this prediction, the mutation of His269 to Ala previously was reported to abrogate both Cif's effect on airway epithelial cells and the ability to hydrolyze the colorimetric substrate S-NEPC (18, 37). However, the crystal structure of Cif revealed that His297 occupies the position of the charge-relay side chain in the Cif catalytic triad. Moreover, His269 is positioned in the middle of a stretch of five Gly residues and is on the surface rather than buried in the active site (Fig. 7A). Its contribution to Cif's catalytic activity therefore was not immediately obvious.
FIG. 7.
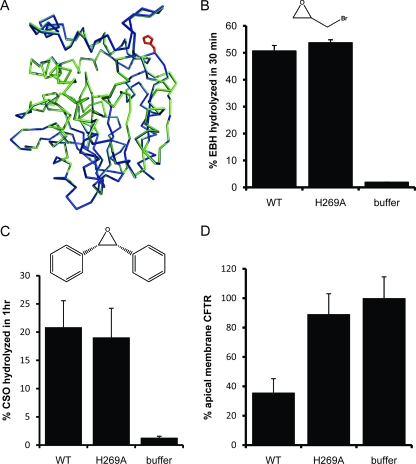
Reassessment of Cif-H269A EH and CFTR inhibitory activity. (A) Cα traces for Cif-WT (green) and Cif-H269A (blue) are shown following superposition, which yielded an rmsd of 0.08 Å. The side chains of residue 269 for both structures are shown in red. (B) Hydrolysis of 10 mM EBH by 20 μM Cif-WT or Cif-H269A. (C) Hydrolysis of radiolabeled CSO (50 μM) by Cif-WT and Cif-H269A (1 μM). (D) Cif-WT decreases the apical membrane abundance of CFTR, while the H269A mutation abrogates this effect. Fifty μg of either Cif-WT or Cif-H269A was added to the apical surface of CFBE WT-CFTR cells and incubated for 60 min, followed by Western blot analysis to determine the apical membrane abundance of CFTR. Samples are scaled in comparison to the buffer control, which was normalized to 100%.
To validate the reported loss of enzymatic activity of Cif-H269A (37), we tested both WT and mutant Cif for the hydrolysis of S-NEPC. While coincubation with either Cif-WT or Cif-H269A generated a colorimetric signal, the specific activities of both were indistinguishable from that of a negative control (see Fig. S7 in the supplemental material). In all cases, S-NEPC hydrolysis correlated with the total amount of protein in solution. It therefore seems likely that the signal generated is caused by the inherent instability of the compound during the extended incubation required to obtain a measurable color change (15-fold longer than that used for sEH) (18). If Cif is able to hydrolyze S-NEPC at all, it does so at a rate slower than that of autolysis.
To assess the enzymatic activity of Cif-H269A, we determined its ability to hydrolyze EBH (Fig. 7B). Consistently with the structural data, we found no significant difference between the hydrolytic activities of Cif-WT and those of Cif-H269A, with Cif-H269A yielding a specific activity of 0.26 U/mg compared to 0.25 U/mg for the wild-type enzyme. Additionally, we compared Cif-WT and -H269A activity on CSO, a more traditional substrate for assessing mammalian EH activity (Fig. 7C). Once again, we found no difference in the enzymatic activity between Cif-WT and Cif-H269A; both yielded a specific activity of 0.005 ± 0.001 U/mg for this experiment. Based on the similar specific activities observed for all substrates tested, we conclude that the H269A mutation does not affect the immediate vicinity of the catalytic triad, although it may have a modest impact on the kinetics of hydrolysis that is not detectable with our assays. Unfortunately, we were unable to purify mutants targeting the catalytic histidine at position 297 (H297A and H297N; data not shown), most likely because the active site is located at the core interface with the cap domain and therefore is structurally sensitive.
Based on the evidence that Cif-H269A has EH activity similar to that of Cif-WT for small, xenobiotic substrates, we decided to reevaluate the ability of Cif-H269A to reduce the apical membrane abundance of CFTR (37). Using aliquots of Cif-H269A that had been confirmed to hydrolyze EBH, we tested CFTR inhibitory activity in triplicate. This allowed us to rule out potential variation in specific activity between different preparations of purified protein. In agreement with previous reports (37), we found that the H269A mutation abrogates the ability of Cif to reduce the levels of CFTR at the apical membrane of airway epithelial cells (Fig. 7D) despite its retention of EH activity. Taken together, our data clearly demonstrate that the Cif hydrolase activity detected by small, artificial substrates is not by itself sufficient to account for the CFTR inhibitory effect.
One possible cause of the loss of CFTR inhibitory activity is if the H269A mutation alters the protein structure away from the catalytic site and thus affects Cif trafficking, localization, or protein interactions. To address this issue, we crystallized and determined the structure of Cif-H269A at 1.5-Å resolution. Molecular replacement was used to obtain phase information from the initial Cif-WT selenomethionine model. The starting model of Cif-H269A was determined by a composite omit strategy (50) to avoid potential model bias. Standard molecular refinement yielded excellent agreement with the diffraction data (Table 1). An alignment of the refined structures of Cif-WT and Cif-H269A (Fig. 7A) gave a Z score of 55.6 and a Cα rmsd value of 0.08 Å, which is less than the maximum-likelihood coordinate error estimates for each structure of 0.19 and 0.16 Å, respectively. We conclude that even at high stringency, there is no measurable conformational difference between the WT and H269A crystal structures.
Like Cif-WT, Cif-H269A crystallizes as a homodimer. However, to test the possibility that the H269A mutation could alter the affinity of Cif dimerization in solution, we repeated the hydrodynamic analyses that were performed for Cif-WT. As seen for Cif-WT, the sedimentation coefficient of the H269A mutant also was 4.3S ± 0.2S, and the protein eluted at the same volume during analytical SEC. Thus, there is no evidence that the mutation affects Cif homodimerization.
These data confirm that Cif-WT and Cif-H269A exhibit the same tertiary and quaternary conformations, and that significant structural changes therefore are not responsible for the effects of this mutation. One limitation of our enzymatic studies is that our test substrates are, at most, 6 Å long. As a result, they effectively probe only the portion of the substrate-binding site closest to the catalytic triad. Longer substrates would fill the narrow access tunnel (Fig. 5B) and emerge near His269 at the dimer interface, roughly 14 Å from Asp129 of the catalytic triad (Fig. 7A). Cif-H269A therefore could act as a KM mutant, affecting interactions with extended candidate endogenous substrates such as epoxides found in physiological pathways.
Conclusion.
The goal of this study was to determine the structure of Cif and characterize its enzymatic activity in order to understand the basis of its action as a CFTR inhibitory virulence factor. We demonstrate here that Cif has bona fide EH activity, despite deviations from otherwise strictly conserved EH motifs. In accordance with these findings, the crystal structure of Cif confirmed that the observed mutations are stereochemically conservative. In contrast, and in agreement with the absence of essential HAD or HLD catalytic motifs, we were unable to detect HAD or HLD activity against standard substrates. Taken together, these data suggest that epoxides are the primary catalytic target of Cif, which is thus the first validated bacterial EH known to interact directly with mammalian cells. The observations that Cif is expressed in bacteria from the lungs of CF patients and is preferentially expressed by nonmucoid strains of P. aeruginosa (37) suggest that Cif is a virulence factor that plays an important role in the early colonization of the lung.
This work lays the foundation for a detailed biochemical dissection of the mechanism of Cif's effect on CFTR plasma membrane abundance. In particular, the active-site stereochemistry of the structure presented here can be used to guide the selection of candidate endogenous substrates. In parallel, it will help us to develop novel active-site mutations to probe the coupling between the EH and CFTR inhibitory activities of Cif.
Supplementary Material
Acknowledgments
Funding support was provided by the NIH (R01-DK075309 to D.R.M., R01-ES002710 and R01-ES013933 to B.D.H., R01-HL074175 to B.A.S., and a training grant predoctoral fellowship from T32-AI007519 to C.D.B.) and the Cystic Fibrosis Foundation (MADDEN08G0 to D.R.M. and STANTO97R0 Research Development Program pilot funds to G.A.O.).
We thank Nobuyoshi Esaki and Tatsuo Kurihara for their generous gift of purified FAc-DEX FA1 protein, Brian Jackson of the Dartmouth Trace Element Analysis Core for inductively coupled plasma mass spectrometry analysis, Daniel P. MacEachran and Gordon W. Gribble for helpful discussions and advice, and Vivian Stojanoff and Jean Jakoncic at NSLS/Brookhaven for assistance with data collection and processing.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.American Thoracic Society and Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 171:388-416. [DOI] [PubMed] [Google Scholar]
- 2.Arand, M., A. Cronin, M. Adamska, and F. Oesch. 2005. Epoxide hydrolases: structure, function, mechanism, and assay. Methods Enzymol. 400:569-588. [DOI] [PubMed] [Google Scholar]
- 3.Bahl, C. D., D. P. MacEachran, G. A. O'Toole, and D. R. Madden. 2010. Purification, crystallization and preliminary X-ray diffraction analysis of Cif, a virulence factor secreted by Pseudomonas aeruginosa. Acta Crystallogr. F66:26-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bebok, Z., J. F. Collawn, J. Wakefield, W. Parker, Y. Li, K. Varga, E. J. Sorscher, and J. P. Clancy. 2005. Failure of cAMP agonists to activate rescued ΔF508 CFTR in CFBE41o− airway epithelial monolayers. J. Physiol. 569:601-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bomberger, J. M., D. P. Maceachran, B. A. Coutermarsh, S. Ye, G. A. O'Toole, and B. A. Stanton. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr, P. D., and D. L. Ollis. 2009. Alpha/beta hydrolase fold: an update. Protein Peptide Lett. 16:1137-1148. [DOI] [PubMed] [Google Scholar]
- 7.Cedrone, F., T. Bhatnagar, and J. C. Baratti. 2005. Colorimetric assays for quantitative analysis and screening of epoxide hydrolase activity. Biotechnol. Lett. 27:1921-1927. [DOI] [PubMed] [Google Scholar]
- 8.Chaloupková, R., J. Sykorova, Z. Prokop, A. Jesenska, M. Monincova, M. Pavlova, M. Tsuda, Y. Nagata, and J. Damborsky. 2003. Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J. Biol. Chem. 278:52622-52628. [DOI] [PubMed] [Google Scholar]
- 9.Chiamvimonvat, N., C. M. Ho, H. J. Tsai, and B. D. Hammock. 2007. The soluble epoxide hydrolase as a pharmaceutical target for hypertension. J. Cardiovasc. Pharmacol. 50:225-237. [DOI] [PubMed] [Google Scholar]
- 10.Chovancova, E., J. Kosinski, J. M. Bujnicki, and J. Damborsky. 2007. Phylogenetic analysis of haloalkane dehalogenases. Proteins 67:305-316. [DOI] [PubMed] [Google Scholar]
- 11.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Cushing, P. R., A. Fellows, D. Villone, P. Boisguerin, and D. R. Madden. 2008. The relative binding affinities of PDZ partners for CFTR: a biochemical basis for efficient endocytic recycling. Biochemistry 47:10084-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dam, J., and P. Schuck. 2004. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 384:185-212. [DOI] [PubMed] [Google Scholar]
- 15.Davies, J. C., and D. Bilton. 2009. Bugs, biofilms, and resistance in cystic fibrosis. Respir. Care 54:628-640. [DOI] [PubMed] [Google Scholar]
- 16.Debernard, S., C. Morisseau, T. F. Severson, L. Feng, H. Wojtasek, G. D. Prestwich, and B. D. Hammock. 1998. Expression and characterization of the recombinant juvenile hormone epoxide hydrolase (JHEH) from Manduca sexta. Insect Biochem. Mol. Biol. 28:409-419. [DOI] [PubMed] [Google Scholar]
- 17.DeLano, W. L. 2008. The PyMOL molecular graphics system. DeLano Scientific LLC, Palo Alto, CA.
- 18.Dietze, E. C., E. Kuwano, and B. D. Hammock. 1994. Spectrophotometric substrates for cytosolic epoxide hydrolase. Anal. Biochem. 216:176-187. [DOI] [PubMed] [Google Scholar]
- 19.Donnelly, C., and C. D. Murphy. 2009. Purification and properties of fluoroacetate dehalogenase from Pseudomonas fluorescens DSM 8341. Biotechnol. Lett. 31:245-250. [DOI] [PubMed] [Google Scholar]
- 20.Eegriwe, E. 1932. Reaktionen und Reagenzien zum Nachweis organischer Verbindungen I. Fresenius Zeitschrift Analytische Chemie 89:121-125. [Google Scholar]
- 21.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 22.Geller, D. E. 2009. Aerosol antibiotics in cystic fibrosis. Respir. Care 54:658-670. [DOI] [PubMed] [Google Scholar]
- 23.Gill, S. S., K. Ota, and B. D. Hammock. 1983. Radiometric assays for mammalian epoxide hydrolases and glutathione S-transferase. Anal. Biochem. 131:273-282. [DOI] [PubMed] [Google Scholar]
- 24.Gribble, G. W. 1973. Fluoroacetate toxicity. J. Chem. Educ. 50:460-462. [DOI] [PubMed] [Google Scholar]
- 25.Hammock, B. D., D. H. Storms, and D. F. Grant. 1997. Epoxide hydrolases, p. 283-305. In F. P. Guengerich (ed.), Comprehensive toxicology, vol. 3. Pergamon, Oxford, United Kingdom. [Google Scholar]
- 26.Hendlich, M., F. Rippmann, and G. Barnickel. 1997. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph Model 15:359-389. [DOI] [PubMed] [Google Scholar]
- 27.Holloway, P., J. T. Trevors, and H. Lee. 1998. A colorimetric assay for detecting haloalkane dehalogenase activity. J. Microbiol. Methods 32:31-36. [Google Scholar]
- 28.Holm, L., S. Kaariainen, P. Rosenstrom, and A. Schenkel. 2008. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24:2780-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jitsumori, K., R. Omi, T. Kurihara, A. Kurata, H. Mihara, I. Miyahara, K. Hirotsu, and N. Esaki. 2009. X-Ray crystallographic and mutational studies of fluoroacetate dehalogenase from Burkholderia sp. strain FA1. J. Bacteriol. 191:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, A. M. 2005. Eradication therapy for early Pseudomonas aeruginosa infection in CF: many questions still unanswered. Eur. Respir. J. 26:373-375. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 32.Kadurugamuwa, J. L., and T. J. Beveridge. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177:3998-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lacourciere, G. M., and R. N. Armstrong. 1994. Microsomal and soluble epoxide hydrolases are members of the same family of C-X bond hydrolase enzymes. Chem. Res. Toxicol. 7:121-124. [DOI] [PubMed] [Google Scholar]
- 34.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer sciences. Royal Society for Chemistry, Cambridge, United Kingdom.
- 35.Leonard, G. A., G. Sainz, M. M. de Backer, and S. McSweeney. 2005. Automatic structure determination based on the single-wavelength anomalous diffraction technique away from an absorption edge. Acta Crystallogr. D 61:388-396. [DOI] [PubMed] [Google Scholar]
- 36.MacEachran, D. P., B. A. Stanton, and G. A. O'Toole. 2008. Cif is negatively regulated by the TetR family repressor CifR. Infect. Immun. 76:3197-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacEachran, D. P., S. Ye, J. M. Bomberger, D. A. Hogan, A. Swiatecka-Urban, B. A. Stanton, and G. A. O'Toole. 2007. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 75:3902-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morisseau, C., and B. D. Hammock. 2005. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu. Rev. Pharmacol. Toxicol. 45:311-333. [DOI] [PubMed] [Google Scholar]
- 39.Morisseau, C., and B. D. Hammock. 2008. Gerry Brooks and epoxide hydrolases: four decades to a pharmaceutical. Pest Manag. Sci. 64:594-609. [DOI] [PubMed] [Google Scholar]
- 40.Morisseau, C., and B. D. Hammock. 2007. Measurement of soluble epoxide hydrolase (sEH) activity. Curr. Protoc. Toxicol. 33:4.23.1-4.23.18. [DOI] [PubMed] [Google Scholar]
- 41.Moyer, B. D., J. Loffing, E. M. Schwiebert, D. Loffing-Cueni, P. A. Halpin, K. H. Karlson, I. I. Ismailov, W. B. Guggino, G. M. Langford, and B. A. Stanton. 1998. Membrane trafficking of the cystic fibrosis gene product, cystic fibrosis transmembrane conductance regulator, tagged with green fluorescent protein in Madin-Darby canine kidney cells. J. Biol. Chem. 273:21759-21768. [DOI] [PubMed] [Google Scholar]
- 42.Murphy, T. F., A. L. Brauer, K. Eschberger, P. Lobbins, L. Grove, X. Cai, and S. Sethi. 2008. Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 177:853-860. [DOI] [PubMed] [Google Scholar]
- 43.Nardini, M., I. S. Ridder, H. J. Rozeboom, K. H. Kalk, R. Rink, D. B. Janssen, and B. W. Dijkstra. 1999. The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides. J. Biol. Chem. 274:14579-14586. [PubMed] [Google Scholar]
- 44.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, J. L. Sussman, K. H. G. Verschueren, and A. Goldman. 1992. The α/β hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 45.Petrek, M., M. Otyepka, P. Banas, P. Kosinova, J. Koca, and J. Damborsky. 2006. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics 7:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian, Z., C. J. Fields, Y. Yu, and S. Lutz. 2007. Recent progress in engineering α/β hydrolase-fold family members. Biotechnol. J. 2:192-200. [DOI] [PubMed] [Google Scholar]
- 47.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 48.Swiatecka-Urban, A., M. Duhaime, B. Coutermarsh, K. H. Karlson, J. Collawn, M. Milewski, G. R. Cutting, W. B. Guggino, G. Langford, and B. A. Stanton. 2002. PDZ domain interaction controls the endocytic recycling of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 277:40099-40105. [DOI] [PubMed] [Google Scholar]
- 49.Swiatecka-Urban, A., S. Moreau-Marquis, D. P. Maceachran, J. P. Connolly, C. R. Stanton, J. R. Su, R. Barnaby, G. A. O'Toole, and B. A. Stanton. 2006. Pseudomonas aeruginosa inhibits endocytic recycling of CFTR in polarized human airway epithelial cells. Am. J. Physiol. Cell Physiol. 290:C862-C872. [DOI] [PubMed] [Google Scholar]
- 50.Terwilliger, T. C., R. W. Grosse-Kunstleve, P. V. Afonine, N. W. Moriarty, P. D. Adams, R. J. Read, P. H. Zwart, and L. W. Hung. 2008. Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr D. 64:515-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Loo, B., J. Kingma, M. Arand, M. G. Wubbolts, and D. B. Janssen. 2006. Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl. Environ. Microbiol. 72:2905-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolff, W. 1893. Ueber ein dinaphtoxanthen (methylendinaphtylenoxyd). Berichte Deutschen Chemischen Gesellschaft 26:83-86. [Google Scholar]
- 54.Wood, D. M., and A. R. Smyth. 2006. Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis. Cochrane Database Syst. Rev. 2006:CD004197. [DOI] [PubMed] [Google Scholar]
- 55.Yamada, T., C. Morisseau, J. E. Maxwell, M. A. Argiriadi, D. W. Christianson, and B. D. Hammock. 2000. Biochemical evidence for the involvement of tyrosine in epoxide activation during the catalytic cycle of epoxide hydrolase. J. Biol. Chem. 275:23082-23088. [DOI] [PubMed] [Google Scholar]
- 56.Ye, S., D. P. MacEachran, J. W. Hamilton, G. A. O'Toole, and B. A. Stanton. 2008. Chemotoxicity of doxorubicin and surface expression of P-glycoprotein (MDR1) is regulated by the Pseudomonas aeruginosa toxin Cif. Am. J. Physiol. Cell Physiol. 295:C807-C818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou, J., B. M. Hallberg, T. Bergfors, F. Oesch, M. Arand, S. L. Mowbray, and T. A. Jones. 2000. Structure of Aspergillus niger epoxide hydrolase at 1.8 Å resolution: implications for the structure and function of the mammalian microsomal class of epoxide hydrolases. Structure 8:111-122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.