Abstract
During brain development, neurons and their nerve fibers are often segregated in specific layers. The hippocampus is a well-suited model system to study lamination in health and aberrant cell/fiber lamination associated with neurological disorders. SRF (serum response factor), a transcription factor, regulates synaptic-activity-induced immediate-early gene (IEG) induction and cytoskeleton-based neuronal motility. Using early postnatal conditional SRF ablation, we uncovered distorted hippocampal lamination, including malpositioning of granule cell neurons and disruption of layer-restricted termination of commissural-associational and mossy fiber axons. Besides axons, dendrite branching and spine morphogenesis in Srf mutants were impaired, offering a first morphological basis for SRF's reported role in learning and memory. Srf mutants resemble mice lacking components of the reelin signaling cascade, a fundamental signaling entity in brain lamination. Our data indicate that reelin signaling and SRF-mediated gene transcription might be connected: reelin induces IEG and cytoskeletal genes in an SRF-dependent manner. Further, reelin-induced neurite motility is blocked in Srf mutants and constitutively active SRF rescues impaired neurite extension in reeler mouse mutants in vitro. In sum, data provided in this report show that SRF contributes to hippocampal layer and nerve fiber organization and point at a link between Srf gene transcription and reelin signaling.
Cells in many tissues are organized into layers. An intriguing example of such cell lamination is the vertebrate brain. During brain development, coordinated cell migration, nerve fiber outgrowth, and guidance ensure layer formation. In the hippocampus, cell bodies are compacted in two layers, the dentate gyrus (DG) granule cell layer (GCL) and the cornu ammonis (CA) pyramidal cell layer (PCL) (14). Besides cell bodies, nerve fibers are bundled in distinct layers, including the mossy fiber axons connecting the GCL with the PCL (see Fig. 1A). Additionally, both hippocampi are connected via commissural/associational (C/A) axons.
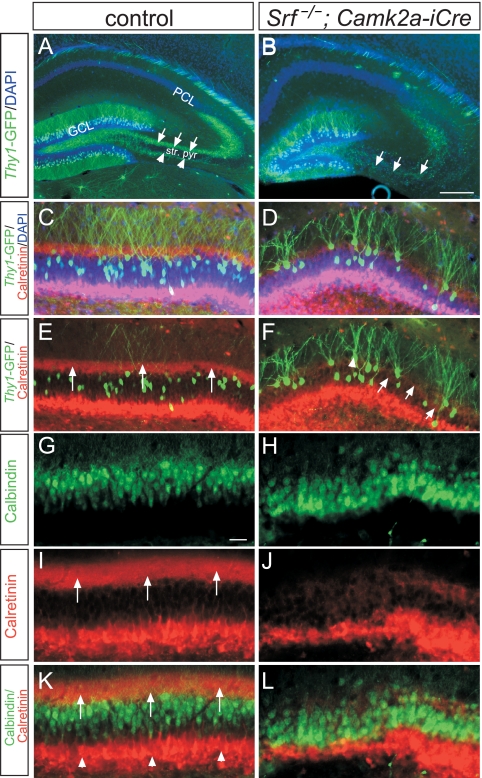
FIG. 1.
SRF controls cell body and fiber lamination in the DG. (A and B) In P14 control (A) and Srf mutant (B) mice, mossy fibers (green) were highlighted by GFP expression via the thy1 promoter of transgenic mice. In controls (A), mossy fibers navigated in either the suprapyramidal (arrows) or infrapyramidal (arrowheads) blade outside the stratum pyramidale (str. pyr.). In Srf mutants (B), mossy fibers navigated inside the stratum pyramidale rather than growing outside. (C to F) Using thy1-GFP mice, a subpopulation of DG granule cells was visualized in control (C and E) and Srf mutant (D and F) mice. C/A fibers were stained red by calretinin. In controls, essentially all of the GFP-positive granule cells were localized underneath the C/A fibers (indicated by arrows in panel E). In contrast, GFP-positive granule cells in Srf mutants were intermingled with C/A fibers (arrows in panel F). C/A fibers in Srf mutants (D and F) did not form a tight bundle, as seen for control mice (C and E). GFP-positive granule cells in Srf mutants were aberrantly positioned above the DAPI-positive GCL (D; arrowhead in panel F). (G to L) All granule cell neurons were visualized with calbindin (green), in parallel with calretinin (red; arrows in panel I). In controls, calbindin-positive cells (green) were embedded by the calretinin-positive C/A fiber layer (arrows in panel K) and neurons in the hilus/subgranular zone (arrowhead in panel K). In contrast, in Srf mutants (J and L), granule cell neurons were more dispersed and were found within or above C/A fibers. Scale bars: A and B, 200 μm; C to L, 50 μm.
In this study, we investigated whether gene expression programs governed by SRF (serum response factor) contribute to lamination in the mouse brain. In neurons, SRF emerged as a paradigmatic transcriptional regulator conveying the immediate-early gene (IEG) response (12, 20, 29). In addition, SRF modulates cytoskeletal dynamics by regulating actin gene transcription (23, 28) and adjusting cofilin activity (1). In neurons (35) and nonneuronal cells (34), actin treadmilling and SRF represent an intimate signaling entity, with actin signaling adjusting SRF activity (20). In conditional Srf mutants, processes relying on the propagation of neuronal activity via IEGs and motility are distorted, including tangential neuronal migration (1), neurite outgrowth, growth cone motility, and axon guidance (19, 35, 38). These deficits in initial brain wiring are associated with impaired learning and memory in the adult (12, 29).
In the present work, we demonstrate hippocampal lamination defects including GCL dispersion, C/A misrouting, aberrant dendritic branching, and a reduced dendritic spine number in Srf mutants.
Phenotypes associated with SRF deficiency resemble those of mutant mice deficient in reelin signaling, an important signaling complex in brain lamination (7, 14, 30). Reelin is an extracellular matrix protein engaging VLDLR (very-low-density lipoprotein receptor) and ApoER2 (apolipoprotein E receptor 2) as receptors (7, 14, 30). In the hippocampi of reelin, Vldlr, and Apoer2 mutant mice, GCL dispersion and C/A fiber misrouting were reported (2, 4, 8, 9, 11, 15, 39). In addition to distorted cell migration, reeler mice display severe axonal and dendritic malformations, including reduced branching and overall neurite length (2, 3, 26, 27). In vitro, reelin enhances neurite length and branch formation (17, 26). Until now, reelin signaling has not been implicated in altering gene expression, except for reelin-activating Egr1 (32), an SRF target gene.
In this study, we explored reelin's potential to modulate gene transcription. Reelin upregulates IEGs and activates cytoskeletal genes in an SRF-dependent manner. We further demonstrate a potential functional interdependence of reelin and SRF signaling. In Srf mutant neurons, reelin-induced neurite branching is blocked. Complementarily, constitutively active SRF can rescue impaired neurite outgrowth in reeler mice.
In sum, our data identify SRF as a regulator of hippocampal lamination and indicate that SRF may operate as a downstream transcriptional target of the reelin signaling cascade.
MATERIALS AND METHODS
Animals.
Srf (flex1neo/flex1neo) and CamK2α-iCre mice were bred to obtain Srf mutants (Srf−/−; Camk2a-iCre) or control littermates (Srf−/−, Srf+/−, or Srf+/−; Camk2a-iCre) (1, 19). Srf mutants were crossed with thy1-green fluorescent protein (GFP) transgenic mice (provided by J. Sanes, Harvard University) (13). Reeler mice were provided by M. Frotscher (Institute for Anatomy and Cell Biology, University of Freiburg). Animal experiments and housing were approved by the local ethics committee (Tübingen University).
Neuronal cell culture.
Postnatal day 1 (P1) hippocampal or embryonic day 17.5 cortical neurons were cultured on poly-l-lysine (100 μg/ml; Sigma)- and laminin (20 μg/ml; Gibco)-coated coverslips (13 mm) as described before (19). Hippocampal cultures from P1-to-P3 reeler mice (5 × 103 cells/13-mm coverslip) were infected 1 day after plating with adenoviral particles expressing either SRF-VP16ΔMADS (final concentration, 4.9 × 107 PFU/ml) or SRF-VP16 (4.6 × 107 PFU/ml; both made by Vector Biolabs). Virus infection efficiency was 60 to 70% of all cells. After an additional 2 days in culture, cells were processed for immunocytochemistry (see below). P1-to-P3 hippocampal cultures from control or Srf mutant mice (8 × 103 cells/13-mm coverslip) were incubated for approximately 24 h in reelin or control supernatant (derived from a GFP-expressing cell line; see below). For quantitative real-time PCR (qRT-PCR), cortical neurons (approximately 106 cells/35-mm dish) were cultured for 3 days in vitro prior to stimulation (see below).
Reelin preparation.
HEK293 cells stably expressing a reelin expression vector (pCrl; a gift from T. Curran, Children's Hospital of Philadelphia) and control HEK293 cells expressing GFP were provided by M. Frotscher (University of Freiburg). HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-GlutaMAX (Invitrogen) with 10% fetal calf serum (PAA Laboratories, Inc.) and 0.5 mg/ml G418 (Geneticin; Roth). The medium was replaced with Opti-MEM (Invitrogen), and the cells were incubated for a further 3 days. Supernatants of control and reelin-expressing cells were centrifuged at 1,000 rpm for 8 min. For stimulation in qRT-PCR experiments (see Fig. 5; see also Fig. S4 in the supplemental material), reelin or control Opti-MEM supernatants (approximately 1.5 ml/35-mm dish) were immediately applied to neurons. To investigate reelin-induced neurite branching (see Fig. 7), supernatants were concentrated 10-fold with Centricon columns (Amicon; Millipore) and diluted in neuronal minimal essential medium-B27 supplement 1:5 in a total volume of 0.5 ml.
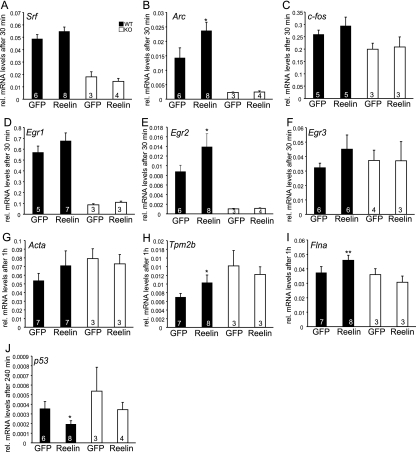
FIG. 5.
Reelin elevated IEG and cytoskeletal mRNA levels through SRF. Cortical cultures derived from embryonic day 17.5 WT or Srf mutant (KO) mice were stimulated with reelin-containing or control supernatant for the indicated times. RNA was isolated, and cDNA was subjected to qRT-PCR using primers to the genes indicated. Reelin increased the mRNA abundance of the IEGs Srf (A), Arc (B), c-fos (C), Egr1 (D), Egr2 (E), and Egr3 (F) to a variable extent. In addition to IEGs, reelin induces mRNA levels of cytoskeletal genes, including Acta (G), Tpm2b (H), and Flna (I). p53 levels are suppressed by reelin in both WT and Srf mutant neurons (J). In Srf mutants, reelin does not upregulate mRNA levels of IEGs and cytoskeletal genes, indicating that SRF is the transcriptional regulator mediating reelin stimulation. Statistical significance was calculated by comparing control values with those obtained with the respective reelin-containing media. The values in the bars are the numbers of animals analyzed.
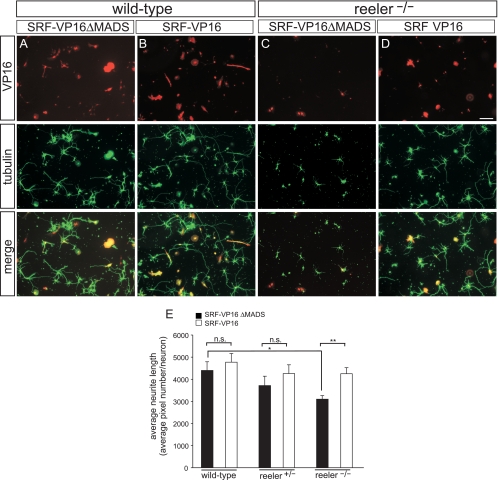
FIG. 7.
SRF-VP16 rescues impaired neurite outgrowth in reeler mutants. Hippocampal neurons derived from P1-to-P3 pups were stained for tubulin (green) and expression of the SRF constructs via antibodies directed to the VP16 domain (red) of the fusion protein. (A and C) WT (A) and hippocampal neurons lacking reelin (C) were infected with adenoviruses expressing the SRF control construct SRF-VP16ΔMADS (lacking DNA binding activity). Typically, WT neurons (A) elaborated more and longer neurites than did neurons lacking reelin (C). (B and D) WT (B) and reeler (D) cultures were infected with constitutively active SRF-VP16. In reelin-deficient neurons, SRF-VP16 strongly increased neurite outgrowth, almost reaching the levels observed for WT cultures. (E) Quantification of neurite outgrowth in WT and reeler hetero- and homozygous cultures. For quantification of neurite outgrowth, neurite length was determined by SlideBook software depicting the numbers of tubulin pixels per DAPI-positive neuron. Scale bars in panels A to D, 100 μm.
qRT-PCR.
Total RNA derived from tissue or culture was isolated with the RNeasy kit (Qiagen). Reverse transcription was performed with 1 μg RNA using reverse transcriptase (Promega) and random hexamers. qRT-PCR was performed on an ABI PRISM 7700 Sequence Detector with the Power PCR SYBR green PCR master mix (Applied Biosystems). Expression was determined in relation to Gapdh RNA levels. Primer sequences can be obtained upon request. Of note, replacement of NMEM/B27 with Opti-MEM of a GFP-expressing HEK293 cell line alone already induced some IEGs and not the cytoskeletal genes. Importantly, IEG gene induction by reelin containing Opti-MEM further enhanced this “baseline” IEG response (see Fig. 5; see also Fig. S4 in the supplemental material). Therefore, we included this Opti-MEM (GFP) control for every individual period of reelin stimulation (30 min to 4 h).
Immunohistochemistry.
Brains were fixed in 4% paraformaldehyde (PFA), and 60-μm sections were prepared on a Vibratome. Slices were blocked for 30 min in 5% normal goat serum-0.1% phosphate-buffered saline (PBS)-Tween 20 and incubated overnight at 4°C with rabbit anticalretinin (1:5,000; Swant), mouse antireelin (1:1,000, G10; Chemicon), mouse anticalbindin (1:500; Swant), mouse anti-Smi32 (1:1,000; Sternberger), rabbit anti-Map2 (1:1,000; Chemicon), and rabbit anti-SRF (1:500; Santa Cruz) primary antibodies. Sections were incubated for 1 h at room temperature with Alexa-conjugated secondary antibodies (1:300), counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and embedded in Mowiol.
Immunocytochemistry.
Cells were fixed for 15 min in 4% PFA-5% sucrose-PBS, permeabilized for 5 min in 0.1% Triton X-100-PBS, and blocked for 30 min in 2% bovine serum albumin-PBS. Mouse anti-β-tubulin (1:5,000; Sigma), mouse anti-class III β-tubulin (1:1,000; Covance), and rabbit anti-VP16 (1:5,000; Abcam) primary antibodies were incubated for 1 h at room temperature. The primary antibodies were detected with Alexa-conjugated secondary antibodies (1:1,000; Molecular Probes). Cells were stained for F-actin with Texas Red-X phalloidin (1:100; Molecular Probes).
Statistics and image acquisition.
The numbers of samples tested are indicated in Results or the figures. For cell culture experiments, at least three independent experiments employing a minimum of three mice per bar (each control and Srf mutant) were analyzed. For neurite branching and length (see Fig. 6 and 7), we analyzed more than 30 neurons/mouse. For spine morphology, more than three GFP-positive neurons (with three apical and basal dendrites/neuron) per mouse were analyzed. Branches and neurite lengths were measured with Axiovision (Zeiss) or SlideBook software (Intelligent Imaging Innovations, Göttingen, Germany). Only neurons (e.g., identified via βIII-tubulin-positive signals) were taken into account. Pictures were further processed using Photoshop software (Adobe). Statistical significance was tested using a two-tailed t test with *, **, and *** representing P ≤ 0.05, P ≤ 0.01, and P ≤ 0.001, respectively, in the graphs. Error bars indicate standard deviations, except in Fig. 5, where they indicate the standard errors of the means.
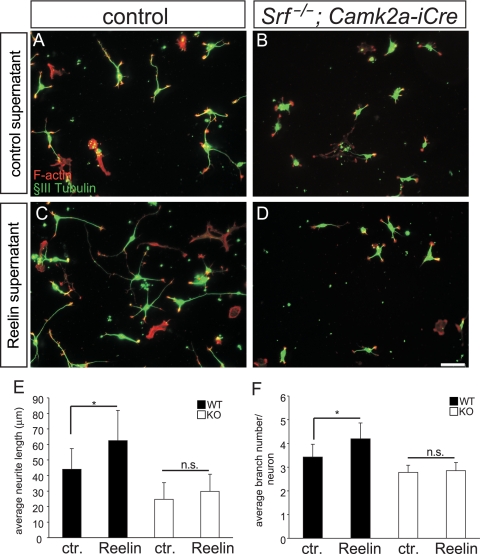
FIG. 6.
Reelin-stimulated neurite elongation and branching requires SRF. (A) WT hippocampal neurons derived from P1-to-P3 pups were stained for F-actin (red) and βIII-tubulin (green). Neurons were incubated with control (ctr.) supernatant derived from GFP-expressing HEK293 cells. (B) Srf mutant neurons—in the presence of control supernatant—are, compared to control-treated WT neurons (A), shorter and contain fewer branches. (C) Addition of reelin to WT neurons increases the average neurite length and number of branches (see quantification in panels E and F). (D) In SRF-deficient cultures, exogenous reelin fails to elevate the neurite length and branch number, indicating that reelin signaling requires SRF activity to modulate neuron morphology. (E and F) Quantification of the average neurite length (E) and the average number of branches per neuron (F) under the various conditions. Scale bars in panels A to D, 50 μm.
RESULTS
SRF regulates lamination of DG neurons and C/A fiber segregation.
To address a role for SRF in hippocampal layering, we analyzed conditional Srf mutants (Srf−/−; Camk2a-iCre). These mice allow early postnatal SRF ablation in forebrain regions such as the hippocampus and cortex commencing at P1 (1, 19). We interbred Srf mutants with thy1-GFP mice, allowing the visualization of individual neuron morphology via GFP distribution (13).
In mice carrying either one or two Srf wild-type (WT) alleles (hereafter referred to as controls), mossy fiber axons emanating from the GCL segregated into the supra- and infrapyramidal branch navigating outside the CA3 stratum pyramidale (arrows and arrowheads in Fig. 1A, respectively). In contrast, in Srf mutants, mossy fiber axons entered the CA3 stratum pyramidale and did not bifurcate (arrows in Fig. 1B). This mossy fiber misrouting in SRF-deficient mice is in line with our previous findings (19).
In addition to mossy fibers (Fig. 1A and B), targeting of C/A axons to GCL neuron dendrites was disturbed in Srf mutants (Fig. 1C to L). In control mice (Fig. 1C and E), C/A fibers labeled with calretinin formed a distinct bundle terminating at proximal parts of GCL dendrites (arrows in Fig. 1E). In contrast, in Srf mutants (Fig. 1D and F), C/A fibers failed to fasciculate and now spread throughout the GCL layer (arrows in Fig. 1F).
We also noted dispersion of GCL neurons in Srf mutants (Fig. 1D and F). In controls (Fig. 1C and E), GFP-positive GCL neurons were situated underneath the calretinin-labeled C/A fibers. In Srf mutants (Fig. 1D and F), many GFP-positive neurons localized ectopically (arrowhead in Fig. 1F) above the DAPI-positive GCL layer (WT, 10% ± 6% mislocalized neurons; knockout [KO], 42% ± 12%; n = 6 mice per genotype; P ≤ 0.001). In a different experimental set (Fig. 1G to L), we colocalized C/A fibers with all calbindin-expressing GCL neurons. As before (Fig. 1C to F), calretinin-positive C/A fibers were misrouted and calbindin-positive GCL neurons were intermingled with C/A fibers in Srf mutants (Fig. 1H, J, and L) but not in controls (Fig. 1G, I, and K; WT, 8.6 ± 4 mislocalized neurons/mm DG; KO, 67 ± 4; n = 3 mice per genotype; P ≤ 0.001).
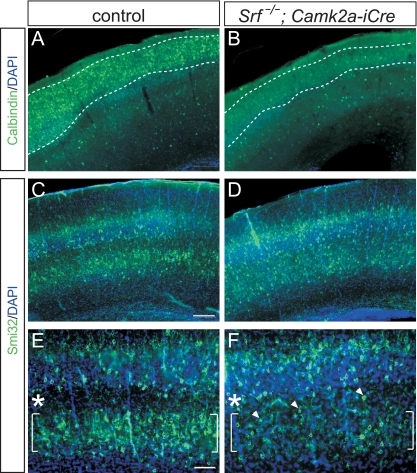
SRF contributes to cortical lamination.
In addition to hippocampal layer formation, we inspected cortical lamination (Fig. 2). We employed calbindin, a marker of layer II/III neurons and interneurons dispersed throughout all cortical layers (Fig. 2A and B). In WT mice, calbindin-positive cells accumulated in layer II/III (Fig. 2A). In contrast, in Srf mutant cortices, numbers of calbindin-positive neurons in layer II/III were reduced (Fig. 2B). In addition, we demonstrated impaired cortical layering in Srf mutants by using Smi-32, a marker for layer III and V cortical neurons (Fig. 2C to F). In WT mice, Smi-32-positive neurons segregated in two layers, with only a few neurons being interspersed between layers III and V (Fig. 2C and E). In the SRF-deficient cortex, layer V contained fewer neurons and Smi-32-positive cells were intermingled between layer III and the remaining cells in layer V (Fig. 2D and F, arrowheads in panel F). In sum, SRF is involved in both hippocampal (Fig. 1) and cortical (Fig. 2) brain layering.
FIG. 2.
Impaired cell layering in the Srf mutant cortex. (A and B) P14 cortices were stained with calbindin, an interneuron and pyramidal cell marker. In the WT (A), calbindin-positive cells localize to the upper cortical layers, whereas fewer cells populate the Srf mutant cortex (B). (C to F) Smi32 labels layer III and layer V pyramidal neurons. In the WT (C and E), Smi32-psoitive neurons segregated into layers III and V (brackets), leaving a Smi32-free zone in between (asterisks). In Srf mutants (D and F), layer V contained fewer Smi32-positive cells and many Smi32-positive neurons gathered between layers III and V (arrowheads). Scale bars: A to D, 200 μm; E and F, 100 μm.
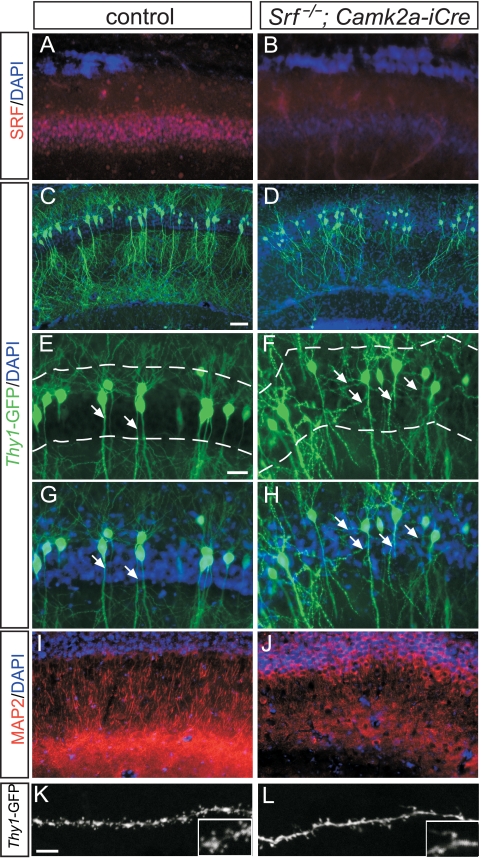
Ectopic dendritic branching and reduced dendritic arbor complexity in Srf mutants.
In addition to the DG, SRF is also deleted in the CA1 stratum pyramidale of Srf mutants (Fig. 3A and B) (11). Besides a lamination defect (Fig. 1), we observed reduced compaction of CA1 pyramidal neurons of Srf mutants (Fig. 3D) compared to controls (Fig. 3C). In thy1-GFP mice used as a reporter for individual neuron morphology, control CA1 pyramidal neurons protruded as two dendritic tufts, a basal tuft localized above and an apical tuft situated underneath the CA1 cell body layer (Fig. 3C, E, and G). Within the CA1 stratum pyramidale of WT mice, dendritic branching is nearly absent (arrows in Fig. 3E and G). On the contrary, in Srf mutants, we noted exuberant dendritic branches within the stratum pyramidale (arrows in Fig. 3F and H; WT, 11.7% ± 10% neurons with ectopic branching; KO, 74% ± 7.6%; n = 8 mice per genotype; P ≤ 0.001). This ectopic dendritic branching (Fig. 3F and H) is accompanied by an overall reduction in the apical dendritic arborization outside the stratum pyramidale of Srf mutants (Fig. 3J). Using the dendrite marker MAP2, we observed a reduction in the CA1 apical dendrite number in Srf mutants (Fig. 3J), compared to the control (Fig. 3I; WT, 75.3 ± 11 dendrites/area; KO, 20 ± 7.5; n = 3 mice per genotype; P ≤ 0.01). This reduction is CA1 specific, as in the entire hippocampus, dendrite arborization was only slightly reduced in Srf mutants (Fig. 1A and B; see Fig. S1 in the supplemental material). In line with SRF loss of function revealing impaired neurite branching (Fig. 3), constitutively active SRF (SRF-VP16) (31) elevated branch formation in vitro (see Fig. S2 in the supplemental material).
FIG. 3.
Aberrant dendrite arborization and spine formation in Srf mutant hippocampi. (A and B) SRF is expressed in WT CA1 neurons (A). Cre-mediated recombination effectively diminished SRF levels in CA1 neurons of Srf mutants (B). (C and D) Dendritic arborizations were visualized in a subpopulation of CA1 pyramidal neurons of P14 thy1-GFP transgenic mice. In the WT (C), CA1 neurons elaborated dendritic arborizations above (basal) and below (apical) the DAPI-positive cell body layer. In Srf mutants (D), the complexity of dendritic arborizations was decreased. Additionally, ectopic dendritic protrusions were observed within the cell body layer (D; see also panels E to H). (E to H) Higher magnifications of results shown in panels C and D. In control mice (E and G), dendrites assembled a basal and an apical tuft. In the WT, dendrites did not protrude within the DAPI-positive cell body layer (arrows in panels E and G; dashed lines label the cell body layer). In Srf mutants, dendrites were ectopically localized within the cell body layer (arrows in panels F and H). (I and J) Dendrites of all CA1 neurons were highlighted with anti-MAP2 antibodies in controls (I) and Srf mutants (J). In Srf mutants (J), dendritic arborization is reduced compared to that in control mice (I). (K and L) Dendritic spine formation was investigated using thy1-GFP mice interbred with WT (K) or Srf mutant mice (L) at P14. In WT mice (K), the dendritic spine number exceeded that of Srf mutants (L). Inserts are higher magnifications. Scale bars: A, B, and E to J, 20 μm; C and D, 50 μm; K and L, 5 μm.
As SRF deficiency impairs neuronal IEG induction (29) and learning and memory (12), we inspected dendritic spine formation (Fig. 3K and L). All dendritic spines, regardless of morphology, were counted, and numbers were normalized to the total dendritic length. Numbers of spines of apical and basal dendrites were reduced in Srf mutants (Fig. 3L) compared to those in controls (Fig. 3K) (WT basal dendrites, 5.3 ± 1.2 spines/10 μm; KO, 3.5 ± 0.9; P ≤ 0.01; WT apical dendrites, 5.7 ± 1.3 spines/10 μm; KO, 3.7 ± 0.4; n = ≥6 mice per genotype; P ≤ 0.01).
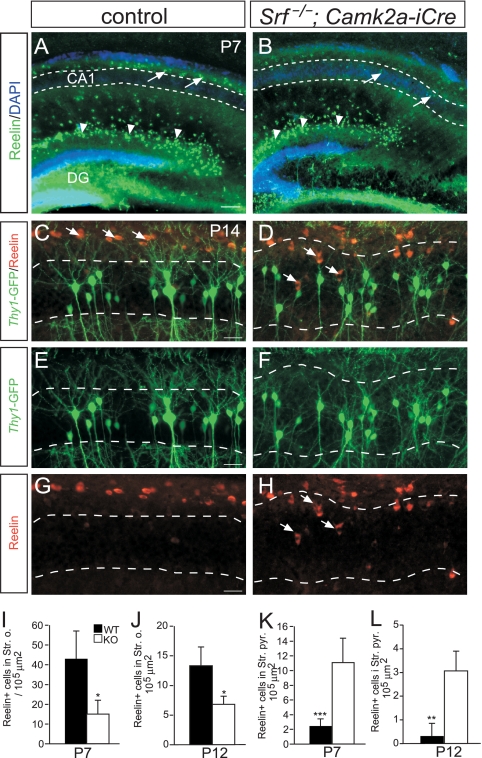
Reelin-expressing cells are displaced in the Srf mutant hippocampus.
In Srf mutants, brain lamination was impaired, including dispersed GCL and cortical lamination (Fig. 1 and 2), C/A misrouting (Fig. 1), and impaired dendritogenesis (Fig. 3). These phenotypes are reminiscent of mutant mice deficient in components of the reelin signaling pathway (see introduction). Lamination defects in SRF-deficient brains are similar to several histological aberrations described for reeler mice. However, the severity of phenotypes is overall weaker in Srf mutant mice. Impaired lamination associated with SRF deficiency more closely resembles that of reelin receptor VLDLR and ApoER2 mutant mice (e.g., see reference 11).
The similarity of the phenotypes caused by impaired SRF or reelin signaling encouraged us to address whether SRF is linked to reelin signaling. We first analyzed whether the positioning of reelin-positive cells is affected by SRF deficiency (Fig. 4; n = ≥3 mice per genotype). Reelin expression in CR cells of the DG molecular layer was not obviously altered by SRF deficiency (arrowheads in Fig. 4A and B). Reelin is also localized in the stratum oriens in close proximity to CA1 pyramidal neurons (arrows in Fig. 4A and C). In controls (Fig. 4A, C, E, and G), reelin surrounded the basal dendritic tuft of CA1 pyramidal neurons (arrows in Fig. 4A and C). In Srf mutants, we found additional reelin-positive cells entering the CA1 cell body layer (arrows in Fig. 4B, D, and H; quantification in panels K and L). Next, we assessed whether SRF operates as a transcriptional regulator of components of the reelin signaling cascade. Using qRT-PCR, we found no mRNA alterations of genes such as Rln, Vldlr, Apoer2, and Dab1 between control and Srf mutant P6 and P14 hippocampi (see Fig. S3 in the supplemental material). This suggests that SRF is not a major transcriptional regulator of “canonical” reelin signaling components. Yet, cytoskeletal genes (Flna, Dcx) whose mutation results in “reeler-like” phenotypes were affected by SRF deficiency (see Fig. S3 in the supplemental material).
FIG. 4.
Reelin-positive cells are misplaced in Srf mutant hippocampi. (A and B) Overview of reelin distribution in the hippocampi of P7 WT (A) and Srf mutant (B) mice. In Cajal-Retzius cells of the DG (arrowheads), no obvious changes in reelin expression were noticed between the genotypes. In Srf mutants, reelin-positive cells entered the CA1 cell body region (arrows in panel B), whereas in WT mice (arrows in panel A), they remained restricted to areas outside CA1 (indicated by the dashed lines in panels A and B). (C to H) In WT P14 mice (C, E, and G), reelin-positive cells were confined to positions above the CA1 stratum pyramidale (indicated by dashed lines). In Srf mutants (D, F, and H), reelin-positive cells enter the CA1 stratum pyramidale (arrows in panels D and H). Thus, in Srf mutants, ectopic dendritic arborizations inside the stratum pyramidale are next to reelin-expressing cells. (I to L) Quantification of reelin localization in the stratum oriens (I and J) and the stratum pyramidale (K and L) at P7 (I and K) and P12 (J and L). Scale bars: A and B, 100 μm; C to H, 20 μm.
Reelin modulates SRF-mediated gene transcription.
To analyze a potential functional interdependence of reelin signaling and SRF-mediated gene expression, we used two experimental strategies, (i) qRT-PCR analysis of endogenous mRNA expression of SRF target genes upon reelin stimulation (Fig. 5; see S4 and S5 in the supplemental material) and (ii) testing the consequence of reelin application in SRF-dependent reporter gene assays of primary neurons (see Fig. S6 in the supplemental material).
In order to test whether reelin stimulation can activate SRF-mediated gene transcription, we stimulated primary cortical neurons with recombinant reelin or BDNF (brain-derived neurotrophic factor), a known SRF-activating stimulus (18, 38). This was followed by qRT-PCR allowing the quantification of mRNA levels of well-known SRF target genes such as IEGs and actin cytoskeletal genes (Fig. 5; see Fig. S4 and S5 in the supplemental material).
In agreement with previous results (32), reelin elevated mRNA levels of the well-known SRF target gene Egr1 (Fig. 5D; see Fig. S4 and S5 in the supplemental material). We extended this study by demonstrating that reelin upregulated various IEG mRNAs; in addition to Egr1, Srf (Fig. 5A), Arc (Fig. 5B), c-fos (Fig. 5C), Egr2 (Fig. 5E), and Egr3 (Fig. 5F) were transiently upregulated by reelin (see also Fig. S4 and S5 in the supplemental material). However, only upregulation of Egr2 and Arc was statistically significant. Reelin also increased mRNA levels of cytoskeletal genes that encode actin isoforms (Acta2) (Fig. 5G), tropomyosin 2b (Tpm2b) (Fig. 5H), filamin A (Flna) (Fig. 5I), and Arc (Fig. 4B) (the last is both an IEG and a cytoskeletal modulator [see also Fig. S4 and S5 in the supplemental material]).
Of note, compared to mRNA levels induced by BDNF stimulation, reelin responses were less pronounced (see Fig. S4 in the supplemental material). However, it should be kept in mind that BDNF is commercially available, highly purified, and added in high concentrations. On the contrary, we employed tissue culture supernatants of HEK293 cells secreting reelin to stimulate neurons with reelin.
Reelin's potential to stimulate SRF-dependent gene expression was further analyzed by reporter gene assays of primary neurons (see Fig. S6 in the supplemental material). In these experiments, reelin augmented SRF-dependent reporter gene activity from a luciferase construct carrying serum response elements derived from the c-fos promoter (see Fig. S6 in the supplemental material).
Our data suggest that reelin- and BDNF-mediated gene transcription requires SRF activity. In Srf mutant neurons, mRNA levels of the genes analyzed were less efficiently induced by either stimulus (Fig. 5; see Fig. S4 in the supplemental material).
We also analyzed the Egr1 target gene p53 (25), whose homolog p73 is coexpressed with reelin in CR cells (22). Interestingly, p53 levels are downregulated after 4 h of reelin stimulation yet, as revealed by Srf mutant cultures, independently of SRF (Fig. 5J; see Fig. S4 in the supplemental material). The implication of this p53 downregulation by reelin remains speculative; however, p53 emerges—besides its role in neuronal apoptosis—as a positive stimulator of neuronal motility (37). Thus, reelin-induced reduction of p53 levels might influence, e.g., cell migration. In sum, we show that, in neurons, reelin increased the mRNA levels of some IEGs and cytoskeletal genes in an SRF-dependent manner.
Reelin-stimulated neuritogenesis requires SRF.
To further elucidate a possible downstream role for SRF in reelin signaling, we asked whether reelin-dependent neuronal motility requires SRF activity (Fig. 6).
Reelin enhances neurite length and branching in primary neuronal culture (17, 26). We stimulated hippocampal neurons by adding reelin to their growth medium. In WT culture, reelin increased the neurite length and branch number (Fig. 6A and C) (n = 12 mice). However, reelin failed to increase the neurite length and branch number in SRF-deficient cultures (Fig. 6B and D; quantification in panels E and F) (n = 9 mice).
Although we cannot completely rule out the possibility that, due to SRF's impact on cytoskeletal dynamics, Srf mutant neurons are intrinsically altered in such a way that they are less responsive to exogenously applied stimuli, this set of experiments suggests that reelin-induced neuronal motility requires nuclear SRF activity.
SRF rescues reeler phenotypes in vitro.
We next asked whether constitutively active SRF could rescue phenotypes evoked by reelin deficiency. To test this, we used reelin-deficient hippocampal neurons in vitro (Fig. 7). Reelin-deficient neurons revealed reduced neurite length and branching (Fig. 7C) (e.g., see reference 26) compared to WT cultures (Fig. 7A) (n = 3 mice). Overexpression of constitutively active SRF-VP16 significantly rescued impaired neurite outgrowth and branch formation evoked by reelin deficiency (Fig. 7D) (n = 4 mice). In contrast to SRF-VP16, SRF-VP16ΔMADS lacking SRF's DNA binding activity was unable to rescue reeler phenotypes in vitro (Fig. 7C). We also investigated heterozygous reeler mice (n = 3). Here, SRF-VP16 expression increased neurite outgrowth almost to WT levels (quantification, Fig. 7E). In sum, both loss- and gain-of-function experiments suggest that reelin and SRF may operate in a shared signaling pathway.
DISCUSSION
SRF controls many aspects of hippocampal development.
In this study, we uncovered SRF as an important regulator of hippocampal development. First, SRF gene activity influences DG and cortical lamina formation and fiber segregation (Fig. 1 and 2). The DG is initially formed by neurons migrating from the ventricular zone (16). As the DG area in Srf mutants is populated similarly to that of the WT (Fig. 1), primary DG formation occurs SRF independently. A secondary postnatal proliferation zone is localized to the hilus, from where granule cells invade the DG. In Srf mutants, many GCL neurons were dispersed (Fig. 1) (1), suggesting that SRF contributes to granule cell radial migration from the hilus to the DG. This finding, together with impaired cell layering in the Srf mutant cortex (Fig. 2), suggests that SRF directs radial cell migration and, as shown before, also tangential migration (1). Besides GCL dispersion, C/A fibers in the DG are displaced in Srf mutants (Fig. 1). The number of commissural axons is unaltered in Srf mutants (36), arguing against extensive fiber loss as the major cause. Instead, our data favor a model where—similar to the situation in reeler mice—C/A fibers are misrouted as a secondary effect to an initial GCL dispersion. In reeler mutants, impaired signaling by guidance cues presented on GCL dendrites, but not on C/A fibers, appears critical for proper C/A fiber targeting (14).
Second, besides axon pathfinding (19), SRF stimulated dendritic branching and the spine number (Fig. 3; see Fig. S2 in the supplemental material). SRF elicits long-term potentiation (29) and long-term depression responses (12) and memory formation while encountering a novel environment (12). Thus, reduced dendrite complexity and spine number in Srf mutants (Fig. 3) might represent a first morphological counterpart underlying these neuronal transmission and behavioral impairments.
Reelin and SRF can operate in a shared signaling pathway.
In this study, we provide data arguing that reelin and SRF operate in a shared signaling pathway. First, Srf mutant phenotypes are similar to those of reelin cascade mutant mice, particularly those of reelin receptor VLDLR and ApoER2 mutants. The phenotypes shared include aberrant DG lamination, C/A fiber misrouting, and reduced dendrite and spine numbers (7, 14, 27, 30). This suggests that reelin and SRF might synergize in the same signaling pathway. Consequently, loss of SRF function prevents coordinate propagation of reelin signaling and thereby results in “reeler-like” phenotypes. Srf mutant phenotypes are not as severe as those of reeler mice. This suggests that SRF is not the only reelin target and that reelin likely signals multiple downstream targets.
Second, SRF deficiency affects the positioning of reelin-positive cells in the hippocampus (Fig. 4). This might suggest the presence of a feedback mechanism by which SRF ensures appropriate localization and thereby signaling of reelin-expressing cells (Fig. 4).
In Srf mutants, ectopic reelin-expressing cells enter the CA1 stratum pyramidale, coinciding with ectopic dendritic branching in this area (Fig. 3 and 4). Reelin stimulates neurite branching (17, 26, 27); thus, mislocalized reelin in Srf mutants might cause unrestrained dendrite formation inside CA1. In agreement, our in vitro data suggest that there is some functional interdependence of reelin and SRF in the regulation of neurite outgrowth: in Srf mutants, reelin fails to stimulate neuronal motility (Fig. 6). Complementarily, gain-of-function experiments employing SRF-VP16 can partially rescue reeler phenotypes (Fig. 7). How might SRF-VP16 rescue reduced neurite outgrowth induced by reelin deficiency? Reelin was recently reported to stabilize F-actin via the LIM kinase-cofilin pathway (5; see also below). As a consequence, actin might become destabilized in reeler mice. We have recently shown that SRF-VP16 increases the abundance of various actin isoforms and the F-actin-stabilizing proteins tropomyosin and calponin (36). Thus, SRF-VP16 could rescue reelin deficiency by reconstituting F-actin stabilization, which might be impaired by reelin ablation alone.
A third mode of reelin-SRF interaction might be activation of SRF-mediated gene expression upon reelin stimulation. So far, reelin has only been reported to upregulate Egr1, yet the transcriptional regulator responsible was not elucidated (32). We extended this study by showing that reelin increased the mRNA levels of various IEGs and cytoskeletal genes (Fig. 5 and 7; see Fig. S6 in the supplemental material). We identified SRF as a transcription factor conveying reelin stimulation. What might be the consequence of a reelin-induced IEG response? IEG upregulation marks neuronal activation upon various physiological and pathological stimuli and has also been associated with apoptosis during brain development (6, 24). As reelin-secreting CR cells are a transiently existing cell population prone to elimination during hippocampal development (33), a reelin-induced IEG response might assist this elimination process. Reelin also upregulated genes that encode actin isoforms (Acta2) or actin binding proteins (filamin, tropomyosin, and Arc) (Fig. 5; see Fig. S4 and S5 in the supplemental material). Previously, reelin was shown to inhibit the actin-severing protein cofilin, resulting in F-actin stabilization (5). Notably, filamin A and tropomyosin likewise stabilize F-actin (10), as does Arc, by maintaining cofilin inhibition (21). Thus, reelin might enhance F-actin stability during cell migration, neurite motility, and synapse maturation, i.e., by elevating Actin, Flna, Tpm2b, and Arc mRNA levels via SRF-dependent gene transcription.
In sum, our data identify SRF as an important regulator of hippocampal development. Additionally, we provide three lines of data arguing for a functional interdependence of SRF and reelin in brain lamination.
Supplementary Material
Acknowledgments
We are grateful to Daniela Sinske for excellent technical support. We thank A. Wizenmann, A. Nordheim, and E. Förster for critically reading the manuscript.
B.K. is supported by the DFG through the Emmy Noether program and SFB 446, the Schram-Stiftung, and young investigator grants from Tübingen University.
We have no conflicting financial interests.
Footnotes
Published ahead of print on 1 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alberti, S., S. M. Krause, O. Kretz, U. Philippar, T. Lemberger, E. Casanova, F. F. Wiebel, H. Schwarz, M. Frotscher, G. Schutz, and A. Nordheim. 2005. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc. Natl. Acad. Sci. U. S. A. 102:6148-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrell, V., J. A. Del Río, S. Alcantara, M. Derer, A. Martinez, G. D'Arcangelo, K. Nakajima, K. Mikoshiba, P. Derer, T. Curran, and E. Soriano. 1999. Reelin regulates the development and synaptogenesis of the layer-specific entorhino-hippocampal connections. J. Neurosci. 19:1345-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrell, V., L. Pujadas, S. Simó, D. Dura, M. Sole, J. A. Cooper, J. A. Del Río, and E. Soriano. 2007. Reelin and mDab1 regulate the development of hippocampal connections. Mol. Cell. Neurosci. 36:158-173. [DOI] [PubMed] [Google Scholar]
- 4.Borrell, V., M. Ruiz, J. A. Del Río, and E. Soriano. 1999. Development of commissural connections in the hippocampus of reeler mice: evidence of an inhibitory influence of Cajal-Retzius cells. Exp. Neurol. 156:268-282. [DOI] [PubMed] [Google Scholar]
- 5.Chai, X., E. Förster, S. Zhao, H. H. Bock, and M. Frotscher. 2009. Reelin stabilizes the actin cytoskeleton of neuronal processes by inducing n-cofilin phosphorylation at serine3. J. Neurosci. 29:288-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, S. C., T. Curran, and J. I. Morgan. 1995. Apoptosis in the nervous system: new revelations. J. Clin. Pathol. 48:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Arcangelo, G. 2006. Reelin mouse mutants as models of cortical development disorders. Epilepsy Behav. 8:81-90. [DOI] [PubMed] [Google Scholar]
- 8.Deller, T., A. Drakew, B. Heimrich, E. Förster, A. Tielsch, and M. Frotscher. 1999. The hippocampus of the reeler mutant mouse: fiber segregation in area CA1 depends on the position of the postsynaptic target cells. Exp. Neurol. 156:254-267. [DOI] [PubMed] [Google Scholar]
- 9.Del Río, J. A., B. Heimrich, V. Borrell, E. Förster, A. Drakew, S. Alcantara, K. Nakajima, T. Miyata, M. Ogawa, K. Mikoshiba, P. Derer, M. Frotscher, and E. Soriano. 1997. A role for Cajal-Retzius cells and reelin in the development of hippocampal connections. Nature 385:70-74. [DOI] [PubMed] [Google Scholar]
- 10.Dent., E. W., and F. B. Gertler. 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40:209-227. [DOI] [PubMed] [Google Scholar]
- 11.Drakew, A., T. Deller, B. Heimrich, C. Gebhardt, D. Del Turco, A. Tielsch, E. Förster, J. Herz, and M. Frotscher. 2002. Dentate granule cells in reeler mutants and VLDLR and ApoER2 knockout mice. Exp. Neurol. 176:12-24. [DOI] [PubMed] [Google Scholar]
- 12.Etkin, A., J. M. Alarcon, S. P. Weisberg, K. Touzani, Y. Y. Huang, A. Nordheim, and E. R. Kandel. 2006. A role in learning for SRF: deletion in the adult forebrain disrupts LTD and the formation of an immediate memory of a novel context. Neuron 50:127-143. [DOI] [PubMed] [Google Scholar]
- 13.Feng, G., R. H. Mellor, M. Bernstein, C. Keller-Peck, Q. T. Nguyen, M. Wallace, J. M. Nerbonne, J. W. Lichtman, and J. R. Sanes. 2000. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28:41-51. [DOI] [PubMed] [Google Scholar]
- 14.Förster, E., S. Zhao, and M. Frotscher. 2006. Laminating the hippocampus. Nat. Rev. Neurosci. 7:259-267. [DOI] [PubMed] [Google Scholar]
- 15.Frotscher, M., C. A. Haas, and E. Förster. 2003. Reelin controls granule cell migration in the dentate gyrus by acting on the radial glial scaffold. Cereb. Cortex 13:634-640. [DOI] [PubMed] [Google Scholar]
- 16.Frotscher, M., S. Zhao, and E. Förster. 2007. Development of cell and fiber layers in the dentate gyrus. Prog. Brain Res. 163:133-142. [DOI] [PubMed] [Google Scholar]
- 17.Jossin, Y., and A. M. Goffinet. 2007. Reelin signals through phosphatidylinositol 3-kinase and Akt to control cortical development and through mTor to regulate dendritic growth. Mol. Cell. Biol. 27:7113-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalita, K., G. Kharebava, J. J. Zheng, and M. Hetman. 2006. Role of megakaryoblastic acute leukemia-1 in ERK1/2-dependent stimulation of serum response factor-driven transcription by BDNF or increased synaptic activity. J. Neurosci. 26:10020-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knöll, B., O. Kretz, C. Fiedler, S. Alberti, G. Schutz, M. Frotscher, and A. Nordheim. 2006. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat. Neurosci. 9:195-204. [DOI] [PubMed] [Google Scholar]
- 20.Knöll, B., and A. Nordheim. 2009. Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci. 32:432-442. [DOI] [PubMed] [Google Scholar]
- 21.Messaoudi, E., T. Kanhema, J. Soule, A. Tiron, G. Dagyte, B. da Silva, and C. R. Bramham. 2007. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J. Neurosci. 27:10445-10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer, G., A. Cabrera Socorro, C. G. Perez Garcia, L. Martinez Millan, N. Walker, and D. Caput. 2004. Developmental roles of p73 in Cajal-Retzius cells and cortical patterning. J. Neurosci. 24:9878-9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miano, J. M., X. Long, and K. Fujiwara. 2007. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 292:C70-C81. [DOI] [PubMed] [Google Scholar]
- 24.Miyashita, T., S. Kubik, G. Lewandowski, and J. F. Guzowski. 2008. Networks of neurons, networks of genes: an integrated view of memory consolidation. Neurobiol. Learn. Mem. 89:269-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair, P., S. Muthukkumar, S. F. Sells, S. S. Han, V. P. Sukhatme, and V. M. Rangnekar. 1997. Early growth response-1-dependent apoptosis is mediated by p53. J. Biol. Chem. 272:20131-20138. [DOI] [PubMed] [Google Scholar]
- 26.Niu, S., A. Renfro, C. C. Quattrocchi, M. Sheldon, and G. D'Arcangelo. 2004. Reelin promotes hippocampal dendrite development through the VLDLR/ApoER2-Dab1 pathway. Neuron 41:71-84. [DOI] [PubMed] [Google Scholar]
- 27.Niu, S., O. Yabut, and G. D'Arcangelo. 2008. The reelin signaling pathway promotes dendritic spine development in hippocampal neurons. J. Neurosci. 28:10339-10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posern, G., and R. Treisman. 2006. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16:588-596. [DOI] [PubMed] [Google Scholar]
- 29.Ramanan, N., Y. Shen, S. Sarsfield, T. Lemberger, G. Schutz, D. J. Linden, and D. D. Ginty. 2005. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci. 8:759-767. [DOI] [PubMed] [Google Scholar]
- 30.Rice, D. S., and T. Curran. 2001. Role of the reelin signaling pathway in central nervous system development. Annu. Rev. Neurosci. 24:1005-1039. [DOI] [PubMed] [Google Scholar]
- 31.Schratt, G., U. Philippar, J. Berger, H. Schwarz, O. Heidenreich, and A. Nordheim. 2002. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 156:737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simó, S., L. Pujadas, M. F. Segura, A. La Torre, J. A. Del Río, J. M. Urena, J. X. Comella, and E. Soriano. 2007. Reelin induces the detachment of postnatal subventricular zone cells and the expression of the Egr-1 through Erk1/2 activation. Cereb. Cortex 17:294-303. [DOI] [PubMed] [Google Scholar]
- 33.Soriano, E., and J. A. Del Río. 2005. The cells of Cajal-Retzius: still a mystery one century after. Neuron 46:389-394. [DOI] [PubMed] [Google Scholar]
- 34.Sotiropoulos, A., D. Gineitis, J. Copeland, and R. Treisman. 1999. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98:159-169. [DOI] [PubMed] [Google Scholar]
- 35.Stern, S., E. Debre, C. Stritt, J. Berger, G. Posern, and B. Knöll. 2009. A nuclear actin function regulates neuronal motility by serum response factor-dependent gene transcription. J. Neurosci. 29:4512-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stritt, C., S. Stern, K. Harting, T. Manke, D. Sinske, H. Schwarz, M. Vingron, A. Nordheim, and B. Knöll. 2009. Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat. Neurosci. 12:418-427. [DOI] [PubMed] [Google Scholar]
- 37.Tedeschi, A., and S. Di Giovanni. 2009. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 10:576-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickramasinghe, S. R., R. S. Alvania, N. Ramanan, J. N. Wood, K. Mandai, and D. D. Ginty. 2008. Serum response factor mediates NGF-dependent target innervation by embryonic DRG sensory neurons. Neuron 58:532-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, S., X. Chai, E. Förster, and M. Frotscher. 2004. Reelin is a positional signal for the lamination of dentate granule cells. Development 131:5117-5125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.