Abstract
The complement regulation protein CD46 is the primary attachment receptor for most species B adenoviruses (Ads). However, significant variability exists in sequence and structure among species B Ads in the CD46-binding regions, correlating with differences in affinity. Here, we report a structure-function analysis of the interaction of the species B Ad21 knob with the two N-terminal repeats SCR1 and SCR2 of CD46, CD46-D2. We have determined the structures of the Ad21 knob in its unliganded form as well as in complex with CD46-D2, and we compare the interactions with those observed for the Ad11 knob-CD46-D2 complex. Surface plasmon resonance measurements demonstrate that the affinity of Ad21 knobs for CD46-D2 is 22-fold lower than that of the Ad11 knob. The superposition of the Ad21 and Ad11 knob structures in complex with CD46-D2 reveals a substantially different binding mode, providing an explanation for the weaker binding affinity of the Ad21 knob for its receptor. A critical difference in both complex structures is that a key interaction point, the DG loop, protrudes more in the Ad21 knob than in the Ad11 knob. Therefore, the protruding DG loop does not allow CD46-D2 to approach the core of the Ad21 knob as closely as in the Ad11 knob-CD46-D2 complex. In addition, the engagement of CD46-D2 induces a conformational change in the DG loop in the Ad21 knob but not in the Ad11 knob. Our results contribute to a more profound understanding of the CD46-binding mechanism of species B Ads and have relevance for the design of more efficient gene delivery vectors.
The 52 human adenovirus (Ad) serotypes are divided into seven species (species A to G) (20). Species B Ads are of interest, as they cause severe infections of the respiratory tract, urinary tract, and kidney as well as multiorgan system failure and death in immunocompromised patients (2, 23, 24). Species B Ads can be further grouped into subspecies B1 (Ad3, Ad7, Ad16, Ad21, and Ad50) and subspecies B2 (Ad11, Ad14, Ad34, and Ad35). Viruses in the two subspecies differ in their tropisms: while most B1 viruses cause ocular and/or acute respiratory tract infections, the B2 viruses primarily cause persistent infections of the urinary tract as well as eye infections, meningitis, and infections of the gastrointestinal tract (10, 11, 43). The subspecies B1 Ad21, which is the subject of this study, recently caused outbreaks of acute respiratory disease (28).
Adenoviruses have a nonenveloped icosahedral capsid with a linear double-stranded DNA (46). The major capsid proteins are the hexon, the penton base, and the fiber. The trimeric fiber protein, which protrudes from each of the 12 capsid vertices, consists of three distinct domains: an N-terminal tail, an elongated shaft, and a globular knob. The knob mediates cellular attachment to the primary receptors CD46 (16, 26, 38), coxsackievirus and adenovirus receptor (CAR) (36), and sialic acid (3). Virus attachment is followed by internalization into the host cell via clathrin-coated endocytosis and macropinocytosis, triggered by αv integrins (17, 27, 45).
The species B adenovirus receptor CD46 is a member of a family of proteins that regulate complement activation and are constructed mainly from short consensus repeat (SCR) domains (25). The extracellular portion of CD46 contains four such domains (SCR1 to SCR4), and structural and functional analyses have established that interactions with Ad knobs require only the SCR1 and SCR2 domains (34). The four SCR domains are followed by a 25-amino-acid sequence that is rich in serine, threonine, and proline (the STP region); a single transmembrane segment; and a short cytoplasmic tail. Structural information is currently limited to the N-terminal CD46 domains SCR1 and SCR2. The crystal structure of a fragment comprising these two domains revealed a pronounced kink between the two repeats and some flexibility at the domain interface (7). The protein is expressed on all human cells with the exception of erythrocytes. CD46 acts as a cofactor for factor I, a serine protease that blocks further recruitment of the membrane attack complex by cleaving C3b and C4b (25, 40).
CD46 is also a receptor for many other pathogens, including measles virus, human herpesvirus 6, Neisseria gonorrhoeae, Neisseria meningitidis, and group A streptococci (8, 13, 22, 30, 37).
The structural analysis of the Ad11 knob in complex with the SCR1-SCR2 fragment of CD46 (CD46-D2) showed that engagement by the knob triggers a conformational change in CD46, producing an elongated, nearly linear conformation that differs substantially from the kinked conformation of the unliganded receptor (34). Furthermore, the structure of this complex provided an explanation for the previously observed critical role of Ad11 knob residue Arg279 in CD46 binding. Earlier mutagenesis studies demonstrated that a mutation of Arg279 to glutamine abolishes binding to CD46-D2 (18). Although the structure of the complex showed that Arg279 does not contact the receptor directly, its side chain lies parallel to that of the CD46-contacting residue Arg280. Stacking interactions between the guanidinium groups of Arg279 and Arg280, resulting in an arginine sandwich, likely play a central role in determining receptor specificity (33), and the mutation of Arg279 is thought to prevent Arg280 from forming contacts with CD46 (18).
Although it also uses CD46 as a receptor, the Ad21 knob does not contain an arginine sandwich, as the residue corresponding to Arg279 in Ad11 is a serine. Therefore, the mode of binding of the Ad21 knob to CD46 is likely distinct from that observed for Ad11, consistent with the observation that previously reported mutational studies failed to determine a central binding motif among species B Ads (32, 44). The Ad21 and Ad11 knobs exhibit low sequence identity, especially at the surface loops that mediate binding to CD46 in Ad11. We therefore used a combination of structural and functional studies to establish the mechanism of Ad21 knob binding to CD46-D2. Our structural analysis reveals substantial differences in the numbers and types of contacts between the complexes of Ad11 and Ad21 with CD46-D2 and also in the relative orientations of CD46-D2 and its contacting knobs. Furthermore, our analysis allows the comparison of structural features of the Ad21 and Ad35 knobs, which are closely related in sequence yet also display significantly different CD46-D2-binding properties as well as different tissue tropisms. Thus, our findings result in a significantly enhanced understanding of the interactions between species B Ads and CD46.
In addition to their role as pathogens, species B Ads serve as widely used gene delivery vectors, as they can transduce a broad range of possible target cells that are normally poorly permissive to other Ads, such as hematopoietic stem cells, dendritic cells, and malignant tumor cells (19, 41). Therefore, our results should also guide efforts to improve the gene delivery properties of these viruses.
MATERIALS AND METHODS
Protein expression and purification.
A DNA fragment encoding amino acids 123 to 323 of the Ad21 knob was cloned into vector pET15b in frame with an N-terminal hexahistidine tag followed by a thrombin cleavage site. Expression in Escherichia coli Rosetta 2 (DE3) cells was induced with 1 mM isopropyl-β-d-thiogalactopyranoside at an optical density at 600 nm of 0.6, followed by growth at 20°C for 14 h. Bacteria were harvested in a solution containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, and 20 mM imidazole and lysed by using a homogenizer. Clarified supernatant was loaded onto a nickel affinity chromatography HisTrap column (GE Healthcare). The target protein was eluted by using a gradient ranging from 10 mM to 500 mM imidazole. The terminal histidine tag was cleaved by incubation with 1 U of thrombin (GE Healthcare) per mg of recombinant protein at 20°C for 12 h. Excess thrombin was removed by size-exclusion chromatography on a Superdex 75 column (GE Healthcare) with 20 mM HEPES (pH 7.4) and 125 mM NaCl. A second nickel affinity chromatography step was used for the removal of uncleaved protein.
CD46-D2 was purified as described previously (7). For complex formation, the purified Ad21 knob was incubated with a 3.3 molar excess of CD46-D2 at 4°C overnight. The CD46-D2 protein carries two N-linked high-mannose carbohydrate moieties (at Asn49 and Asn80), which hampered crystallization efforts. Therefore, the Ad21 knob-CD46-D2 complex was incubated with 20 U endoglycosidase Hf (New England Biolabs)/μg of Ad21 knob-CD46-D2 complex for 20 h at 25°C. This procedure resulted in the quantitative cleavage of the two glycans, leaving only a terminal N-acetylglucosamine residue at each of the two asparagine residues (data not shown). The deglycosylated complex was further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare) with 20 mM HEPES and 125 mM NaCl (pH 7.4).
Crystallization and structure determination.
All crystals were grown by using the sitting-drop vapor diffusion technique by mixing protein and precipitant solution in a 1:1 ratio, followed by incubation over precipitant solution in a sealed compartment. Unliganded Ad21 knob crystals were grown at 20°C with a reservoir solution containing 30% polyethylene glycol 3000, 100 mM Tris buffer, and 200 mM NaCl (pH 7.1). The crystals belong to space group P2 and contain four Ad21 knob trimers in their asymmetric unit. The crystals were flash-frozen for X-ray data collection at 100 K by using 20% glycerol as a cryoprotectant. Diffraction data were collected at the Swiss Light Source (Villigen, Switzerland), beam line X06SA, by using a Pilatus 6 M detector and were processed with XDS (21). Initial phases were obtained by molecular replacement using a modified Ad3 knob structure (14) as a search model with PHASER (35). Modifications of the Ad3 structure included the deletion of surface loops and exchanges of amino acids to reflect the Ad21 knob sequence. The structure was then refined by using rigid body refinement and simulated annealing with CNS (6). Subsequent refinement was carried out by alternating rounds of model building with Coot (15) and restrained refinement using 12-fold noncrystallographic symmetry (NCS) restraints with Refmac (29).
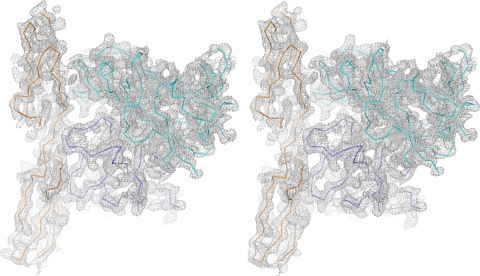
Crystals of the Ad21 knob in complex with CD46-D2 were grown at 20°C by using a reservoir solution consisting of 6% polyethylene glycol 8000, 100 mM citrate buffer, and 200 mM LiCl (pH 5.6). The crystals belong to space group P1 and contain four Ad21 knob trimers, each complexed with three CD46-D2 molecules, in their unit cell. Crystals appeared in 6 to 10 months. They were flash-frozen for X-ray data collection at 100K by using 28% glycerol as a cryoprotectant. Diffraction data were collected at the European Synchrotron Radiation Facility (Grenoble, France) and processed with XDS (21). Initial phases were determined by molecular replacement with PHASER, using only the unliganded Ad21 knob structure as a search model (35). Clear solutions could be obtained for all four Ad21 knob trimers, and these were then refined by using rigid body refinement with PHENIX (1). A difference electron density map [Fobs − Fcalc exp(iαcalc)], calculated with Fcalc amplitudes and phases derived from the four Ad21 knob trimer coordinates, revealed clear electron density for the SCR1 and SCR2 domains. The CD46 SCR1 and SCR2 domains were placed separately into the electron density, followed by rigid body and simulated annealing refinement with PHENIX (1). Subsequent refinement was then carried out by alternating rounds of model building with Coot (15) and restrained refinement with PHENIX (1). For each complex, 3-fold NCS restraints were applied separately to restrain the three Ad21 monomers in the knob, the three SCR1 domains, and the three SCR2 domains. The high level of noncrystallographic symmetry allowed us to average the electron density and aided in the convergence of the refinement. Thus, although the resolution of the structure is intermediate, the refinement statistics are nevertheless good (Table 1). The asymmetric unit comprises four trimeric Ad21 knobs and 12 monomeric CD46-D2 molecules. Figure 1 shows that the electron density map is remarkably clear for the complex shown. The other complexes all have similar electron densities, with the exception of two CD46-D2 SCR1 domains, chains Q and S, which are less well ordered. Data statistics for both the unliganded Ad21 knob and the Ad21 knob-CD46-D2 structures are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
| Statistic | Value |
|
|---|---|---|
| Ad21 knob | Ad21 knob- CD46-D2 | |
| Data collection | ||
| Space group | P2 | P1 |
| No. of monomers in the asymmetric unit | 12 | 12 |
| Cell dimensions | ||
| a, b, c (Å) | 85.0, 63.9, 209.5 | 94.5, 107.7, 154.1 |
| α, β, γ (°) | 90.0, 91.7, 90.0 | 90.0, 90.1, 104.7 |
| Resolution range (Å)a | 49.5-2.5 (2.68-2.5) | 48.0-3.5 (3.62-3.5) |
| Rmergea,b | 11.0 (20.3) | 17.9 (35.5) |
| I/σIa | 8.83 (4.17) | 8.93 (3.61) |
| Completeness (%)a | 91.6 (78.8) | 96.2 (74.9) |
| Redundancya | 3.0 (2.1) | 3.8 (2.7) |
| Refinement | ||
| Resolution (Å)a | 49.5-2.5 (2.68-2.5) | 48.0-3.5 (3.62-3.5) |
| No. of reflections | 71,875 | 71,363 |
| Rwork/Rfreec | 23.4 (25.9) | 20.4 (23.9) |
| No. of atoms | ||
| Protein | 17,249 | 28,961 |
| Water | 67 | |
| Carbohydrate | 56 | |
| B factors (Å2) | ||
| Ad21 knob | 20.4 | 51.2 |
| CD46-D2 | 69.1 | |
| Carbohydrate | 63.5 | |
| Water | 22.1 | |
| RMS deviations | ||
| Bond length (Å) | 0.004 | 0.005 |
| Bond angle (°) | 0.594 | 0.812 |
Data for the highest-resolution shell are shown in parentheses.
Rmerge = ΣhklI − <I>/ΣhklI, where I is the intensity of reflection hkl and <I> is the average over symmetry-related observations of hkl.
Rwork = Rfree = ΣhklFobs − Fcalc/ΣhklFobs, where Fobs and Fcalc are the observed and calculated structure factors, respectively. The free set contains 5% of reflections, which were omitted from the refinement.
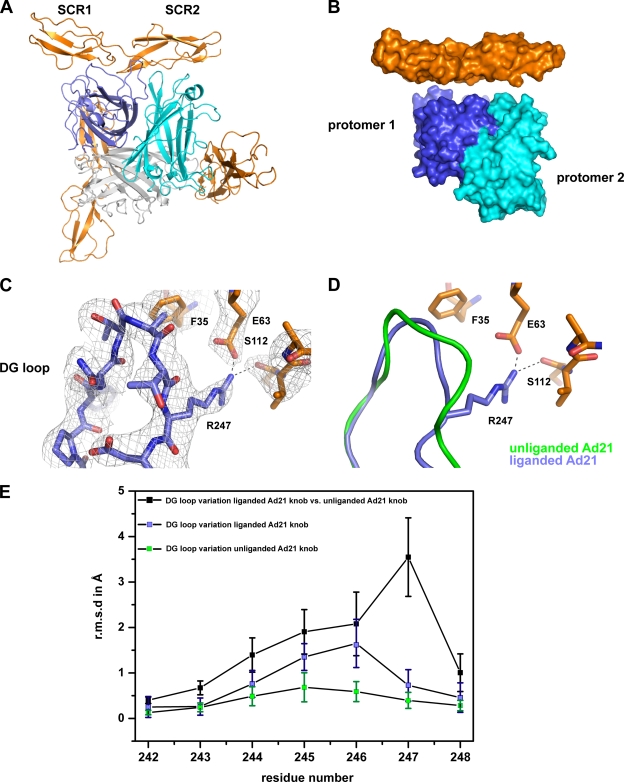
FIG. 1.
Electron density map for the Ad21 knob-CD46-D2 structure. Shown is the stereo view of the final, nonaveraged 2Fo-Fc electron density map for the Ad21 knob-CD46-D2 complex, contoured at 1.0 σ. Ribbon tracings for two Ad21 knob protomers (blue and cyan) and one CD46-D2 molecule (orange) are shown.
Determination of binding parameters by use of surface plasmon resonance (SPR).
Kinetic and steady-state interaction experiments with the Ad21, Ad35, and Ad11 knobs and their monomeric binding partner CD46-D2 were performed at 25°C by use of a Biacore 2000 instrument and data collection rates ranging from 2.5 Hz (Ad11 and Ad35) to 5 Hz (Ad21). One knob can bind up to three copies of CD46-D2. To avoid influencing the binding parameters through avidity effects, the Ad21, Ad11, and Ad35 knobs were immobilized onto the biosensor surface in all experiments. Furthermore, the CD46-D2 protein from the same batch was used for all measurements to eliminate variations in protein quality. The stock concentration of this CD46-D2 sample was 385 μM, calculated by using absorbance values at 280 nm and theoretical extinction coefficients. CM5 chips, an amine-coupling kit, and surfactant P20 were all purchased from GE Healthcare. Buffer containing 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.005% (vol/vol) surfactant P20 (pH 7.4) was used as a running buffer for the CM5 chips. Kinetic experiments were performed with two consecutive flow cells on a CM5 sensor chip. The Ad21, Ad35, and Ad11 knobs were covalently immobilized onto the surface of the downstream experimental flow cell via amine-coupling chemistry according to the manufacturer's instructions. The surface of the upstream flow cell was subjected to the same coupling reaction in the absence of knobs and used as a reference.
Kinetic experiments.
The analyte (CD46-D2) was serially diluted in running buffer and injected in series over the reference and experimental biosensor surfaces at a flow rate of 75 μl/min. A sample containing only running buffer was injected under the same conditions. Sensorgrams of each sample were double referenced by the subtraction of sensorgrams obtained from the reference surface of each cycle to remove bulk effects, followed by a further subtraction of the referenced sensorgram obtained from the running buffer sample injection to remove drift and system noise. The association rate (on-rate) constant (ka) and dissociation rate (off-rate) constant (kd) were determined simultaneously by globally fitting double-referenced sensorgrams of the entire titration series to a “1:1 binding with mass transfer, refractive index (RI) effect = 0” model with BIAevaluation software 4.1 (Biacore). The dissociation constant (KD) was calculated by the following equation: KD = kd/ka.
Steady-state experiments.
When the sensorgram of a kinetic sensorgram series reached the equilibrium binding level, which is represented by the plateau phase of a sensorgram, during each individual injection of analyte, the averaged (AVG) response in this area (Eq-Response AVG) was plotted against the injected CD46 concentration. By fitting these data to a “1:1 Langmuir isotherm” model, a KD value can be obtained that is not calculated from kinetic rate constants and that can therefore be used to cross-validate the KD value obtained from a corresponding kinetic analysis. In all SPR experiments, surface regeneration was not necessary, since all interactions showed either very rapid or moderate dissociation rates.
Preparation of figures.
Amino acid sequences of knobs were aligned by using the program ClustalW2 (9). Structures were aligned by the secondary-structure-matching superposition of knob structures with the program Coot (15). Structural figures (Fig. 1 and see Fig. 3 to 6) were generated with PyMOL (DeLano Scientific LLC) (12).
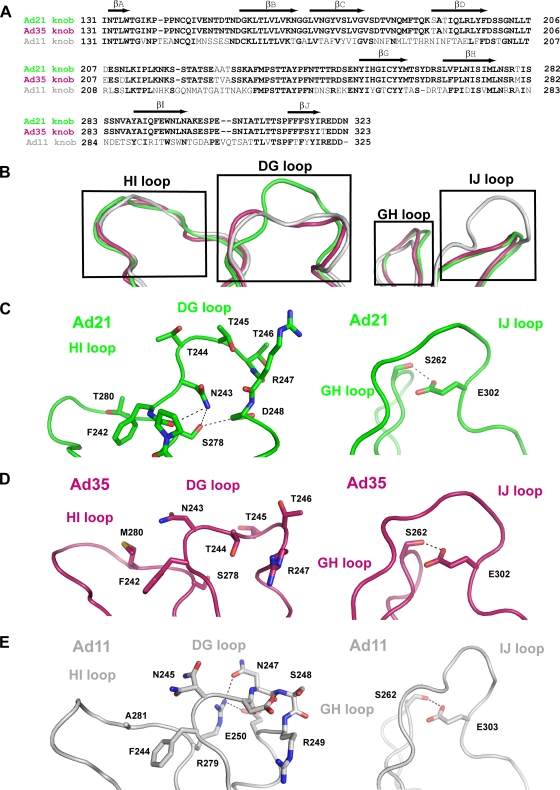
FIG. 3.
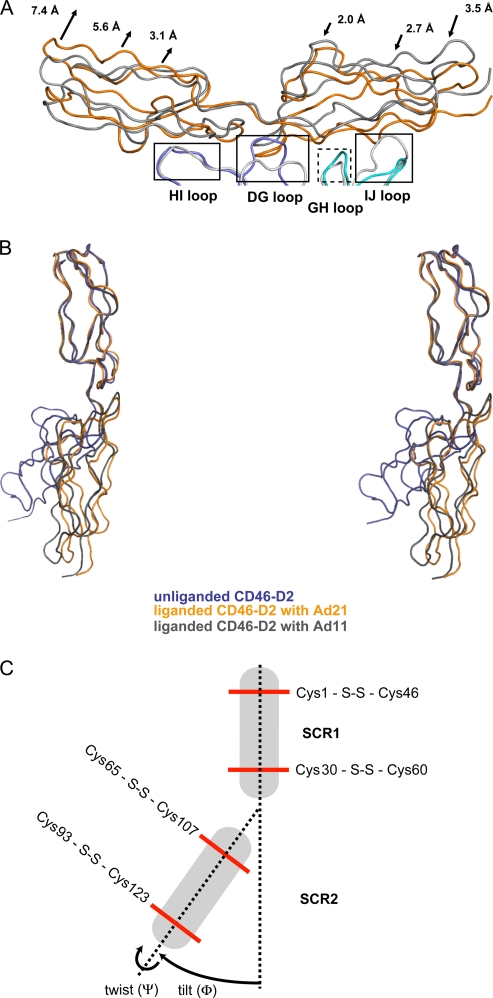
Comparison of the unliganded Ad11, Ad21, and Ad35 knob structures. (A) Alignment of the Ad21, Ad35, and Ad11 knob sequences performed with ClustalW2. The positions of β-strands are indicated above the alignment with arrows. (B) Superposition of the unliganded Ad21, Ad35, and Ad11 knobs showing the surface-exposed HI, DG, GH and IJ loops. The Ad21 knob is green, the Ad35 knob is ruby, and the Ad11 knob is gray. (C) Close-up views of the Ad21 DG and HI loops (left) and the Ad21 GH and IJ loops (right). (D) Close-up view of the corresponding regions in the Ad35 knob. (E) Close-up view of the corresponding regions in the Ad11 knob. The Ad21 knob, Ad35 knob, and Ad11 knob loops are colored as described above (B). Hydrogen bonds are represented with dashed lines.
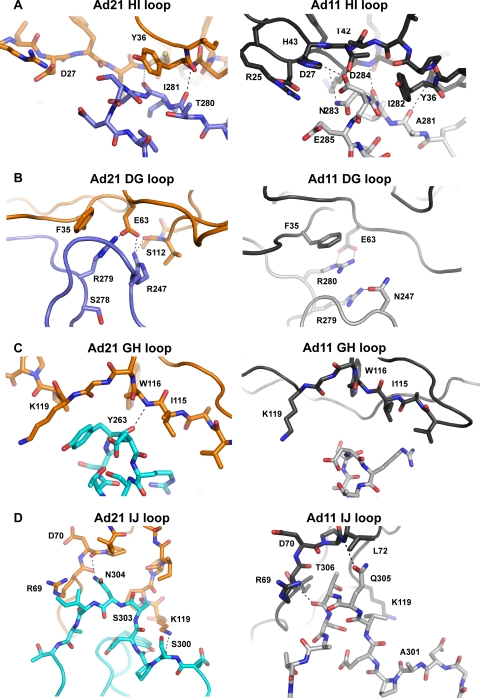
FIG. 6.
Comparison of interactions of CD46 with the Ad21 and Ad11 knobs. The close-up views show the four contact regions of the Ad21 knob-CD46-D2 interaction (centered at loops HI, DG, GH, and IJ, respectively) and compare them with the respective interactions in the Ad11 knob-CD46 complex. The color scheme is the same as that described in the legend of Fig. 5A. Hydrogen bonds and salt bridges are represented with dashed lines.
Protein structure accession numbers.
The structures reported here have been deposited with the Protein Data Bank (http://www.rcsb.org/) under accession numbers 3L88 (Ad21 knob) and 3L89 (Ad21 knob in complex with CD46-D2).
RESULTS
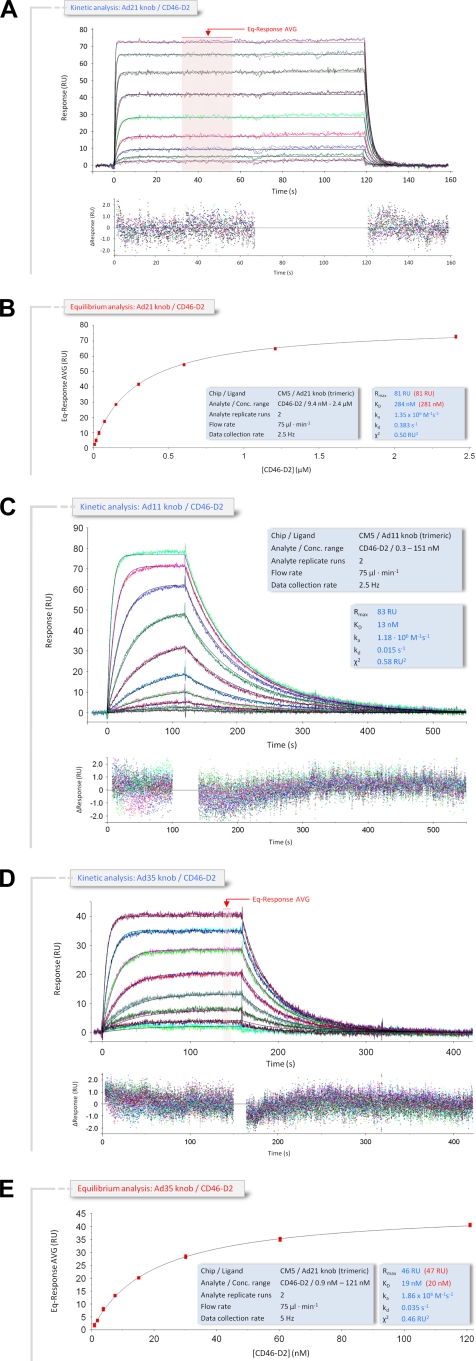
The affinity of Ad21 knobs for CD46-D2 is significantly lower than those of the Ad35 and Ad11 knobs.
The subspecies B1 Ad21 and the subspecies B2 Ad35 and Ad11 knobs all use CD46 as their primary receptor for infection (26). With 93.2% sequence identity, the Ad21 and Ad35 knobs are highly homologous and exhibit an almost complete conservation of surface loop residues. In contrast, the Ad35 and Ad21 knobs each share only 49.5% identical residues with the Ad11 knob, and residues in surface loops are poorly conserved. Our SPR measurements show that the affinities of the Ad11 and Ad35 knobs for CD46-D2 are almost identical (13 nM and 19 nM, respectively) despite these differences in sequence (Table 2) (33). In order to determine the corresponding affinity values for the Ad21 knob under identical conditions, kinetic and steady-state SPR studies were performed to determine the binding parameters of the interaction between the immobilized Ad21 knob and soluble CD46-D2 (Fig. 2A and B). Unexpectedly, we found that the KD value for the Ad21 knob-CD46-D2 interaction is 284 nM and, therefore, 22-fold lower than the binding affinity of Ad11 and 20-fold lower than the binding affinity of Ad35 for the same receptor. The sensorgram profile of the kinetics of the binding of the Ad21 knob to CD46-D2 shows a curve progression that is typical for rapid dissociation, with an almost square-wave-like shape (Fig. 2B). A quantitative evaluation of the kinetic data shows that the on-rate constant (ka) values are fairly stable for all three interactions (Table 2) (33), while the off-rate constant (kd) values are significantly higher for the Ad21 knob-CD46-D2 interaction (0.383 s−1) than for the corresponding Ad11 knob-CD46-D2 (0.015 s−1) and Ad35 knob-CD46-D2 (0.035 s−1) interactions. This increase of the Ad21 knob off-rate constant is clearly the main reason for the reduced affinity (Table 2) and provides evidence that the binding stability of the Ad21 knob-CD46-D2 complex is significantly weaker than the corresponding Ad11 and Ad35 knob-CD46-D2 interactions. The corresponding half-lives (t1/2 = 0.693·kd−1) of the three complexes also reflect this. The Ad21 knob-CD46-D2 complex exhibits a half-life of only 1.8 s, whereas the Ad11 knob-CD46-D2 and Ad35 knob-CD46-D2 complexes have considerably longer half-lives of 46.2 s and 19.8 s, respectively (Table 2). Fitting the data with a “1:1 binding with mass transfer, RI = 0” model with BIAevaluation 4.1 (Biacore) yields excellent residual plots with no obvious deviations of the sensorgram data from the fit. As the measurements were performed in the exact same manner for all three knobs, and by using the same CD46-D2 protein, we conclude that the lower affinity of the Ad21 knob for CD46-D2 is not due to a variation in experimental conditions but rather is a result of differences in stabilizing interactions.
TABLE 2.
Interaction data from SPR
| Protein interaction | KD (M) | ka (M−1 s−1) | kd (s−1) | t1/2 (s)a |
|---|---|---|---|---|
| Ad21 knob-CD46-D2 | 284 × 10−9 | 1.35 × 106 | 0.383 | 1.8 |
| Ad11 knob-CD46-D2 | 13 × 10−9 | 1.18 × 106 | 0.015 | 46.2 |
| Ad35 knob-CD46-D2 | 19 × 10−9 | 1.86 × 106 | 0.035 | 19.8 |
t1/2 = 0.693·kd−1.
FIG. 2.
Kinetic and equilibrium SPR analyses of the CD46-D2 interaction with the Ad21, Ad11, and Ad35 knobs. (A) Ad21 knob interaction with CD46-D2. Double-referenced sensorgrams of a kinetic SPR titration series (shown in color) are overlaid with fits of a “1:1 binding with mass transfer, RI = 0” model (black lines) on top of the corresponding residual values showing the kinetic-fit range and absolute deviation (Δ) of data points from curve fit values. The arrow indicates data used to determine averaged (AVG) equilibrium response values (Eq-Response AVG) for equilibrium analysis. (B) Equilibrium analysis of CD46-D2 binding to the Ad21 knob. Averaged equilibrium response values (red squares) obtained from data in A were plotted against the corresponding CD46-D2 concentration and fitted to a “1:1 Langmuir isotherm” model (black line). The light-blue-shaded boxes show setup details and measured parameters of the kinetic (blue font) and equilibrium (red font) analyses. (C) Kinetic SPR analysis of CD46-D2 binding to the Ad11 knob. The analysis was carried out as described above (A). (D) Kinetic SPR analysis sensorgram of CD46-D2 binding to the Ad35 knob. The analysis was carried out as described above (A). (E) Equilibrium analysis of CD46-D2 binding to the Ad35 knob. The analysis was carried out as described above (B). RU, resonance units.
Comparison of the Ad21, Ad11, and Ad35 knob structures reveals conformational differences in surface loops.
Next, we investigated whether the lower CD46-D2 binding affinity of the Ad21 knob is linked to structural features unique to this protein. Crystal structures of the Ad11 and Ad35 knobs are available (31, 33, 44). In order to provide a basis for their comparison with the Ad21 knob, we determined the crystal structure of the unliganded Ad21 knob to a 2.5-Å resolution. Not surprisingly, the overall fold and trimeric organization of the Ad21 knob are very similar to those seen for other knobs. However, the Ad21 knob structure reveals interesting differences in the conformation of three surface loops that participate in the interaction with CD46-D2 in the Ad11 knob: the HI loop, the DG loop, and the IJ loop (34). An additional loop, termed the GH loop, is also in close proximity and was therefore included in our discussion, although residues in the Ad11 variant of this loop do not directly contact CD46-D2. A sequence alignment of all three knobs is given in Fig. 3A, and a superposition of the loop regions of the unliganded Ad21, Ad35, and Ad11 knob structures is shown in Fig. 3B. From this comparison, it is apparent that despite differences in sequence, the HI and GH loops have almost identical conformations in all three structures. The DG and IJ loops, however, differ significantly, which has implications for the interactions with CD46-D2.
The DG loop has a narrow shape and protrudes from the Ad21 knob core structure, whereas in Ad35 and Ad11, it is wider and less protruding. A closer inspection suggests an explanation for the observed conformational DG loop differences between the Ad21 and Ad11 knobs (Fig. 3C and E). A conserved asparagine residue, which is located at one end of the DG loop, performs drastically different functions in the two proteins. In the Ad21 knob, the side chain of Asn243 faces toward the DG loop center, with its amide group forming hydrogen bonds with the hydroxyl group of Ser278 and the backbone carbonyl of Pro241. The corresponding Ad11 asparagine (Asn245) cannot perform similar interactions and instead faces toward the solvent. In the Ad11 knob, Ser278 is replaced with an arginine (Arg279). The larger Arg279 side chain fills much of the center of the Ad11 knob DG loop. As it forms hydrogen bonds with the side chain of loop residues Asn247 and Glu250, it appears that Arg279 is responsible for forcing the DG loop into a wider, more oval conformation. The observed differences in loop structure are unrelated to crystal packing effects. The Ad21 knob structure contains 12 noncrystallographically linked copies of the monomer, which all have closely similar conformations of their DG loops. The overall root mean square (RMS) deviation for Cα atoms among the 12 DG loops is 0.401 Å. While the Ad11 knob structure contains only one monomer in the asymmetric unit, the crystal structure of the Ad11 knob in complex with CD46-D2 contains two copies of the Ad11 knob DG loop. All three Ad11 knob DG loops adopt the same conformation.
Upon first inspection, the conformation of the DG loop in the Ad35 knob resembles that seen for the Ad11 knob (Fig. 3B, D, and E). However, the sequence of that loop is identical in Ad21 but quite different from the sequence in Ad11. Thus, the sequence in the DG loop does not correlate directly with its conformation. The adjacent regions of the DG loops in Ad21 and Ad35 knob structures differ only in the HI loop residue Met280 in Ad35, which is replaced with Thr280 in the Ad21 knob. This sequence difference could be an explanation for the observed loop structure differences between Ad35 and Ad21 knobs, as the Ad35 Met280 is closer to Phe242 than the Ad21 Thr280 (3.4 Å compared to 4.4 Å) (Fig. 3A, C, and D). Furthermore, the Ad35 knob structures have been determined to 2.0-Å and 2.7-Å resolutions (31, 44), and in both structures, the DG loop exhibits high mobility, as judged from its elevated-temperature factors, indicating multiple possible conformations.
The IJ loop also differs substantially between the Ad21 and Ad11 knobs (Fig. 3B, C, and D). In all three structures, including the Ad35 knob, the IJ loop is stabilized through a hydrogen bond between a conserved glutamic acid side chain (Glu302 in Ad21) and the hydroxyl group of a conserved serine (Ser262 in Ad21) contributed by the GH loop (Fig. 3C). In all three structures, the IJ loop exhibits little mobility and low-temperature factors. An inspection of the sequence shows that the Ad11 IJ loop contains two additional amino acids, Val304 and Gln305 (Fig. 3A), that are inserted into the tip of the loop, accounting for its more protruding conformation.
Structure determination of the Ad21 knob in complex with CD46-D2.
The crystal structure of the complex between the Ad11 knob and CD46-D2 demonstrated that the DG, HI, and IJ loops of the Ad11 knob mediate key contacts with CD46-D2 and also revealed a striking shape complementarity of the Ad11 knob and the CD46-D2 surfaces (34). As the conformations of the DG and IJ loops differ significantly among the Ad21, Ad11, and Ad35 knobs (Fig. 3), it is unlikely that the Ad21 knob engages CD46-D2 in a manner identical to that seen for the Ad11 knob. In order to provide a structural basis for Ad21 knob binding to CD46-D2, we crystallized a complex of the Ad21 knob with the exact same CD46-D2 fragment that was also used to generate the Ad11 knob-CD46-D2 complex. The obtained crystals were sensitive to radiation and diffracted to only a 3.5-Å resolution at synchrotron sources. Initial phases were obtained by molecular replacement using only the Ad21 knob trimer as a search model. The resulting 2Fo-Fc and Fo-Fc electron density maps clearly showed the location and orientation of the SCR1 and SCR2 domains of the CD46-D2 protein at this stage. The crystals contain 4 noncrystallographically related copies of Ad21 knobs. Thus, we were able to apply NCS restraints to aid crystallographic refinement. All noncrystallographic symmetry operators were calculated separately for the Ad21 knob, the CD46-D2 SCR1 domain, and the CD46-D2 SCR2 domain in order to allow for differences in the relative orientations of these units. The symmetry operators were also updated after each refinement cycle. Using this strategy, we were able to obtain a model for the complex that agrees well with the crystallographic data and has good geometry (Table 1). The Ad21 knob and CD46-D2 backbone chains are well defined by electron density (Fig. 1). Many side chains, especially at the CD46-D2-Ad21 knob interface, are also visible. Despite its moderate resolution, our model therefore provides a solid basis for an understanding of the intermolecular interactions and allows a meaningful comparison with the previously determined structure of the Ad11 knob-CD46-D2 complex. Four of the 12 CD46-D2 molecules are involved in crystal contacts in our structure. Their orientation is therefore somewhat influenced by packing forces, resulting in small differences in the relative orientation of one binding loop region, the DG loop, as well as CD46-D2 domains. The remaining eight CD46-D2 monomers lack crystal contacts near the knob-receptor interface. As they all make very similar contacts with the Ad21 knob, we will use a representative of this group for the presentation and discussion of the interactions.
Overall structure of the Ad21 knob in complex with CD46-D2.
As expected, each trimeric Ad21 knob engages three CD46-D2 molecules (Fig. 4A). At the interface with one CD46-D2 molecule, two Ad21 protomers are involved in complex stabilization: one protomer interacts via residues in the DG and HI loops, and a neighboring protomer interacts with the GH and IJ loops (Fig. 4A). The interactions bury an area of 2,320 Å2 from solvent and feature high surface complementarity (Fig. 4B). The overall conformations of most of the Ad21 knob residues, including those located in the HI, GH, and IJ loops, are similar in both the unliganded and the CD46-D2-bound states, with an overall RMS deviation of 0.47 Å. However, the electron density clearly shows that the DG loop in the liganded Ad21 knob has a different conformation (Fig. 4C) compared to that of the unliganded Ad21 knob (Fig. 4C and D). Variations in DG loop structures among all unliganded Ad21 knobs, among all liganded Ad21 knobs, and between the liganded and unliganded Ad21 knobs are shown in Fig. 4E. The local variations in DG loops of the unliganded and liganded Ad21 knobs were calculated by separately superposing the eight liganded Ad21 knob protomers onto the 12 unliganded protomers. The average of these 96 superpositions is 1.6 Å. However, a large deviation, about 3.6 Å, was seen at residue Arg247, which has different conformations in the unliganded and liganded Ad21 knobs. In the unliganded Ad21 knob, the Arg247 main chain occupies a similar position, while its side chain is flexible and adopts several orientations among the 12 monomers in the asymmetric unit. In all copies of the liganded Ad21 knob, however, the Arg247 main chain shifted, and its side chain formed a salt bridge with CD46-D2 residue Glu63 as well as a hydrogen bond with the backbone carbonyl atom of CD46-D2 residue Ser112. We therefore propose that contacts involving Arg247 are primarily responsible for the conformational change in the DG loop upon the engagement of CD46-D2. The rearrangement of an Ad knob DG loop is unique to the Ad21 knob-CD46-D2 interaction, as changes in the main chain conformation were not observed for the Ad11 knob structure upon the engagement of CD46-D2 (33, 34).
FIG. 4.
Structure of the Ad21 knob in complex with CD46-D2. (A) Ribbon drawing of the trimeric Ad21 knob in complex with three CD46-D2 molecules. The three protomers that form the knob are shown in blue, cyan, and gray, and the CD46-D2 ligands are shown in orange. One Ad21 knob-CD46-D2 interface is shown from the front, whereas two other Ad21 knob-CD46-D2 interfaces are shown from behind. (B) Surface representation of two Ad21 knob protomers (labeled protomer 1 and protomer 2) bound to one CD46-D2 ligand. The color code used is described above (A). (C) CD46-D2-bound Ad21 knob DG loop with the final nonaveraged 2Fo-Fc electron density map. (D) Superposition of the DG loop of the unliganded Ad21 knob (green) and the CD46-D2-bound Ad21 knob (blue). The relevant portion of CD46-D2 is shown in orange. The hydrogen bond and salt bridge are shown with dashed lines. (E) Comparison of DG loop conformations (residues 242 to 248) among unliganded Ad21 knob structures (green squares), among liganded Ad21 knob structures (blue squares), and between the liganded and unliganded structures (black squares). The average RMS deviation values for the Cα atoms of each residue are plotted as a function of the residue number. The bars show the standard deviations of the measured RMS deviation values for the Cα atoms.
The overall organization of the complex resembles the Ad11 knob-CD46-D2 structure (34). However, a comparison of the buried surface area in the interactions (1,681 Å2 for the Ad11 knob-CD46-D2 complex and 2,320 Å2 for the Ad21 knob-CD46-D2 complex) shows that the Ad21 knob-CD46-D2 contact area is larger. Moreover, a superposition of the two complexes reveals that the location of the CD46-D2 molecules relative to the knob clearly differs in both cases (Fig. 5A). Compared with the Ad11 knob-CD46-D2 complex, the SCR1 and SCR2 domains are shifted by distances that range from 2 Å to more than 7 Å in the complex with the Ad21 knob. The overall shift of CD46-D2 can be attributed, at least in part, to the profoundly different conformation of the DG loop in the Ad21 knob, which would sterically interfere with CD46-D2 and prevent it from binding in the orientation observed for the complex with the Ad11 knob. The protruding DG loop in the Ad21 knob thus displaces the SCR1 domain, which was previously shown to comprise the major determinants in species B adenovirus binding (34, 44). In addition, the shorter IJ loop in Ad21 allows SCR2 to approach the Ad21 knob more closely.
FIG. 5.
Comparison of the CD46-D2 structure in different states of ligation. (A) Superposition of the Ad21 knob-CD46-D2 complex (Ad21 knob protomers are in cyan and blue, and CD46-D2 is in orange) with the Ad11 knob-CD46-D2 complex (Ad11 knob protomers are in light gray, and CD46-D2 is in dark gray). Common binding regions are boxed. The dashed box indicates a binding region that mediates interactions only in the Ad21 knob. The arrows depict the displacement of the Ad21 knob-bound CD46-D2 regions compared to their positions in the Ad11 knob-CD46-D2 complex. (B) Superposition, based on residues in the SCR1 domain, of the three CD46-D2 structures known to date, shown in stereo. Unliganded CD46-D2 is shown in dark blue, CD46-D2 as seen in the complex with the Ad21 knob is shown in orange, and CD46-D2 as seen in the complex with the Ad11 knob is shown in dark gray. (C) Schematic view of unliganded CD46-D2. Disulfide bonds are represented with red lines. Two dashed lines link the disulfide bonds in SCR1 and SCR2. The variations in domain orientation are expressed in tilt angles (Φ) and twist angles (Ψ), as indicated with arrows (5).
A comparison of the overall conformation of the CD46-D2 molecules bound to Ad11 and Ad21 shows that although CD46-D2 adopts an elongated conformation in both complex structures, the elongated structures are not identical (Fig. 5B). To compare the overall conformations of the two-domain CD46-D2 protein in different states of ligation, tilt (Φ) and twist (Ψ) angles were calculated, as previously described (Table 3) (5). The tilt angle differs by 8° and the twist angle differs by 10° in the two complexes, indicating that the relative orientations of SCR1 and SCR2 are not identical. The CD46-D2 protein therefore possesses some flexibility at its interdomain interface, which allows it to adapt to subtly varying surfaces provided by different Ad knobs. Both interdomain angles differ substantially from those of the unliganded CD46-D2 conformation (Fig. 5B and Table 3).
TABLE 3.
Interdomain orientations of CD46-D2
| Conformation of CD46-D2 | Angle (°) |
|
|---|---|---|
| Φ | ψ | |
| Unligandeda | 116 | 70 |
| Bound to Ad21 knobb | 161 | 152 |
| Bound to Ad11 knobc | 153 | 162 |
Tilt angles vary by 15° among the six copies of the molecule in the asymmetric unit (7).
Twist angles vary by 12° among the 12 copies of the molecule in the asymmetric unit. There are no significant differences in tilt angles.
There are no significant differences in the tilt and the twist angles between the two copies of the molecule in the asymmetric unit.
Contacts at the Ad21 knob-CD46-D2 interface and comparison with Ad11 knob contacts.
The Ad21 knob contacts CD46-D2 via the HI, DG, GH, and IJ loops. The HI and DG loops are contributed by one protomer (blue in Fig. 5A and B), whereas the GH and IJ loops belong to the second protomer (cyan in Fig. 5A and B). Contacts involving the HI loop are limited to the SCR1 domain of CD46-D2 and are centered at Tyr36 and neighboring residues in CD46-D2 (Fig. 6A and B). Compared to the Ad11 knob-CD46-D2 complex, contacts in Ad21 are less extensive (Fig. 6A). A hydrogen bond tethering the Tyr36 backbone to the Ad knob HI loop backbone is conserved in both cases, but several additional hydrogen bonds and polar interactions between the side-chain and main-chain atoms are present only in the Ad11 knob-CD46-D2 complex.
The second knob-CD46-D2 interface involves the HI and DG loops (Fig. 6B). This region features a conserved salt bridge between an arginine side chain (Arg279 in Ad21 and Arg280 in Ad11) and Glu63 of CD46-D2. In both cases, the arginine guanidium group is stacked against the phenyl ring of Phe35. The salt bridge and the stacking of the guanidinium group against the aromatic ring are critical for allowing CD46 to bind in an extended conformation, as they determine interactions with SCR1 residue Phe35 and SCR2 residue Glu63. Furthermore, the Phe35 side chain is buried in the SCR1-SCR2 interface in unliganded CD46-D2 and becomes solvent exposed upon Ad knob engagement (7, 34). In Ad11, the Arg280 side chain stacks against that of the neighboring Arg279. Mutagenesis experiments with Ad11 have previously shown that the presence of Arg279 is critical for the engagement of CD46-D2 (18). The most likely reasons for this are favorable π-π interactions between the two guanidinium groups of Arg279 and Arg280 as well as the stabilization of the HI and DG loop conformations through a hydrogen bond with Asn247 in the DG loop (34). In Ad21, residue Arg279 is replaced with a serine (Ser278) that is involved in neither contacts with CD46-D2 nor interactions that would stabilize the conformations of the HI and DG loops. The replacement of Arg279 with serine would therefore be expected to render the DG loop more flexible, and this explains our observation that the DG loop rearranges upon the engagement of the receptor in the Ad21 knob but not in the Ad11 knob. An additional contact in the Ad21 knob-CD46-D2 complex is formed by a salt bridge between Glu63 and Arg247 (Fig. 6B).
The GH loop forms contacts with CD46-D2 only in the Ad21 knob (Fig. 6C). The interactions are centered at Ad21 knob residue Tyr263, which stacks against the hydrophobic portion of Lys119 in SCR2. A main-chain hydrogen bond between the Tyr263 carbonyl oxygen and the Trp116 nitrogen atom further stabilizes the interaction. In the Ad11 knob, the GH loop is not involved in CD46-D2 contacts (Fig. 6C).
The Ad knob IJ loop mediates extensive interactions in both complexes (Fig. 6D). In the Ad11 knob-CD46-D2 complex, the interface is stabilized by two hydrogen bonds between knob residues Gln305 and Thr306 and the CD46 main chain. The corresponding Ad21 knob-CD46-D2 complex also features two hydrogen bonds at this position, located between the Ad21 knob Ser300 main chain and the Lys119 side chain of SCR2 and between the Ad21 knob Asn304 main chain and the side chain of Asp70 in SCR2. Several additional polar interactions also stabilize this interface.
DISCUSSION
A number of species B Ads use CD46 as a cellular receptor (26), and the crystal structure of the Ad11 knob in complex with CD46-D2 has provided a structural basis for this interaction (34). By performing a structure-function analysis of the Ad21 knob and its interaction with CD46-D2, we establish here the determinants of receptor binding for a second species B Ad knob and compare them with those of the Ad11 knob-CD46-D2 interaction. Our analysis reveals a generally conserved binding mode, with key differences in intermolecular contacts that can be linked to differences in affinity.
Compared with the Ad11 knob, the Ad21 knob features a substantially more-protruding DG loop that creates a “hump” at the center of the CD46-D2-binding surface and a shorter IJ loop. The protruding DG loop of the unliganded Ad21 knob is ordered and well defined by electron density, and its loop conformation is also stabilized by interactions involving loop side chains. The structure of the Ad16 knob, which was solved recently, also features a protruding DG loop (called FG loop in that serotype), and this loop protrudes even further due to a 2-amino-acid insertion (32). According to the conformation of this loop, the CD46-binding Ad knobs can be grouped into two classes: knobs with a retracted DG loop and knobs with a protruding DG loop (14, 31, 32, 33, 44). The Ad11 knob features a retracted variant of this loop, which does not change its conformation upon ligand binding but rather serves as a rigid docking platform for CD46-D2. The conformation of the loop is stabilized by a complex network of interactions centered at the Arg279 side chain. However, the DG loop of the Ad21 knob rearranges significantly upon CD46 binding. Moreover, the receptor in the Ad21 knob complex is shifted compared to its location in the Ad11 knob complex. The Ad knob DG loop thus emerges as the key determinant of differences in CD46-D2 engagements.
The observed retracted and protruding conformations of the DG loop also correlate well with CD46-D2 binding affinities that have been determined for several Ad knobs. The Ad11 knob binds CD46-D2 with a 13 nM affinity, whereas the Ad21 and Ad16 knobs bind CD46-D2 with 284 nM and 437 nM affinities, respectively (32). As the length of the DG loop determines the position of CD46-D2 relative to the knob, a protruding DG loop does not allow the residues at the base of SCR1 and the SCR1-SCR2 interface to approach the core of the knob as closely as is the case for a knob with a retracted loop. The SCR1 domain and its contacts with the HI loop have been shown to be a major determinant in species B adenovirus binding (18, 34, 44). The total buried surface area in the Ad21 knob-CD46-D2 complex is larger (2,320 Å2) than that in the Ad11 knob-CD46-D2 complex (1,681 Å2). However, the buried surface area between SCR1 and the Ad21 knob HI loop (624 Å2) is smaller than the surface area buried in the contact between SCR1 and the Ad11 knob HI loop (676 Å2). It is thus tempting to speculate that altered contacts at the base of SCR1 and the SCR1-SCR2 interface result in weaker binding.
An additional determinant of CD46-D2 engagement by Ad knobs is the IJ loop, which interacts with the SCR2 domain. The shorter IJ loop in the Ad21 knob than in the Ad11 knob allows an SCR2 domain shift toward the knob in order to form contacts.
The structure determination of the species B Ad11 knob in complex with the two N-terminal CD46 domains revealed key determinants of this interaction, such as a central salt bridge between an Ad11 knob Arg280 and CD46 residue Glu63. This contact is augmented by cation-π interactions between the aromatic ring of CD46 residue Phe35 and the guanidinium group of Arg280. As Phe35 and Glu63 are located at the CD46 SCR1-SCR2 domain interface, their engagement by the Ad11 knob leads to a conformational rearrangement of the CD46-D2 fragment, straightening the protein into an almost linear conformation. Additional contacts involving the surface-exposed DG, HI, and IJ loops at the interface of two adjacent Ad11 knob protomers cement the interaction, providing a basis for a high-affinity interaction.
Interestingly, Ad21, Ad34, Ad35, and Ad50 all use CD46 as a receptor, and all of them feature similar sequences in their knob regions. However, their loop sequences differ substantially from those of the Ad11 knob. Moreover, the closest Ad11 relatives, Ad7 and Ad14, bind CD46 rather poorly and likely do not use CD46 as a receptor at all (26, 33, 34).
The interaction between a virus and a host cell receptor is the first step in a complex process that leads to the infection of the cell. Not only must viruses be able to specifically attach to cells in order to gain entry into the cell, but their newly formed progeny must also be able to release themselves from the cell membrane after an infection. Therefore, attachment and release processes depend on precisely regulated contacts and affinities between viral proteins and their cognate receptor molecules on the cell surface. Subtle modifications in the virus coat proteins can have drastic consequences, leading to the emergence of a new pathogen with altered infectivity, tissue tropism, or host range. We cannot exclude the possibility that other, postentry mechanisms contribute to the differences in tropism that were observed for species B Ads. However, it is tempting to speculate that the affinity for CD46, to a certain extent, contributes to the observed differences in tropism: high affinity (Ad11 and Ad35) correlates with urinary tract tropism, and low affinity (Ad7, Ad14, Ad16, and Ad21) correlates with ocular and/or respiratory tract tropism (10, 11, 43). Furthermore, Ad8, Ad19, and Ad37, which cause epidemic keratoconjunctivitis, are the only Ads that use sialic acid as a receptor and thus show a clear link to tropism and receptor usage (3).
Several well-resolved structures of viruses or viral proteins in complex with cellular receptors are available. In very few cases, however, has one been able to compare structures of different strains or serotypes of the same virus in complex with the same receptor (4, 39, 42). The structure-function analysis of the Ad21 knob interaction with CD46-D2 presented here exemplifies such a case, advancing our understanding of how subtle differences at the receptor interface can lead to altered binding properties of the same receptor.
Acknowledgments
We thank Ulrike Scheu for purification of the CD46-D2 construct and the staff at beam lines X06SA of the Swiss Light Source (Villigen, Switzerland) and ID14-4 of the European Synchrotron Radiation Facility (Grenoble, France) for beam time and support.
We gratefully acknowledge financial support from German Research Foundation grants STE-1463 and SFB-685 (T.S.) and from Swedish Research Council grant 521-2007-3402 (N.A.).
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, H., T. Kurosu, C. Sakashita, T. Inoue, S. Mori, K. Ohashi, S. Tanikawa, H. Sakamaki, Y. Onozawa, Q. Chen, H. Zheng, and T. Kitamura. 2001. Adenovirus is a key pathogen in hemorrhagic cystitis associated with bone marrow transplantation. Clin. Infect. Dis. 32:1325-1330. [DOI] [PubMed] [Google Scholar]
- 3.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 4.Bewley, M. C., K. Springer, Y. B. Zhang, P. P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 5.Bouma, B., P. G. de Groot, J. M. van den Elsen, R. B. Ravelli, A. Schouten, M. J. Simmelink, R. H. Derksen, J. Kroon, and P. Gros. 1999. Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure. EMBO J. 18:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Casasnovas, J. M., M. Larvie, and T. Stehle. 1999. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 18:2911-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cattaneo, R. 2004. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens' magnet. J. Virol. 78:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darougar, S., R. Pearce, J. A. Gibson, and D. A. McSwiggan. 1978. Adenovirus type 21 keratoconjunctivitis. Br. J. Ophthalmol. 62:836-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Azevedo, J. P., L. R. Nascimento, M. C. Cortinovis, S. S. Oliveira, E. V. da Costa, and E. E. da Silva. 2004. Characterization of species B adenoviruses isolated from fecal specimens taken from poliomyelitis-suspected cases. J. Clin. Virol. 31:248-252. [DOI] [PubMed] [Google Scholar]
- 12.DeLano, W. E. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 13.Dorig, R. E., A. Marcil, A. Chopra, and C. D. Richardson. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295-305. [DOI] [PubMed] [Google Scholar]
- 14.Durmort, C., C. Stehlin, G. Schoehn, A. Mitraki, E. Drouet, S. Cusack, and W. P. Burmeister. 2001. Structure of the fiber head of Ad3, a non-CAR-binding serotype of adenovirus. Virology 285:302-312. [DOI] [PubMed] [Google Scholar]
- 15.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 16.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 17.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 18.Gustafsson, D. J., A. Segerman, K. Lindman, Y. F. Mei, and G. Wadell. 2006. The Arg279Gln substitution in the adenovirus type 11p (Ad11p) fiber knob abolishes EDTA-resistant binding to A549 and CHO-CD46 cells, converting the phenotype to that of Ad7p. J. Virol. 80:1897-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havenga, M. J., A. A. Lemckert, O. J. Ophorst, M. van Meijer, W. T. Germeraad, J. Grimbergen, M. A. van Den Doel, R. Vogels, J. van Deutekom, A. A. Janson, J. D. de Bruijn, F. Uytdehaag, P. H. Quax, T. Logtenberg, M. Mehtali, and A. Bout. 2002. Exploiting the natural diversity in adenovirus tropism for therapy and prevention of disease. J. Virol. 76:4612-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, M. S., II, B. Harrach, R. D. Ganac, M. M. Gozum, W. P. Dela Cruz, B. Riedel, C. Pan, E. L. Delwart, and D. P. Schnurr. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 22.Kallstrom, H., M. K. Liszewski, J. P. Atkinson, and A. B. Jonsson. 1997. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol. Microbiol. 25:639-647. [DOI] [PubMed] [Google Scholar]
- 23.Leen, A. M., and C. M. Rooney. 2005. Adenovirus as an emerging pathogen in immunocompromised patients. Br. J. Haematol. 128:135-144. [DOI] [PubMed] [Google Scholar]
- 24.Liles, W. C., H. Cushing, S. Holt, C. Bryan, and R. C. Hackman. 1993. Severe adenoviral nephritis following bone marrow transplantation: successful treatment with intravenous ribavirin. Bone Marrow Transplant. 12:409-412. [PubMed] [Google Scholar]
- 25.Liszewski, M. K., T. W. Post, and J. P. Atkinson. 1991. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu. Rev. Immunol. 9:431-455. [DOI] [PubMed] [Google Scholar]
- 26.Marttila, M., D. Persson, D. Gustafsson, M. K. Liszewski, J. P. Atkinson, G. Wadell, and N. Arnberg. 2005. CD46 is a cellular receptor for all species B adenoviruses except types 3 and 7. J. Virol. 79:14429-14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier, O., K. Boucke, S. V. Hammer, S. Keller, R. P. Stidwill, S. Hemmi, and U. F. Greber. 2002. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 158:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzgar, D., M. Osuna, A. E. Kajon, A. W. Hawksworth, M. Irvine, and K. L. Russell. 2007. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J. Infect. Dis. 196:1465-1473. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 30.Naniche, D., G. Varior-Krishnan, F. Cervoni, T. F. Wild, B. Rossi, C. Rabourdin-Combe, and D. Gerlier. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pache, L., S. Venkataraman, G. R. Nemerow, and V. S. Reddy. 2008. Conservation of fiber structure and CD46 usage by subgroup B2 adenoviruses. Virology 375:573-579. [DOI] [PubMed] [Google Scholar]
- 32.Pache, L., S. Venkataraman, V. S. Reddy, and G. R. Nemerow. 2008. Structural variations in species B adenovirus fibers impact CD46 association. J. Virol. 82:7923-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persson, B. D., S. Muller, D. M. Reiter, B. B. Schmitt, M. Marttila, C. V. Sumowski, S. Schweizer, U. Scheu, C. Ochsenfeld, N. Arnberg, and T. Stehle. 2009. An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46. J. Virol. 83:673-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persson, B. D., D. M. Reiter, M. Marttila, Y. F. Mei, J. M. Casasnovas, N. Arnberg, and T. Stehle. 2007. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat. Struct. Mol. Biol. 14:164-166. [DOI] [PubMed] [Google Scholar]
- 35.Read, R. J. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57:1373-1382. [DOI] [PubMed] [Google Scholar]
- 36.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santoro, F., P. E. Kennedy, G. Locatelli, M. S. Malnati, E. A. Berger, and P. Lusso. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817-827. [DOI] [PubMed] [Google Scholar]
- 38.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seiradake, E., H. Lortat-Jacob, O. Billet, E. J. Kremer, and S. Cusack. 2006. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J. Biol. Chem. 281:33704-33716. [DOI] [PubMed] [Google Scholar]
- 40.Seya, T., J. R. Turner, and J. P. Atkinson. 1986. Purification and characterization of a membrane protein (gp45-70) that is a cofactor for cleavage of C3b and C4b. J. Exp. Med. 163:837-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stehle, T., and J. M. Casasnovas. 2009. Specificity switching in virus-receptor complexes. Curr. Opin. Struct. Biol. 19:181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Avoort, H. G., T. Adrian, R. Wigand, A. G. Wermenbol, T. P. Zomerdijk, and J. C. de Jong. 1986. Molecular epidemiology of adenovirus type 21 in The Netherlands and the Federal Republic of Germany from 1960 to 1985. J. Clin. Microbiol. 24:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, H., Y. C. Liaw, D. Stone, O. Kalyuzhniy, I. Amiraslanov, S. Tuve, C. L. Verlinde, D. Shayakhmetov, T. Stehle, S. Roffler, and A. Lieber. 2007. Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob. J. Virol. 81:12785-12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 46.Wold, W. S., and M. S. Horwitz. 2007. Adenoviruses, p. 2395-2436. In D. M. Knipe et al. (ed.), Field's virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]