Abstract
In Drosophila species, molecular asymmetries guiding embryonic development are established maternally. Vasa, a DEAD-box RNA helicase, accumulates in the posterior pole plasm, where it is required for embryonic germ cell specification. Maintenance of Vasa at the posterior pole requires the deubiquitinating enzyme Fat facets, which protects Vasa from degradation. Here, we found that Gustavus (Gus) and Fsn, two ubiquitin Cullin-RING E3 ligase specificity receptors, bind to the same motif on Vasa through their paralogous B30.2/SPRY domains. Both Gus and Fsn accumulate in the pole plasm in a Vasa-dependent manner. Posterior Vasa accumulation is precocious in Fsn mutant oocytes; Fsn overexpression reduces ovarian Vasa levels, and embryos from Fsn-overexpressing females form fewer primordial germ cells (PGCs); thus, Fsn destabilizes Vasa. In contrast, endogenous Gus may promote Vasa activity in the pole plasm, as gus females produce embryos with fewer PGCs, and posterior accumulation of Vas is delayed in gus mutant oocytes that also lack one copy of cullin-5. We propose that Fsn- and Gus-containing E3 ligase complexes contribute to establishing a fine-tuned steady state of Vasa ubiquitination that influences the kinetics of posterior Vasa deployment.
Establishment and maintenance of polarity is essential for multicellular development. Asymmetric distribution of proteins within cells is often realized by localizing specific mRNAs to distinct positions and by tightly regulating their translation (1). During Drosophila oogenesis, polarized deployment of key mRNAs is crucial for the maternal determination of the embryonic body axes (17). However, although asymmetric mRNA localization within cells is widespread (1, 22), some proteins localize directly. An example is Vasa (Vas), which accumulates in a highly polarized fashion in Drosophila oocytes from a uniformly distributed mRNA that is not believed to be under translational control (8, 20, 21). Vas accumulates in the pole plasm of the oocyte, where it is necessary for embryonic posterior patterning and primordial germ cell (PGC) formation (26). Accumulation of high levels of Vas in the pole plasm requires the deubiquitinating enzyme (DUB) Fat facets (Faf) (25). In faf mutants, levels of posterior Vas are reduced, and polyubiquitinated forms of Vas accumulate. This indicates that Vas stability in the pole plasm is regulated by ubiquitin-dependent pathways.
Ubiquitination culminates in the E3 ligase-catalyzed formation of a covalent bond between the C terminus of ubiquitin and a lysine residue of the ubiquitinated protein (12). Target proteins can be ubiquitinated simultaneously and/or sequentially on different lysine residues, and the presence of seven internal lysine residues in ubiquitin itself allows for the formation of topologically distinct polyubiquitin chains (9, 12, 27). Different forms of ubiquitination usually produce different effects on the target protein, and modulation of the steady-state dynamics of ubiquitin conjugation can strongly influence a target's activity and/or stability. The regulatory logic governing the steady state of target ubiquitination can consist of nonlinear pathways that involve feedback mechanisms, responses to cellular stimuli such as phosphorylation, and complex cross-regulation between individual components of the ubiquitin conjugation machinery and corresponding DUBs.
Cullin-RING ubiquitin E3 ligases (CRLs) comprise the largest class of ubiquitin E3 ligases (30). CRLs contain a substrate specificity receptor that binds the ubiquitinated target and a RING protein that is involved in recruiting an E2-conjugating enzyme, which catalyzes transfer of ubiquitin to the associated substrate through the E3 ligase. RING proteins and particular substrate specificity receptors are brought together by scaffold proteins called Cullins, often through small adaptor proteins that link the Cullin with the receptor. Cullin-1 (Cul-1) CRLs recruit their substrate through F-box proteins, with a Skp family adaptor protein forming a bridge between the Cullin and the F-box. In contrast, CRLs containing Cullin-5 (Cul-5) recognize their substrates through receptor proteins that contain a SOCS-box, which are linked to the Cullin by the Elongin B/Elongin C (EloBC) adaptor complex.
In this study we identified the F-box protein Fsn and the SOCS-box protein Gus as in vivo regulators of Vas. Fsn and its Caenorhabditis elegans orthologue are required for normal synaptic development and associate with RING proteins encoded by highwire and rpm-1, respectively (24, 46). Gus was previously shown to interact with Vas and was implicated in its posterior localization (37). Gus features a B30.2/SPRY domain through which it directly interacts with a five-amino-acid motif on Vas (DINNN), and it can be cocrystallized with the EloBC complex (44, 45). Fsn has a B30.2/SPRY domain that is very similar to that of Gus (40% identity). We show experimentally that Fsn also binds the DINNN motif and that Gus associates more stably with Vas than does Fsn. Using genetic methods, we investigated the contributions of Gus and Fsn to regulating Vas activity and deployment.
MATERIALS AND METHODS
Structural predictions.
The amino acid sequences of the B30.2/SPRY domains of Gus and Fsn were aligned using ClustalW (19) and introduced into the MODELLER program (32). The B30.2/SPRY domain of Fsn was modeled using the previously solved structures of Gus bound to the Vas peptide (protein database [PDB] code 2IHS) (Fig. 1A to D) (45) and of Gus in complex with EloBC (PDB code 2FNJ) (44 and data not shown).
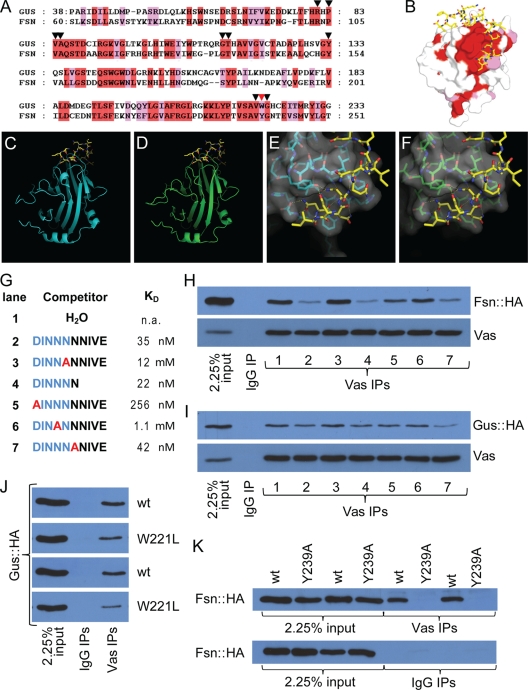
FIG. 1.
Gus and Fsn are Vas-interacting proteins. (A) Sequence alignment of the SPRY domains of Gus and Fsn. Identical and similar residues are highlighted in red and pink, respectively. Inverted triangles highlight the residues that form the Vas binding pocket. The red inverted triangle marks the residue W221 of Gus and Y239 of Fsn. (B) Surface view of a Gus crystal structure (PDB code 2IHS) (44). Surface residues conserved between Gus and Fsn are colored as described for panel A. (C to F) The B30.2/SPRY domains of Gus and Fsn are likely to form highly similar 3D structures. Panels C and E show crystal structures obtained from Gus (cyan) in complex with a Vas fragment (yellow) (PDB code 2IHS) (45). Panels D and F are structural predictions of the B30.2/SPRY domain of Fsn onto the crystals shown in panels C and E, respectively. The surface topology of the predicted Vas-binding site on Fsn (F) is almost identical to that of Gus (E). (G) Peptides used for the competition experiments shown in panels H and I. Kd values (apparent dissociation constants) for the interactions between Gus and peptides (45) are shown. (H) Fsn::HA and Vas coimmunoprecipitate. The interaction is sensitive to competition with peptides containing an intact DINNN motif. (I) GusL::HA and Vas coimmunoprecipitate. While the interaction is sensitive to peptide competition, the effects are much less pronounced than is the case for Fsn. (J) Both the wild type and W221L mutant Gus::HA associate with Vas in coimmunoprecipitation experiments; however, the amino acid substitution reduces the affinity of the interaction. All transgenic lines expressing GusL::HAW221L produced low levels of protein, possibly indicating a destabilizing effect of the substitution. Two transgenic lines are shown for Gus::HAW221L (second and fourth panels from the top), both compared to the same Gus::HA strain that expressed at a similar level to the mutant protein (first and third panels). (K) Unlike wild-type Fsn::HA, transgenic Fsn containing the Y239A mutation does not detectably coimmunoprecipitate with Vas. Two independent transgenic lines for each protein are shown.
Transgenic fly strains.
Transgenes encoding green fluorescent protein (GFP)-tagged Vas (GFP::Vas) were generated as described previously (10), except that site-directed mutagenesis was performed using GeneTailor (Invitrogen). Fsn and Gus fusion proteins were produced by PCR amplification of the full-length open reading frames (ORFs) of Fsn, GusL, and GusS from the Berkeley Drosophila Genome Project Drosophila gene collection (BDGP DGC) cDNA clones LD47425 and LD34464, respectively, and then cloning the products into pENTR/D-TOPO (Invitrogen). When applicable, mutations were introduced at this step using GeneTailor (Invitrogen). Positive clones were then recombined with pPVW and pPWH (Drosophila Gateway collection, provided by the Murphy lab) to produce Venus::Fsn (V::Fsn), Venus::GusL (V::GusL), Venus::GusS (V::GusS), Fsn::hemagglutinin (Fsn::HA), GusL::HA, and GusS::HA. Standard procedures were used to transform the constructs into flies and to map the insertion chromosomes. Expression of the transgenic proteins was achieved by crossing to the appropriate Gal4 driver strains.
Fly stocks.
gusf04367, cul-5EY32463, Df(2L)nap1, and Df(2L)Exel7124 were provided by the Bloomington Stock Center, Fsnf06595, gusf07073, and guse00456 were from the Exelixis Collection (Harvard Medical School).
Hatching assays and PGC counts.
Virgin females were collected for 3 days and then mated to Oregon-R males for 24 h. Subsequently, embryos were repeatedly collected, and the number of hatched and unhatched embryos was determined 36 to 48 h after each egg lay. For PGC counts, embryos were fixed and immunostained with anti-Vas antibodies. PGCs were counted in stage 10 embryos at ×200 magnification.
Immunoprecipitations.
Immunoprecipitations were performed as described in reference 16, using agarose-coupled mouse monoclonal anti-HA or rabbit anti-Vas antibodies. HA-tagged transgenic proteins were expressed in the ovaries under the control of the nanos-Gal4::VP16 driver (41). Rabbit or agarose-coupled mouse immunoglobulin G (IgG) (Santa Cruz) was used as control. For peptide competition assays, 10 μl water or 10 μl of a 1-mg/ml aqueous solution of the indicated peptides (45) was added to each immunoprecipitation reaction mixture. Western blots of the immunoprecipitates were probed with rabbit or rat anti-Vas antibodies (1:10,000), horseradish peroxidase (HRP)-coupled rat anti-HA antibodies (Roche; 1:2,000), rabbit anti-Cullin-1 antibodies (Zymed; 1:250), or rabbit anti-Cullin-5 antibodies (1:200) (31).
Immunocytochemistry.
Immunocytochemistry was performed using standard procedures. Rabbit and rat anti-Vas antibodies were used at 1:2,000 and detected with AlexaFluor-488 or AlexaFluor-555 after mounting in Antifade (Molecular Probes). Ovaries expressing GFP or Venus fusion transgenes were treated likewise. Images were taken on LSM510meta confocal microscopes (Zeiss); analysis and processing were done with ImageJ, LSM image browser, and Adobe Photoshop. Embryos derived from mothers expressing GFP-Vas in a vas mutant background were collected, dechorionated with 50% bleach for 2 min, and imaged live.
Preparation of ovarian extracts for Western blot analysis.
Ovaries were dissected into phosphate-buffered saline (PBS) and frozen in liquid nitrogen. Samples were homogenized in cold hybridization buffer (HB) (1× PBS with 10× Roche EDTA-free protease inhibitor cocktail) and cleared by centrifugation at 20,000 × g for 10 min. The supernatant was recovered, its protein concentration was determined by a Bradford assay (Bio-Rad), and the remaining sample was boiled with an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer (33). Samples were then diluted with 1× SDS loading buffer to the desired final concentration and boiled again before being loaded onto polyacrylamide gel electrophoresis (PAGE) gels. We dissected 15 to 35 ovaries from 24- to 48-h-old females per sample. We noted that Df(2R)Exel7124/Fsnf06595 ovaries were often smaller and contained fewer late-stage egg chambers than those of other genotypes of comparable age. Therefore, to ensure comparability we chose only well-developed Fsn mutant ovaries that most closely resembled those of other genotypes for analysis.
Quantitative Western blot analysis.
Ovarian extracts were diluted to a final concentration of typically 0.33 mg/ml (in our initial experiments we performed serial dilutions to determine and ensure the linear range of our detection method), and 15 μl was loaded per lane onto 6% E-PAGE gels (Invitrogen). We tested six genotypes per 96-well gel, such that each extract was loaded in eight replicates with eight adjacent wild-type control extracts in a sextant of the gel. We ran at least three independent extracts, generating ≥32 sample/control data points, per tested genotype (except for vasPH165/vasPD, only one extract was used for negative control purposes). After running the extracts, gels were equilibrated in transfer buffer (2.5 mM Tris, 19.2 mM glycine, 20% MeOH) for 30 min of shaking and then wet transferred onto activated Hybond-LFP membranes at 25 V at 4°C overnight. Membranes were then developed at room temperature. Following blocking with 2% ECL-Advance blocking agent (GE Healthcare) in PBST (1× PBS, 0.2% Tween 20) for 1 h, membranes were rinsed two times in PBST, washed two times with PBST for 5 min, and incubated with duplexed primary antibodies in blocking solution for 1.5 h. Primary antibodies were rabbit anti-Vas antibodies (rabbit 3005) at 1:4,000 and mouse anti-α-tubulin antibodies (Sigma, clone DM1A) at 1:2,500. Membranes were then rinsed two times, washed two times with PBST for at least 5 min each, and subsequently incubated with duplexed Cy3-conjugated anti-mouse and Cy5-conjugated anti-rabbit antibodies (GE Healthcare; 1:1,250) for 1 h. Membranes were then rinsed three times with PBST, washed with PBST four times for at least 5 min each, rinsed three times with PBS, and dried in the dark overnight at room temperature or for 1 h at 37°C. Blots were scanned with a Typhoon Trio and analyzed using ImageQuant. For each sextant, the Cy5 and Cy3 signal of one wild-type lane was set arbitrarily to 100%. Subsequently, we calculated the relative amount of Vas, standardized to α-tubulin, in our test genotype in comparison to its control by using the following equation:
 |
where I is the normalized fluorescence intensity of a given band and n is the number of paired control/test lanes. The graph presents the average of all pooled data points obtained for each genotype; error bars denote the standard deviations (±SD).
To quantitate GFP::Vas levels, we produced Western blots of ovarian extracts which were separated on SDS mini gels in a dilution series. Subsequent to probing the membranes as described above, we normalized each GFP::Vas/α-tubulin signal ratio of the transgene of GFP::Vas(5xAla) (a GFP-Vas fusion with a DINNN motif substituted by five Ala residues) to that of GFP::Vas(wt) (the wild-type GFP-Vas fusion). Four independent extracts were compared in four dilution steps each.
Quantitative reverse transcription-PCR (qRT-PCR).
To assess the effect of the PBac insertions on the expression of gus and Fsn, ovarian RNA was isolated from 15 females using the RNeasy kit (Qiagen) and reverse transcribed using SuperScript II (Invitrogen) following the supplier's instructions. Intron-spanning primers were used in PCR analyses with Taq polymerase (Qiagen) according to the manufacturer's recommendations.
Expression levels of GFP::Vas transgenes were assessed as follows: ovarian RNA was isolated with RNeasy, including on-column digestion of contaminating DNA with the RNase-free DNase kit (Qiagen). Total RNA (0.8 μg) was used for reverse transcription and quantitative PCR (qPCR) using the DyNAmo cDNA synthesis kit and qPCR kit (Finnzymes), following the supplier's recommendations. Transgenic RNA was quantified on a Bio-Rad CFX98 system using a GFP-specific and a vas-specific primer. Rp49-, β-tubulin-, and cul-1-specific primer pairs were used for normalization. Data were analyzed using the Bio-Rad CFX Manager software. All reactions were performed in triplicates of two independently isolated RNA samples per genotype.
RESULTS
Structural predictions suggest a similar mode of interaction of Gus and Fsn with Vas.
The SPRY domain of Gus forms a rigid surface pocket which embeds the DINNN motif of Vas (42, 43). Cocrystalization studies of SSB proteins, the mammalian orthologues of Gus, with a Vas peptide revealed that their SPRY domains feature very similar Vas-binding pockets (see Fig. S1A in the supplemental material) (15). Drosophila Fsn contains a SPRY domain that is also very similar to those of Gus (Fig. 1A and B) and the SSB family members (see Fig. S1B in the supplemental material). We modeled the B30.2/SPRY domain of Fsn onto the existing Gus structures that were derived by X-ray crystallography (Fig. 1C to F and data not shown) (44, 45). As predicted by the sequence similarity, the overall organization of the predicted B30.2/SPRY domain structure of Fsn is almost identical to that of Gus. Moreover, the surface topology as well as the position and identity of most of the amino acids implicated in Gus-Vas interaction are conserved in the predicted Fsn structure. As expected, the predicted Fsn structure is also highly similar to the structure of the SSB proteins (see Fig. S1C in the supplemental material). These predictions suggest that Fsn would associate with Vas in a manner that is structurally similar to Gus, which we proceeded to test experimentally.
Gus and Fsn interact with a common site on Vas.
To directly test whether Fsn is a Vas-interacting protein and to further characterize the Vas-Gus interaction in vivo, we generated transgenic flies that express HA-tagged versions of Gus (GusL::HA; see below) or Fsn (Fsn::HA) upon Gal4 induction. We then immunoprecipitated Vas from ovarian extracts expressing either transgene in the germ line and found that both Fsn::HA and GusL::HA efficiently copurified with Vas (Fig. 1H and I, lane 1). Vas also copurified with GusL::HA and Fsn::HA in the reciprocal experiment using anti-HA resin (data not shown). The interactions persist even in modified radioimmunoprecipitation (mRIPA) buffers with high concentrations of detergents (see Materials and Methods). Thus, both Gus and Fsn can form protein complexes with Vas in ovaries.
Short peptides derived from Vas that contain an intact DINNN motif bind to Gus (45). Similar peptides (Fig. 1G) efficiently competed with Vas for interaction with Fsn::HA (Fig. 1H, lanes 2 and 4), indicating that the Fsn-Vas interaction likely occurs through direct binding to the DINNN motif. To confirm the relevance of the DINNN motif of Vas for binding to Gus or Fsn, we assessed peptides with single-amino-acid substitutions within or outside the DINNN motif for their ability to compete with these interactions (Fig. 1G). Substitution of either of two DINNN residues (N187 or N188) with Ala greatly reduced affinity of Vas for Gus (45), and peptides with these substitutions failed to compete with the Vas-Fsn::HA interactions (Fig. 1H, lanes 3 and 6). Substitution of D184, another residue included in the DINNN motif, with Ala more modestly reduced the affinity of Vas for Gus (45), and this peptide acted more weakly as a competitor with Vas for binding to Fsn::HA (Fig. 1F, lane 5). In contrast, substitution of N189, a residue outside the DINNN motif, with Ala had no impact on the affinity of the mutant peptide for Gus (45) nor on the efficiency of Vas-Fsn::HA coimmunoprecipitation (Fig. 1H, lane 7). Taken together, these results show that the DINNN motif in Vas is important for its interaction with the B30.2/SPRY domain of Fsn.
In equivalent experiments with GusL::HA we observed qualitatively similar but significantly weaker inhibitory effects for these peptides (Fig. 1I). This confirms previous observations (44, 45) that the DINNN motif is important for the interaction between Gus and Vas, but, importantly, it also indicates that Gus, unlike Fsn, may also associate with Vas through additional contacts. As full-length Vas and a DINNN motif containing peptide bind to Gus equally well in vitro (44), these additional contacts are likely stabilized in vivo by other molecules.
We also investigated the role of the B30.2/SPRY domains of Gus and Fsn on Vas interaction by examining the effect of mutating W221 of Gus, a residue of the B30.2/SPRY domain, which when altered to Leu greatly reduced the interaction of Gus with a Vas-derived peptide containing the DINNN motif (44). Consistent with this, GusL::HA carrying the W221L substitution (GusL::HAW221L) coimmunoprecipitated less efficiently with Vas than wild-type GusL::HA. However, this mutation did not fully abrogate association with Vas (Fig. 1J), again suggesting that interactions other than those between the B30.2/SPRY domain of Gus and the DINNN motif of Vas contribute to the interactions of the two full-length proteins. We performed the equivalent experiment for Fsn by substituting Y239 (45) with Ala, and found that Fsn::HAY239A did not immunoprecipitate with Vas at detectable levels (Fig. 1K). This indicated that the interaction between the B30.2/SPRY domain of Fsn and the DINNN motif of Vas are largely responsible for the Fsn::HA-Gus association.
Molecular genetics of gus and Fsn.
We next addressed the function of Gus and Fsn during oogenesis. Based on the phenotype of a mutant allele (gusZ409), gus was previously implicated in localizing Vas to the posterior of the oocyte (37). We and others, however, found during subsequent work that the gusZ409 chromosome, which was recovered from a chemical mutagenesis screen (14), contains at least two additional mutations that affect female fertility and embryonic patterning, including an allele of Bicaudal-C. Because of the confounding effects of these additional mutations, we obtained fully independent gus alleles for this study.
Expressed sequence tag (EST) evidence predicts six gus mRNAs, five transcribed from a common proximal promoter (gus-RA and gus-RC through gus-RF; the “RA class”) and one (gus-RB) from a second promoter approximately 10 kb upstream (FlyBase) (Fig. 2A). We used RT-PCR to show that both the RA class and gus-RB are expressed in ovaries (Fig. 2B). All annotated gus mRNAs possess an ORF of 281 amino acids whose start codon resides in an exon common to all transcripts, but all except gus-RF also contain an upstream in-frame AUG and could thus encode longer protein isoforms with different N-terminal regions. We raised two polyclonal antibodies against recombinant Gus to test which of the putative isoforms are expressed in ovaries. When interrogating Western blots with these antibodies, we detected a band of approximately 30 kDa that is reduced in ovaries bearing the gusf07073 allele (see below), which likely corresponds to the 281-amino-acid protein (Fig. 2C). We did not reproducibly detect any bands that correspond to a longer Gus isoform. This is consistent with our previous observation that an antibody raised against the unique N-terminal part of Gus did not show immunoreactivity on Western blots (37). However, these negative data cannot rule out that small amounts of one or more longer isoforms of Gus are expressed.
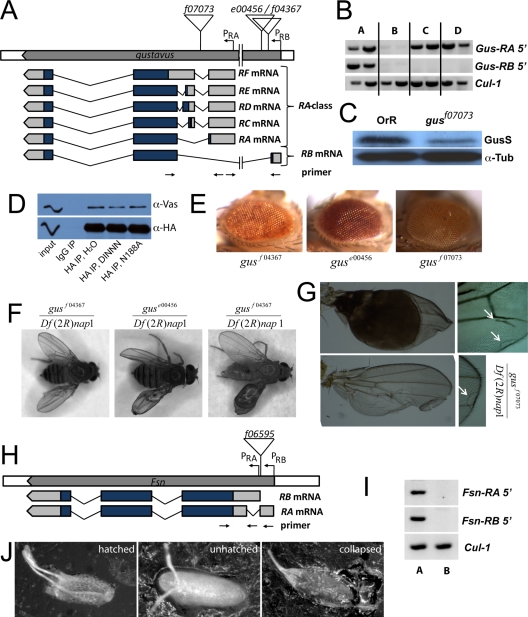
FIG. 2.
Molecular genetics of gus and Fsn. (A) Schematic representation of the gus locus and its transcripts (not to scale). Insertion sites of the PBac elements used in this study are indicated. (B) RT-PCR amplifications specific for the RA class and gus-RB transcripts indicate that both mRNAs are expressed in wild-type ovaries (lane A). gusf07073 impairs expression of all tested transcripts (lane B), while guse00456 in trans to gusf07073 (lane C) or homozygous (lane D) affects only gus-RB. Positions of the primer pairs used are indicated in panel A; exact sequences are available upon request. A fragment of cul-1 mRNA was amplified as a control. (C) An antibody raised against recombinant Gus recognizes a ∼30-kDa protein that is reduced in gusf07073 ovaries, corresponding most likely to the 281-amino-acid-long isoform of Gus (GusS). The blot was also probed with anti-α-tubulin antibodies (α-Tub) as a loading control. (D) Transgenic GusS::HA efficiently coimmunoprecipitates Vas. A peptide containing an intact DINNN motif moderately interferes with this reaction, while a corresponding peptide bearing the N188A substitution within the DINNN motif does not have this effect. (E) Eyes from flies bearing the gusf07073, guse00456, and gusf04367 insertions. The mini white marker that tags the PBac shows position effect variegation in its expression. (F) In trans to the deficiency Df(2R)nap1, the gusf07073, guse00456, and gusf04367 mutations cause defects in the wing, including blisters, folded wings and ectopic vein material. (G) Hoyer's embedded wings derived from Df(2R)nap1/gusf07073 flies. (H) Schematic representation of the Fsn locus (not to scale). The insertion site of PBac Fsnf06595 is indicated. (I) RT-PCR amplifications specific for the Fsn-RA and Fsn-RB transcripts indicating expression of both isoforms in wild-type ovaries (lane A). In Df(2R)Exel7124/Fsnf06595 ovaries (lane B) these mRNAs are not expressed at detectable levels. (J) Eggs produced by gusf07073/gusf07073 mothers. Many of these eggs collapse.
For this reason, we prepared HA-tagged transgenic proteins corresponding to the 281-amino-acid Gus protein (GusS::HA) as well as to the 349-amino-acid protein (GusL::HA) that could be produced by gus-RA using its upstream AUG for functional studies. Notably, both GusL::HA (Fig. 1) and GusS::HA (Fig. 2D) associated with Vas in coimmunoprecipitation experiments. The interactions of either isoform with Vas were similarly sensitive to competition by a peptide bearing an intact DINNN motif and insensitive to the N188A-containing peptide (Fig. 1H and 2D). Therefore, the short and predicted long isoforms of Gus appear to have a similar capability to interact with Vas.
Several alleles of gus were recovered from a large-scale genetic screen (39). gusf07073 is a PiggyBac (PBac) insertion into the first intron of all annotated gus transcripts, which largely abrogates expression of all gus mRNAs (Fig. 2B). gusf04367 and guse00456 are PBac insertions proximal to the first exon of gus-RB. guse00456 reduces expression of gus-RB but not of RA class-specific products (Fig. 2B). We did not quantitate gus expression levels in gusf04367 mutant flies. We verified the PBac insertion sites in these alleles by genomic PCR experiments using combinations of PBac and gene-specific primers (not shown) and noted that the mini-white marker of PBac showed variegated expression (Fig. 2E), consistent with the position of gus and thus the PBac insertion sites, near the euchromatin-heterochromatin junction (36). Finally, all three insertions cause obvious and similar wing morphology phenotypes, including blistering, formation of ectopic vein material, and loss of material at the margin (Fig. 2F and G), usually affecting only one wing. These phenotypes occur at low penetrance in guse00456/Df(2R)nap1, gusf04367/Df(2R)nap1, and gusf07073/gusf07073 flies but are frequent in gusf07073/Df(2R)nap1 flies [Df(2R)nap1 is a deletion mutation of a chromosomal region that includes gus]. Taken together, these data indicate that all three mutant strains contain bona fide mutations of gus.
The Fsn locus is less complex. We confirmed expression of two mRNAs, Fsn-RA and Fsn-RB, in ovaries. These mRNAs differ slightly in their 5′ untranslated regions (UTRs) but encode identical proteins of 255 amino acids (Fig. 2H and I). The PBac insertion Fsnf06595 is inserted in a sequence common to the 5′ UTRs of the two transcripts and abrogates expression of both mRNAs (Fig. 2I) (46).
gus is a maternal effect gene.
We tested whether gus and Fsn mutant mothers produce defective eggs or embryos. gusf07073/gusf07073 or gusf07073/Df(2R)nap1 embryos (for simplicity we refer to embryos by their maternal genotype) hatched only rarely (Table 1), and most viable embryos were produced by young females. guse00456/guse00456 embryos also rarely hatched; however, in trans to stronger alleles [Df(2R)nap1 or gusf07073] this allele has little effect on embryonic viability. This indicates either that guse00456 acts as a gain-of-function allele or that the chromosome carries a second site mutation. gusf04367 can be maintained as a homozygous stock, and most embryos hatch into viable larvae; thus, it is a very weak allele. For further work we concentrated on gusf07073, as both molecular and phenotypic evidence indicate that it most closely approximates a simple null allele.
TABLE 1.
Gus maternally affects embryonic viability
| Maternal genotype | Total no. of embryos scored for hatching frequency | % Hatched embryos | Total no. of embryos scored for collapsed phenotype | % Collapsed embryos |
|---|---|---|---|---|
| Oregon-R | 6,851 | 92.8 | 3,597 | 0 |
| +/gusf07073 | 2,561 | 94.3 | 1,214 | 0 |
| +/guse00456 | 2,679 | 92.4 | 1,236 | 0 |
| +/gusf04367 | 1,193 | 97.4 | 1,193 | 0 |
| +/Df(2R)nap1 | 1,892 | 85.6 | NDa | |
| gusf07073/guse00456 | 2,890 | 74.8 | 1,529 | 0.2 |
| guse00456/gusf07073 | 1,522 | 80.6 | 1,522 | 0.6 |
| gusf07073/gusf07073 | 4,543 | 2.3 | 3,127 | 68.1 |
| guse00456/guse00456 | 4,071 | 5.5 | 2,495 | 2.7 |
| gusf07073/Df(2R)nap1 | 2,710 | 9.2 | 1,413 | 76.9 |
| guse00456/Df(R2)nap1 | 1,536 | 73.8 | 1,536 | 1.2 |
| gusf04367/Df(2R)nap1 | 2,109 | 70.4 | 2,109 | 1.1 |
| gusf07073/gusf07073; actGal4/TM3Sb | 526 | 0.6 | 526 | 88.4 |
| gusf07073/gusf07073; TM3Sb/Gus(S)::HA | 853 | 0.9 | 853 | 86.3 |
| gusf07073/gusf07073; Gus(S)::HA/TM3Sb | 799 | 0.3 | 799 | 91.9 |
| gusf07073/gusf07073; Gus(S)::HA/nosGal4 | 1,801 | 0.6 | 1,801 | 93.7 |
| gusf07073/gusf07073; actGal4/Gus(S)::HA | 1,670 | 20.1 | 1,670 | 2.8 |
| gusf07073/gusf07073; Fsn::HA/TM3Sb | 87 | 1.1 | 87 | 81.6 |
| gusf07073/gusf07073; Fsn::HA/actGal4 | 1,135 | 14.2 | 1,135 | 47.3 |
| Fsnf06595/+ | 394 | 88.5 | 394 | 0 |
| +/Df(2R)Exel7124 | 393 | 93.2 | 393 | 0 |
| Fsnf06595/Df(2R)Exel7124 | 657 | 76.7 | 657 | 0 |
| +/cul-5EY21463 | 877 | 90.3 | 877 | 1.0 |
| cul-5EY21463/cul-5EY21463 | 1,805 | 5.9 | 1,805 | 62.3 |
ND, not determined.
Most gusf07073/gusf07073 or gusf07073/Df(2R)nap1 embryos desiccate and collapse soon after egg laying (Fig. 2J; Table 1). This phenotype was nearly fully rescued by maternal overexpression of GusS::HA using the ubiquitous Act5C-Gal4 driver, but not when the germ line-specific nanosGal4::VP16 (nosGal4) driver was used (41). This most likely reflects a requirement for gus in somatic follicle cells, although effects from the promoters driving different amounts of gus expression in germ line cells cannot be entirely ruled out. Unlike GusS::HA, GusL::HA did not rescue the collapsed-egg phenotype; in fact, gusf07073/gusf07073 flies containing both the GusL::HA transgene and the Act5C-Gal4 driver were never recovered, suggesting that GusL::HA, expressed at high levels in gusf07073/gusf07073 somatic cells, is lethal. Df(2R)Exel7124/Fsnf06595 [Df(2R)Exel7124 is a chromosomal deletion that includes Fsn] embryos have nearly wild-type viability. Intriguingly, Fsn::HA driven by Act5C-Gal4 could partially rescue the collapsed-egg gusf07073 phenotype.
Fsn and Gus can modulate Vas levels in ovaries and interact with Cul-1 and Cul-5, respectively.
The predicted function of Fsn and Gus as E3 ligase receptor subunits suggests that they may modulate Vas abundance by recruiting it to Cul-1 and Cul-5-containing CRL complexes, respectively. To accurately compare Vas levels in ovarian extracts from different genotypes, we produced Western blots from 96-well E-PAGE gels incubated in duplex with primary rabbit anti-Vas and mouse anti-α-tubulin antibodies, which we then detected with fluorescently labeled secondary antibodies. This allowed us to simultaneously test multiple replicates of each extract on the same gel under identical conditions. We took care to use only flies of comparable age, with ovaries of similar size and a similar distribution of egg chamber stages, for these comparisons (see Materials and Methods for details).
Using these methods, we found a consistent and statistically significant reduction of Vas levels to 58.2% ± 16.2% of that of the wild type upon nosGal4-driven overexpression of Fsn::HA (Fig. 3A). Importantly, this requires direct interaction between Fsn::HA and Vas, as the effect was much smaller and not statistically significant (83.7% ± 27.1%) when we overexpressed Fsn::HAY239A. We observed similar reductions in Vas levels upon nosGal4-driven overexpression of GusL::HA or GusS::HA (51.6% ± 13.9% and 56.8% ± 15.7% of wild-type control levels, respectively). Thus, we conclude that overexpression of either Fsn or Gus destabilizes Vas. We next analyzed extracts prepared from gus or Fsn mutant ovaries. Consistent with our overexpression data, we observed a statistically significant increase in overall Vas levels to 123.2% ± 22.6% in Df(2R)Exel7124/Fsnf06595 ovaries. We did not observe a statistically significant change in Vas levels in gusf07073/gusf07073 ovaries (92.1% ± 22.8%). However, Vas levels were further reduced in gusf07073/gusf07073 ovaries overexpressing Fsn::HA (41.5% ± 6.6% versus 58.2% ± 16.2%; P < 0.0001).
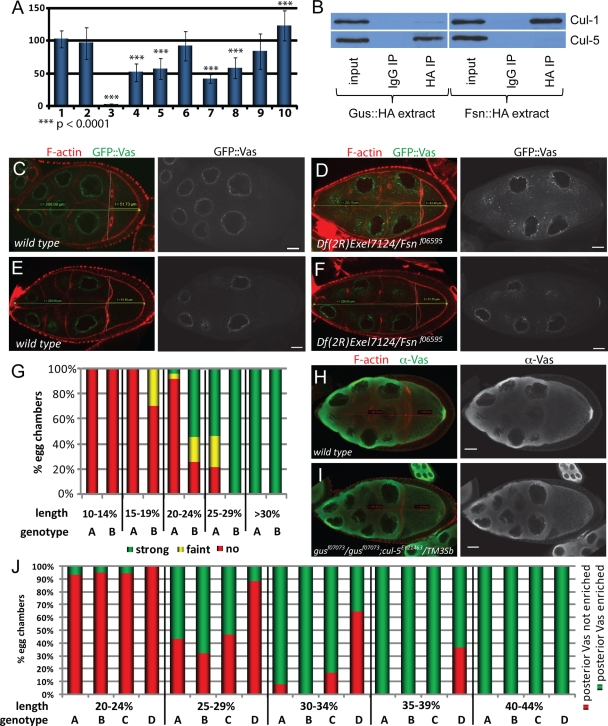
FIG. 3.
Effects of manipulations of gus- and Fsn-dependent pathways on Vas in ovaries. (A) Relative amount of Vas in ovaries normalized to the α-tubulin signal. Bar 1 shows a comparison of an Oregon-R wild-type extract to itself and bar 2 shows a comparison of an Oregon-R extract to three independently prepared Oregon-R wild-type extracts. Bars 3 to 10 show comparisons of extracts from the following genotypes to Oregon-R: vasPD/vasPH165 (bar 3), GusL::HA/nosGal4 (bar 4), GusS::HA/nosGal4 (bar 5), gusf07073/gusf07073 (bar 6), gusf07073/gusf07073; Fsn::HA/nosGal4 (bar 7), Fsn::HA/nosGal4 (bar 8), Fsn::HAY239A/nosGal4 (bar 9), and Fsnf06595/Df(2R)Exel7124 (bar 10). (B) Cul-1 is found in Fsn::HA but not Gus::HA immunoprecipitates. Conversely, Cul-5 associates with Gus::HA but not with Fsn::HA. (C to F) Examples in which posterior GFP-Vas is not apparent in wild-type oocytes (C and E) but is faint (D) or obvious (F) in smaller, earlier-stage Fsn mutant oocytes. (G) Correlation between posterior GFP::Vas accumulation and the ratio of oocyte to germ line cyst length. Posterior GFP::Vas is detectable in smaller Df(2R)Exel7124/Fsnf06595 oocytes (columns B) than in the wild type (columns A) (n = 74 for the wild type; n = 90 for Fsn). (H and I) Stage 10 oocytes of wild-type females and gusf07073/gusf07073; cul-5EY21463/TM3Sb females immunostained for Vas. F-actin is labeled with rhodamine phalloidin. No posterior enrichment of Vas is evident in the mutant oocyte. (J) Correlation between posterior Vas enrichment and the ratio of oocyte to germ line cyst length. Genotypes represented by the columns are Oregon-R (A), gusf07073/gusf07073 (B), cul-5EY21463/TM3Sb (C), and gusf07073/gusf07073; cul-5EY21463/TM3Sb (D). Posterior Vas accumulation is delayed in gusf07073 homozygotes sensitized by introduction of a cul-5 P-element insertion (n = 63 for the wild type; n = 66 for gusf07073/gusf07073; n = 55 for cul-5EY21463/TM3Sb; n = 55 for gusf07073/gusf07073; cul-5EY21463/TM3Sb).
To directly test whether Fsn is associated with ubiquitin E3 ligase complexes, we probed immunoprecipitates of Fsn::HA and Gus::HA for Cul-1. Cul-1 associated with Fsn::HA, but not with Gus::HA (Fig. 3B), corroborating the idea that Fsn acts as a substrate recognition subunit of a Cul-1 E3 ligase. Notably, we observed this interaction only in low-stringency buffers and not in mRIPA buffers (see Materials and Methods). Fsn interacts with Skp proteins in yeast two-hybrid assays (7, 35), further supporting the conclusion that Fsn is a subunit of a Cul-1 CRL.
We next probed immunoprecipitates of Fsn::Ha and Gus::HA for the presence of Cul-5. As expected, we found that Cul-5 associates with Gus::HA, but not with Fsn::HA (Fig. 3B), indicating that Gus most likely acts as a substrate receptor for a Cul-5-containing ubiquitin E3 ligase complex. Furthermore, Gus and the EloBC complex associate stably through several purification steps and have been cocrystallized (44). They also interact in yeast two-hybrid screens (7, 35). Human SSB1, a likely orthologue of Gus that is 69% identical in sequence, interacts with Cul-5 (11). Lastly, we found that females homozygous for the cul-5EY21463 mutation, a viable P-element insertion that reduces cul-5 mRNA and Cul-5 protein levels (31), produce mostly collapsed embryos which resemble those produced by gusf07073/gusf07073 mutants (Table 1), further supporting the idea that Gus and Cul-5 act together.
Endogenous Fsn and Gus fine-tune posterior Vas deployment.
We next asked whether genetic manipulations of Fsn- and gus-dependent pathways affect Vas localization. Immunostainings revealed that Vas accumulates both in the nurse cell perinuclear nuage and oocyte pole plasm in gus and Fsn mutants and upon Gus or Fsn overexpression (see Fig. S2 in the supplemental material). Notably, however, from individual images we observed that posterior accumulation of GFP::Vas appeared premature in Fsn mutant oocytes (Fig. 3C to F). To address this quantitatively, we scored the percentage of wild-type and Fsn oocytes that showed posterior GFP::Vas accumulation as a function of the anterior-posterior length of the oocyte divided by the length of the germ line cyst (Fig. 3G). We found obvious posterior GFP::Vas accumulation in 55% of the Fsn mutant oocytes that were between 20 and 24% cyst length (corresponding to stage 8 [34]), while we observed this in only 4.3% of equally staged wild-type egg chambers. As Vas and Fsn directly interact, this, taken together with the immunoblotting data discussed above, indicates that Vas is stabilized and accumulates prematurely in the pole plasm when Fsn activity is reduced.
We observed little effect of gusf07073 on posterior Vas deployment. This is contrary to previous results indicating that Vas failed to stably accumulate in the pole plasm of gusZ409 mutants (37). While posterior Vas does appear reduced in some gusf07073 oocytes, variability among individual oocytes, and ambiguity in strictly delineating the pole plasm for quantitation purposes, prevented us from drawing a firm conclusion in this regard. However, when we sensitized the gusf07073/gusf07073 mutant background by introduction of a single copy of the cul-5EY21463 allele we observed a notable delay in posterior accumulation of Vas (Fig. 3H and I). Nearly 40% of gusf07073/gusf07073; cul-5EY21463/TM3 oocytes that were between 35 and 39% of the cyst length had not accumulated posterior Vas, while all control oocytes (wild type, gusf07073/gusf07073, or cul-5EY21463/TM3) of a similar stage had done so (Fig. 3J). We attempted to extend this analysis to gusf07073/gusf07073; cul-5EY21463/cul-5EY21463 individuals, but found that these females produce mostly aberrant egg chambers with numerous defects (18), precluding the possibility of comparison.
Maternal manipulation of Gus- and Fsn-dependent pathways influences PGC formation.
Primordial germ cell (PGC) counts are a sensitive bioassay for Vas activity (2, 10, 21). Consistent with our observation that ovaries overexpressing Fsn::HA have clearly reduced Vas levels, we found that embryos from Fsn::HA-overexpressing females form fewer PGCs than do wild-type embryos (an average of 17 versus 34) (Table 2). This effect was enhanced when one genetic dose of vas was removed; embryos from vasPD/+ or vasPH165/+ females have 32 PGCs on average, but they have only 10 to 12 when Fsn::HA is overexpressed. Importantly, overexpression of the Fsn::HAY239A mutant has a much less dramatic impact; such embryos form an average of 27 PGCs, despite an expression level similar to that of Fsn::HA (Fig. 1K). Reducing endogenous Fsn by introducing Fsnf06595 or Df(2R)Exel7124 into the maternal Fsn::HA overexpression background suppresses the loss of PGCs in a statistically significant manner (average of 22 PGCs versus 17). Introducing both Fsn mutant chromosomes causes an even stronger suppression (25 PGCs). These results support the argument that endogenous Fsn contributes to the overexpression phenotypes and, taken together with the protein quantitation and affinity data described above, strongly support the idea that endogenous Fsn regulates Vas stability during oogenesis, destabilizing it through a direct interaction.
TABLE 2.
Average no. of PGCs per embryo derived from mothers of the indicated genotypesii
| Maternal genotype | Total no. of embryos | Avg no. of PGCs/embryo | ±SD |
P |
|
|---|---|---|---|---|---|
| Oregon-Ra | Transgeneb | ||||
| Oregon-R | 60 | 34.33 | 4.02 | NA | |
| +/vasPD | 50 | 32.58 | 4.29 | <0.05 | |
| +/vasPH165 | 50 | 31.68 | 4.47 | <0.05 | |
| +/+; GFP::Vas(wt1)/GFP::Vas(wt2) | 47 | 32.30 | 3.89 | <0.05 | |
| nosGal4/Fsn::HA(6.11) | 195 | 17.01 | 3.57 | <0.0001 | |
| nosGal4/Fsn::HA(6.15) | 69 | 16.81 | 3.43 | <0.0001 | |
| nosGal4/Fsn::HA(6.18) | 130 | 17.52 | 3.96 | <0.0001 | |
| +/vasPD; nosGal4/Fsn::HA(6.15) | 64 | 11.97 | 3.05 | <0.0001 | |
| +/vasPH165; nosGal4/Fsn::HA(6.15) | 63 | 10.67 | 3.98 | <0.0001 | |
| nosGal4/Fsn::HAY239A(24.7) | 53 | 27.72 | 3.75 | <0.0001 | <0.0001c |
| nosGal4/Fsn::HAY239A(24.10) | 53 | 27.49 | 4.13 | <0.0001 | <0.0001c |
| Fsnf06595/+; nosGal4/Fsn::HA(6.11) | 123 | 21.72 | 4.11 | <0.0001 | <0.0001c |
| +/Df(2R)Exel7124; nosGal4/Fsn::HA(6.11) | 64 | 21.94 | 3.03 | <0.0001 | <0.0001c |
| Fsnf06595/Df(2R)Exel7124; nosGal4/Fsn::HA(6.11) | 64 | 24.95 | 3.63 | <0.0001 | <0.0001c |
| Fsnf06595/Df(2R)Exel7124 | 51 | 34.33 | 5.77 | NSd | |
| +/Df(2R)Exel7124; nosGal4/Fsn::HA(6.15) | 63 | 22.76 | 3.57 | <0.0001 | <0.0001e |
| Fsnf06595/Df(2R)Exel7124; nosGal4/Fsn::HA(6.18) | 44 | 25.39 | 5.37 | <0.0001 | <0.0001f |
| +/gusf07073 | 55 | 35.44 | 4.73 | NSd | |
| gusf07073/gusf07073 | 83 | 21.12 | 8.49 | <0.0001 | |
| cul-5EY21463/cul-5EY21463 | 49 | 25.00 | 5.52 | <0.0001 | |
| nosGal4/Gus(S)::HA(5.3) | 63 | 20.40 | 4.05 | <0.0001 | |
| nosGal4/Gus(S)::HA(5.5) | 110 | 19.22 | 3.97 | <0.0001 | |
| nosGal4/Gus(L)::HA(4.6) | 147 | 22.35 | 4.65 | <0.0001 | |
| nosGal4/V::GusS(2.11) | 41 | 24.54 | 4.32 | <0.0001 | |
| nosGal4/Gus(L)::HAW221L(17.1) | 56 | 28.34 | 4.34 | <0.0001 | <0.0001g |
| nosGal4/Gus(L)::HAW221L(17.4) | 52 | 28.63 | 3.71 | <0.0001 | <0.0001g |
| gusf07073/CyO; GusS::HA/act5C-Gal4 | 50 | 31.80 | 5.16 | <0.01 | |
| gusf07073/gusf07073; GusS::HA/act5C-Gal4 | 55 | 22.64 | 4.14 | <0.0001 | NSh |
P value calculated in comparison to the Oregon-R sample.
P values calculated in comparison to the relevant reference transgene as indicated below.
Compared to nosGal4/Fsn::HA(6.11).
Difference not statistically significant in comparison to Oregon-R.
Compared to nosGal4/Fsn::HA(6.15).
Compared to nosGal4/Fsn::HA(6.18).
Compared to nosGal4/GusL::HA(4.6).
Difference not statistically significant in comparison to gusf07073/gusf07073.
Numbers in parentheses after genotypes denote independent transgene insertions. NA, not applicable; NS, not statistically significant.
Fsn mutant embryos did not form extra PGCs (34 versus 34) (Table 2). Consistent with this, overexpression of GFP::Vas from transgenes that rescue vas mutant phenotypes also did not lead to the formation of additional PGCs. This indicates that while reducing maternal vas reduces the number of PGCs, additional Vas is not sufficient to cause elevated PGC numbers, or the threshold of additional Vas for doing so was not reached in our experiments.
In similar experiments with Gus, we found that maternal overexpression of either GusL::HA or GusS::HA caused a significant reduction in the number of PGCs, consistent with the reduced amount of Vas we observed in the corresponding ovaries (19 to 22 PGCs versus 34) (Table 2). The majority of Gus:HA-overexpressing embryos developed normally and had only rare displaced Vas-positive cells, as are also observed in wild-type embryos. However, some eggs produced from Gus:HA-overexpressing females were unfertilized, and some embryos developed aberrantly and had scattered Vas-positive cells in ectopic sites. We only scored PGCs from the class that apparently developed normally. gusf07073/gusf07073 embryos that did not collapse (see above) were frequently unfertilized or sometimes developed aberrantly with scattered Vas-positive cells; those that were morphologically normal and could be scored also displayed a significant reduction of PGCs in comparison to wild-type or +/gusf07073 control embryos (21 versus 34 or 35, respectively) (Table 2). Expression of Gus under the control of Act5C-Gal4 rescued the collapsed-egg phenotype and had little effect on the number of PGCs formed by +/gusf07073 embryos (32 versus 35). However, gusf07073/gusf07073; GusS::HA/Act5C-Gal4 embryos formed only 23 PGCs on average, corroborating the idea that gus is required maternally in the germ line, not the soma, for efficient PGC formation.
The reduced number of PGCs in gus mutant embryos is consistent with a positive regulatory function of Gus for Vas in the pole plasm, as is the delay in posterior Vas deployment in gusf07073/gusf07073; cul-5EY21463/TM3 oocytes. However, this conclusion is inconsistent with the Gus overexpression data. We propose that while ectopic, overexpressed Gus can promote ubiquitin-mediated Vas degradation, in the pole plasm endogenous Gus functions in a stabilizing role or as a positive regulator of Vas activity.
Like gusf07073/gusf07073 eggs, cul-5EY21463/cul-5EY21463 eggs often collapsed and were not fertilized, and some cul-5EY21463/cul-5EY21463 embryos developed aberrantly with scattered Vas-positive cells. Similar to gusf07073/gusf07073 embryos, those that were morphologically normal averaged only 25 PGCs (Table 2). The similarity between gus and cul-5 mutant phenotypes further corroborates the idea that Gus and Cul-5 act together during oogenesis to promote PGC formation.
Gus and Fsn accumulate in overlapping but distinct patterns.
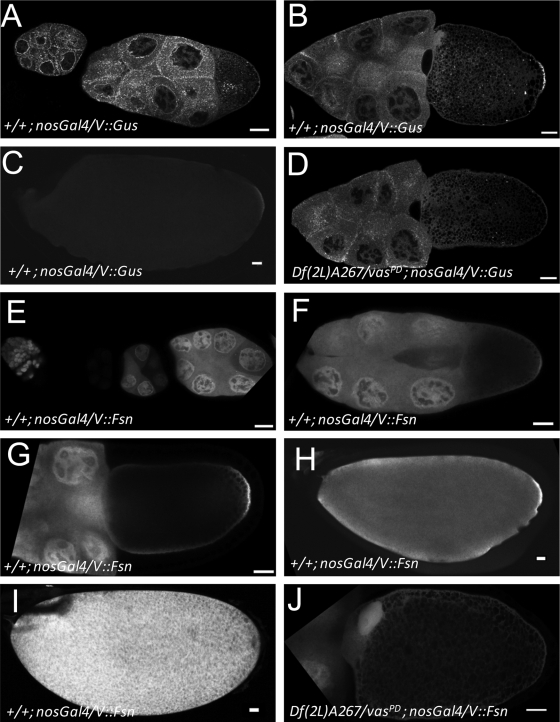
To assess the subcellular distribution of Fsn and Gus in ovaries, we prepared flies that express Venus-tagged forms of Gus and Fsn (V::GusS; V::GusL and V::Fsn, respectively) under nosGal4 control. The expression pattern of V::GusS essentially recapitulates earlier results for endogenous Gus (37). In the nurse cells through stage 8, V::GusS accumulates at high levels surrounding the nurse cell nuclei and in punctate aggregates throughout the cytoplasm (Fig. 4A). We also observed some cortical enrichment of these aggregates. In early oocytes, V::GusS forms a particulate pattern throughout the ooplasm and is also cortically enriched (Fig. 4A). We also noted some nuclear V::GusS in both nurse cells and the oocyte. At stage 10, the signal surrounding the nurse cell nuclei becomes weaker and the particulate nature of the cytoplasmic aggregates becomes less pronounced (Fig. 4B). Importantly, V::GusS is enriched in the posterior of the oocyte at stage 10, indicating that Gus is indeed a pole plasm component. In later stages, posterior V::GusS diminishes (Fig. 4C) and is not detectable in early embryos. Importantly, posterior V::GusS accumulation requires vas. In vasPD/Df(2L)A267 [Df(2L)A267 is a chromosomal deletion that includes vas] mutant egg chambers, cortical V::GusS remains detectable, but the posterior enrichment is clearly lost (Fig. 4D). The distribution of V::GusL is indistinguishable from that of V::GusS (data not shown).
FIG. 4.
V::Gus and V::Fsn accumulate in the pole plasm in a vas-dependent manner. (A to C) In a wild-type background, V::Gus driven by nosGal4 is expressed at low levels in germ line nuclei but accumulates to high levels in the cytoplasm surrounding the nurse cell nuclei. In the germ line cytoplasm, V::Gus is ubiquitous, but it is enriched in numerous luminous foci, particularly during the earlier stages of oogenesis. In the oocyte, V::Gus is enriched in the cortex but accumulates in the pole plasm during stage 10. In late oocytes, posterior V::Gus levels decrease. (D) Pole plasm accumulation of V::Gus is lost in vas mutants (compare with panel B). (E to I) V::Fsn driven by nosGal4 in a wild-type background during early oogenesis (notice that transgene expression follows the expression pattern of the driver) (E), stage 9 (F), stage 10 (G), stage 13 (H), and stage 14 (I). V::Fsn accumulates in nuclei and is uniformly distributed throughout the nurse cell cytoplasm. In the oocyte, the protein appears cortically enriched, and from stage 10 on, V::Fsn is enriched in the pole plasm. Posterior enrichment is not evident in mature eggs. (J) In a vas mutant background, pole plasm accumulation of V::Fsn is lost; however, a general cortical stain is still observed. Bars, 20 mm.
When expressed with the nosGal4 driver, V::Fsn is enriched in the nuclei of both nurse cells and oocytes (Fig. 4E to G). Unlike V::Gus, V::Fsn does not accumulate in the perinuclear region of nurse cells, although we observed some cortical accumulation in mid- to late-stage oocytes. V::Fsn is recruited to the pole plasm of mid- to late-stage oocytes (Fig. 4F to H); however, this enrichment is later lost (Fig. 4I). In mature eggs V::Fsn is present at high levels throughout the ooplasm (Fig. 4I). Posterior accumulation of V::Fsn also requires vas (Fig. 4J).
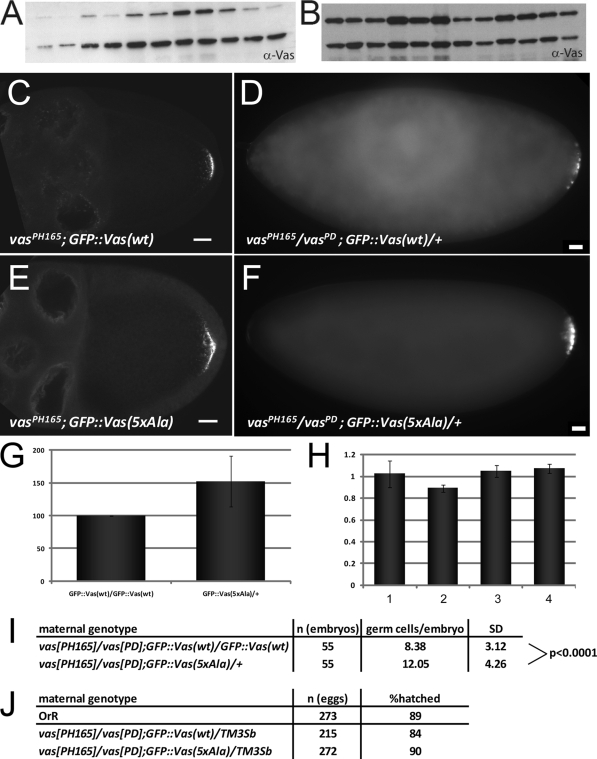
Vas lacking the DINNN motif accumulates to high levels and efficiently rescues vas phenotypes.
To test how Vas would be affected by a mutation that impairs its association with Gus and Fsn, we compared the distribution of a wild-type GFP-Vas fusion with a counterpart that had the DINNN motif substituted by five Ala residues [GFP::Vas(wt) and GFP::Vas(5xAla), respectively]. Many similar constructs with the same promoter (vas) that produced wild-type or other mutant forms of Vas (Fig. 5A and data not shown) (10) express protein at levels much lower than endogenous Vas. In contrast, many independent transformant lines for GFP::Vas(5xAla) expressed the protein at very high levels (Fig. 5B), suggesting that the substitution of five Ala residues might stabilize Vas. The distribution of GFP::Vas(5xAla) reproduces the wild type, although it clearly accumulates at higher levels in the nuage, pole plasm, and PGCs (Fig. 5C to F). We next quantitated transgenic protein and RNA levels through quantitative Western blot analyses and qRT-PCR experiments (Fig. 5G and H, respectively, and data not shown). When comparable levels of the two transgenic mRNAs were expressed, the mutant form of GFP::Vas accumulated to about 1.5 times the level of the corresponding wild-type protein. This suggests that the substitution of five Ala residues has a stabilizing effect on Vas, GFP::Vas(5xAla) rescues PGC formation in vasPD/vasPH165 embryos more efficiently than GFP::Vas(wt) (Fig. 5I), and either transgene rescues viability of vasPD/vasPH165 embryos (Fig. 5J). We conclude that the substitution of five Ala residues enhances Vas stability and increases its biological activity. As our competition experiments indicate that the binding of Fsn to Vas is more sensitive to the DINNN motif than that of Gus, this is consistent with the conclusion that Fsn, but not Gus, is a negative regulator of Vas.
FIG. 5.
GFP::Vas lacking the DINNN motif accumulates to high levels, localizes normally, and rescues vas phenotypes. (A and B) Western blots of ovarian extracts showing expression levels of individually established lines for GFP::Vas(wt) (A) and GFP::Vas(5xAla) (B) transgenes (upper band) and endogenous Vas (lower band). (C to F) Wild-type GFP::Vas (C) and a mutant form lacking the DINNN motif (E) localize similarly during oogenesis. Both transgenic proteins rescue PGC formation in vas mutant embryos. Bars, 20 mm. (G and H) GFP::Vas(5xAla) accumulates to higher levels than GFP::Vas(wt). (G) Normalized expression levels of the mutant protein in comparison to wild type. (H) RNA expression levels assayed by qRT-PCR. Bars 1 and 2 represent two independently prepared samples of vasPH165/vasPD; GFP::Vas(wt)/GFP::Vas(wt). Bars 3 and 4 represent two independent samples of vasPH165/vasPD; GFP::Vas(5xAla)/+. One copy of the mutant transgene expresses an RNA amount comparable to two copies of the wild-type transgene. (I) One copy of the GFP::Vas(5xAla) transgene permits the formation of more PGCs in vasPH165/vasPD embryos than two copies of GFP::Vas(wt). (J) Both constructs rescue the ability of vas embryos to hatch.
DISCUSSION
Full accumulation of Vas in the pole plasm requires the activity of Faf, a DUB, indicating that Vas is regulated in the oocyte posterior through ubiquitin-dependent pathways (25). Our study identifies two CRL receptors, Fsn and Gus, as additional regulators of Vas. Our biochemical and genetic data suggest that Fsn and Gus act by directly recruiting Vas to their respective CRL complexes, and both proteins accumulate together with Vas in the pole plasm.
Fsn is clearly a negative regulator of Vas, presumably targeting it via Cul-1 for proteasomal-mediated degradation. In Fsn mutant ovaries, we observed increased Vas, and Fsn overexpression produces the reciprocal effect. Furthermore, reduction of endogenous Fsn suppresses the Fsn overexpression phenotype, indicating that the endogenous protein acts in the same direction as the one we expressed ectopically. We also observed precocious accumulation of Vas expression in the posterior of Fsn mutant oocytes, demonstrating that endogenous Fsn antagonizes Vas accumulation. Taken together, these findings are consistent with a role for endogenous Fsn in promoting Vas degradation.
The function of Gus in regulating Vas, however, is more complex, as its overexpression and mutant phenotypes are pleiotropic and not reciprocal. In a gus mutant background where cul-5 dosage is also reduced, posterior Vas accumulation is delayed, the opposite of what we observed in Fsn mutant ovaries. Consistent with this, reduction of gus enhances the effects of Fsn overexpression on Vas levels. Taken together with our molecular experiments, which indicate that Gus and Fsn cannot bind simultaneously to the DINNN motif of Vas, these data support a conclusion that endogenous Gus is a positive regulator of Vas, acting antagonistically with Fsn to establish a steady state of Vas ubiquitination that allows timely posterior Vas accumulation and full activity. We believe that these loss-of-function data more accurately reflect the function of endogenous gus and that Gus overexpression might alter its activity, perhaps by favoring conjugation of destructive long ubiquitin chains rather than that of short activating ones and/or by impacting other regulators of Vas, as E3 ligases and DUBs often cross-regulate each other (28).
Gus and Fsn could catalyze different ubiquitination events, with Fsn promoting Vas degradation and endogenous Gus in the pole plasm promoting Vas activity while antagonizing Fsn-dependent destabilization by rendering the DINNN motif less accessible. Further modulation of Vas regulation may be accomplished through Gus-Vas interactions that are independent of the DINNN motif, likely mediated in vivo by additional proteins that stabilize the Gus-Vas complex through associations with other domains of Vas or that provide a bridging mechanism to indirectly recruit Gus to Vas. Such interactions would explain why Vas accumulates in the pole plasm despite the presence there of Fsn.
Fsn may also be counter-balanced by Faf, which has previously been shown to stabilize Vas in the pole plasm (25). A dynamic equilibrium among Gus, Fsn, and Faf may establish and maintain a steady state of ubiquitination that allows stable Vas expression and Vas activity. Excess Gus or Fsn (this study) or loss of Faf (25) would shift this dynamic equilibrium to a state of ubiquitination that promotes Vas degradation while, in the absence of Gus, Vas may be more prone to Fsn-dependent degradation.
Additional layers of complexity in the regulatory logic governing Vas ubiquitination are likely. For example, Vas can be copurified from embryonic extracts with the E3 ligase purity of essence (Poe) (25, 40), and vasPH165/poe03420 females produce embryos with substantially fewer PGCs than vas or poe heterozygotes (22.1 ± 4.5 versus 30.1 ± 3.2 for poe03420/CyO; N. Liu and P. Lasko, unpublished observation). poe is expressed during oogenesis (36), and Vas distribution is aberrant in eggs produced by females bearing certain poe alleles (C. Bazinet, personal communication).
Proteins often have to be phosphorylated on particular motifs called phosphodegrons to be modified by the ubiquitination machinery (42). A C. elegans Vas homolog, GLH-1, features a functional phosphodegron that, when phosphorylated by the Jun N-terminal kinase (JNK) KGB-1, targets it for proteasomal degradation (29). The KGB-1 binding site and phosphodegron are conserved in Drosophila Vas, which can associate with KGB-1 in vitro, suggesting that JNK-dependent regulation of Vas might occur in flies as well (29). In C. elegans, KGB-1-dependent GLH-1 degradation is antagonized by the Cop9 signalosome subunit CSN-5, which regulates CRL activity by inactivating the Cullin subunit (30). In Drosophila species, maternal csn-5 mutants produce embryonic patterning defects that resemble those of vas mutants, and Vas shows altered electrophoretic mobility (4). However, similar shifts in Vas mobility also result from mutations that affect a meiotic DNA damage response checkpoint in Drosophila oogenesis (3, 6, 13). It is believed that this pathway negatively regulates Vas activity, at least with respect to its function in activating Grk translation (6, 38), but it is unclear whether it influences Vas abundance. These considerations might help explain the relatively small effects of genetic manipulation of Gus and Fsn on overall Vas levels, as the fraction of Vas that can be modified through ubiquitination might be limited through other pathways that affect its phosphorylation state.
gus mutant phenotypes imply that Vas is not its only target, as vas is required neither in follicle cells nor for wing development. Recognition of a target by both Gus and Fsn implies a B30.2/SPRY domain-DINNN motif interaction; consistently, several Gus-interacting proteins recovered through genome-wide yeast two-hybrid screens (7, 35) contain a DINNN motif (45).
Both mammals and sea urchins possess Gus orthologues (11, 43, 47). Intriguingly, in these organisms Gus appears to be expressed primarily in cells that do not express Vas but are adjacent to Vas-expressing cells, leading to a hypothesis that Gus may be involved in removing Vas from cells where it might be detrimental (43). However, this interesting idea has not been tested experimentally. In zebrafish, gus orthologues are, conversely, expressed together with Vas in the germ line in both adult testes and ovaries (23). Fsn orthologues are also present throughout animal evolution. In the synapse they are believed to interact with Highwire to form a ubiquitin E3 ligase that downregulates a protein kinase, Wallenda, which in turn restrains synaptic terminal growth (46). The C. elegans Fsn orthologue, FSN-1, is not only involved in synaptogenesis (24) but has also been recently identified as a negative regulator of germ line apoptosis that is dependent on the p53 orthologue CEP-1 (5). It will be interesting to unravel whether the Gus and Fsn orthologues in these and other species also interact with Vas in the same cell and to elucidate how recruitment of Vas to different E3 ligase complexes can influence Vas dynamics.
Supplementary Material
Acknowledgments
We thank Bob Duronio, Niankun Liu, and Hong Han for antibodies, Beili Hu for the transgene injections, Maria Kilfoil for help with statistical analyses, the Murphy lab for the Drosophila gateway collection, and Chris Bazinet, Niankun Liu, and Paul MacDonald for sharing results prior to publication.
This work was supported by NICHD (R01HD036631) to P.L. and by the GRL Program (K20815000001) of KICOS/KMEST to B.-H.O.
Footnotes
Published ahead of print on 1 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Besse, F., and A. Ephrussi. 2008. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9:971-980. [DOI] [PubMed] [Google Scholar]
- 2.Carrera, P., O. Johnstone, A. Nakamura, J. Casanova, H. Jackle, and P. Lasko. 2000. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell 5:181-187. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y., A. Pane, and T. Schüpbach. 2007. cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr. Biol. 17:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doronkin, S., I. Djagaeva, and S. K. Beckendorf. 2002. CSN5/Jab1 mutations affect axis formation in the Drosophila oocyte by activating a meiotic checkpoint. Development 129:5053-5064. [DOI] [PubMed] [Google Scholar]
- 5.Gao, M. X., E. H. Liao, B. Yu, Y. Wang, M. Zhen, and W. B. Derry. 2008. The SCF FSN-1 ubiquitin ligase controls germline apoptosis through CEP-1/p53 in C. elegans. Cell Death Differ. 15:1054-1062. [DOI] [PubMed] [Google Scholar]
- 6.Ghabrial, A., and T. Schüpbach. 1999. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1:354-357. [DOI] [PubMed] [Google Scholar]
- 7.Giot, L., J. S. Bader, C. Brouwer, A. Chaudhuri, B. Kuang, Y. Li, Y. L. Hao, C. E. Ooi, B. Godwin, E. Vitols, G. Vijayadamodar, P. Pochart, H. Machineni, M. Welsh, Y. Kong, B. Zerhusen, R. Malcolm, Z. Varrone, A. Collis, M. Minto, S. Burgess, L. McDaniel, E. Stimpson, F. Spriggs, J. Williams, K. Neurath, N. Ioime, M. Agee, E. Voss, K. Furtak, R. Renzulli, N. Aanensen, S. Carrolla, E. Bickelhaupt, Y. Lazovatsky, A. DaSilva, J. Zhong, C. A. Stanyon, R. L. Finley, Jr., K. P. White, M. Braverman, T. Jarvie, S. Gold, M. Leach, J. Knight, R. A. Shimkets, M. P. McKenna, J. Chant, and J. M. Rothberg. 2003. A protein interaction map of Drosophila melanogaster. Science 302:1727-1736. [DOI] [PubMed] [Google Scholar]
- 8.Hay, B., L. Y. Jan, and Y. N. Jan. 1988. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55:577-587. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser, M. 2006. Lingering mysteries of ubiquitin-chain assembly. Cell 124:27-34. [DOI] [PubMed] [Google Scholar]
- 10.Johnstone, O., and P. Lasko. 2004. Interaction with eIF5B is essential for Vasa function during development. Development 131:4167-4178. [DOI] [PubMed] [Google Scholar]
- 11.Kamura, T., K. Maenaka, S. Kotoshiba, M. Matsumoto, D. Kohda, R. C. Conaway, J. W. Conaway, and K. I. Nakayama. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159-180. [DOI] [PubMed] [Google Scholar]
- 13.Klattenhoff, C., D. P. Bratu, N. McGinnis-Schultz, B. S. Koppetsch, H. A. Cook, and W. E. Theurkauf. 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12:45-55. [DOI] [PubMed] [Google Scholar]
- 14.Koundakjian, E. J., D. M. Cowan, R. W. Hardy, and A. H. Becker. 2004. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167:203-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuang, Z., S. Yao, Y. Xu, R. S. Lewis, A. Low, S. L. Masters, T. A. Willson, T. B. Kolesnik, S. E. Nicholson, T. J. Garrett, and R. S. Norton. 2009. SPRY domain-containing SOCS box protein 2: crystal structure and residues critical for protein binding. J. Mol. Biol. 386:662-674. [DOI] [PubMed] [Google Scholar]
- 16.Kugler, J. M., J. Chicoine, and P. Lasko. 2009. Bicaudal-C associates with a Trailer Hitch/Me31B complex and is required for efficient Gurken secretion. Dev. Biol. 328:160-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kugler, J. M., and P. Lasko. 2009. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly (Austin) 3:15-28. [DOI] [PubMed] [Google Scholar]
- 18.Kugler, J. M., C. Lem, and P. Lasko. 2010. Reduced cul-5 activity causes aberrant follicular morphogenesis and germ cell loss in Drosophila. PLoS One 5:e9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 20.Lasko, P. F., and M. Ashburner. 1988. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 335:611-617. [DOI] [PubMed] [Google Scholar]
- 21.Lasko, P. F., and M. Ashburner. 1990. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 4:905-921. [DOI] [PubMed] [Google Scholar]
- 22.Lécuyer, E., H. Yoshida, N. Parthasarathy, C. Alm, T. Babak, T. Cerovina, T. R. Hughes, P. Tomancak, and H. M. Krause. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131:174-187. [DOI] [PubMed] [Google Scholar]
- 23.Li, J. Z., Y. P. Zhou, Y. Zhen, Y. Xu, P. X. Cheng, H. N. Wang, and F. J. Deng. 2009. Cloning and characterization of the SSB-1 and SSB-4 genes expressed in zebrafish gonads. Biochem. Genet. 47:179-190. [DOI] [PubMed] [Google Scholar]
- 24.Liao, E. H., W. Hung, B. Abrams, and M. Zhen. 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430:345-350. [DOI] [PubMed] [Google Scholar]
- 25.Liu, N., D. A. Dansereau, and P. Lasko. 2003. Fat facets interacts with Vasa in the Drosophila pole plasm and protects it from degradation. Curr. Biol. 13:1905-1909. [DOI] [PubMed] [Google Scholar]
- 26.Mahowald, A. P. 2001. Assembly of the Drosophila germ plasm. Int. Rev. Cytol. 203:187-213. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay, D., and H. Riezman. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201-205. [DOI] [PubMed] [Google Scholar]
- 28.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773-786. [DOI] [PubMed] [Google Scholar]
- 29.Orsborn, A. M., W. Li, T. J. McEwen, T. Mizuno, E. Kuzmin, K. Matsumoto, and K. L. Bennett. 2007. GLH-1, the C. elegans P granule protein, is controlled by the JNK KGB-1 and by the COP9 subunit CSN-5. Development 134:3383-3392. [DOI] [PubMed] [Google Scholar]
- 30.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds, P. J., J. R. Simms, and R. J. Duronio. 2008. Identifying determinants of cullin binding specificity among the three functionally different Drosophila melanogaster Roc proteins via domain swapping. PLoS One 3:e2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 34.Spradling, A. C. 1993. Developmental genetics of oogenesis, p. 1-70. In M. Bate and A. Martinez-Arias (ed.), The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 35.Stanyon, C. A., G. Liu, B. A. Mangiola, N. Patel, L. Giot, B. Kuang, H. Zhang, J. Zhong, and R. L. Finley, Jr. 2004. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 5:R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stapleton, M., G. Liao, P. Brokstein, L. Hong, P. Carninci, T. Shiraki, Y. Hayashizaki, M. Champe, J. Pacleb, K. Wan, C. Yu, J. Carlson, R. George, S. Celniker, and G. M. Rubin. 2002. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12:1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Styhler, S., A. Nakamura, and P. Lasko. 2002. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev. Cell 3:865-876. [DOI] [PubMed] [Google Scholar]
- 38.Styhler, S., A. Nakamura, A. Swan, B. Suter, and P. Lasko. 1998. vasa is required for GURKEN accumulation in the oocyte and is involved in oocyte differentiation and germline cyst development. Development 125:1569-1578. [DOI] [PubMed] [Google Scholar]
- 39.Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe, C. M. Singh, R. Buchholz, M. Demsky, R. Fawcett, H. L. Francis-Lang, L. Ryner, L. M. Cheung, A. Chong, C. Erickson, W. W. Fisher, K. Greer, S. R. Hartouni, E. Howie, L. Jakkula, D. Joo, K. Killpack, A. Laufer, J. Mazzotta, R. D. Smith, L. M. Stevens, C. Stuber, L. R. Tan, R. Ventura, A. Woo, I. Zakrajsek, L. Zhao, F. Chen, C. Swimmer, C. Kopczynski, G. Duyk, M. L. Winberg, and J. Margolis. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and PiggyBac. Nat. Genet. 36:283-287. [DOI] [PubMed] [Google Scholar]
- 40.Thomson, T., N. Liu, A. Arkov, R. Lehmann, and P. Lasko. 2008. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 125:865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Doren, M., A. L. Williamson, and R. Lehmann. 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8:243-246. [DOI] [PubMed] [Google Scholar]
- 42.Varshavsky, A. 2005. Regulated protein degradation. Trends Biochem. Sci. 30:283-286. [DOI] [PubMed] [Google Scholar]
- 43.Voronina, E., M. Lopez, C. E. Juliano, E. Gustafson, J. L. Song, C. Extavour, S. George, P. Oliveri, D. McClay, and G. Wessel. 2008. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev. Biol. 314:276-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woo, J.-S., J. H. Imm, C. K. Min, K. J. Kim, S. S. Cha, and B.-H. Oh. 2006. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 25:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo, J.-S., H. Y. Suh, S. Y. Park, and B.-H. Oh. 2006. Structural basis for protein recognition by B30.2/SPRY domains. Mol. Cell 24:967-976. [DOI] [PubMed] [Google Scholar]
- 46.Wu, C., R. W. Daniels, and A. DiAntonio. 2007. DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Dev. 2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing, Y., R. Gosden, P. Lasko, and H. Clarke. 2006. Murine homologues of the Drosophila gustavus gene are expressed in ovarian granulosa cells. Reproduction 131:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.