Abstract
Bioactive compounds have been invaluable for dissecting the mechanisms, regulation, and functions of cellular processes. However, very few such reagents have been described for pre-mRNA splicing. To facilitate their systematic discovery, we developed a high-throughput cell-based assay that measures pre-mRNA splicing by utilizing a quantitative reporter system with advantageous features. The reporter, consisting of a destabilized, intron-containing luciferase expressed from a short-lived mRNA, allows rapid screens (<4 h), thereby obviating the potential toxicity of splicing inhibitors. We describe three inhibitors (out of >23,000 screened), all pharmacologically active: clotrimazole, flunarizine, and chlorhexidine. Interestingly, none was a general splicing inhibitor. Rather, each caused distinct splicing changes of numerous genes. We further discovered the target of action of chlorhexidine and show that it is a selective inhibitor of specific Cdc2-like kinases (Clks) that phosphorylate serine-arginine-rich (SR) protein splicing factors. Our findings reveal unexpected activities of clinically used drugs in splicing and uncover differential regulation of constitutively spliced introns.
Extensive posttranscriptional processing of eukaryotic pre-mRNA is required for the biogenesis of mRNAs, including 7-methylguanosine cap addition at the 5′ end, cleavage and polyadenylation at the 3′ end, and splicing of introns. The vast majority of genes in complex eukaryotes contain multiple introns that need to be spliced out by the spliceosome with high fidelity in order to generate the open reading frame (ORF) and the 5′ and 3′ untranslated regions (UTR) carried by the exons. Splicing depends on the recognition of short consensus sequences at the intron-exon boundaries and within introns by a set of small nuclear ribonucleoprotein (snRNP) complexes (consisting of snRNPs U1, U2, U4, U5, U6, U11, U12, U4atac, and U6atac) and a large number of proteins, including spliceosomal proteins and positively as well as negatively acting splicing modulators (12, 59). Serine-arginine-rich (SR)-domain-containing proteins (36) generally serve to promote constitutive splicing. They also modulate alternative splicing by binding to intronic or exonic splicing enhancer (ISE or ESE, respectively) sequences (5, 20). Other pre-mRNA binding proteins, such as hnRNPs, that lack SR domains regulate splicing by binding to intronic or exonic splicing suppressor (ISS or ESS, respectively) sites and also act as general splicing modulators (14, 62).
Alternative splicing allows for a single gene to express different isoforms of mRNA, thus playing a major role in contributing to the cellular complexity in higher eukaryotes without the need to expand the genome (6). Global surveying of the human transcriptome estimates that up to 95% of multiexon genes undergo alternative splicing (46, 60). Importantly, these events are highly regulated by numerous splicing factors in a tissue type-, developmental stage-, and signal-dependent manner. Aberrations in splicing due to mutations in the pre-mRNA are responsible for up to 15% of inherited diseases (32). In addition, defects in the splicing machinery itself due to the loss/gain of function of splicing factors or their relative stoichiometry are causes of a wide range of human ailments, from cancer to neurodegenerative diseases (12, 18, 35, 57). It has been established that splicing is subject to regulation by upstream signaling pathways. However, the details of the underlying mechanisms or the specific proteins involved in such regulation remain largely unclear. The significant link between splicing defects and human diseases underscores the paramount importance of understanding the mechanisms of splicing, including the signaling pathways that regulate general splicing as well as splicing of specific subsets of transcripts.
Small molecules have been essential in uncovering the mechanisms, regulations, and functions of many cellular processes, including DNA replication, transcription, and translation. While several recent reports have described screens for effectors of splicing, only a small number of constitutive or alternative splicing inhibitors have been identified (24, 31, 34, 42, 47, 53-55). Here, we describe a cell-based reporter system with advantageous properties that can be used for systematic discovery of modulators of splicing and splicing-dependent processes. Screening a collection of >23,000 compounds identified small molecules that modulate both constitutive and alternative splicing. We describe here three of the most active compounds, all of which are in wide clinical use. While none was a general splicing inhibitor, we show that, unexpectedly, clotrimazole and flunarizine each inhibited a different set of constitutively spliced introns. Chlorhexidine predominantly modulated alternative splicing by selectively inhibiting Cdc2-like kinase (Clk) SR protein kinases (SRPKs). Our data underscore the capacity of this rapid-response screen to identify a broad range of modulators of the splicing machinery, which shed new insights into regulation of both constitutive and alternative splicing in cells.
MATERIALS AND METHODS
Reporter construction and generation of stable cell lines.
An intron-containing luciferase reporter was generated by inserting a 132-nucleotide chimeric β-globin/immunoglobulin intron (Promega) at nucleotide position 1344 of the firefly luciferase gene. The luciferase protein is destabilized using both a PEST protein degradation sequence and a CL1 sequence. In order to destabilize the mRNA, five tandem AUUUA repeats were introduced into the 3′ UTR of the reporter luciferase gene. Both intron-containing and intronless luciferases were transcribed from a cytomegalovirus (CMV) promoter. To generate stable cell lines, transfected HeLa cells were selected for integration using hygromycin for 2 to 3 weeks. The luciferase assay was done according to the manufacturer's recommendation (Promega).
Chemical compounds and high-throughput screen (HTS).
A collection of >23,000 small molecules, including known bioactive compounds and FDA-approved drugs, was assembled from several commercial sources (Microsource Diversity, Sigma-Aldrich, BioMol, Tocris, Lopac, Prestwick, Maybridge, and Chembridge) and maintained at a 2 mM stock concentration in dimethyl sulfoxide (DMSO). For confirmation studies, clotrimazole, flunarizine, and chlorhexidine were purchased from Sigma-Aldrich.
For HTS, 2,000 cells in 4 μl medium per well were seeded on white 1,536-well plates (Corning) by using an automated dispenser (Deerac Fluidics) and allowed to grow overnight. For each plate, the first two and last two columns were controls treated with DMSO and assigned an activity of 100%. Compounds were administered to the central wells in duplicates. Briefly, 70 nl of compounds diluted in growth medium (final concentration of 20 μM) was added to wells by using a Pintool (V&P Scientific) attached to a Biomek FX workstation (Beckman Coulter), and the plates were incubated for an additional 4 h. Subsequently, 4 μl of One-Glo reagent (Promega) was added, and luminescence signals were measured using an EnVision microplate reader (Perkin Elmer) with standard luminescence settings.
RNA extraction and reverse transcription-PCR (RT-PCR).
To analyze the effect of the splicing inhibitors on reporter RNA and endogenous RNA splicing, HeLa cells were seeded in 12-well plates for 24 h, followed by treatment with the various compounds for the indicated times. Total RNA was extracted using an RNeasy kit (Qiagen) in accordance with the manufacturer's instructions, followed by cDNA synthesis from 1 μg total RNA. Nonquantitative PCR was performed using 5 μl of diluted cDNA and Pfx DNA polymerase (Invitrogen). Quantitative real-time PCR using 5 μl of the same cDNA and relative quantification analysis were performed with an Applied Biosystems 7500 fast system by using SYBR green dye chemistry in accordance with the manufacturer's recommendations. All primers are listed in Table S2 in the supplemental material.
In vitro splicing.
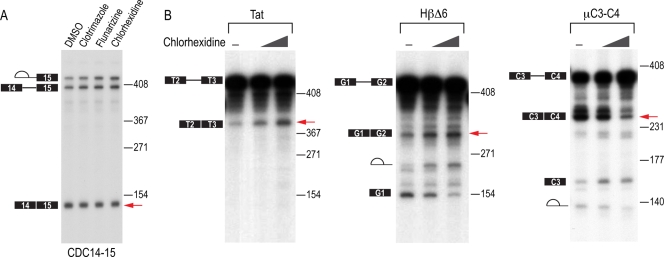
The pre-mRNA minigenes CDC14-15 (chicken delta crystalline), HIV Tat2-3, HβΔ6 (Globin1-2), and μC3-C4 and their splicing conditions were previously described (27, 37, 38). Briefly, the pre-mRNAs were labeled with [α-32P]UTP by in vitro transcription with SP6 polymerase, followed by gel purification. Splicing reactions were performed using HEK 293T total (CDC14-15) or HeLa nuclear (Tat2-3, μC3-C4, and HβΔ6) extract in the presence of DMSO or the specified compounds and incubated for 90 to 120 min at 30°C. Splicing products were purified by Trizol for CDC14-15 or by proteinase K digestion, followed by phenol-chloroform extraction for HIV Tat2-3, μC3-C4, and HβΔ6, and resolved on denaturing polyacrylamide gel electrophoresis (PAGE) gel.
Quantitative Western blotting.
Protein samples were resolved by SDS-PAGE and transferred to a 0.2-μm nitrocellulose membrane, which was blocked in Li-COR blocking buffer. Primary antibodies were diluted in Li-COR blocking buffer plus 0.2% Tween 20. The membranes were then washed 4 times, 15 min each, with phosphate-buffered saline (PBS) containing 0.1% Tween 20. Secondary antibodies (IRDye 800; Rockland) were used at a 1:5,000 dilution. The membranes were washed 4 times as described above. Detection of signal and quantification were done using an Odyssey infrared imaging system. Anti-phospho-SR protein (1H4), anti-SF2/ASF, anti-SRm160, and anti-SRp20 antibodies were purchased from Abcam. Anti-eIF4A3 (3F1) and anti-Magoh (18G12) were previously described (10, 28).
In vitro kinase profiling.
Activated recombinant kinases, including Cdc2-like kinases 1 to 4 (Clk1-4), SR protein kinases 1 and 2 (SRPK1-2), Jun N-terminal protein kinase (JNK), and extracellular signal-related kinase (ERK), were incubated with a synthetic SR-rich substrate peptide in the presence of 50 μM ATP and [α-32P]ATP cocktail in kinase assay buffer (5 mM MOPS, pH 7.2, 2.5 mM β-glycerol-phosphate, 5 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA, 0.05 mM dithiothreitol [DTT]) for 20 min. Either DMSO or chlorhexidine was added at various concentrations. The assay was terminated by spotting 10 μl of the reaction mixture onto a phosphocellulose P81 plate, followed by washing 3 times for 15 min each in 1% phosphoric acid solution. The radioactivity on the P81 plate was counted with scintillation fluid in a TriLux scintillation counter.
Exon microarray target preparation, array hybridization, and data analysis.
Biotinylated sense-strand DNA targets were prepared using the Affymetrix GeneChip whole-transcript (WT) sense target labeling assay in accordance with the manufacturer's directions. One microgram of total RNA from cells treated with chlorhexidine (n = 3), clotrimazole (n = 3), flunarizine (n = 3), and DMSO (n = 6) was used as an input for rRNA reduction with an Invitrogen RiboMinus transcriptome isolation kit in accordance with the manufacturer's directions. The total resulting volume was used for the first round of amplification. Ten micrograms of the resulting cRNA was used to proceed to the second round of amplification. A hybridization cocktail including 5.5 μg of fragmented, end-labeled single-stranded DNA (ssDNA) was applied to GeneChip Human Exon 1.0 ST arrays. Hybridization was performed using F450-001 fluidics wash and stain script on an Affymetrix GeneChip Fluidics Station 450. Arrays were scanned using an Affymetrix GCS 3000 7G and GeneChip operating software (GCOS) to produce CEL intensity files.
Probe set intensities were calculated from the CEL files of the 15 samples by using the robust multiarray average (RMA) algorithm with default settings at both the gene level and the probe set level in Partek Genomic Suite 6.4, using the core probe sets as defined by Affymetrix. Probe sets with a maximum RMA intensity of 3 across all samples were excluded to eliminate probe sets with low expression levels. Alternative splicing multiway analysis of variance (ANOVA) was applied using Partek defaults with terms (probe set identification number and group) not only reflecting experimental conditions but also allowing detection of alternative splicing events that differ between the treated samples and their appropriate controls. Step-up false discovery rates (FDR) were calculated, and genes with FDR values of <0.01 for differential expression or alternative splicing were considered. One-way ANOVA was also applied at the exon level to determine differential expression of exons. These data were used to generate heat maps by using SpotFire DecisionSite.
Microarray data accession number.
The CEL files were deposited in the NCBI Gene Expression Omnibus repository under accession number GSE19891.
RESULTS
Rapid-response splicing reporter system for discovery of modulators of splicing.
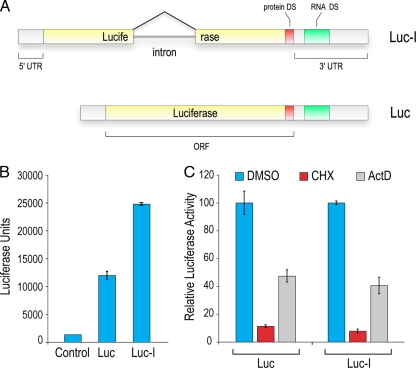
In order to identify small molecules that regulate the splicing machinery or splicing-dependent processes, we generated a reporter in which the ORF of firefly (Photinus pyralis) luciferase is interrupted by a chimeric β-globin/immunoglobulin intron (Luc-I) that has been optimized to splice with high efficiency (51) (Fig. 1A). In cases where this intron is not spliced out, several stop codons appear in frame, resulting in a truncated luciferase protein that has no enzymatic activity. To characterize the splicing reporters, we transfected cells with either intronless (Luc) or intron-containing luciferase reporters. Consistent with previous reports (13, 43, 63), splicing conferred an advantage to gene expression, as equal amounts of transfected DNA generated 2- to 3-fold more luciferase signal from Luc-I than from Luc (Fig. 1B).
FIG. 1.
Characterization of the splicing reporter. (A) Schematic diagram of the intron-containing (Luc-I) and intronless (Luc) luciferase reporters. Solid yellow bars denote exons and the black line depicts the inserted intron. 5′ and 3′ untranslated regions (UTR) are indicated. The destabilizing sequences (DS) are CL1 and PEST for protein DS and five tandem repeats of the AUUUA sequence for RNA. “ORF” represents the open reading frame of luciferase. (B) Expression of the luciferase reporters in cells. The luciferase assay was performed 24 h after transfection of HeLa cells. Error bars denote the standard deviations for three independent experiments. (C) Half-life measurement of reporter protein and mRNA. Luc and Luc-I stably expressing cells were treated with cycloheximide (CHX) or actinomycin D (ActD) for 4 h, followed by a luciferase assay. Data are presented as percent activities compared to the level for DMSO-treated cells. Error bars denote the standard deviations for three independent experiments.
Since splicing is an essential process in mammalian cells, we reasoned that splicing inhibitors would be toxic to cells. To circumvent this, we designed a reporter system that enables the readout of signals in a short screening time (<4 h) so that the time during which cells are exposed to small molecules is minimal, and no global toxic effects are observed, but is long enough for the reporter to sense changes in splicing. To achieve this, the splicing reporter needs to be highly sensitive and fast responding. Thus, we designed the reporters with elements to shorten the half-life of the luciferase protein as well as the luciferase mRNA. We utilized destabilizing sequences (DS) that were added to the C terminus of the protein (protein DS CL1 and PEST) as well as five consecutive AUUUA elements to the 3′ UTR (RNA DS ARE) for rapid mRNA turnover (Fig. 1A). We confirmed that these elements confer a fast response by treatment of Luc- or Luc-I-transfected cells with either cycloheximide or actinomycin D. As shown in Fig. 1C, 4 h of treatment with cycloheximide, which blocks protein synthesis, leads to the loss of 90% of the luciferase signal, indicating that the reporter protein did not accumulate in cells and that this protein is highly sensitive to inhibitors of gene expression. Blocking transcription with actinomycin D for the same time period caused 60% reduction in the signal, suggesting that the mRNA has a half-life of around 4 h. Importantly, both reporters showed similar responses to regulation of gene expression, eliminating the need to adjust for potentially differential stability conferred by splicing of the Luc-I reporter. We concluded that the reporter should be able to detect changes in splicing, especially inhibition, within 4 h of drug treatment of cells and thus performed all further analyses of the luciferase reporter within that time frame.
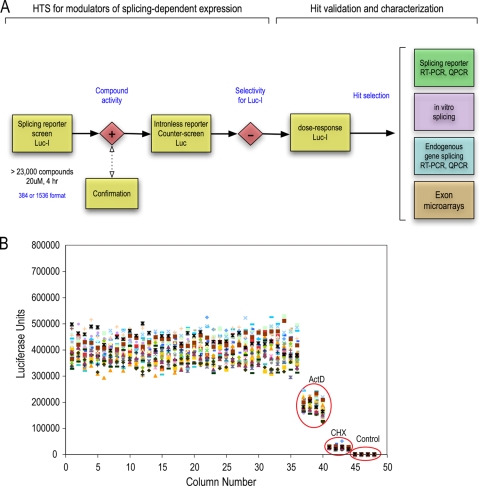
High-throughput screening identifies splicing inhibitors.
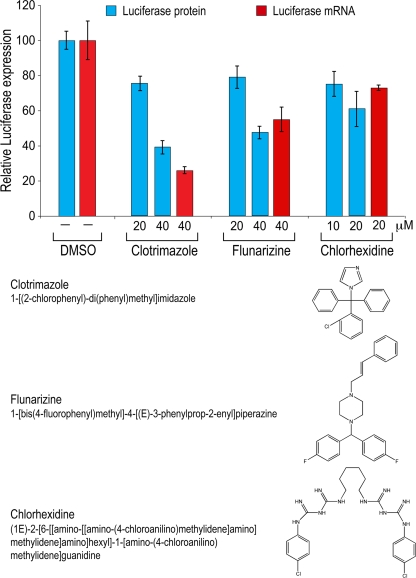
We next generated HeLa cell lines harboring the Luc or Luc-I luciferase reporter. Analysis of RNA processed from these cells revealed that the majority of detectable Luc-I mRNA from the stable lines is spliced (data not shown). To identify modulators of splicing, we performed an ultra-high-throughput screen (1,536-well format) utilizing these cell lines. The strategy of the screen, including the validation and characterization assays, is summarized in a workflow diagram in Fig. 2A. Shown in Fig. 2B are data from a representative plate treated with DMSO, actinomycin D, or cycloheximide. Analysis of this plate shows a high signal-to-noise ratio (more than 100-fold), a coefficient of variance (CV) of 11 to 15%, and a Z′ factor of at least 0.6, indicating that this cell-based assay is highly robust and reliable. The short assay time made it unnecessary to normalize the signal-to-cell number. A collection of >23,000 chemically and functionally diverse compounds assembled from commercial sources was first screened on the Luc-I cell line. Approximately 250 compounds showed an effect of more than 3 times the standard deviation (SD) in comparison to the level for the DMSO control wells on the same plate. However, compounds that affect firefly luciferase enzymatic activity or protein stability could be mistakenly identified as gene expression modulators if counterscreened on Renilla luciferase or other reporters (2). We therefore counterscreened the same compounds in cells expressing an intronless firefly luciferase (Luc). This also allows us to filter away compounds that modulate general processes other than splicing, such as transcription or translation. Indeed, a large fraction of compounds modulated Luc in the same manner as Luc-I and were therefore eliminated. The remaining compounds that showed an intron-dependent effect were studied further (see Table S3 in the supplemental material). Three of these, clotrimazole {1-[(2-chlorophenyl)-di(phenyl)methyl]imidazole}, flunarizine {1-[bis(4-fluorophenyl)methyl]- 4-[(E)-3-phenylprop-2-enyl]piperazine}, and chlorhexidine {(1E)-2-[6-[[amino-[[amino-(4-chloroanilino)methylidene]amino]methylidene]amino]hexyl]-1-[amino-(4-chloro-anilino)methylidene]guanidine}, which are compounds in wide clinical use and have not previously been shown to affect splicing, are described here in detail. Measurements of a wide range of concentrations showed that clotrimazole and flunarizine had a dose-dependent inhibition of Luc-I, with 50% inhibitory concentrations (IC50s) between 40 and 50 μM, while chlorhexidine had a smaller effect on Luc-I at concentrations up to 20 μM (Fig. 3; see also Fig. S1 in the supplemental material). Higher concentrations of chlorhexidine were toxic to these cells even at the 4-hour treatment time point. To verify that the change in Luc-I signal is due to splicing modulation, we used real-time PCR to measure the levels of spliced luciferase mRNA in cells treated with the three compounds at their IC50s. Both clotrimazole and flunarizine strongly decreased spliced Luc-I mRNA levels, while chlorhexidine had a modest but reproducible effect (Fig. 3). The spliced mRNA measurements were normalized to those of intronless Luc in order to eliminate effects on transcription or mRNA stability.
FIG. 2.
High-throughput screen for splicing modulators. (A) Workflow for HTS. Cells stably expressing Luc-I were screened with a collection of >23,000 compounds for 4 h (see Materials and Methods). Hits were counterscreened on Luc cells, and compounds that passed the selectivity step and showed a well-behaved dose response were further characterized as illustrated in the scheme. QPCR, quantitative PCR. (B) Scatter plot of a representative control plate of Luc-I cells in a 1,536-well format. Four columns on the plate were treated with ActD or CHX. The last four columns (Control) did not contain any cells and reflect the background/noise of the assay. The calculated Z′ value was 0.6, and the CV was 11%.
FIG. 3.
Splicing inhibitors identified from HTS inhibit luciferase protein and mRNA expression. Luc-I cells were treated with various concentrations of clotrimazole, flunarizine, and chlorhexidine for 4 h. Luciferase activity, measured in light units, reflects the amount of luciferase protein produced (blue bars). To quantify the amount of spliced luciferase mRNA, real-time qPCR was performed at the IC50 of each compound (red bars). The spliced mRNA measurements were normalized to those of intronless Luc in order to eliminate effects on transcription or mRNA stability. Data are presented as percentages of the level for DMSO-treated cells, and error bars denote the standard deviations for three independent experiments.
Effect of inhibitors on splicing of endogenous genes in vivo.
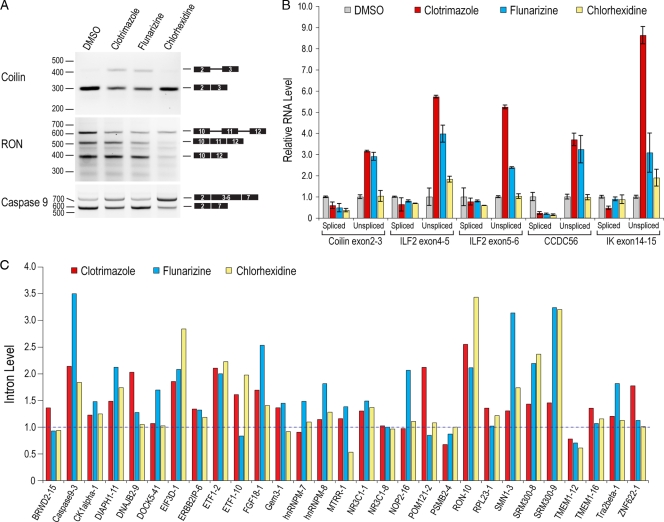
To determine the effect of the compounds on splicing of endogenous genes, we analyzed constitutive and alternative splicing patterns of several genes by RT-PCR. Interestingly, none of the compounds inhibited every splicing event; rather, each compound showed a unique pattern of inhibition and induced distinct alternative splicing changes (Fig. 4A and data not shown). For example, both clotrimazole and flunarizine inhibited splicing of coilin intron 2, whereas chlorhexidine, which had no effect on this intron, modulated the splicing of several alternatively spliced exons, including SR protein-regulated RON exon 11 and caspase 9 (Fig. 4A). To obtain a quantitative measure of the differential effects of these compounds, we performed detailed real-time PCR for spliced and unspliced mRNAs of several major (U2-dependent) and minor (U12-dependent) introns. As shown in Fig. 4B, splicing of both major (coilin and ILF2) and minor (CCDC56 and IK) introns was strongly inhibited by clotrimazole and flunarizine, albeit to different extents, while chlorhexidine only slightly inhibited the splicing of ILF2 intron 4 and IK intron 14 (Fig. 4B). We further extended this analysis to more than 30 constitutively spliced introns that had a wide range of sizes and have not previously been shown to be regulated (Fig. 4C). These data indicated that clotrimazole and flunarizine are not general splicing inhibitors. Instead, each inhibited a subset of these introns, whereas chlorhexidine inhibited a smaller and distinct set of constitutively spliced introns (Fig. 4C), suggesting that these compounds regulate splicing by different mechanisms. Bioinformatic analysis of the affected introns did not show distinctive features such as splice site strength or common motifs. Nevertheless, these data uncover an unexpected differential regulation of constitutively spliced introns in cells.
FIG. 4.
Splicing modulators have distinct effects on constitutive and alternative splicing of endogenous genes in cells. (A) RT-PCR analysis of total RNA extracted from HeLa cells treated with clotrimazole, flunarizine, and chlorhexidine for 6 h. Results are shown for constitutive pre-mRNA splicing of endogenous coilin intron 2 and SR protein-regulated alternative splicing of exon 11 of RON and exons 3 to 6 of caspase 9. (B) Real-time qPCR analysis of the samples shown in panel A. For each endogenous gene, a set of primers was designed to distinguish between exon-exon junction (spliced) and exon-intron junction (unspliced). Coilin intron 2 and ILF2 introns 4 and 5 are major (U2 dependent), whereas CCDC56 intron 1 and IK intron 14 are minor (U12 dependent). Three independent replicates were used for each treatment, and data are presented relative to the level for DMSO, which was set to 1. (C) Real-time qPCR analysis of introns from various endogenous genes, as described for panel A. For each transcript, the intron tested is indicated as a number after the gene name. Each bar represents the average of results from three measurements, and data are presented relative to the level for DMSO, which was set to 1. All qPCR measurements were normalized to the level for β-actin.
Chlorhexidine modulates SR protein-regulated splicing by inhibiting the Clk family of SR protein kinases.
We next tested the effect of the compounds on constitutive and SR protein-regulated splicing in vitro. CDC14-15 pre-mRNA is constitutively spliced in HeLa nuclear extract, and none of the compounds inhibited its splicing in vitro (Fig. 5A). However, the splicing of the same intron in the context of the luciferase reporter was modulated by the three compounds when tested with intact cells (data not shown). This suggests that the compounds do not target the basal splicing machinery but rather modulate its activity in vivo. Interestingly, chlorhexidine modulated the in vitro splicing of the three SR protein-regulated pre-mRNAs that were tested (Tat, μC3-C4, and HβΔ6), suggesting that it can modulate SR protein-regulated splicing both in vitro and in vivo (Fig. 5B and 4A).
FIG. 5.
In vitro effects of splicing modulators. (A) Constitutive splicing of CDC14-15 pre-mRNA was analyzed in the presence of 50 μM clotrimazole, flunarizine, or chlorhexidine for 90 min. Splicing intermediates are depicted to the left of the gel, and molecular size markers are indicated to the right of the gel. Fully spliced mRNA is indicated with a red arrow and corresponds to 140 nucleotides. (B) SR protein-regulated splicing of HIV Tat2-3, HβΔ6, and μC3-C4 pre-mRNAs was analyzed in the presence of DMSO or 50 and 100 μM chlorhexidine in vitro. Splicing intermediates are depicted to the left of the gels, and molecular size markers are indicated to the right of the gels. Red arrows point to the altered levels of the spliced mRNA, which are 371, 367, and 271 nucleotides for Tat2-3, HβΔ6, and μC3-C4, respectively.
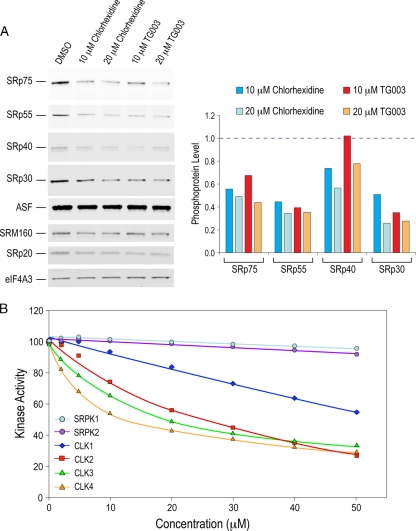
We therefore investigated the effect of chlorhexidine on SR protein phosphorylation, which is a major determinant of splicing regulation. Western blotting using antibodies specific for phosphorylated SR proteins revealed that chlorhexidine decreased the phosphorylation of several SR proteins, including SRp75, SRp55, SRp40, and SRp30, at concentrations as low as 10 μM (Fig. 6A). These effects were comparable to those observed for TG003, a previously described regulator of SR protein phosphorylation (42). Other SR and SR-like proteins, such as SF2/ASF, SRp20, and SRm160, were not significantly affected at these concentrations. For reference, the levels of other proteins involved in mRNA biogenesis were not affected (Fig. 6A).
FIG. 6.
Chlorhexidine inhibits SR protein phosphorylation. (A) Total protein extracts from cells treated with 0, 10, and 20 μM chlorhexidine or TG003 for 6 h were separated by SDS-PAGE, and phosphorylated SR proteins were detected by Western blotting using the SR protein phospho-specific monoclonal antibody (1H4). Quantification of the phosphorylated SR proteins is depicted in the histogram. Values are presented relative to the level for DMSO, which was set to 1 (dashed line). (B) The ability of known recombinant SR protein kinases to phosphorylate an SR-rich substrate was tested in vitro in the presence of either DMSO or increasing concentrations of chlorhexidine. Activity in the presence of DMSO was set to 100%.
In light of the pronounced effect of chlorhexidine on SR protein-regulated splicing and SR protein phosphorylation, we tested its effect on several kinases known to target SR proteins. With the use of recombinant kinases and an SR-rich peptide derived from SF2/ASF as a substrate, chlorhexidine showed specific inhibition of the Cdc2-like kinase (Clk) family of SR protein kinases. No inhibition by chlorhexidine of other known SR protein kinases (SRPK1 and SRPK2) as well as other unrelated kinases (ERK1 and JNK1) (data not shown) was detected (Fig. 6B). Moreover, chlorhexidine showed selectivity for different members of the Clk family. Clk4 and Clk3 were most sensitive to chlorhexidine treatment, with IC50s of 10 and 15 μM, respectively, whereas Clk2 and Clk1 were less affected, with IC50s of 25 and >50 μM, respectively.
Differential effects of the compounds on alternative splicing.
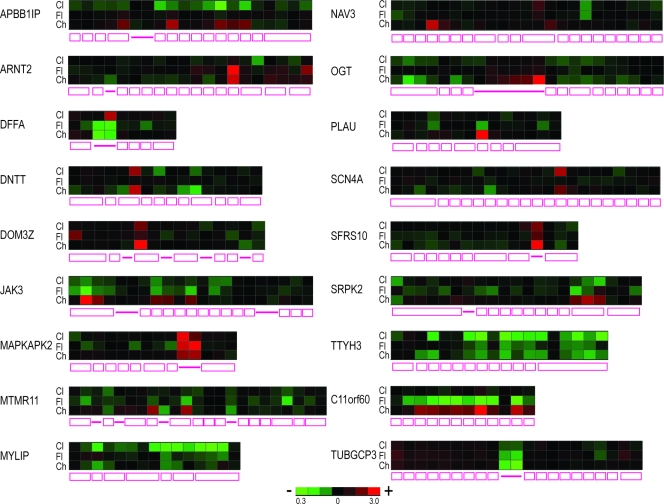
Given the effect of chlorhexidine on several alternatively spliced exons, we wished to assess its effect, as well as those of clotrimazole and flunarizine, on splicing at a genome-wide level. RNAs processed from cells treated with clotrimazole (40 μM), flunarizine (40 μM), and chlorhexidine (10 μM) were analyzed by exon arrays. For chlorhexidine, the data showed a large number of alternative splicing changes (1,444 transcripts at FDR values of <0.01), compared to only 191 genes affected at the whole-transcript level, namely, at transcriptional or mRNA stability levels, indicating high selectivity of chlorhexidine for alternative splicing regulation (see Table S1 in the supplemental material). Clotrimazole and flunarizine were found to cause fewer alternative splicing changes when the same statistical cutoff was used (874 and 326 genes, respectively). Confirmation of a select number of the affected transcripts is presented in Fig. S2 in the supplemental material, and the overlap among these transcripts and the breakdown of the splicing alterations are shown in Fig. S3 in the supplemental material. We note that transcripts that were affected by at least two of the compounds frequently showed different profiles at the exon level (Fig. 7). For example, some exons were increased or decreased in all three treatments (DOM3Z, MYLIP, and TTYH3). Others were affected by two of the three compounds (ARNT2, SCN4A, DNTT, and NAV3), whereas some showed opposing effects in different treatments (APBB1IP, JAK3, MTMR11, C11orf60, PLAU, and SRPK2). In addition, some transcripts with intronic probes showed unique effects for each of the three compounds (DFFA, OGT, MTMR11, MAPKAPK2, TUBGCP3, and SFRS10). Thus, clotrimazole, flunarizine, and chlorhexidine have differential effects on constitutively spliced introns and on alternative splicing in cells.
FIG. 7.
Differential effects of splicing modulators on alternative splicing. Heat maps of representative transcripts from the exon array are shown, with the gene structure depicted below the heat map.
DISCUSSION
The splicing reporter system that we describe here has several advantageous properties that make it a powerful tool for the discovery of inhibitors and modulators of splicing and splicing-dependent processes in cells. Our assay system was designed to overcome two major potential problems. First, given the essential role of splicing in gene expression, prolonged exposure to splicing inhibitors would be toxic to cells and preclude their discovery. We therefore reasoned that the reporter needed to be highly sensitive and robust and provide real-time readout of changes in splicing before general toxic effects interfere. We achieved this by incorporating destabilizing elements in both the luciferase protein and the mRNA encoding it, generating a rapid-response reporter with a half-life of less than 4 h. Second, many compounds could affect the enzymatic activity or stability of the reporter or steps in its expression other than splicing, such as transcription and translation. To address this, we constructed a firefly luciferase reporter gene that was the same except for its lack of an intron and eliminated compounds that showed an effect on both Luc and Luc-I. The assay is suitable for large-scale screens for chemical and genetic modifiers of splicing in 384- and 1,536-well formats. This strategy opens up the possibility of discovering splicing inhibitors that would likely not be identified by other recently described cell-based assays (34, 44, 54). Furthermore, the short time frame of the assay helps minimize secondary effects of compounds and increases the likelihood of capturing direct effects on splicing.
Screening with this assay, using a strong intron commonly used for constitutive splicing studies, identified several inhibitors. Importantly, each showed differential effects on constitutive and alternative splicing. Both clotrimazole and flunarizine inhibited splicing of many introns but had distinct profiles. Of the introns tested, each compound inhibited some that the other did not; some were inhibited by both, and some showed different degrees of inhibition for the two compounds. This suggests that although both clotrimazole and flunarizine inhibit the splicing of the reporter, they do so by different modes of action. We note that neither compound inhibited splicing in vitro of several test introns. Thus, unlike spliceostatin A and pladienolide (24, 31), clotrimazole and flunarizine do not inhibit components of the basal splicing machinery such as U2 snRNP, which is surprising because the β-globin/immunoglobulin intron used in the screen is considered to be independent of splicing factors due to its strong (constitutive) splice sites (51). These findings also underscore the capacity of cell-based screens to reveal regulatory mechanisms that are not recapitulated in an in vitro system. Our observations suggest that entirely constitutive splicing may not exist in cells and that all introns are regulated. The compounds that we identified should make it possible to discover the pathways and factors involved in this unexpected aspect of regulation of constitutively spliced introns.
Clotrimazole is widely used without prescription for treatment of fungal infections. In mammalian cells, it inhibits cytochrome P450, interferes with cellular calcium homeostasis (1, 4, 58), and inhibits proliferation of cancer and vascular endothelial cells (4, 29, 56). Flunarizine is a calcium channel blocker and vasodilator that is used to treat neurological disorders, including vertigo, migraine, and epilepsy (22, 49). It has a protective effect on neurons from serum and nerve growth factor deprivation, oxidative stress, and axotomy (25). Clotrimazole and flunarizine have not previously been shown to have an effect on splicing. The mechanism by which these compounds inhibit splicing is not presently known, and it is thus not clear whether this is related to their clinical efficacy. It is noted, however, that both affect calcium physiology. Several anticancer drugs have been recently shown to affect alternative splicing of a group of apoptosis-related test genes (52). However, the drug treatments were for 24 h, and thus, the effects on splicing could be an adaptive response rather than a direct effect. Some of these were included in the library of compounds that we screened but did not show specific activity in our assay.
Our results demonstrate that chlorhexidine had a weaker effect on constitutive splicing than clotrimazole and flunarizine. However, it was highly effective in modulating alternative splicing. Chlorhexidine is a widely used, prescription-free disinfectant and topical anti-infective agent, most commonly used in mouthwash. In addition to its broad-spectrum antibacterial activity, chlorhexidine is active against yeast and some lipid-enveloped viruses, including HIV (40). It has also been reported to induce an inflammatory reaction (45, 65) and tissue necrosis (16). Several mechanisms have been suggested for this induced toxicity, including inhibition of mitochondrial activity, protein and DNA synthesis, endoplasmic reticulum stress, an increase in intracellular calcium, and induction of oxidative stress (19, 23). However, chlorhexidine has not previously been known to have an effect on splicing.
Because our data demonstrated that chlorhexidine affected alternative splicing of many exons, including those of RON, caspase 9, and HIV Tat2-3, which are known to be regulated by SR proteins, we studied its effect on this major class of splicing factors. SR proteins play critical roles in both constitutive and alternative splicing (5, 7, 20, 36). Phosphorylation of the RS domain of SR proteins regulates their activity in alternative splicing by modulating their protein-protein interactions, RNA binding, cellular localization, and stability (8, 9, 15, 41, 64). We found marked reductions in the phosphorylation states of SRp75, SRp55, and SRp30 after chlorhexidine treatment. Other SR proteins, such as SRp20, were not affected. It has been shown that the relative stoichiometry of SR proteins on a given pre-mRNA is complex and depends on their phosphorylation state (50) and that disruption of this stoichiometry leads to not only negative but also positive outcomes on splicing (17, 26, 39, 61). Since chlorhexidine did not affect the phosphorylation of all SR proteins to the same level, it is impossible to predict the outcome of chlorhexidine treatment on a given splicing event. For example, while inhibition of μC3-C4 splicing could be due to the inability of a dephosphorylated SR protein(s), such as ASF/SF2, to bind and enhance splicing, the increased splicing efficiency of Tat2-3 could be explained in light of previous data showing that SC35 can bind to an exonic splicing silencer and repress splicing of Tat2-3 (38). Thus, the release of SC35 from this pre-mRNA or the disruption of the balance between SC35 and ASF/SF2 could lead to enhanced splicing of Tat2-3 pre-mRNA. Several kinases that phosphorylate SR proteins and regulate their activity have been described, particularly SRPKs (21, 33) and Clks (3, 11). Other potential SR protein kinases have also been reported but remain less characterized, including pre-mRNA processing mutant 4 (PRP4) (30) and topoisomerase I (48). Our studies show that chlorhexidine is a selective inhibitor of Clk4 and Clk3 and to a lesser extent Clk2. Clk1, on the other hand, is only weakly inhibited by chlorhexidine. This activity profile is different from that of a previously described Clk inhibitor, TG003, which mainly targets Clk1 and Clk4 (42). While all Clks can phosphorylate any SR protein in vitro, it was not previously known whether the individual Clks have activities toward different SR proteins in cells. Our findings suggest that by inhibiting Clk4 and Clk3, chlorhexidine decreases or alters the phosphorylation signature of a subset of SR proteins, resulting in a change in the splicing pattern of a distinct group of exons. Further characterization of these splicing events should provide a better understanding of the functional relationship between phosphorylation of specific SR proteins by different Clks and their role in regulating alternative splicing. Chlorhexidine is therefore a valuable tool with which to dissect these mechanisms.
Exon array analysis of clotrimazole-, flunarizine-, or chlorhexidine-treated cells showed that each compound affects a distinct set of splicing events. Closer analysis of the transcripts that were affected by at least two compounds showed that a large fraction of these transcripts were affected on different exons or in opposite directions, indicating that each compound regulates splicing by a unique mechanism. The effect of the clinically used drugs clotrimazole, flunarizine, and chlorhexidine on splicing is unexpected and off target at concentration ranges in which they show pharmacologic activity. This effect may provide a molecular basis for their toxicity as well as their activity and should help design more-specific next-generation drugs. The screening method that we describe will provide a wealth of additional reagents for studying the mechanism and regulation of splicing. With slight modifications, the same reporter can be used to screen for effectors of specific introns and of other splicing-dependent processes.
Supplementary Material
Acknowledgments
We are grateful to members of our laboratory, especially Mumtaz Kasim, Jennifer Bachorik, Jeongsik Yong, and Lili Wan, for stimulating discussions and comments on the manuscript.
This work was supported by the Association Française Contre les Myopathies (AFM). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 1 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Alvarez, J., M. Montero, and J. Garcia-Sancho. 1992. High affinity inhibition of Ca(2+)-dependent K+ channels by cytochrome P-450 inhibitors. J. Biol. Chem. 267:11789-11793. [PubMed] [Google Scholar]
- 2.Auld, D. S., N. Thorne, W. F. Maguire, and J. Inglese. 2009. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc. Natl. Acad. Sci. U. S. A. 106:3585-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-David, Y., K. Letwin, L. Tannock, A. Bernstein, and T. Pawson. 1991. A mammalian protein kinase with potential for serine/threonine and tyrosine phosphorylation is related to cell cycle regulators. EMBO J. 10:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benzaquen, L. R., C. Brugnara, H. R. Byers, S. Gatton-Celli, and J. A. Halperin. 1995. Clotrimazole inhibits cell proliferation in vitro and in vivo. Nat. Med. 1:534-540. [DOI] [PubMed] [Google Scholar]
- 5.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 6.Blencowe, B. J. 2006. Alternative splicing: new insights from global analyses. Cell 126:37-47. [DOI] [PubMed] [Google Scholar]
- 7.Bourgeois, C. F., F. Lejeune, and J. Stevenin. 2004. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 78:37-88. [DOI] [PubMed] [Google Scholar]
- 8.Caceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, W., S. F. Jamison, and M. A. Garcia-Blanco. 1997. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3:1456-1467. [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, C. C., J. Dostie, M. D. Diem, W. Feng, M. Mann, J. Rappsilber, and G. Dreyfuss. 2004. eIF4A3 is a novel component of the exon junction complex. RNA 10:200-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colwill, K., T. Pawson, B. Andrews, J. Prasad, J. L. Manley, J. C. Bell, and P. I. Duncan. 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15:265-275. [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, T. A., L. Wan, and G. Dreyfuss. 2009. RNA and disease. Cell 136:777-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diem, M. D., C. C. Chan, I. Younis, and G. Dreyfuss. 2007. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 14:1173-1179. [DOI] [PubMed] [Google Scholar]
- 14.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, P. I., D. F. Stojdl, R. M. Marius, and J. C. Bell. 1997. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol. Cell. Biol. 17:5996-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faria, G., C. R. Cardoso, R. E. Larson, J. S. Silva, and M. A. Rossi. 2009. Chlorhexidine-induced apoptosis or necrosis in L929 fibroblasts: a role for endoplasmic reticulum stress. Toxicol. Appl. Pharmacol. 234:256-265. [DOI] [PubMed] [Google Scholar]
- 17.Gallego, M. E., R. Gattoni, J. Stevenin, J. Marie, and A. Expert-Bezancon. 1997. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 16:1772-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Blanco, M. A., A. P. Baraniak, and E. L. Lasda. 2004. Alternative splicing in disease and therapy. Nat. Biotechnol. 22:535-546. [DOI] [PubMed] [Google Scholar]
- 19.Giannelli, M., F. Chellini, M. Margheri, P. Tonelli, and A. Tani. 2008. Effect of chlorhexidine digluconate on different cell types: a molecular and ultrastructural Invest. Toxicol. In Vitro 22:308-317. [DOI] [PubMed] [Google Scholar]
- 20.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gui, J. F., W. S. Lane, and X. D. Fu. 1994. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369:678-682. [DOI] [PubMed] [Google Scholar]
- 22.Hamalainen, M. L. 2006. Migraine in children and adolescents: a guide to drug treatment. CNS Drugs 20:813-820. [DOI] [PubMed] [Google Scholar]
- 23.Hidalgo, E., and C. Dominguez. 2001. Mechanisms underlying chlorhexidine-induced cytotoxicity. Toxicol. In Vitro 15:271-276. [DOI] [PubMed] [Google Scholar]
- 24.Kaida, D., H. Motoyoshi, E. Tashiro, T. Nojima, M. Hagiwara, K. Ishigami, H. Watanabe, T. Kitahara, T. Yoshida, H. Nakajima, T. Tani, S. Horinouchi, and M. Yoshida. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3:576-583. [DOI] [PubMed] [Google Scholar]
- 25.Kaminski Schierle, G. S., O. Hansson, and P. Brundin. 1999. Flunarizine improves the survival of grafted dopaminergic neurons. Neuroscience 94: 17-20. [DOI] [PubMed] [Google Scholar]
- 26.Kanopka, A., O. Muhlemann, and G. Akusjarvi. 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381:535-538. [DOI] [PubMed] [Google Scholar]
- 27.Kataoka, N., and G. Dreyfuss. 2004. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J. Biol. Chem. 279:7009-7013. [DOI] [PubMed] [Google Scholar]
- 28.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673-682. [DOI] [PubMed] [Google Scholar]
- 29.Khalid, M. H., Y. Tokunaga, A. J. Caputy, and E. Walters. 2005. Inhibition of tumor growth and prolonged survival of rats with intracranial gliomas following administration of clotrimazole. J. Neurosurg. 103:79-86. [DOI] [PubMed] [Google Scholar]
- 30.Kojima, T., T. Zama, K. Wada, H. Onogi, and M. Hagiwara. 2001. Cloning of human PRP4 reveals interaction with Clk1. J. Biol. Chem. 276:32247-32256. [DOI] [PubMed] [Google Scholar]
- 31.Kotake, Y., K. Sagane, T. Owa, Y. Mimori-Kiyosue, H. Shimizu, M. Uesugi, Y. Ishihama, M. Iwata, and Y. Mizui. 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3:570-575. [DOI] [PubMed] [Google Scholar]
- 32.Krawczak, M., J. Reiss, and D. N. Cooper. 1992. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 90:41-54. [DOI] [PubMed] [Google Scholar]
- 33.Kuroyanagi, N., H. Onogi, T. Wakabayashi, and M. Hagiwara. 1998. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem. Biophys. Res. Commun. 242:357-364. [DOI] [PubMed] [Google Scholar]
- 34.Levinson, N., R. Hinman, A. Patil, C. R. Stephenson, S. Werner, G. H. Woo, J. Xiao, P. Wipf, and K. W. Lynch. 2006. Use of transcriptional synergy to augment sensitivity of a splicing reporter assay. RNA 12:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Licatalosi, D. D., and R. B. Darnell. 2006. Splicing regulation in neurologic disease. Neuron 52:93-101. [DOI] [PubMed] [Google Scholar]
- 36.Manley, J. L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 37.Mayeda, A., and Y. Ohshima. 1990. Beta-globin transcripts carrying a single intron with three adjacent nucleotides of 5′ exon are efficiently spliced in vitro irrespective of intron position or surrounding exon sequences. Nucleic Acids Res. 18:4671-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayeda, A., G. R. Screaton, S. D. Chandler, X. D. Fu, and A. R. Krainer. 1999. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol. 19:1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNally, L. M., and M. T. McNally. 1996. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 70:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milstone, A. M., C. L. Passaretti, and T. M. Perl. 2008. Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin. Infect. Dis. 46:274-281. [DOI] [PubMed] [Google Scholar]
- 41.Misteli, T., J. F. Caceres, J. Q. Clement, A. R. Krainer, M. F. Wilkinson, and D. L. Spector. 1998. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143:297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muraki, M., B. Ohkawara, T. Hosoya, H. Onogi, J. Koizumi, T. Koizumi, K. Sumi, J. Yomoda, M. V. Murray, H. Kimura, K. Furuichi, H. Shibuya, A. R. Krainer, M. Suzuki, and M. Hagiwara. 2004. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 279:24246-24254. [DOI] [PubMed] [Google Scholar]
- 43.Nott, A., H. Le Hir, and M. J. Moore. 2004. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 18:210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Brien, K., A. J. Matlin, A. M. Lowell, and M. J. Moore. 2008. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J. Biol. Chem. 283:33147-33154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oncag, O., M. Hosgor, S. Hilmioglu, O. Zekioglu, C. Eronat, and D. Burhanoglu. 2003. Comparison of antibacterial and toxic effects of various root canal irrigants. Int. Endod. J. 36:423-432. [DOI] [PubMed] [Google Scholar]
- 46.Pan, Q., O. Shai, L. J. Lee, B. J. Frey, and B. J. Blencowe. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40:1413-1415. [DOI] [PubMed] [Google Scholar]
- 47.Pilch, B., E. Allemand, M. Facompre, C. Bailly, J. F. Riou, J. Soret, and J. Tazi. 2001. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 61:6876-6884. [PubMed] [Google Scholar]
- 48.Rossi, F., E. Labourier, T. Forne, G. Divita, J. Derancourt, J. F. Riou, E. Antoine, G. Cathala, C. Brunel, and J. Tazi. 1996. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature 381:80-82. [DOI] [PubMed] [Google Scholar]
- 49.Santi, C. M., F. S. Cayabyab, K. G. Sutton, J. E. McRory, J. Mezeyova, K. S. Hamming, D. Parker, A. Stea, and T. P. Snutch. 2002. Differential inhibition of T-type calcium channels by neuroleptics. J. Neurosci. 22:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sapra, A. K., M. L. Anko, I. Grishina, M. Lorenz, M. Pabis, I. Poser, J. Rollins, E. M. Weiland, and K. M. Neugebauer. 2009. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol. Cell 34:179-190. [DOI] [PubMed] [Google Scholar]
- 51.Senapathy, P., M. B. Shapiro, and N. L. Harris. 1990. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 183:252-278. [DOI] [PubMed] [Google Scholar]
- 52.Shkreta, L., U. Froehlich, E. R. Paquet, J. Toutant, S. A. Elela, and B. Chabot. 2008. Anticancer drugs affect the alternative splicing of Bcl-x and other human apoptotic genes. Mol. Cancer Ther. 7:1398-1409. [DOI] [PubMed] [Google Scholar]
- 53.Soret, J., N. Bakkour, S. Maire, S. Durand, L. Zekri, M. Gabut, W. Fic, G. Divita, C. Rivalle, D. Dauzonne, C. H. Nguyen, P. Jeanteur, and J. Tazi. 2005. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc. Natl. Acad. Sci. U. S. A. 102:8764-8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoilov, P., C. H. Lin, R. Damoiseaux, J. Nikolic, and D. L. Black. 2008. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl. Acad. Sci. U. S. A. 105:11218-11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sumanasekera, C., D. S. Watt, and S. Stamm. 2008. Substances that can change alternative splice-site selection. Biochem. Soc. Trans. 36:483-490. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi, H., M. Abe, T. Sugawara, K. Tanaka, Y. Saito, S. Fujimura, M. Shibuya, and Y. Sato. 1998. Clotrimazole, an imidazole antimycotic, is a potent inhibitor of angiogenesis. Jpn. J. Cancer Res. 89:445-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venables, J. P. 2004. Aberrant and alternative splicing in cancer. Cancer Res. 64:7647-7654. [DOI] [PubMed] [Google Scholar]
- 58.Villalobos, C., R. Fonteriz, M. G. Lopez, A. G. Garcia, and J. Garcia-Sancho. 1992. Inhibition of voltage-gated Ca2+ entry into GH3 and chromaffin cells by imidazole antimycotics and other cytochrome P450 blockers. FASEB J. 6:2742-2747. [DOI] [PubMed] [Google Scholar]
- 59.Wahl, M. C., C. L. Will, and R. Luhrmann. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701-718. [DOI] [PubMed] [Google Scholar]
- 60.Wang, E. T., R. Sandberg, S. Luo, I. Khrebtukova, L. Zhang, C. Mayr, S. F. Kingsmore, G. P. Schroth, and C. B. Burge. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, J., and J. L. Manley. 1995. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1:335-346. [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Z., and C. B. Burge. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14:802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiegand, H. L., S. Lu, and B. R. Cullen. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. U. S. A. 100:11327-11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao, S. H., and J. L. Manley. 1997. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 11:334-344. [DOI] [PubMed] [Google Scholar]
- 65.Yesilsoy, C., E. Whitaker, D. Cleveland, E. Phillips, and M. Trope. 1995. Antimicrobial and toxic effects of established and potential root canal irrigants. J. Endod. 21:513-515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.