Abstract
Identification of interacting proteins in stable complexes is essential to understand the mechanisms that regulate cellular processes at the molecular level. Transcription initiation in prokaryotes requires coordinated protein-protein and protein-DNA interactions that often involve one or more transcription factors in addition to RNA polymerase (RNAP) subunits. The RNAP α subunit (RNAPα) is a key regulatory element in gene transcription and functions through direct interaction with other proteins to control all stages of this process. A clear description of the RNAPα protein partners should greatly increase our understanding of transcription modulation. A functional proteomics approach was employed to investigate protein components that specifically interact with RNAPα. A tagged form of Escherichia coli RNAPα was used as bait to determine the molecular partners of this subunit in a whole-cell extract. Among other interacting proteins, 50S ribosomal protein L2 (RPL2) was clearly identified by mass spectrometry. The direct interaction between RNAPα and RPL2 was confirmed both in vivo and in vitro by performing coimmunoprecipitation and bacterial two-hybrid experiments. The functional role of this interaction was also investigated in the presence of a ribosomal promoter by using a β-galactosidase gene reporter assay. The results clearly demonstrated that RPL2 was able to increase β-galactosidase expression only in the presence of a specific ribosomal promoter, whereas it was inactive when it was assayed with an unrelated promoter. Interestingly, other ribosomal proteins (L1, L3, L20, and L27) did not have any effect on rRNA expression. The findings reported here strongly suggest that in addition to its role in ribosome assembly the highly conserved RPL2 protein plays a specific and direct role in regulation of transcription.
Understanding the mechanism and regulation of transcription in bacteria requires dissection of the specific roles of the individual components of the multiprotein transcription complex, including the multisubunit enzyme RNA polymerase (RNAP). It is well known, in fact, that one of the major factors ensuring correct gene expression in microorganisms is the efficiency with which RNAP recognizes the specific promoters of different genes.
The single form of RNAP in Escherichia coli consists of a tetrameric core enzyme (ββ′α2) that is capable of RNA synthesis and factor-independent termination and a σ subunit that is responsible for recognition of specific transcription initiation sites. However, although the RNAP σ subunit is the main determinant for promoter recognition, the α subunit also plays a key role in the stability of the transcription complex (3, 10).
The E. coli RNAP α subunit (RNAPα) consists of two domains, an amino-terminal domain (NTD) and a carboxy-terminal domain (CTD), that are connected by a flexible linker that allows the CTD to interact with promoter elements located different distances from the RNAP binding site (2). The α subunit has at least two functions during gene transcription; it is involved in both assembly of the core enzyme and regulation of transcription initiation. The role of the two domains is clear; the NTD is needed for dimerization and interaction with the β and β′ subunits, whereas the CTD is important for both DNA binding and protein-protein interactions.
Transcription initiation requires coordinated protein-protein and protein-DNA interactions that often involve one or more transcription factors in addition to RNAP subunits (19). Transcription factors can interact with nearly all RNAP components, although the αCTD is the most frequent target. Class I activators bind to an upstream site and contact the αCTD, thereby recruiting RNAP to the promoter (6, 20). The α subunit is thus a key regulatory element in gene transcription and functions through direct interaction with other proteins to control all stages of the process. A clear description of the α-subunit protein partners should greatly increase our understanding of modulation of transcription.
Recently, functional proteomics approaches essentially based on pull-down or immunoprecipitation assays (17) have been used to investigate protein-protein interactions both in vitro and in vivo. Association of specific proteins with molecular partners belonging to protein complexes involved in particular mechanisms can contribute substantially to description of the cellular processes at the molecular level (9, 12).
In this work, a six-His-tagged recombinant of the α subunit of E. coli RNAP (RNAPα) was used as bait to determine the molecular partners of this subunit in a whole-cell extract. Among other interacting proteins, 50S ribosomal protein L2 (RPL2) was clearly identified by mass spectrometry. The direct interaction between RNAPα and RPL2 was further confirmed in vivo and in vitro by performing coimmunoprecipitation and two-hybrid experiments. The functional role of this interaction was investigated by performing in vivo transcription assays using lacZ as the reporter gene. RPL2 was shown to increase β-galactosidase expression only when a specific ribosomal promoter was used in this assay, whereas it was inactive in the presence of an unrelated promoter. Interestingly, other ribosomal proteins (L1, L3, L20, and L27) did not have any effect on rRNA expression. Findings reported here strongly suggest that the highly conserved RPL2 protein plays a specific and direct role in regulation of transcription in addition to its role in ribosome assembly.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli K-12 | Wild type | DSMZ |
| C41(DE3) | Derived from BL21 [F−ompT hsdSB(rB− mB−) gal dcm (DE3)]; has at least one uncharacterized mutation that prevents cell death associated with expression of many toxic recombinant proteins | 15 |
| R721 | 71/18 glp O-P434/p22lacZ | 5 |
| Plasmids | ||
| pET22b(+) | N-terminal pelB signal sequence plus an optional C-terminal His-tagged sequence; ampicillin resistant | Novagen |
| pET28a(+) | N-terminal His tag-thrombin-T7 tag configuration plus optional C-terminal His tag sequence; kanamycin resistance | Novagen |
| pET22b-αpol | pET22b(+) Δ(NdeI-XhoI) Ω(rpoA) | This study |
| pET22b-c-Myc | pET22b(+) Δ(NdeI-BamHI) Ω(c-myc tag) | This study |
| pET22b-c-Myc-L2 | pET22b-c-Myc Δ(BamHI-XhoI) Ω(rplB) | This study |
| pET22b-c-Myc-Thioredoxin | pET22 b-c-Myc Δ(BamHI-XhoI) Ω(trxC) | This study |
| pET28a-αpol | pET28a Δ(BamHI-XhoI) Ω(rpoA) | This study |
| pcIP22 | pC132 derivative carrying N-terminal end of p22 repressor; ampicillin resistance | 5 |
| pcI434 | pACYC177 derivative carrying N-terminal end of 434 repressor; chloramphenicol resistance | 5 |
| pcIP22-RNAPα | pcIP22Δ(SalI-BamHI) Ω(rpoA) | This study |
| pcI434-RNAPa | pcI434Δ(SalI-BamHI) Ω(rpoA) | This study |
| pcI434-RPL2 | pcI434Δ(SalI-BamHI) Ω(rplB) | This study |
| pcI-L2 | pcI434 Δ(NdelI-BamHI) Ω(rplB) | This study |
| pcI-L1 | pcI434Δ(NdelI-BamHI) Ω(rplA) | This study |
| pcI-L3 | pcI434Δ(NdelI-BamHI) Ω(rplC) | This study |
| pcI-L20 | pcI434Δ(NdelI-BamHI) Ω(rplT) | This study |
| pcI-L27 | pcI434Δ(NdelI-BamHI) Ω(rpmA) | This study |
| pET22b-PrrnD | pET22b Δ(SphI-HindIII) Ω(rrsD promoter) | This study |
| pET22b-PaidB | pET22b Δ(SphI-HindIII) Ω(aidB promoter) | This study |
| pET22b-PrrnD-lacZ | pET22b-PrrnDΔ(HindIII-XhoI) Ω(lacZ) | This study |
| pET22b-PaidB-lacZ | pET22b-PaidBΔ(HindIII-XhoI) Ω(lacZ) | This study |
Media and chemicals.
Luria-Bertani (LB) or nutrient broth (for bacterial cultures and plating) and suspension medium (for bacterial dilutions) were used as described by Miller (14). Ampicillin, kanamycin, and chloramphenicol (Sigma) were used at concentrations of 100, 50, and 34 μg/ml, respectively.
Construction of pET22b-αpol, pET28a-αpol, pET22b-c-Myc-L2, and pET22b-c-Myc-Thioredoxin.
The pET22b-αpol vector was constructed by cloning the rpoA gene (PCR amplified from E. coli genomic DNA using the rpoA-pET22b Fw and rpoA-pET22b Rv primers [Table 2] and digested with NdeI and XhoI) into expression vector pET22b (Novagen), which was linearized with the NdeI and XhoI restriction enzymes. For coimmunoprecipitation experiments, the rpoA gene (PCR amplified using primers rpoA-pET28a Fw and rpoA-pET28a Rv and digested with BamHI and XhoI) was cloned into expression vector pET28a (Novagen), which was linearized with the BamHI and XhoI restriction enzymes.
TABLE 2.
Oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| rpoA-pET22b Fw | 5′ ATACATATGATGCAGGGTTCTGTGACAG 3′ |
| rpoA-pET22b Rv | 5′ TAACCTGTGCTCGAGTAACTCGTCAGCG 3′ |
| c-myc Fw | 5′ATTCATATGGAACAAAAACTCATCTCAGAAGAGGATCTGAATGGGGCCGCAGGATCCTAT 3′ |
| c-myc Rv | 5′ATAGGATCCTGCGGCCCCATTCAGATCCTCTTCTGAGATGAGTTTTTGTTCCATATGAAT 3′ |
| rplB-pET22b Fw | 5′ TATGGATCCATGGCAGTTGTTAAATG 3′ |
| rplB-pET22b Rv | 5′ ATACTCGAGTAATTTGCTACGGCGAC 3′ |
| trxC-pET22b Fw | 5′ CCGGATCCATGAATACCGTTTGTACCCATTG 3′ |
| trxC-pET22b Rv | 5′ CCCTCGAGTAAAAGAGATTCGTTCAGCCAG 3′ |
| rpoA-pET28a Fw | 5′ GAGGGATCCATGCAGGGTTCTGTGACAG 3′ |
| rpoA-pET28a Rv | 5′ GTGCTCGAGTTACTCGTCAGCGATGC 3′ |
| rpoA-pcI434/P22 Fw | 5′ TATCTAGAGCGTCGACCATGCAGGGTTCTGTGACAG 3′ |
| rpoA-pcI434/P22 Rv | 5′ ATACTCGAGCGGGATCCTTACTCGTCAGCGATGCTT 3′ |
| rplB-pcI434 Fw | 5′ TATCTAGAGCGTCGACCATGGCAGTTGTTAAATGTA 3′ |
| rplB-pcI434 Rv | 5′ ATACTCGAGCGGGATCCTTATTTGCTACGGCGACG 3′ |
| rplB-pcI Fw | 5′ TAACATATGATGGCAGTTGTTAAATGTAA 3′ |
| rplB-pcI Rv | 5′ TTAGGATCCTTATTTGCTACGGCGACG 3′ |
| rplA-pcI Fw | 5′ GAGCCATATGCATGGCTAAACTGACCAAGCG 3′ |
| rplA-pcI Rv | 5′ GCGGGATCCTTAGTTTACAGAAGCGCTCAGG 3′ |
| rplC-pcI Fw | 5′ GAGCCATATGCATGATTGGTTTAGTCGGTAAA 3′ |
| rplC-pcI Rv | 5′ GCGGGATCCTTACGCCTTCACAGCTGGTTT 3′ |
| rplT-pcI Fw | 5′ GAGCCATATGCATGGCTCGCGTAAAACGTGG 3′ |
| rplT-pcI Rv | 5′ GCGGGATCCTTATGCCAGAGCTGCTTTCGC 3′ |
| rpmA-pcI Fw | 5′ GAGCCATATGCATGGCACATAAAAAGGCTGG 3′ |
| rpmA-pcI Rv | 5′ GCGGGATCCTTATTCAGCTTCGATGCTGATAAA 3′ |
| PrrnD-pET22b Fw | 5′ GTGCGCATGCACAGAAAAAAAGATC 3′ |
| PrrnD-pET22b Rv | 5′ TCGAAGCTTCGGAGGCGCATTATAG 3′ |
| PaidB-pET22b Fw | 5′ GTGCGCATGCATAAGAATGTTTTAGC 3′ |
| PaidB-pET22b Rv | 5′ TCGAAGCTTCACCATTAGTATGGTC 3′ |
| lacZ-pET22b Fw | 5′ TGTAAGCTTATAACAATTTCACACAGGAA 3′ |
| lacZ-pET22b Rv | 5′ CGGCTCGAGTTATTTTTGACACCAGAC 3′ |
To obtain pET22b-c-Myc, an NdeI/BamHI-digested fragment corresponding to the c-Myc epitope was inserted into the pET22b(+) expression plasmid (Novagen) linearized with NdeI and BamHI. pET22b-c-Myc-L2 and pET22b-c-Myc-Thioredoxin were constructed by cloning the rplB gene (PCR amplified using the rplB-pET22b Fw and rplB-pET22b Rv primers [Table 2]) and the trxC gene (PCR amplified using primers trxC-pET22b Fw and trxC-pET22b Rv) into pET22b-c-Myc digested with BamHI and XhoI. The resulting plasmids, designated as shown in Table 1, were verified by automated DNA sequencing.
Production and purification of recombinant proteins.
The pET22b-αpol, pET22b-c-Myc-L2, pET22b-c-Myc-Thioredoxin, and pET28a-αpol constructs (Table 1) were individually transformed into E. coli strain C41(DE3). For RNAPα and thioredoxin production, the recombinant cells were grown in LB medium at 37°C without induction until the optical density at 600 nm (OD600) was 3.0. The rplB gene was expressed as follows. Recombinant cells were grown at 37°C to an optical density at 600 nm of ∼0.9, and then 0.05 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) was added and the cultures were grown until the OD600 was 3.0. Cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C, resuspended in buffer A (50 mM Na2HPO4 [pH 7.4], 0.3 M NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride [PMSF]), disrupted by passage through a French press, and centrifuged at 14,000 × g for 30 min at 4°C.
The recombinant proteins were purified by affinity chromatography on a His-Select nickel affinity gel (Sigma) using a slight modification of the manufacturer's instructions. The lysate was loaded onto a His-Select nickel affinity gel equilibrated with equilibration buffer (50 mM Na2HPO4 [pH 7.4], 0.3 M NaCl, 10 mM imidazole). After 1 min of incubation at 4°C, the matrix was collected by centrifugation at 11,000 × g for 1 min and washed three times with washing buffer (50 mM Na2HPO4 [pH 7.4], 0.8 M NaCl, 10 mM imidazole). The recombinant proteins were eluted with buffer containing 250 mM imidazole in 50 mM Na2HPO4 (pH 7.4), 0.3 M NaCl.
Protein concentrations were estimated using the Bradford assay (1), and protein contents were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Fishing for partners.
One hundred microliters of a His-Select nickel affinity gel (Sigma) was incubated twice with 10 mg of bacterial protein extract for 2 h at 4°C as a preclearing step. The unbound protein extract was then incubated with six-His-tagged RNAPα linked to agarose beads by His-Ni2+ interactions for 2 h at 4°C. The matrix was collected by centrifugation at 5,000 × g for 10 min and washed four times with washing buffer. The retained proteins were eluted with 100 μl of Laemmli buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue) containing 0.1 M dithiothreitol (DTT) (4). The eluate was fractionated by one-dimensional 10% SDS-PAGE and stained with colloidal Coomassie brilliant blue G (Sigma).
In situ digestion.
Coomassie blue-stained protein bands were excised from SDS-PAGE gels and washed three times in deionized MilliQ-grade water (10 min each time). Then the excised spots were washed first with acetonitrile (ACN) and then three times with 0.1 M ammonium bicarbonate (15 min each time). Protein bands were then digested in the gel as previously described (16).
MALDI MS analyses.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectra were recorded using an Applied Biosystems Voyager DE STR instrument for mass spectrometry (MS). A mixture of analyte and matrix solution (10 mg/ml α-cyanohydroxycinnamic acid in 66% ACN-10 mM citric acid in MilliQ water) was applied to the metallic sample plate and dried at room temperature. Mass calibration was performed using external peptide standards. Raw data were analyzed using the computer software provided by the manufacturer and were expressed as monoisotopic masses. The peptide masses of tryptic fragments from each digested protein were used to search protein databases using an in-house version of the Mascot software (Matrix Science).
Nano-LC-ESI-MS/MS.
Tryptic peptide mixtures obtained as previously described were also analyzed by nano-liquid chromatography-electrospray ionization-tandem mass spectrometry (nano-LC-ESI-MS/MS) online using a linear ion trap instrument (4000Q-trap; Applied Biosystems). The proteolytic digest was fractionated using an HP 1100 nano-high-performance liquid chromatography (nano-HPLC) apparatus (Hewlett-Packard, Palo Alto, CA) and a capillary C18 column (75 μm by 150 mm; 300 Å; Agilent Technologies, Torrance, CA) with 0.1% (vol/vol) formic acid and 2% (vol/vol) ACN in H2O (solvent A) and 0.1% (vol/vol) formic acid and 2% (vol/vol) H2O in ACN (solvent B); a linear gradient from 5 to 70% solvent B for 60 min at a flow rate of 0.2 μl/min was used. The column was directly connected to the ion source through the nanospray probe, and both ESI-MS and ESI-MS/MS spectra were acquired throughout the analysis by dependent data scanning and monitoring the five most intense ions.
Coimmunoprecipitation and Western blotting.
For coimmunoprecipitation, E. coli strain C41(DE3) was cotransformed with pET22b-c-Myc-L2 and pET28a-αpol and with pET22b-c-Myc-Thioredoxin and pET28a-αpol as a control. Portions (2 ml) of cultures that were grown overnight were inoculated into 200 ml of LB medium containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. After expression of the recombinant genes without induction, cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C, resuspended in 50 mM Na2HPO4 (pH 7.4), 1 mM PMSF, disrupted by passage through a French press, and centrifuged at 14,000 × g for 30 min at 4°C. The protein contents were checked by Western blotting. The supernatants were then used for the coimmunoprecipitation experiments. Cell lysates were incubated with agarose-linked T7 antibody (Bethyl) and with agarose beads alone (control) at 4°C overnight. The beads were then collected by centrifugation and washed; the bound proteins were eluted with 1× SDS-PAGE sample buffer (2% SDS, 10% glycerol, 62.5 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 0.01% bromophenol blue) and subjected to SDS-PAGE followed by Western blotting, which was performed by using anti-T7 mouse antibody (Novagen) and anti-c-Myc mouse antibody (Calbiochem) as the primary antibodies and anti-mouse IgG conjugated to peroxidase as the secondary antibody (Calbiochem).
Two-hybrid system: vector construction, growth conditions, and dimerization assay.
The rpoA gene was amplified from E. coli genomic DNA by PCR using the rpoA-pcI434/P22 Fw and rpoA-pcI434/P22 Rv primers (Table 2). The amplified fragment was digested with SalI and BamHI and cloned into the pcI434 and pcIP22 plasmids (containing the N-terminal domain of phage 434 and the P22 repressor, respectively), which were linearized with the same restriction enzymes. The rplB gene was amplified from E. coli genomic DNA using the rplB-pcI434 Fw and rplB-pcI434 Rv primers (Table 2). The amplified fragment was digested with SalI and BamHI and cloned into the pcI434 plasmid. The resulting plasmids, pcIP22-RNAPα, pcI434-RNAPα, and pcI434-RPL2 (Table 1), were verified by automated DNA sequencing.
The pcIP22, pcI434, pcIP22-RNAPα, pcI434-RNAPα, and pcI434-RPL2 plasmids were introduced into R721 competent cells, which were then inoculated into 0.5 ml LB medium, diluted 1:1,000 in 10 ml LB medium supplemented with 1 × 10−4 M IPTG, and grown with aeration at 34°C for about 5 h. After this time the OD600 should have been between 0.3 and 0.4 (5).
The β-galactosidase activity assay was performed as described by Miller (14). The activity was expressed in Miller units and was calculated as follows: activity (Miller units) = 1,000 × A420/(time [min] × culture volume × optical density at 600 nm).
Construction of pET22b-PrrnD-lacZ and pET22b-PaidB-lacZ for transcription assays.
The rrnD P1 and the aidB promoters were amplified from DNA genomic of E. coli by PCR using the primers shown in Table 2. The amplified products were digested with SphI and HindIII and cloned into the pET22b(+) vector, which was linearized with the same restriction enzymes. Then the lacZ gene was positioned downstream of the rrnD P1 and aidB promoters using HindIII and XhoI. Plasmid construction was verified by automated DNA sequencing.
In vivo transcription assays.
C41 cells were either transformed with pET22b-PrrnD-lacZ or cotransformed with pET22b-PrrnD-lacZ and pcI-L2. The pcI-L2 plasmid was constructed by the following procedure. The pcI434 plasmid was digested with NdeI and BamHI to remove the 434 repressor N-terminal fragment; the rplB gene was amplified from E. coli genomic DNA using primers rplB-pcI Fw and rplB-pcIRv (Table 2) and digested with NdeI/BamHI, and the rplB fragment was cloned into the corresponding sites of pcI434, generating the pcI-L2 vector (Table 1) that could express native L2 lacking the N-terminal domain of the phage 434 repressor.
Recombinant C41 cells were grown overnight in LB medium at 37°C and diluted 1:100 in fresh medium, and then selective antibiotics were added. C41 cells transformed with pET22b-PaidB-lacZ and C41 cells cotransformed with pET22b-PaidB-lacZ and pcI-L2 were used as controls. At an optical density at 600 nm of 0.9, IPTG was added (final concentration, 0.05 mM), and cell pellets were collected when the culture OD600 was 1.5 (late exponential phase). The cells were resuspended in 50 mM Na2HPO4 (pH 7.4), disrupted by passage through a French press, and centrifuged at 14,000 × g for 30 min at 4°C. The supernatant was collected, and the protein concentration was determined by the Bio-Rad protein assay (1), using bovine serum albumin as the standard. β-Galactosidase activity was determined by measuring o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolysis, as described by Miller (14).
Overexpressed L2 was detected under test conditions; an aliquot of a bacterial culture overexpressing L2 was collected, and the soluble protein content was analyzed by SDS gel electrophoresis as a control. The presence of an intense Coomassie blue-stained protein band at about 30 kDa demonstrated that a significant amount of L2 was present.
The putative effects of other ribosomal proteins (L1, L3, L20, and L27) on rRNA transcription were also examined by performing in vivo transcription assays. To do this, C41 cells were cotransformed with the following constructs: pET22b-PrrnD-lacZ and pcI-L1, pET22b-PrrnD-lacZ and pcI-L3, pET22b-PrrnD-lacZ and pcI-L20, and pET22b-PrrnD-lacZ and pcI-L27. The β-galactosidase activity was monitored during the late exponential growth phase. C41 cells cotransformed with pET22b-PaidB-lacZ and pcI-L1, pcI-L3, pcI-L20, or pcI-L27 were used as controls. The pcI-L1, pcI-L3, pcI-L20, and pcI-L27plasmids were constructed by the following procedure. The rplA, rplC, rplT, and rpmA genes were amplified from E. coli genomic DNA by PCR using the specific primers shown in Table 2. The amplified fragments were digested with NdeI and BamHI and cloned into the pcI434 plasmid, which was digested with NdeI and BamHI to remove the 434 repressor N-terminal fragment. The resulting pcI-L1, pcI-L3, pcI-L20, and pcI-L27 vectors (Table 1) were able to produce the native L1, L3, L20, and L27 ribosomal proteins without the N-terminal domain of the phage 434 repressor.
Transcription experiments with the control ribosomal proteins were carried out under the conditions described above (i.e., C41 recombinant cell growth and induction) for the in vivo assays in the presence of recombinant L2 protein. All of the ribosomal proteins were stably expressed. Aliquots of bacterial cultures individually expressing L1, L3, L20, and L27 were collected, and the soluble protein contents were analyzed by SDS gel electrophoresis. Intense Coomassie blue-stained protein bands corresponding to the individual recombinant ribosomal proteins in the cell extracts were clearly detected.
RESULTS AND DISCUSSION
Identification of novel proteins associated with the RNAP α subunit.
Understanding protein functions and unraveling cellular mechanisms at the molecular level are major challenges in modern biology. Both of these goals can be addressed through identification of interacting protein partners in vivo. The association of an unknown protein with components belonging to a specific protein complex involved in a particular mechanism would strongly suggest the biological function of the protein (11, 13, 17). Furthermore, detailed description of the cellular signaling pathways might benefit greatly from elucidation of protein-protein interactions in the cell (7, 8).
Identification of interacting proteins in stable complexes in a cellular system by a functional proteomics approach is accomplished essentially by using affinity-based procedures. The basic idea is to express the protein of interest with a suitable tag and use it as bait to determine the specific partners of the protein in a whole-cell extract. Individual components of the multiprotein complex are then fractionated by SDS-PAGE and identified by mass spectrometry methods.
An investigation of the regulatory transcriptional network in E. coli was carried out by identifying transcriptional modulators interacting with the RNAP α subunit, using a “fishing-for-partners” strategy combined with mass spectrometry procedures. Recombinant plasmid pET22b-αpol was introduced into E. coli cells, and the resulting expressed six-His-tagged RNAP α subunit (6HisRNAPα) was purified by affinity chromatography on a His-Select nickel affinity gel. From 1 liter of bacterial growth about 20 mg of pure 6HisRNAPα protein was obtained.
In a classical pull-down experiment, the 6HisRNAPα was linked to agarose beads by His-Ni2+ interactions, and the clear bacterial extract was incubated with the bait. The immobilized protein formed stable noncovalent interactions with specific partners in the E. coli extract that were retained on the insoluble matrix, while the unbound proteins were eluted by repeated washing. After mild washes to eliminate nonspecific interactions, the protein components specifically recognized by the bait were eluted in Laemmli buffer and fractionated by one-dimensional 10% SDS-PAGE. The same amount of bacterial extract was incubated with His-Select nickel affinity gel beads lacking RNAPα under the same experimental conditions as a negative control.
Figure 1 shows an SDS-PAGE gel following colloidal Coomassie blue staining; lanes 4 and 5 contained the negative control and the sample, respectively. Due to the complexity of gel patterns and the low resolution of one-dimensional electrophoresis, several proteins can occur in the same gel band. Therefore, protein bands specifically present in a sample lane and not present in a control lane cannot be identified by simply comparing the two gel profiles. Thus, the entire lanes containing the sample and the control were cut into slices (38 slices each), as shown in Fig. 1, and each gel slice was subjected to the identification procedure. The proteins in the slices were reduced, alkylated, and digested in situ with trypsin. The resulting peptide mixtures were analyzed directly by mass spectrometry using both the peptide mass fingerprinting procedure and LC-MS/MS (12, 17). A comparison of the sample and the control based on the proteins effectively identified in each gel slice was then performed. Proteins that were identified in both the control and sample slices were discarded, and only the proteins identified in the sample slices but not in the control slices were selected as putative proteins that interact with RNAPα. Using this procedure, the RNAPα protein bait was identified only in sample slice C19 at an apparent molecular mass of about 40 kDa, whereas it was not present in corresponding control slice 19 and was not detected in any gel slice from the control lane.
FIG. 1.
Fractionation of proteins interacting with RNAPα by one-dimensional SDS-PAGE. After a preclearing step, the bacterial extract was incubated with 6HisRNAPα linked to agarose beads, and the protein components specifically recognized by the bait were eluted in Laemmli buffer. Lane 1, molecular markers; lane 2, total bacterial extract; lane 3, unbound proteins not retained by the bait; lane 4, protein extract retained by the His-Select nickel affinity gel alone and eluted with Laemmli buffer (control); lane 5, proteins specifically retained by the bait. Control lane 4 and sample lane 5 were cut into 38 slices, as indicated by boxes, and each gel slice from lane 4 (samples 1 to 38) and from lane 5 (samples C1 to C38) was subjected to mass spectrometry analysis.
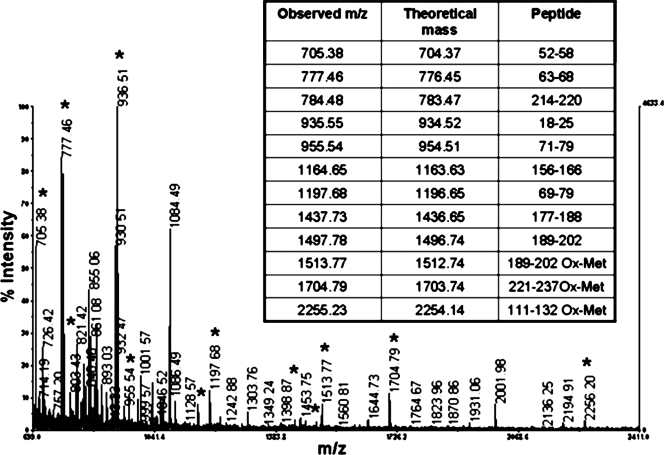
Among many other proteins, we focused on the protein in band C25 at about 30 kDa (Fig. 1, lane 5). The MALDI MS spectrum obtained for an aliquot of the tryptic digest of band C25 is shown in Fig. 2, and some peaks (peaks indicated by an asterisk) were used for a nonredundant sequence database search with an in-house version of the MASCOT software. The query returned a highly significant match (score, 160) with ribosomal protein L2, whose predicted molecular mass (30 kDa) corresponded to the observed electrophoretic mobility of band C25. The mass spectral analyses showed sequence coverage for RPL2 that was greater than 50%.
FIG. 2.
MALDI MS analysis of the tryptic digest of band C25. Band C25 was digested in situ with trypsin, and the resulting peptide mixture was extracted from the gel and directly analyzed by MALDI MS. Peaks indicated by an asterisk were used for a nonredundant sequence database search using the MASCOT software, which led to identification of RPL2. Some of these peaks corresponded to L2 peptides, as shown in the table indicating the measured and theoretical masses and the corresponding peptides in the L2 sequence.
The remaining portion of the tryptic digest of slice C25 was analyzed by LC-MS/MS. Peptides were fractionated by nano-HPLC by direct insertion into the nanoelectrospray source, and their masses were accurately determined. Peptide ions were simultaneously isolated and fragmented in the mass spectrometer, which produced daughter ion spectra from which sequence information could be inferred. Several sequence stretches were obtained that matched peptide fragments in the RPL2 sequence, which confirmed previous MALDI data and indicated that RPL2 is a putative protein that interacts RNAPα. No trace of RPL2 was observed in any gel slice from the control lane.
Verification of RNAPα-RPL2 interaction by coimmunoprecipitation.
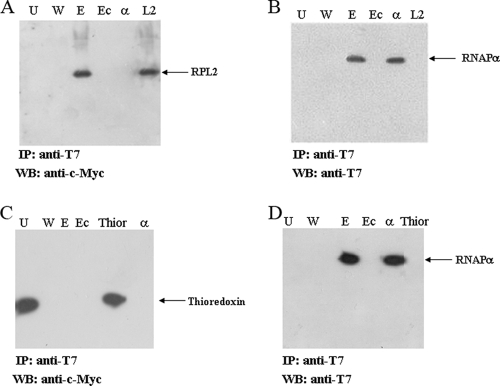
The interaction between RNAPα and RPL2 suggested by the results of the functional proteomics analysis had to be confirmed by both in vitro and in vivo investigations. First, the interaction was verified by a totally independent approach based on coimmunoprecipitation experiments. T7-tagged RNAPα and c-Myc-labeled RPL2 were coexpressed in E. coli C41 cells under the control of an IPTG-inducible promoter. However, since it is well known that pET vectors always express a small amount of the recombinant protein even in the absence of induction, stimulation of the recombinant cells with IPTG was omitted to avoid unnecessary overproduction of the two proteins. The protein extract from recombinant cells was fractionated by SDS-PAGE and stained with colloidal Coomassie blue as a control. The protein bands corresponding to RNAPα and RPL2 could not be detected, thus ruling out the possibility that the two proteins were overexpressed. The total protein extract was immunoprecipitated by anti-T7 antibody linked to agarose beads and underivatized agarose beads as a control. Figure 3A shows a Western blot of the T7-RNAPα immunoprecipitate developed with anti-c-Myc antibodies to highlight the presence of RPL2. A protein band positive for anti-c-Myc antibody that corresponded to the anti-T7 immunoprecipitate was clearly detected in lane E. A pure c-Myc-tagged RPL2 sample was loaded in lane L2 as a control, and it was positively stained by the antibody, whereas pure T7-RNAPα was not recognized, as expected (lane α).
FIG. 3.
Coimmunoprecipitation experiments. Total protein extracts of E. coli strain C41 cotransformed with pET28a-αpol and pET22b-c-Myc-L2 and with pET28a-αpol and pET22b-c-Myc-Thioredoxin were subjected to immunoprecipitation with agarose-linked T7 antibody and with agarose beads alone as a control, followed by immunoblotting with anti-c-Myc antibody (A and C) and with anti-T7 antibody (B and D). In this experiment the recombinant RNAPα contained a T7 tag. In all immunoblots, lanes U, W, and E contained the unbound, wash, and elution fractions when anti-T7 antibody beads were used, respectively, while lane Ec contained the fraction eluted from agarose beads alone. (A and B) T7-RNAPα-c-Myc-L2 coimmunoprecipitation. Lane α contained the purified T7-tagged form of RNAPα, and lane L2 contained the purified c-Myc-tagged form of RPL2. (C and D) No T7-RNAPα-c-Myc-thioredoxin coimmunoprecipitation. Lane Thior contained the purified c-Myc-tagged form of thioredoxin, and lane α contained the purified T7-tagged form of RNAPα. IP, immunoprecipitation; WB, Western blotting.
The presence of T7-RNAPα in the immunoprecipitate was verified by developing the Western blot with anti-T7 antibodies (Fig. 3B). A protein band at about 45 kDa was clearly detected by the antibody in lane E, and this band corresponded perfectly to pure T7-RNAPα loaded in lane α as a control. As expected, pure c-Myc-labeled RPL2 was not recognized by the anti-T7 antibody (lane L2), ruling out the possibility of any cross-reactivity in the experiment. No signal was generated in the fraction eluted from agarose beads alone (Fig. 3A and B, lane Ec).
A coimmunoprecipitation experiment using T7-tagged RNAPα and an unrelated protein, c-Myc-labeled thioredoxin, was performed as a control to rule out any possible artifact in the coimmunoprecipitation of RNAPα and RPL2 and to confirm the physiological relevance of the interaction. Thioredoxin is a cytosolic oxidoreductase that is involved in the reduction of other proteins by cysteine thiol-disulfide exchange, and it is completely unrelated to the transcription mechanism. The recombinant proteins were produced in E. coli C41 cells using the experimental conditions described above, and the total protein extract was immunoprecipitated with anti-T7 agarose-conjugated antibody and underivatized agarose beads as a control. As shown in Fig. 3D, when the Western blot was developed with anti-T7 antibodies, a protein band corresponding to pure T7-RNAPα loaded in lane α as a control was clearly visible in lane E containing the fraction eluted from anti-T7 antibody beads. In contrast, Fig. 3C shows that when the anti-c-Myc antibody was used, a protein band corresponding to pure thioredoxin (loaded in lane Thior as a control) was detected only in lane U containing the unbound material. No trace of this band was observed in the immunoprecipitation lane (Fig. 3C, lane E). These results demonstrated that thioredoxin did not interact with RNAPα, indicating that, at least under the conditions used, no nonspecific aggregation of the expressed proteins occurred and ruling out the possibility that there was an artifact in the coimmunoprecipitation experiments. A biologically significant interaction seemed to take place between RNAPα and RPL2, confirming previous proteomics data.
Investigation of the RNAPα-RPL2 interaction.
Another question was whether the RNAPα and RPL2 proteins interact directly or just belong to the same multiprotein complex. To address this question, we designed a biochemical experiment based on the bacterial two-hybrid system (5).
The genes coding for RNAPα and RPL2 were cloned in the plasmid vectors pcIP22 and pcI434, which contained the N-terminal domain of phage 434 and the P22 repressor, respectively. The recombinant plasmids obtained were used to transform E. coli strain R721, which harbors the 434-P22 chimeric operator located upstream of the lacZ gene. If RNAPα and RPL2 directly interact, a functional repressor would have been produced and the β-galactosidase activity expressed by the lacZ gene would have been decreased.
The ability of the reconstituted repressor to bind to the chimeric operator and to affect enzymatic activity was tested by measuring the residual β-galactosidase activity following induction of bacterial cells with 0.1 mM IPTG. The results are shown in Table 3. When plasmids pcIP22-RNAPα and pcI434-RPL2 were both expressed in E. coli R721 cells, there was a considerable decrease in the β-galactosidase activity; the residual enzymatic activity decreased to 35% of the initial value.
TABLE 3.
Two-hybrid assay results
| pcI plasmid(s) | β-Galactosidase activity (Miller units) | Residual β-galactosidase activity (%) |
|---|---|---|
| Binding to chimeric operatora | ||
| pcI434 | 2,350 ± 2.5 | 95 |
| pcIP22 | 2,517 ± 3.4 | 100 |
| pcIP22+ pcI434 | 2,373 ± 1.5 | 94.1 |
| pcI434-434 | 1,886 ± 2.6 | 75 |
| pcIP22-434 | 2,113 ± 3.1 | 86 |
| pcI434-434 + pcIP22-434 | 351 ± 2.7 | 14 |
| Interaction between RNAPα and RPL2b | ||
| pcIP22-RNAPα | 1,915 ± 1.8 | 76.2 |
| pcI434-RPL2 | 1,829 ± 2.5 | 72.7 |
| pcI434-RNAPα | 2,200 ± 3.4 | 88 |
| pcIP22-RNAPα + pcI434-RPL2 | 880 ± 2.9 | 35 |
| pcIP22-RNAPα + pcI434-RNAPα | 401 ± 3.2 | 16 |
The pcI plasmids were inserted into E. coli strain R721 by transformation, and the ability of each reconstituted repressor to bind to the chimeric operator was then determined by measuring the residual β-galactosidase activity.
A two-hybrid assay was performed to investigate the interaction between RNAPα and RPL2. Plasmids pcIP22-RNAPα and pcI434-L2 were both expressed in E. coli R721 cells, and the residual β-galactosidase activity was recorded. In the absence of any pcI plasmid, strain R721 produced 2,500 Miller units of β-galactosidase activity (mean of five independent experiments). The values are the means of nine independent experiments.
Since RNAPα is known to form a functional dimer, plasmids pcIP22-RNAPα and pcI434-RNAPα were also coexpressed in R721 cells as a control. As expected, when RNAPα formed the dimeric structure, a functional repressor was produced, and the β-galactosidase activity decreased to 16% of the initial value.
The results of the bacterial two-hybrid assay demonstrated that there is a direct interaction between RNAPα and RPL2 in the multiprotein complex.
RPL2 is a transcriptional modulator.
Finally, we examined whether the binding of RPL2 to RNAPα might have a functional role in modulating the transcription mechanism in E. coli. Since RPL2 is essential for growth and survival of E. coli, we could not design a knockout gene experiment. The putative transcriptional role of the interaction between RNAPα and RPL2 was investigated by performing in vivo transcription experiments using a classic gene reporter assay.
The lacZ gene coding for E. coli β-galactosidase was cloned into the pET22b(+) expression vector under the control of the ribosomal promoter rrnD P1, resulting in the recombinant expression vector pET22b-PrrnD-lacZ. The rrnD promoter P1 used contains the DNA sequence from position −61 to position 1, including the core promoter plus the upstream element (Table 2). The gene coding for RPL2 was cloned into the pcI434 plasmid, generating the recombinant expression vector pcI-L2, which was able to express native RPL2. The two compatible expression vectors were then used to cotransform E. coli C41 cells. The same cells were transformed with the pET22b-PrrnD-lacZ vector alone as a control.
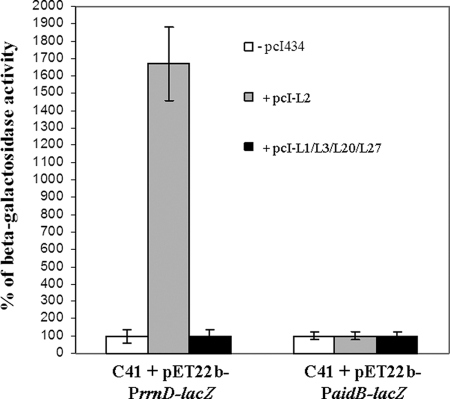
E. coli cells were allowed to grow, and the β-galactosidase activity was monitored in both cell cultures during the late exponential phase. Figure 4 shows that a large increase in the β-galactosidase activity clearly occurred in the E. coli cells producing recombinant RPL2 compared to the control. These finding suggested that the ribosomal protein might act as a transcriptional activator of the ribosomal operon rrnD.
FIG. 4.
In vivo transcription assays. E. coli C41 cells were transformed with pET22b-PrrnD-lacZ or with the pET22b-PaidB-lacZ vector, and the β-galactosidase specific activity was monitored in the absence and in the presence of recombinant proteins L1, L2, L3, L20, and L27. The β-galactosidase activity was evaluated at log phase. Open bars, β-galactosidase activity in the absence of recombinant protein production; gray bars, β-galactosidase activity when recombinant L2 was overproduced; black bars, β-galactosidase activity in the presence of L1, L3, L20, or L27. Means and standard deviations were calculated from the results of three independent assays. The β-galactosidase activities for the PrrnD-lacZ fusion in the presence and in the absence of L2 expression were 52,700 and 3,160 Miller units, respectively; and the β-galactosidase activities for the PaidB-lacZ fusion in the presence and in the absence of L2 expression were 357 and 355 Miller units, respectively. The β-galactosidase activities for the two promoter-lacZ fusions in the presence of L1, L3, L20, or L27 and in the absence of recombinant protein expression were about 360 Miller units. The background activity for the lacZ fusion vector without any promoter insert was 62 Miller units.
The specificity of RPL2 for the rrnD P1 promoter was tested by performing an in vivo transcription assay using the β-galactosidase gene as the reporter gene under control of an unrelated promoter, the aidB promoter. The PaidB promoter used contains the DNA sequence from position −61 to position 1, including the core promoter plus the upstream element (Table 2). As shown in Fig. 4, recombinant RPL2 did not affect β-galactosidase expression when the aidB promoter was used in the assay, as the enzymatic activity in cell cultures transformed with the pET22b-PaidB-lacZ and pcI-L2 constructs was not changed. These data confirmed previous results showing that RPL2 is a transcriptional regulator specific for the ribosomal promoter rrnD P1.
In addition, to ensure that the effect on rrnD P1 could be attributed specifically to RPL2, the putative effects of other ribosomal proteins on rRNA transcription were tested. The following four proteins of the bacterial ribosome were used as controls: L1 and L3, which are relatively large ribosomal proteins, like L2; L20, a ribosomal protein that participates in the early steps during assembly of the 50S ribosomal subunit (18); and L27, which is a relatively late assembly protein (23). As clearly shown in Fig. 4, the β-galactosidase activity was unchanged in the presence of the ribosomal proteins, showing that L1, L3, L20 and L27 were unable to affect lacZ expression driven by both rrnD P1 and PaidB, thus demonstrating the specific and direct effect of RPL2 on rRNA expression.
In addition to their role in ribosome assembly, several ribosome proteins have been shown to have other biological functions in the cell. One of the best-characterized functions is the ability of some E. coli ribosomal proteins to regulate translation of their own multicistronic mRNAs (25). Other E. coli ribosomal proteins have been demonstrated to play roles in transcriptional antitermination (S4 and S10) (21) or to be involved in DNA repair and replication (S9 and L14) (22, 24).
Data reported in this paper showed that the highly conserved RPL2 protein is involved in regulation of transcription by acting as an activator of ribosomal operon transcription. Moreover, our functional proteomics approach indicated that RPL2 belongs to a large multiprotein transcription complex that gathers at the RNAP α subunit. Finally, we demonstrated that the transcriptional activator function of RPL2 is carried out through direct and specific binding of the ribosomal protein to the α subunit of the RNA polymerase.
Acknowledgments
We are indebted to L. Paolozzi and G. Di Lallo of the Università Tor Vergata di Roma for the bacterial two-hybrid system.
This work was supported by grants from the Ministero dell'Università e della Ricerca Scientifica (Progetti di Rilevante Interesse Nazionale 2004, 2005, 2006; FIRB 2001), from the Programma Nazionale di Ricerche in Antartide 2004, and from Regione Campania L.R. 05/03. Support from the National Center of Excellence in Molecular Medicine (MIUR, Rome, Italy) and from the Regional Center of Competence (CRdC ATIBB, Regione Campania, Naples, Italy), Istituto Nazionale Biomolecole e Biosistemi (INBB), is gratefully acknowledged.
Footnotes
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 3.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., C. Röcken, C. Lofton-Day, H. U. Schulz, O. Müller, N. Kutzner, P. Malfertheiner, and M. P. Ebert. 2005. Molecular analysis of APC promoter methylation and protein expression in colorectal cancer metastasis. Carcinogenesis 26:37-43. [DOI] [PubMed] [Google Scholar]
- 5.Di Lallo, G., L. Castagnoli, P. Gherardini, and L. Paolozzi. 2001. A two-hybrid system based on chimeric operator recognition for studying protein homo/heterodimerization in Escherichia coli. Microbiology 147:1651-1656. [DOI] [PubMed] [Google Scholar]
- 6.Ebright, R. H. 1993. Transcription activation at class I CAP-dependent promoters. Mol. Microbiol. 8:797-802. [DOI] [PubMed] [Google Scholar]
- 7.Fraldi, A., E. Zito, F. Annunziata, A. Lombardi, M. Cozzolino, M. Monti, C. Spampanato, A. Ballabio, P. Pucci, R. Sitia, and M. P. Cosma. 2008. Multistep, sequential control of the trafficking and function of the multiple sulfatase deficiency gene product, SUMF1 by PDI, ERGIC-53 and ERp44. Hum. Mol. Genet. 17:2610-2621. [DOI] [PubMed] [Google Scholar]
- 8.Gavin, A. C., M. Bösche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Höfert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 9.Gingras, A. C., M. Gstaiger, B. Raught, and R. Aebersold. 2007. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8:645-654. [DOI] [PubMed] [Google Scholar]
- 10.Gourse, R. L., W. Ross, and T. Gaal. 2000. Ups and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 11.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, P. K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sørensen, J. Matthiesen, R. C. Hendrickson, R. C. F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 12.Köcher, T., and G. Superti-Furga. 2007. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4:807-815. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, T. S., J. B. Hunt, L. D. Aveline, K. R. Jonscher, D. F. Louie, J. M. Yeh, T. S. Nahreini, K. A. Resing, and N. G. Ahn. 2000. Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell 6:1343-1354. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 16.Monti, G., L. De Napoli, P. Mainolfi, R. Barone, M. Guida, G. Marino, and A. Amoresano. 2005. Monitoring food quality by microfluidic electrophoresis, gas chromatography, and mass spectrometry techniques: effects of aquaculture on the sea bass (Dicentrarchus labrax). Anal. Chem. 77:2587-2594. [DOI] [PubMed] [Google Scholar]
- 17.Monti, M., S. Orrù, D. Pagnozzi, and P. Pucci. 2005. Interaction proteomics. Biosci. Rep. 25:45-56. [DOI] [PubMed] [Google Scholar]
- 18.Raibaud, S., I. Lebars, M. Guillier, C. Chiaruttini, F. Bontems, A. Rak, M. Garber, F. Allemand, M. Springer, and F. Dardel. 2002. NMR structure of bacterial ribosomal protein L20: implications for ribosome assembly and translational control. J. Mol. Biol. 323:143-151. [DOI] [PubMed] [Google Scholar]
- 19.Rojo, F. 1999. Repression of transcription in bacteria. J. Bacteriol. 181:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savery, N., V. Rhodius, and S. Busby. 1996. Protein-protein interactions during transcription activation: the case of Escherichia coli cyclic AMP receptor protein. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351:543-550. [DOI] [PubMed] [Google Scholar]
- 21.Torres, M., C. Condon, J. M. Balada, C. Squires, and C. L. Squires. 2001. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 14:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodgate, R., M. Rajagopalan, C. Lu, and H. Echols. 1989. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc. Natl. Acad. Sci. U. S. A. 86:7301-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wower, I. K., J. Wower, and R. A. Zimmermann. 1998. Ribosomal protein L27 participates in both 50S subunit assembly and the peptidyl transferase reaction. J. Biol. Chem. 273:19847-19852. [DOI] [PubMed] [Google Scholar]
- 24.Yancey, J. E., and S. W. Matson. 1991. The DNA unwinding reaction catalyzed by Rep protein is facilitated by an RHSP-DNA interaction. Nucleic Acids Res. 19:3943-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zengel, J. M., and L. Lindahl. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 47:331-370. [DOI] [PubMed] [Google Scholar]