Abstract
We recently reported that the phosphotyrosine-binding (PTB) domain of Anks family proteins binds to EphA8, thereby positively regulating EphA8-mediated signaling pathways. In the current study, we identified a potential role for the SAM domains of Anks family proteins in EphA signaling. We found that SAM domains of Anks family proteins directly bind to ubiquitin, suggesting that Anks proteins regulate the degradation of ubiquitinated EphA receptors. Consistent with the role of Cbl ubiquitin ligases in the degradation of Eph receptors, our results revealed that the ubiquitin ligase c-Cbl induced the ubiquitination and degradation of EphA8 upon ligand binding. Ubiquitinated EphA8 also bound to the SAM domains of Odin, a member of the Anks family proteins. More importantly, the overexpression of wild-type Odin protected EphA8 and EphA2 from undergoing degradation following ligand stimulation and promoted EphA-mediated inhibition of cell migration. In contrast, a SAM domain deletion mutant of Odin strongly impaired the function of endogenous Odin, suggesting that the mutant functions in a dominant-negative manner. An analysis of Odin-deficient primary embryonic fibroblasts indicated that Odin levels play a critical role in regulating the stability of EphA2 in response to ligand stimulation. Taken together, our studies suggest that the SAM domains of Anks family proteins play a pivotal role in enhancing the stability of EphA receptors by modulating the ubiquitination process.
Activation of Eph receptor tyrosine kinases (RTKs) by ephrin ligands stimulates intracellular signaling pathways that regulate diverse cell behaviors such as axon guidance, cell adhesion, and cell migration (1). Activated Eph receptors also initiate negative signaling events that counteract or alter positive signals, thereby modulating biological outcomes. Negative signaling events associated with Eph RTKs include metalloprotease-mediated cleavage of ephrins and trans endocytosis of Eph-ephrin complexes (9, 15, 24). These negative regulatory mechanisms may be important in the repulsive mechanism responsible for retraction of cellular processes. Some studies suggest that c-Cbl, a RING finger E3 ligase, participates in activated Eph receptor signal termination. Ligand stimulation induces the tyrosine phosphorylation of c-Cbl and facilitates the degradation of Eph receptors (19, 23). More recent studies have shown that the E3 ligase activity of c-Cbl is activated through tyrosine phosphorylation by Src family kinases and that c-Cbl is recruited to activated Eph receptors and induces the ubiquitination and degradation of the receptors (6, 14). These studies point to an important role for Cbl family ubiquitin (Ub) ligases in mediating the ubiquitination of activated Eph RTKs and in fine-tuning Eph receptor signaling pathways.
Emerging evidence points to a critical role for Eph receptors in human diseases such as diabetes and cancer (2, 13, 17). For example, EphA2 overexpression has been found in many types of malignant tumors. Overexpression of EphA2 in nontransformed epithelial cells enhances tumorigenic and metastatic potential, whereas downregulation of EphA2 expression suppresses tumor growth and metastasis (4). In addition, either soluble ephrin-A ligand or a monoclonal antibody that activates and degrades EphA2 has been shown to inhibit the growth of human tumor xenografts in nude mice (5, 12). More recent evidence reveals that EphA2 cooperates with Erb2 (also known as Neu) to promote tumor progression in mice (3). These findings strongly suggest that EphA2 and possibly other Eph receptors function in tumor progression in the context of either specific oncogenes or tumor suppressors. In this respect, understanding the negative regulation of Eph receptors, such as their degradation, may have important implications in the design of effective antitumor therapeutics.
Recently, we showed that Anks family proteins act as key scaffolding molecules in EphA8-mediated signaling pathways (20). Anks family proteins contain six ankyrin repeats at their N terminus, two SAM domains, and a phosphotyrosine-binding (PTB) domain at their C terminus (22). Odin and AβPP intracellular domain-associated protein 1b (AIDA-1b) belong to this protein family. Several isoforms of AIDA-1b have been described, and the regions encoding the PTB domain and the two SAM domains are very well conserved among all isoforms (7). Interestingly, AIDA-1 has been implicated in reducing AβPP processing through the inhibition of γ-secretase activity (7) and in increasing the global protein biosynthetic capacity in response to long-term neuronal stimulation through the regulation of nucleolar assembly (10). Functions attributed to Odin have been limited to its negative role in platelet-derived growth factor (PDGF)-mediated cell proliferation (16). In contrast to AIDA-1 proteins, Odin appears to be abundantly and ubiquitously expressed in many different mammalian cell lines, and its expression is restricted to the mouse embryonic brain rather than the adult brain (20). We recently reported that the PTB domains of Anks family proteins are crucial for the association of these proteins with the juxtamembrane (JM) domain of EphA8; however, an as-yet-unidentified motif in Anks family proteins also contributes to stable complex formation between these two proteins (20).
While the SAM domains of Anks family proteins are highly conserved among all isoforms, the function of this domain is not well understood. In the current study, we identified a potential role for SAM domains in EphA signaling. We showed that while the ubiquitin ligase c-Cbl mediates the ubiquitination and degradation of EphA8 upon ligand binding, the SAM domains of Anks family proteins associate with ubiquitinated EphA8 receptor and are critically involved in inhibiting the degradation of EphA2 and EphA8 receptors. These results suggest that the fine-tuning of EphA RTK signaling is regulated by a delicate balance between the activity of c-Cbl E3 ligase and Anks family proteins.
MATERIALS AND METHODS
Plasmids, reagents, and antibodies.
The pcDNA3 expression vectors for EphA8, AIDA-1b, and Odin have been previously described (20). Expression vectors for Ub and c-Cbl were provided by Keun Il Kim (Sookmyung Women's University, Seoul, South Korea) and Pan Gil Suh (Pohang University of Science and Technology, Pohang, South Korea), respectively. The expression vector for EphA8-Ub was constructed by inserting a PCR-amplified fragment encoding Ub (76 residues) at the C terminus of the mouse EphA8 cDNA sequence.
The yeast vectors for EphA8-TKD and -JM have been previously described (20). The other yeast bait vectors were constructed by subcloning PCR-amplified DNA fragments encoding the following amino acid residues into pBHA: for AIDA1-SAM, residues 808 to 963 of human AIDA-1b; for AIDA1-Ank, residues 46 to 247 of human AIDA-1b; for AIDA1-SAM1, residues 814 to 877 of human AIDA-1b; for AIDA1-SAM2, residues 885 to 944 of human AIDA-1b; for Odin-SAM, residues 714 to 845 of mouse Odin; and for EphA8-SAM, residues 926 to 991 of mouse ephA8. UBC42-1 and UBC45-1 are two different yeast clones identified by screening a human fetal brain cDNA library. UBC42-1 contained amino acid residues 158 to 685 of human UBC (GenBank accession no. NM_021009), whereas UBC45-1 contained a single Ub repeat corresponding to residues 616 to 685 of human UBC. For the glutathione S-transferase (GST) fusion constructs, the same regions described for the yeast bait vectors were subcloned into pGEX-5X-1. To construct the SAM domain deletion mutant of Odin, we used an overlapping PCR method to delete residues 687 to 874 of mouse Odin and subcloned the amplified sequence into the full-length Odin cDNA. Oligonucleotide sequences of the primers used in this study are available upon request.
Wild-type ubiquitin, Ub-agarose, cycloheximide, and MG132 were purchased from Sigma-Aldrich Chemical Co. Bafilomycin was purchased from Calbiochem. The human Odin small interfering RNA (siRNA) was previously described (20). Purified E1 (Ub-activating enzyme) and E2 (Ub-conjugating enzyme 6) were purchased from Calbiochem. The anti-Odin antibody was purchased from Calbiochem. The anti-AIDA and horseradish peroxidase-conjugated anti-rabbit IgG antibodies were from Zymed. The monoclonal anti-pTyr antibody (4G10) was obtained from Upstate. Polyclonal rabbit anti-Cbl, monoclonal anti-Ub, and anti-EphA2 antibodies were purchased from Santa Cruz Biotechnology. Antiactin and horseradish peroxidase-conjugated goat anti-mouse IgG antibodies were obtained from Sigma-Aldrich Chemical Co. Goat anti-human IgG was acquired from Jackson ImmunoResearch Laboratories, Inc.
Yeast two-hybrid assay, cell culture, cell transfection, and cell migration assays.
Yeast two-hybrid screening of a human cDNA library was performed essentially as described previously (20). Both HEK293T cells and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), as previously described (20). Inhibitors were diluted to the following concentrations in dimethyl sulfoxide: 10 mM for MG132 (Z-Leu-Leu-Leu-al) and 0.5 mM for bafilomycin. Inhibitors or vehicle was added to culture media at a concentration of 0.1% (vol/vol).
Transwell migration assays were performed essentially as described previously, except that cells were allowed to migrate for 4 h (20). For wound-healing assays, 1 × 105 cells were seeded onto culture dishes (24 wells), grown for 2 days, and wounded by scratching with a sterile micropipette tip (11). After wounding, cells were treated with 5 μg/ml preclustered ephrin-A5-Fc or control Fc and then allowed to migrate for 6 h. Images were captured at 0 and 6 h after wounding using a DP70 (Olympus) camera attached to an IX71 inverted microscope (Olympus).
Immunoprecipitation, WB, and in vitro ubiquitination assays.
Immunoprecipitation and Western blotting (WB) were performed essentially as described previously (20). To detect ubiquitinated proteins, we immediately lysed cells in boiling lysis buffer (2% SDS, 1 mM EDTA, and 50 mM NaF supplemented with phosphatase inhibitor cocktail II and protease inhibitor cocktail). After being boiled for 10 min, the lysates were diluted with 4 volumes of dilution buffer (2.5% Triton X-100, 12.5 mM Tris [pH 7.5], 187.5 mM NaCl, phosphatase inhibitor cocktail II, and protease inhibitor cocktail) and precleared by centrifugation. For Ub-agarose pulldown assays, 500 μl of cell extract was incubated with free ubiquitin in ubiquitin buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2) for 30 min at 4°C and then incubated with 40 μl Ub-agarose (Sigma-Aldrich) for 60 min. The mixture was pelleted, washed three times in ubiquitin buffer, and then analyzed by WB. GST pulldown assays were performed essentially as described previously (20).
For in vitro ubiquitination assays, cell lysates were prepared using a different lysis buffer (0.5% Igepal CA-630 [Sigma], 20 mM HEPES [pH 7.2], 50 mM sodium fluoride, 1 mM sodium vanadate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride). Cell lysate (approximately 2 mg) was subjected to immunoprecipitation using an anti-EphA8 antibody, and immune complexes were successively washed with phosphate-buffered saline (PBS) two times and reaction buffer (20 mM HEPES [pH 7.2], 10 mM MgCl2, 1 mM ATP, 1 mM dithiothreitol) two times. Cell extracts containing c-Cbl E3-ligase were prepared by sonication (two cycles of 30 s each) in a different buffer (20 mM HEPES [pH 7.2], 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol, 20 μM MG132, and the protease inhibitors described above). Ubiquitination reaction mixtures (150 μl) contained EphA8 immune complexes and 300 μg of Cbl cell extract in reaction buffer supplemented with 30 mM creatine phosphate, 0.1 mg/ml creatine kinase, 25 μM MG132, and 7.5 μg of GST-Ub. Reaction mixtures were incubated at 30°C for 2 h. Immune complex beads were then washed with 0.5% Igepal CA-630 in PBS three times and analyzed by WB.
Generation of Odin−/− mice and isolation of mouse embryonic fibroblasts (MEFs).
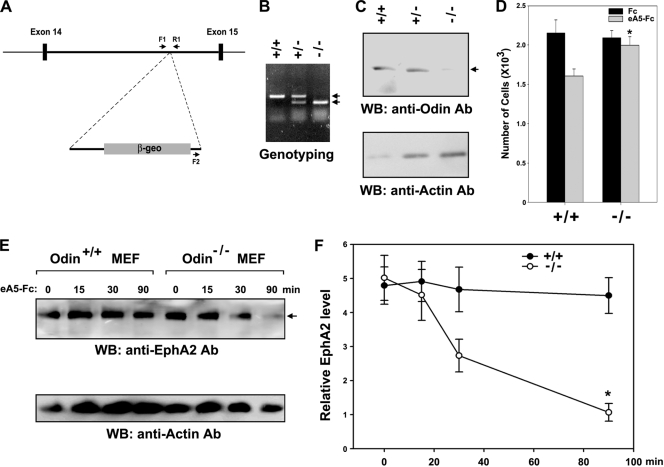
Embryonic stem (ES) cells harboring a gene trap vector insert (pGT0lxr) in the Odin locus (cell line ID CF0537) were purchased from the Mutant Mouse Regional Resource Center, and the location of the gene trap insertion was determined by sequence analysis. The ES cells were microinjected into C57BL/6 blastocysts, which were then transferred into foster mother (ICR) females to generate chimeric animals. Chimeric males were mated with 129/SvJ females, and F1 agouti pups were analyzed for the presence of the transgene by PCR analysis of tail genomic DNA using the following primers to detect mutant and wild-type Odin: 5′-TGAAGGCACATGACCCTGAG-3′ (F1), 5′-ATGTCATAGCTGTTTCCTGT-3′ (F2), and 5′-ACAGCGTTTGCATCTTGCTG-3′ (R1). All mice were generated and maintained in accordance with institutional guidelines approved by the Sookmyung Women's University Animal Care and Use Committee.
For isolation of MEF cells, embryos (13.5 days) were minced and incubated in 0.25% trypsin-EDTA for 20 min. DMEM supplemented with 7% FBS, penicillin, and streptomycin was added to the cell suspension. The cell pellet was then resuspended in DMEM supplemented with 7% FBS, grown in this medium, and passaged by trypsinization. Third-passage cells were used for the cell migration and protein stability assays.
RESULTS
The SAM domains of Anks family proteins bind to ubiquitin.
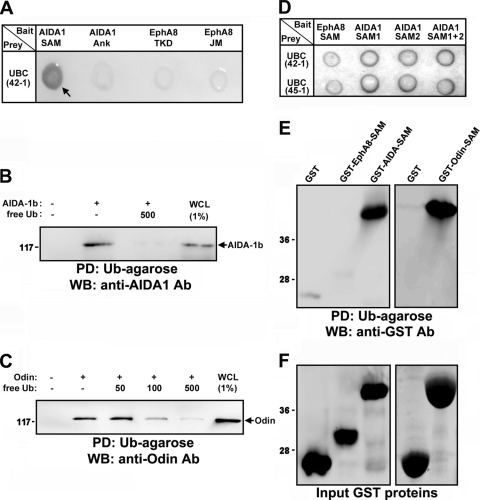
The SAM domains of Anks proteins are well conserved among isoforms, suggesting that SAM domains are critically involved in EphA-mediated signaling pathways. We carried out a yeast two-hybrid screen for binding partners of Anks family SAM domains using the SAM domains of AIDA-1b as bait. This screen identified ubiquitin as a potential binding partner. As shown in Fig. 1A, ubiquitin specifically bound to the SAM domains of AIDA-1 in the yeast two-hybrid assay (lane 1), whereas it did not bind to other motifs present in AIDA-1 or EphA8 (lanes 2 to 4). Transient transfection of HEK293T cells followed by in vitro protein association (pulldown) assays using ubiquitin (Ub)-agarose revealed that both AIDA-1b and Odin bound to ubiquitin (Fig. 1B and C, lane 2), and free ubiquitin effectively competed with Ub-agarose for binding to Anks family proteins (Fig. 1B, lane 3, and C, lanes 3 to 5). The isolated SAM domains of AIDA-1b (SAM1 and SAM2) readily bound to ubiquitin in the yeast two-hybrid assay (Fig. 1D, lanes 2 and 3), whereas the SAM domain of EphA8 did not (Fig. 1D, lane 1). Similarly, in a glutathione S-transferase (GST) pulldown assay, GST fusion proteins of the SAM domains of AIDA-1b or Odin bound to ubiquitin (Fig. 1E, lanes 3 and 5), whereas the SAM domain of EphA8 did not (Fig. 1E, lane 2). AIDA-1b SAM1 bound to ubiquitin to a similar extent as the SAM1 plus SAM2 domains of AIDA-1b or Odin (data not shown). We were unable to analyze isolated SAM2, as it was insoluble when expressed alone (data not shown). These results suggest that the SAM domains of Anks family proteins contain unique structural features that are critical for binding to ubiquitin.
FIG. 1.
Yeast two-hybrid screening showing that ubiquitin interacts with AIDA1-SAM domains. (A) Analysis of the binding of AIDA1-SAM domains to ubiquitin by X-Gal staining. As negative controls, the ankyrin (Ank) repeats of AIDA-1b, the tyrosine kinase domain (TKD) of EphA8, and the JM domain of EphA8 served as bait. UBC42-1 is a clone identified through library screening that contained part of human ubiquitin chain gene (UBC). (B) Pulldown (PD) assay using Ub-agarose beads to probe lysates of HEK293 cells expressing full-length AIDA-1b. Free ubiquitin (500 nM) was used as a competitor for binding to Ub-agarose. WB analysis was carried out to determine the level of AIDA1 bound to Ub-agarose. Ab, antibody. (C) Experiments were performed as described for panel B, except that cell lysates from Odin-expressing cells were used. WCL, whole-cell lysate. (D) A yeast two-hybrid assay was performed as described for panel A. SAM1 and SAM2 indicate the isolated SAM domains of AIDA-1. UBC45-1 represents a second UBC clone identified through library screening. (E) Ub-agarose pulldown assay of SAM domains fused to GST. (F) The samples from panel E were examined by Coomassie blue staining to determine the levels of GST fusion protein.
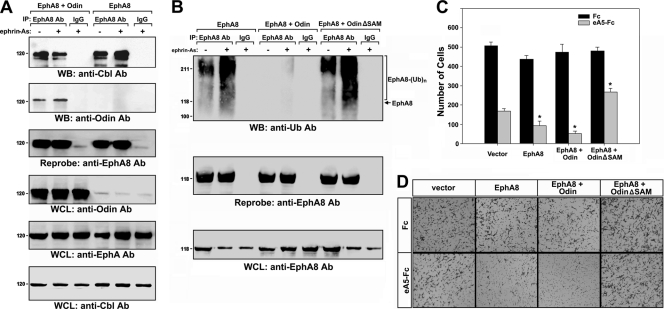
Ubiquitination and degradation of EphA8 are mediated by Cbl E3 ligase.
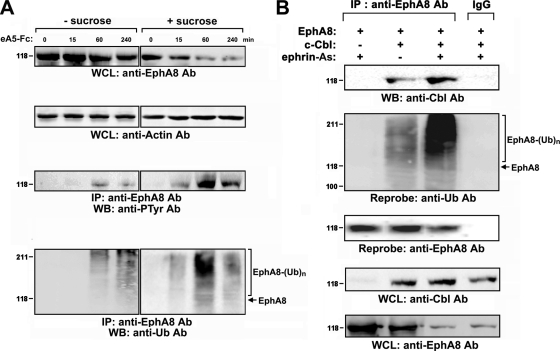
Recent studies have shed new light on ligand-induced ubiquitination and degradation of Eph receptors (6, 14). To determine whether the SAM domains of Anks family proteins bind to ubiquitinated EphA8 with stronger affinity, we examined EphA8 ubiquitination and degradation in response to ephrin-A5. EphA8 ubiquitination and degradation were not detected until 1 h after ligand stimulation (Fig. 2A, first and fourth panels, lanes 1 to 4). At 4 h after ligand treatment, a significant level of EphA8 was reduced, together with the increased level of ubiquitinated EphA8 (Fig. 2A, first and fourth panels, lane 4). This ligand-induced degradation of EphA8 was inhibited by MG132 or bafilomycin (data not shown), suggesting that both proteasomal and lysosomal mechanisms of degradation are involved in the regulation of intact EphA8. It is not clear why the ligand-induced EphA8 degradation and ubiquitination are detectable only after long-term treatment with ligand. One possibility is that rapid endocytosis of EphA8 in response to ephrin-A5 stimulation interferes with ubiquitination and degradation of EphA8 at the cell surface. Consistent with this possibility, when endocytosis of ligand-bound EphA8 complexes was blocked by sucrose, both the degradation and ubiquitination of EphA8 were readily detected within 60 min (Fig. 2A, first and fourth panels, lanes 5 to 8). In addition, tyrosine phosphorylation of EphA8 correlated well with the extent of ubiquitination (Fig. 2A, third and fourth panels, lanes 5 to 8). These results suggest that EphA8 at the cell surface is more readily tyrosine phosphorylated in response to ligand stimulation, making it more susceptible to being targeted by the ubiquitination machinery. Furthermore, the proteasomal inhibitor MG132 efficiently blocked the ligand-induced degradation of EphA8 following sucrose treatment (data not shown), suggesting that ubiquitinated EphA8 at the cell surface is degraded predominantly by a proteosomal mechanism.
FIG. 2.
Cbl E3 ligase mediates ubiquitination and degradation of EphA8. (A) HEK293T cells were transfected with an expression vector for EphA8 for 24 h and then treated with sucrose for 30 min. Cells were stimulated with preclustered ephrin-A5-Fc (2 μg/ml) for the indicated times. Cell extracts were subjected to immunoprecipitation using an anti-EphA8 antibody, and immune complexes were analyzed by WB using antiphosphotyrosine (PTyr) or antiubiquitin (Ub) antibodies, as indicated. Whole-cell lysates (WCL) were directly examined by WB to determine the levels of the indicated antigens. (B) HEK293T cells were cotransfected with an expression vector for EphA8 (1 μg) and an expression vector for c-Cbl (2 μg) and then treated with NIH 3T3 cells (express endogenous ephrin-A ligands) for 30 min.
Next, we performed in vitro ubiquitination assays to investigate the identity of the ubiquitin ligase involved in the ubiquitination and degradation of EphA8. Incubation of EphA8 with c-Cbl induced pronounced ubiquitination of EphA8 (data not shown). Ligand-induced degradation of EphA8 was more rapid in the presence of increasing levels of c-Cbl (data now shown). In cells that coexpressed ectopic c-Cbl and EphA8, ligand stimulation promoted the association of c-Cbl with EphA8 (Fig. 2B, first panel, lanes 2 and 3) and the tyrosine phosphorylation of c-Cbl (data not shown). Consistent with these observations, ubiquitination of EphA8 was readily detected in c-Cbl-overexpressing cells and was enhanced by ligand stimulation (Fig. 2B, second panel, lanes 2 and 3). These results suggest that c-Cbl plays a critical role in the ubiquitination and degradation of EphA8 in response to ligand stimulation.
The SAM domains of Anks family proteins interact with the ubiquitinated EphA8 receptor.
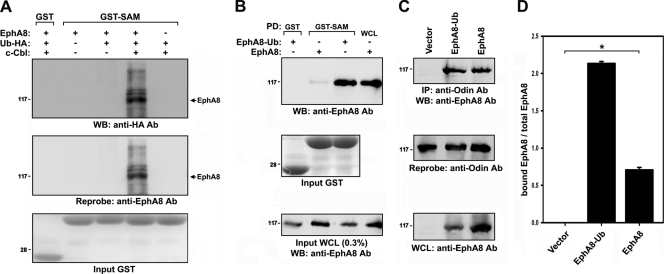
To determine whether the SAM domains of Anks family proteins associate with ubiquitinated EphA8, we performed a GST pulldown assay using cell extracts from Cbl-overexpressing cells. Cells were also transfected with an expression vector for hemagglutinin-tagged ubiquitin (HA-Ub) to label ubiquitinated proteins. The coexpression of c-Cbl strongly induced the ubiquitination of EphA8, and ubiquitinated receptors readily associated with the SAM domains of AIDA-1b (Fig. 3A, lane 4). To determine whether the SAM domains of Anks family proteins associate with ubiquitinated EphA8, the EphA8 receptor was tagged with a single ubiquitin moiety at its C terminus (EphA8-Ub). The expression level of EphA8-Ub was more than 5-fold lower than that of wild-type EphA8 (data not shown). Furthermore, in a pulldown assay, the affinity of the SAM domains of AIDA-1b for EphA8-Ub was greater than that for wild-type EphA8 (Fig. 3B, top panel, lanes 2 and 3).
FIG. 3.
The SAM domains of Anks family proteins associate with the ubiquitinated EphA8 receptor. (A) GST pulldown assay of ubiquitinated EphA8 using AIDA-1 SAM domains. Transfections were performed as described for Fig. 2B, except that an expression vector for HA-tagged ubiquitin (1 μg) was used. The arrow indicates the position of nonubiquitinated EphA8. (B) GST pulldown assay of EphA8-Ub using AIDA-1 SAM domains. Note that expression of EphA8-Ub was lower than that of wild-type EphA8 (third panel). (C) HEK293T cells were transfected with the indicated expression constructs. Cell lysates were subjected to immunoprecipitation using an anti-Odin antibody, and immune complexes were analyzed by WB using an anti-EphA8 antibody (top panel). The blot was reprobed to determine the levels of endogenous Odin (second panel). WCL were directly probed by WB to determine the levels of EphA8 and EphA8-Ub (third panel). (D) The data in panel C were quantitated, and the amount of EphA8 in association with Odin (panel C, top panel) was normalized to total EphA8 (panel C, third panel). Data represents the means ± standard errors (SE) from three independent experiments. EphA8-Ub associated with Odin more strongly than wild-type EphA8 (*, P < 0.001 by analysis of variance [ANOVA]).
AIDA-1b was expressed predominantly in the adult mouse brain (data not shown). In contrast, Odin was abundantly expressed in the embryonic brain and in many types of mammalian cells that also expressed Eph receptors (data not shown). These results suggest that Odin is a physiologically relevant partner of EphA8. We examined the association of EphA8-Ub with full-length, endogenous Odin in HEK293T cells. Similar to earlier results (20), wild-type EphA8 specifically coimmunoprecipitated with Odin (Fig. 3C, first panel, lane 3). Of note, while the expression level of ectopic EphA8-Ub was much lower than that of wild-type EphA8 in HEK293T cells (Fig. 3C, third panel, lane 2), a significant amount of EphA8-Ub coimmunoprecipitated with Odin (first panel, lane 2), indicating that EphA8-Ub has a stronger association with Odin (at least 4-fold stronger) than wild-type EphA8 (Fig. 3D). These results demonstrate that the SAM domains of Anks family proteins are critically involved in the recognition of ubiquitinated EphA8.
The SAM domains of Odin are critical for inhibiting the degradation of EphA8.
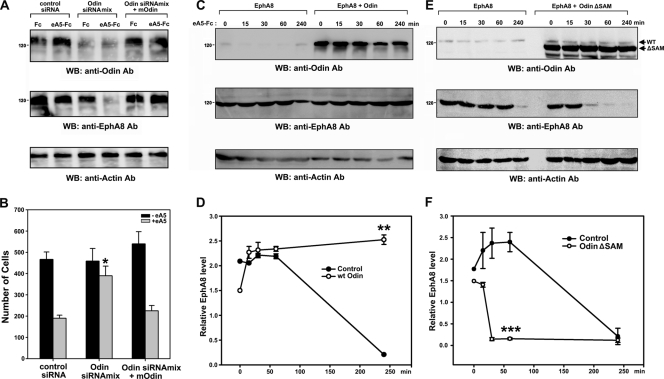
To explore the biological significance of Anks-SAM domain binding to ubiquitinated EphA8, we transfected HEK293T cells expressing EphA8 with Odin siRNAs to downregulate endogenous Odin expression. Following transfection, cells were treated with ephrin-A5 and cycloheximide, an inhibitor of de novo protein synthesis, for 1 h. As shown in Fig. 4A, ligand treatment induced a rapid decay of EphA8 within 1 h in cells in which Odin expression was downregulated with SMART pool siRNAs (lane 4), suggesting that Odin protects the EphA8 receptor from ligand-induced degradation. Similar results were also observed for a specific Odin siRNA, which effectively downregulated Odin expression and attenuated EphA8-mediated inhibition of cell migration in response to ephrin-A5 stimulation (data not shown). Consistent with a previous publication (20), we also found that introduction of a mouse Odin rescue plasmid into siRNA-transfected cells was capable of restoring EphA8 stability and the cell migration-inhibitory effect of EphA8 after Odin knockdown (Fig. 4A and B, lanes 5 and 6).
FIG. 4.
The SAM domains of Odin are required to protect EphA8 from degradation. (A) HEK293T cells were transfected with an expression vector for EphA8 and then retransfected with control siRNA or human Odin siRNA (SMART pool). Mouse Odin cDNA expression vector was also cotransfected with human Odin siRNA for the rescue experiment. At 48 hours after transfection, cells were treated with cycloheximide for 30 min and then incubated with preclustered ephrin-A5-Fc for 1 h. WCL were analyzed by WB using antibodies specific for the indicated antigens. (B) Inhibition of cell migration by ephrin-A5-stimulated EphA8 requires Odin. HEK293T cells were transfected as described for panel A. Cells were then allowed to migrate toward the lower compartment of a Boyden chamber for 4 h. Data represent the mean ± SE from three independent experiments. Cell migration under different conditions was compared with that under the control siRNA-transfected and ephrin-A5-stimulated conditions (*, P < 0.001 by ANOVA). (C) HEK293T cells were cotransfected with an expression vector for EphA8 and a control or Odin expression vector for 48 h. Cells were treated with cycloheximide for 30 min and then stimulated with preclustered ephrin-A5-Fc for the indicated times. (D) The data in panel C were quantitated, and the levels of EphA8 were normalized to the actin content. Data represent the mean ± SE from three independent experiments. The x axis represents the stimulation time. **, P < 0.01 compared to cells expressing EphA8 alone at the 4-h time point (Student's t test). (E and F) Experiments were carried out as described for panels C and D, except that an expression vector for a SAM deletion mutant of Odin, not wild-type Odin, was used. ***, P < 0.01 compared to cells expressing EphA8 alone at the 1-h time point (Student's t test).
The role of Odin in the regulation of EphA8 stability was also investigated using transient-transfection assays. In the absence of Odin overexpression, EphA8 levels were constant up to 1 h after ephrin-A5 treatment and then decreased up to 5-fold at 4 h after treatment (Fig. 4C, second panel, lanes 1 to 5; Fig. 4D). The half-life of EphA8 was estimated to be approximately 2 h under these experimental conditions; however, the results were highly variable, so we did not include the 2-h and 3-h poststimulation time points (data not shown). In contrast, when Odin was overexpressed, the levels of EphA8 were constant for up to 4 h after stimulation (Fig. 4C, second panel, lanes 6 to 10; Fig. 4D). To determine whether the SAM domains of Odin are involved in regulating the stability of EphA8, we constructed a SAM domain deletion mutant of Odin. The SAM domain deletion mutant of Odin specifically coimmunoprecipitated with EphA8 (data not shown). Similar to earlier results, in cells that expressed EphA8 alone, EphA8 decay was not detected until after 1 h of ephrin-A5 treatment (Fig. 4E, second panel, lanes 1 to 5; Fig. 4F). Strikingly, overexpression of the SAM domain deletion mutant of Odin resulted in a rapid decay of EphA8 upon ligand stimulation, suggesting that this mutant functions in a dominant-negative manner (Fig. 4E, second panel, lanes 6 to 10; Fig. 4F). These results strongly suggest that Odin is critically involved in strong binding to ubiquitinated EphA8 receptor and in preventing receptor degradation and that this activity is mediated by the Odin SAM domains.
Odin interferes with EphA8 ubiquitination and prolongs receptor signaling.
Next, we investigated whether Odin affects the binding of Cbl to EphA8. As shown in Fig. 5A, in the presence of endogenous Odin, the association of Cbl with EphA8 increased upon ligand stimulation (first panel, lanes 4 and 5). In contrast, the association of Cbl with EphA8 in response to ligand stimulation was significantly reduced when Odin was overexpressed (first panel, lanes 1 and 2), suggesting that the strong association between Odin and EphA8 interferes with access or binding of Cbl to this complex. The overexpression of Odin also attenuated the ubiquitination of EphA8 in response to ligand stimulation (Fig. 5B, top panel, lanes 4 and 5). On the other hand, overexpression of the SAM domain deletion mutant of Odin enhanced ubiquitination of EphA8 in response to ligand stimulation (Fig. 5B, top panel, lanes 7 and 8). These results suggest that the SAM domain-mediated binding of Odin to EphA8 interferes with the ubiquitination of EphA8.
FIG. 5.
The SAM domain of Odin interferes with the ubiquitination of EphA8. (A) HEK293T cells were transfected with an expression vector for EphA8 together with a control or Odin expression vector for 48 h and then treated with NIH 3T3 cells for 4 h. Cell lysates were subjected to immunoprecipitation using an anti-EphA8 antibody, and immune complexes were analyzed by WB using antibodies specific for Cbl (top panel) or Odin (second panel). Increasing the amount of cell lysate used for immunoprecipitation (lanes 4 and 5) resulted in detectable EphA8 in complex with endogenous Odin (data not shown). WCL were also analyzed by WB using antibodies for the indicated antigens. (B) Experiments were performed as described for Fig. 5A, except that immune complexes were analyzed by WB using an anti-Ub antibody. (C and D) Inhibition of cell migration by ephrin-A5-stimulated EphA8 requires the SAM domains of Odin. HEK293T cells were transfected as described for panel A. Cells were then allowed to migrate toward the lower compartment of a Boyden chamber for 4 h. Data represents the means ± SE from three independent experiments. The inhibitory effect of ephrin-A5 on cell migration under different conditions was compared with that in the control vector-transfected condition (*, P < 0.01 by ANOVA).
To determine the effect of Odin on EphA8-mediated signaling pathways, we transiently transfected HEK293T cells with expression vectors for EphA8 and Odin. As seen in Fig. 5C, control (vector alone) HEK293T cells exhibited reduced cell migration (approximately 65%) in response to ephrin-A5 (bars 1 and 2), most likely through the activity of endogenous Eph receptors. When cells were transfected with an expression vector for EphA8, the inhibitory effect of ephrin-A5 on cell migration increased to 80% (bars 3 and 4). In cells that coexpressed both EphA8 and Odin, the inhibitory effect on cell migration was increased even further to 90% (bars 5 and 6). On the other hand, coexpression of the SAM domain deletion mutant of Odin significantly decreased the inhibitory effect of ephrin-A5 on cell migration (approximately 40%) (bars 7 and 8). A similar trend was obtained in a wound-healing assay (data not shown). These results strongly suggest that Odin SAM domains positively regulate signaling from the EphA8 receptor by promoting receptor stability.
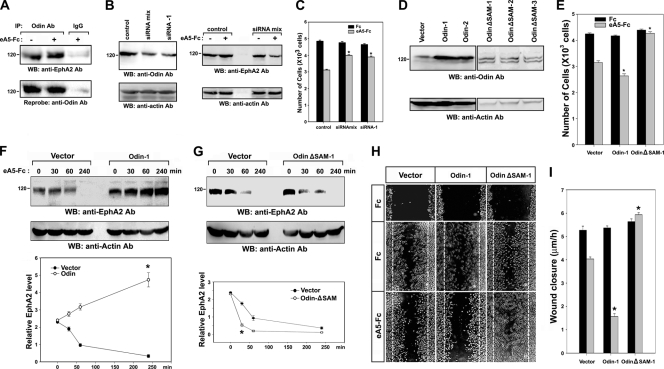
The SAM domains of Odin are critical for inhibiting the degradation of EphA2 in MDA-MB-231 cells.
Next, MDA-MB-231 cultured human breast carcinoma cells were used to investigate whether Odin plays a role in regulating the stability of other EphA receptors and their signaling effects. Western blot analysis revealed that both Odin and EphA2 were endogenously expressed in this cell line, and immunoprecipitations performed on extracts from this cell line using an anti-Odin antibody, but not a control antibody, specifically recovered EphA2, irrespective of ephrin-A5 treatment (Fig. 6A). To verify that the EphA2-Odin interaction is critical for EphA2 stability, we knocked down Odin in MDA-MB-231 cells through transfection of SMART pool siRNAs or a specific siRNA. WB analysis revealed that endogenous Odin expression was effectively downregulated (Fig. 6B, left panels). As expected, ligand treatment induced a rapid decay of EphA2 within 30 min in cells in which Odin expression was downregulated by SMART pool siRNAs (Fig. 6B, right panels, lane 4), consistent with the ability of Odin to protect the EphA8 receptor from ligand-induced degradation in HEK293T cells (Fig. 4A). Similar findings were also observed using a specific Odin siRNA (data not shown). Moreover, like HEK293T cells, MDA-MB-231 cells exhibited reduced cell migration in response to ephrin-A5 (Fig. 6C, bars 1 and 2), and Odin siRNAs significantly attenuated the inhibitory effect of ephrin-A5 on migration of these cells (bars 3 to 6).
FIG. 6.
The SAM domains of Odin are required to protect EphA2 from degradation in MDA-MB-231 cells. (A) MDA-MB-231 cell lysates were subjected to immunoprecipitation using an anti-Odin antibody, and immune complexes were analyzed by WB using an anti-EphA2 antibody (top panel). The blot was reprobed to determine the levels of endogenous Odin (second panel). (B) MDA-MB-231 cells were transfected with a control siRNA, SMART pool siRNAs (siRNA mix), or a specific siRNA. At 48 hours after transfection, 20% of cells were directly analyzed by WB using antibodies specific for the indicated antigens (left panels), whereas the rest of cells were treated with cycloheximide for 30 min and then incubated with preclustered ephrin-A5-Fc for 1 h (right panels). (C) MDA-MB-231 cells were transfected as described for panel B. Cells were then allowed to migrate toward the lower compartment of a Boyden chamber for 4 h. Data represents the means ± SE from three independent experiments. Cell migration under different conditions was compared with that under the control siRNA-transfected and ephrin-A5-stimulated condition (*, P < 0.001 by ANOVA). (D) MDA-MB-231 cells were stably transfected with an expression vector for wild-type Odin or a SAM deletion mutant of Odin. Cell lysates were directly analyzed by WB using antibodies specific for Odin (top panels) or actin (bottom panels). (E) Cell migration experiments were performed as described for panel C. Cell migration under different conditions was compared with that in the vector-transfected and ephrin-A5-stimulated condition (*, P < 0.01 by ANOVA). (F) Experiments were performed as described for Fig. 4C, except MDA-MB-231 cell lysates were analyzed by WB using an anti-EphA2 antibody. *, P < 0.01 compared to cells expressing vector alone at the 4-h time point (Student's t test). (G) Experiments were performed as described for Fig. 4E, except MDA-MB-231 cell lysates were analyzed by WB using an anti-EphA2 antibody. *, P < 0.05 compared to cells expressing vector alone at the 30-min time point (Student's t test). (H and I) Wound-healing assays were carried out using cells that were prepared as described for panels F and G. The wound closure rate (y axis) was calculated from the average distance that cells at the wound edge migrated from their starting point over a period of 6 h. Data represent the means ± SE from three independent experiments. Wound closure rates under different conditions were compared with those in the vector-transfected and ephrin-A5-stimulated conditions (*, P < 0.01 by ANOVA).
To further investigate the role of Odin in the regulation of EphA2 stability, we generated MDA-MB-231 cells stably expressing wild-type Odin or the SAM domain deletion mutant of Odin (Fig. 6D). Both cell migration assays and wound-healing assays revealed that wild-type Odin promoted the inhibitory effect of ephrin-A5 on cell migration, whereas expression of the SAM domain deletion mutant of Odin attenuated this inhibitory effect (Fig. 6E, H, and I). In control MDA-MB-231 cells treated with cycloheximide, EphA2 levels remained constant up to 30 min after ephrin-A5 treatment and then decreased up to 5-fold at 4 h after treatment (Fig. 6F and G, lanes 1 to 4). The half-life of EphA2 was estimated to be approximately 1 h under these experimental conditions. When Odin was overexpressed in MDA-MB-231 cells, the EphA2 levels were rather increased up to 4 h poststimulation (Fig. 6F, lanes 6 to 9). In contrast, expression of the SAM domain deletion mutant of Odin resulted in a rapid decay of EphA2 upon ligand stimulation (Fig. 6G, lanes 6 to 9), similar to its effect on the stability of EphA8 as shown in Fig. 4E. These results were reproducibly observed in different cell lines stably expressing wild-type Odin or the SAM domain deletion mutant of Odin (data not shown). Taken together, these results strongly suggest that Odin is critically involved in preventing EphA2 receptor degradation and that this activity is mediated by the Odin SAM domains.
The level of Odin is critical for regulating EphA2 stability in MEFs.
To further investigate the physiological role of Odin in controlling the stability of EphA receptors, we generated Odin-deficient mice carrying a gene trap insertion in intron 14 of the Odin gene (Fig. 7A). The mice were genotyped by PCR with primers located at the 3′ side of the gene trap and either side of the vector insertion site in intron 14 (Fig. 7B). In general, survival, body size, sex ratio, and life expectancy were indistinguishable between knockout mice and their wild-type littermates (data not shown). We cultured 13.5-day MEFs to analyze the effect of Odin deficiency on EphA-mediated cell migration and EphA protein stability. As shown in Fig. 7C, WB analysis revealed that Odin was barely detectable in Odin−/− mice compared with Odin+/+ and Odin+/− mice (lane 3). Nevertheless, no abnormal morphology or proliferative behaviors were observed in Odin−/− MEF cells (data not shown). WB analysis using various EphA antibodies revealed that EphA2 was highly expressed in MEFs (Fig. 7E, top panel) and that EphA2 was tightly associated with Odin in MEFs irrespective of ephrin-A5 stimulation (data not shown). Consistent with Odin knockdown results in MDA-MD-231 cells, Odin deficiency in MEFs attenuated the inhibitory effect of ephrin-A5 on cell migration (Fig. 7D). In addition, protein stability assays using cycloheximide revealed that EphA2 levels remained constant up to 90 min after stimulation in Odin+/+ MEF cells, whereas the absence of Odin resulted in a rapid decay of EphA2 upon ligand stimulation in Odin−/− MEF cells (Fig. 7E and F). Taken together, these results strongly support our hypothesis that Odin plays an important role in promoting the stability of EphA receptors.
FIG. 7.
Odin is required to protect EphA2 from degradation in MEFs. (A) Schematic representation of the vector integration site within the 14th intron of the mouse Odin gene in the ES clone CF0537. Exons are shown as black boxes, and the locations of the PCR primers (F1, F2, and R1) used in genotyping are indicated. (B) A typical genotyping analysis showing the 479-bp PCR fragment for the wild-type allele and the 310-bp product for the mutant allele. (C) Fibroblasts were derived from 13.5-day embryos, and the expression of Odin was analyzed by WB. (D) MEFs derived from Odin−/− knockout and Odin+/+ wild-type embryos were allowed to migrate toward the lower compartment of a Boyden chamber for 2 h. Data represents the means ± SE from three independent experiments. *, P < 0.01 compared to Odin+/+ cells (Student's t test). (E and F) Experiments were performed as described for Fig. 4C and E, except MEF lysates were analyzed by WB using an anti-EphA2 antibody. *, P < 0.01 compared to Odin+/+ cells at the 90-min time point (Student's t test).
DISCUSSION
Here we report that EphA8-mediated signaling is modulated by the opposing actions of c-Cbl and Anks family proteins. Ligand stimulation of EphA8 leads to the activation of c-Cbl, thereby inducing the ubiquitination and degradation of EphA8. EphA8 also interacts with Odin, a member of the Anks family of proteins. Odin not only binds to EphA8 through its PTB domain (20) but also binds ubiquitinated EphA8 receptors with a higher affinity through its SAM domains, thereby protecting EphA8 from degradation. More importantly, Odin plays an important role in protecting EphA2 from degradation in response to ephrin-A5 stimulation in MDA-MB-231 cells and MEFs. Therefore, our findings suggest that the delicate balance between c-Cbl E3 ligase activity and Anks family proteins is a critical determinant of the levels of active EphA receptors within a cell.
SAM domains are typically found in large multidomain proteins and have been shown to have diverse functions (18). Unlike some protein modules, SAM domains are not easily categorized into subgroups, because close homologues can have different functions. Our finding that the SAM domains of Anks family proteins bind to ubiquitin points to another novel binding property of SAM domains but also to a new biological function for these motifs in regulating protein degradation and stability. The number of ubiquitin-binding domains has expanded over recent years and now is at least 16 domains, including UBM and UIM (8). Similar to other ubiquitin-binding domains, SAM domains contain a high α-helical content, based on secondary structural predictions. Whether SAM domains are similar to other ubiquitin-binding domains and interact with a single region of ubiquitin (i.e., the Ile 44 hydrophobic patch) remains to be determined. Although the current study centers on the biological significance of the Anks-SAM domains in the context of EphA receptors, investigating whether these domains play a role in interacting with other ubiquitinated proteins such as epidermal growth factor (EGF) and PDGF receptors would also be interesting.
What mechanism underlies the ability of the Odin SAM domains to prevent EphA8 from undergoing degradation after ephrin-A5 stimulation? Our intensive biochemical studies with HEK293T cells suggest the following scenario. Upon ligand binding, Cbl E3 ligase rapidly induces the ubiquitination of the EphA8 receptor at the cell surface, leading to rapid degradation of EphA8. On the other hand, Odin targets and binds to ubiquitinated EphA8 more strongly than nonubiquitinated EphA8 through its SAM domains, thereby inhibiting additional Cbl-mediated ubiquitination processes. It is possible that the association of ubiquitinated EphA8 with Odin interferes with access to the complex by Cbl or decreases the binding affinity of Cbl for EphA8. Alternatively, Odin may somehow interact with deubiquitination machinery to promote the stability of EphA8. Further experiments will be needed to dissect the role of Odin in inhibiting Cbl-mediated ubiquitination of EphA8.
The next important question is whether the SAM domains of Odin also regulate the stability of other EphA receptors via a similar mechanism. We found that Odin is constitutively associated with EphA2 and EphA4 mainly through its PTB domain (unpublished results). We also observed that EphA2 is ubiquitinated in response to ephrin-A5 stimulation in MDA-MB-231 cells and that the SAM domains of Odin associate with ubiquitinated EphA2 in these cells (unpublished results). However, the level of ubiquitinated EphA2 in MDA-MB-231 cells or MEFs appears to be very low due to the unstable properties of these species, and we had technical difficulties investigating whether expression of wild-type Odin or the SAM domain deletion Odin mutant significantly influences the ubiquitination of EphA2 in these cells. Nevertheless, in both MDA-MB-231 cells and MEFs, the stability of EphA2 after ephrin-A5 stimulation was strongly influenced by the level of Odin. Moreover, the SAM domains of Odin were shown to be critical for inhibiting the degradation of EphA2 in MDA-MB-231 cells. These results support our hypothesis that the SAM domains of Odin play a role in maintaining the stability of multiple EphA receptors, possibly through binding to ubiquitin moieties. However, we cannot rule out the possibility that Odin regulates the stability of EphA receptors through a different mechanism, such as interfering with the activity of proteases. Interestingly, EphB2 can be cleaved by presenilin-dependent γ-secretase activity (14). AIDA-1 has also been implicated in the reduction of AβPP processing through the inhibition of γ-secretase activity (7). In this respect, it will be interesting to investigate whether Anks family proteins also regulate the proteolytic system involved in the processing of Eph receptors.
Previous studies suggest that the activated EphA8 receptor induces inhibition of cell migration through Odin-dependent signaling functions. Consistent with this finding, the activated EphA2 receptor also induces inhibition of cell migration in an Odin-dependent manner in both MDA-MB-231 cells and MEFs. It is not clear why Odin is required for the inhibitory effect of ephrin-A5 on the migration of EphA-expressing cells. One possibility is that Odin is the key adaptor protein that regulates the EphA receptor signaling pathway leading to inhibition of cell migration by interacting with other signaling components downstream of EphA receptors. However, we have not yet observed that Odin interacts with the well-known signaling proteins downstream of EphA receptors, such as small GTP-binding proteins and their exchange factors (unpublished results). Alternatively, Odin may maintain the level of EphA receptors by preventing further degradation of these receptors after ligand stimulation, in this way sustaining EphA-mediated signaling and leading to inhibition of cell migration. This model is more consistent with our current hypothesis that the SAM domains of Odin play a pivotal role in maintaining the stability of EphA receptors, although we cannot exclude the possibility that Odin is a key adaptor protein linking EphA receptors with other downstream signaling components.
To further address the physiological role of Odin in regulating the stability of EphA receptors, we generated Odin-deficient mice harboring a gene trap in intron 14 of the Odin gene. The gene trap leads to the fusion of the first 253 amino acids of Odin to LacZ. The resulting protein likely has no biological function because it lacks both the SAM and PTB domains. We have not observed any overt anatomical or behavioral phenotypes in Odin-deficient mice, which reach adulthood and are fertile. Nevertheless, our current study using MEFs from wild-type and Odin knockout mice strongly supports that the notion that the level of Odin plays a critical role in protecting EphA receptors from undergoing degradation after ligand stimulation. We further determined the developmental expression of Odin by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) histochemistry in mice heterozygous (+/−) and homozygous (−/−) for the targeted allele (unpublished results). Interestingly, we found that a significant level of Odin expression was observed in the cells lining the lateral ventricular region of the telencephalon. A recent study indicates that during cortical development, EphA and ephrin-A signaling plays an important role in regulating the lateral dispersion of migrating neurons originating from the proliferative units at the ventricular surface (21). This circumstantial evidence suggests that Odin may play a role in regulating radial neuronal cell migration to properly form the cortical columns, possibly by modulating the stability of EphA receptors. In addition, Odin knockout mice will be a useful in vivo model for exploring the function of Odin in well-known EphA signaling pathways, such as those involved in retinocollicular topographic map formation.
Acknowledgments
This work was supported by a grant (no. 2009-0083149) from the Korea Science and Engineering Foundation (KOSEF) and by a grant (no. 2009K001246) from the Brain Research Center of the 21st Century Frontier Research Program funded by the Ministry of Education, Science and Technology, Republic of Korea.
Footnotes
Published ahead of print on 25 January 2010.
REFERENCES
- 1.Arvanitis, D., and A. Davy. 2008. Eph/ephrin signaling: networks. Genes Dev. 22:416-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batlle, E., J. Bacani, H. Begthel, S. Jonkheer, A. Gregorieff, M. van de Born, N. Malats, E. Sancho, E. Boon, T. Pawson, S. Gallinger, S. Pals, and H. Clevers. 2005. EphB receptor activity suppresses colorectal cancer progression. Nature 435:1126-1130. [DOI] [PubMed] [Google Scholar]
- 3.Brantley-Sieders, D. M., G. Zhuang, D. Hicks, W. B. Fang, Y. Hwang, J. M. Cates, K. Coffman, D. Jackson, E. Bruckheimer, R. S. Muraoka-Cook, and J. Chen. 2008. The receptor tyrosine kinase EphA2 promotes mammary adenocarcinoma tumorigenesis and metastatic progression in mice by amplifying ErbB2 signaling. J. Clin. Invest. 118:64-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., G. Zhuang, L. Frieden, and W. Debinski. 2008. Eph receptors and Ephrins in cancer: common themes and controversies. Cancer Res. 68:10031-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, N., D. M. Brantley, H. Liu, Q. Lin, M. Enriquez, N. Gale, G. Yancopoulos, D. P. Cerretti, T. O. Daniel, and J. Chen. 2002. Blockade of EphA receptor tyrosine kinase activation inhibits vascular endothelial cell growth factor-induced angiogenesis. Mol. Cancer Res. 1:2-11. [PubMed] [Google Scholar]
- 6.Fasen, K., D. P. Cerretti, and U. Huynh-Do. 2008. Ligand binding induces Cbl-dependent EphB1 receptor degradation through the lysosomal pathway. Traffic 9:251-266. [DOI] [PubMed] [Google Scholar]
- 7.Ghersi, E., C. Noviello, and L. D'Adamio. 2004. Amyloid-beta protein precursor (AbetaPP) intracellular domain-associated protein-1 proteins bind to AbetaPP and modulate its processing in an isoform-specific manner. J. Biol. Chem. 279:49105-49112. [DOI] [PubMed] [Google Scholar]
- 8.Hicke, L., H. L. Schubert, and C. P. Hill. 2005. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6:610-621. [DOI] [PubMed] [Google Scholar]
- 9.Janes, P. W., N. Saha, W. A. Barton, M. V. Kolev, S. H. Wimmer-Kleikamp, E. Nievergall, C. P. Blobel, J. P. Himanen, M. Lackmann, and D. B. Nikolov. 2005. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123:291-304. [DOI] [PubMed] [Google Scholar]
- 10.Jordan, B. A., B. D. Fernholz, L. Khatri, and E. B. Ziff. 2007. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat. Neurosci. 10:427-435. [DOI] [PubMed] [Google Scholar]
- 11.Katoh, H., K. Hiramoto, and M. Negishi. 2006. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 119:56-65. [DOI] [PubMed] [Google Scholar]
- 12.Kiewlich, D., J. Zhang, C. Gross, W. Xia, B. Larsen, R. R. Cobb, S. Biroc, J. M. Gu, T. Sato, D. R. Light, T. Heitner, J. Willuda, D. Vogel, F. Monteclaro, A. Citkowicz, S. R. Roffler, and D. A. Zajchowski. 2006. Anti-EphA2 antibodies decrease EphA2 protein levels in murine CT26 colorectal and human MDA-231 breast tumors but do not inhibit tumor growth. Neoplasia 8:18-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konstantinova, I., G. Nikolova, M. Ohara-Imaizumi, P. Meda, T. Kucera, K. Zarbalis, W. Wurst, S. Nagamatsu, and E. Lammert. 2007. EphA-Ephrin-A-mediated beta cell communication regulates insulin secretion from pancreatic islets. Cell 129:359-370. [DOI] [PubMed] [Google Scholar]
- 14.Litterst, C., A. Georgakopoulos, J. Shioi, E. Ghersi, T. Wisniewski, R. Wang, A. Ludwig, and N. K. Robakis. 2007. Ligand binding and calcium influx induce distinct ectodomain/gamma-secretase-processing pathways of EphB2 receptor. J. Biol. Chem. 282:16155-16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marston, D. J., S. Dickinson, and C. D. Nobes. 2003. Rac-dependent trans-endocytosis of ephrinBs regulates Eph-ephrin contact repulsion. Nat. Cell Biol. 5:879-888. [DOI] [PubMed] [Google Scholar]
- 16.Pandey, A., B. Blagoev, I. Kratchmarova, M. Fernandez, M. Nielsen, T. Z. Kristiansen, O. Ohara, A. V. Podtelejnikov, S. Roche, H. F. Lodish, and M. Mann. 2002. Cloning of a novel phosphotyrosine binding domain containing molecule, Odin, involved in signaling by receptor tyrosine kinases. Oncogene 21:8029-8036. [DOI] [PubMed] [Google Scholar]
- 17.Pasquale, E. B. 2008. Eph-ephrin bidirectional signaling in physiology and disease. Cell 133:38-52. [DOI] [PubMed] [Google Scholar]
- 18.Qiao, F., and J. U. Bowie. 2005. The many faces of SAM. Sci. STKE 2005:re7. [DOI] [PubMed] [Google Scholar]
- 19.Sharfe, N., A. Freywald, A. Toro, and C. M. Roifman. 2003. Ephrin-A1 induces c-Cbl phosphorylation and EphA receptor down-regulation in T cells. J. Immunol. 170:6024-6032. [DOI] [PubMed] [Google Scholar]
- 20.Shin, J., C. Gu, E. Park, and S. Park. 2007. Identification of phosphotyrosine binding domain-containing proteins as novel downstream targets of the EphA8 signaling function. Mol. Cell. Biol. 27:8113-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torii, M., K. Hashimoto-Torii, P. Levitt, and P. Rakic. 2009. Integration of neuronal clones in the radial cortical columns by EphA and ephrin-A signalling. Nature 461:524-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlik, M. T., B. Temple, S. Bencharit, A. J. Kimple, D. P. Siderovski, and G. L. Johnson. 2005. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 345:1-20. [DOI] [PubMed] [Google Scholar]
- 23.Wang, Y., S. Ota, H. Kataoka, M. Kanamori, Z. Li, H. Band, M. Tanaka, and H. Sugimura. 2002. Negative regulation of EphA2 receptor by Cbl. Biochem. Biophys. Res. Commun. 296:214-220. [DOI] [PubMed] [Google Scholar]
- 24.Zimmer, M., A. Palmer, J. Kohler, and R. Klein. 2003. EphB-ephrinB bi-directional endocytosis terminates adhesion allowing contact mediated repulsion. Nat. Cell Biol. 5:869-878. [DOI] [PubMed] [Google Scholar]