Abstract
Human cytomegalovirus (HCMV) virion assembly takes place in the nucleus and cytoplasm of infected cells. The HCMV virion tegument protein pp150 (ppUL32) is an essential protein of HCMV and has been suggested to play a role in the cytoplasmic phase of HCMV assembly. To further define its role in viral assembly and to identify host cell proteins that interact with pp150 during viral assembly, we utilized yeast two-hybrid analyses to detect an interaction between pp150 and Bicaudal D1 (BicD1), a protein thought to play a role in trafficking within the secretory pathway. BicD1 is known to interact with the dynein motor complex and the Rab6 GTPase. The interaction between pp150 and BicD1 was confirmed by coimmunoprecipitation and fluorescence resonance energy transfer. Depletion of BicD1 with short hairpin RNA (shRNA) caused decreased virus yield and a defect in trafficking of pp150 to the cytoplasmic viral assembly compartment (AC), without altering trafficking to the AC of another essential tegument protein, pp28, or the viral glycoprotein complex gM/gN. The C terminus of BicD1 has been previously shown to interact with the GTPase Rab6, suggesting a potential role for Rab6-mediated vesicular trafficking in HCMV assembly. Finally, overexpression of the N terminus of truncated BicD1 acts in a dominant-negative manner and leads to disruption of the AC and a decrease in the assembly of infectious virus. This phenotype was similar to that observed following overexpression of dynamitin (p50) and provided additional evidence that morphogenesis of the AC and virus assembly were dynein dependent.
Human cytomegalovirus (HCMV) (human herpesvirus 5 [HHV-5]), the prototypical betaherpesvirus, is ubiquitous in humans and establishes a persistent infection in the host (19). HCMV also reinfects healthy seropositive individuals, suggesting another mechanism for maintaining persistence in a population (9). Intrauterine transmission and HCMV infection of the developing fetus constitute a leading viral cause of birth defects (32). HCMV is also a leading cause of opportunistic infections in immunocompromised patients, including transplant recipients and patients with AIDS (10, 20). HCMV infection has also been implicated as a cofactor in such diverse diseases as atherosclerosis and cancer (8, 17, 33, 66).
HCMV replicates its genome in the nucleus, and acquisition of the final tegument and envelope is thought to occur in the cytoplasm of infected cells (73, 77). Envelopment of HCMV has been reported to occur by budding into cytoplasmic vacuoles that are composed of HCMV glycoproteins required for the assembly of infectious virions (37). The fully mature virus is released from the cell through either exocytosis or, possibly, lysis of the infected cells (56). The nucleic acid-containing capsid is embedded in a proteinaceous tegument layer that occupies the space between the nucleocapsid and the envelope. The tegument contains approximately 40% of the virion protein mass and approximately 20 to 25 known virion proteins, most of which are phosphorylated (40, 44). The assembly pathway and protein interactions required for formation of the tegument layer and the role of individual tegument proteins in the replication and assembly of infectious HCMV remain poorly understood. Deletion of viral genes encoding some tegument proteins results in varying levels of impairment in virus production (11-13, 35, 43, 45, 53, 68). Some tegument proteins, such as pp28 (pUL99) and ppUL25, are expressed only in the cytoplasm of infected cells during HCMV replication, whereas others, such as ppUL53 and pp65 (pUL83), are expressed in the nuclei of cells early in infection but are localized predominantly in the cytoplasm late in infection (68). Others, such as the tegument protein ppUL69, are expressed only in the nuclei of infected cells. Finally, the intracellular localization of other tegument proteins, such as pp150 (pUL32), is less well defined in that both nuclear and cytoplasmic localizations have been described (34, 68).
HCMV pp150 (basic phosphoprotein [BPP], pUL32) is the 1,048-amino-acid product of the UL32 gene of HCMV and an abundant constituent of the HCMV virion. Homologues of pp150 are found in other betaherpesviruses, including chimpanzee CMV, rat CMV, mouse CMV, HHV-6, and HHV-7, but not in alpha- or gammaherpesviruses (2). It is expressed late in HCMV infection (15, 68). It comprises 9.1% of infectious virion mass and 2% of the mass of dense bodies, suggesting that it is preferentially incorporated into virions (87). It has an estimated molecular mass of 113 kDa and is posttranslationally modified by phosphorylation and glycosylation, resulting in a molecular mass of 150 kDa in purified virus preparations analyzed by SDS-PAGE (41, 42, 65). pp150 has been classified as a tegument protein based on its presence in virion preparation, noninfectious enveloped particles, and cytoplasmic nucleocapsids but not in immature nuclear capsids (27, 28, 40). It has been suggested that pp150 contacts the capsids through the distal end of the capsomeres or through the triplex subunits that interlink them (16, 86). It has been reported to bind HCMV capsids in vitro through its amino one-third (6). We have also noted association of pp150 with the virion capsid by cryo-immunoelectron microscopy (W. Britt and H. Zhou, UCLA, Los Angeles, CA, unpublished findings). In primary human foreskin fibroblast (HFF) cells infected with HCMV, pp150 accumulates in a juxtanuclear structure that is termed the assembly compartment (AC), which colocalizes with markers of the distal secretory pathway and with other tegument proteins, including pp28 and pp65 and envelope glycoproteins gB, gH, and gM/gN (68). The virus-induced AC appears to overlap with microtubules emanating from the microtubule-organizing center (MTOC) and is proposed to be a cytoplasmic site of virion assembly (37, 68).
The function of pp150 is unknown, although its close association with the nucleocapsid suggests potential involvement in nuclear targeting during entry and in nuclear targeting of the encapsidated viral DNA, capsid tegumentation, and/or envelopment late in infection. It is essential for production of infectious virus, since the deletion of the UL32 open reading frame (ORF) leads to loss of virus replication and has been reported to be important in cytoplasmic maturation of HCMV, especially in viral egress (2, 22, 84, 91, 92). In cells infected with ΔUL32 virus, which lacks pp150, fewer virus particles accumulated in the cytoplasm, although nuclear steps in virus assembly were not affected (84). It was also observed that in the absence of pp150, nucleocapsids were present in the viral assembly compartment but failed to proceed further to vesicle transport-associated release (84). These observations, together with pp150 abundance in the virion, suggest a primary contribution for this structural protein in the morphogenesis and/or cytoplasmic transport of progeny virion particles to sites of virion envelopment.
Since pp150 has no predicted intracellular trafficking signals, its localization to the AC in virus-infected cells has been postulated to be dependent on interactions with cellular and/or viral proteins. Using yeast two-hybrid (Y2H) screening experiments we identified the cellular protein Bicaudal D1 (BicD1) as an interacting cellular protein. Bicaudal D was originally defined as a Drosophila protein that is involved in establishing the asymmetric cytoplasm in the developing oocyte (82, 89). Two homologues of Bicaudal D, BicD1 and BicD2, have been reported in humans, and these proteins have been reported to be involved in dynein-mediated microtubule transport as well as in COPI-independent Golgi-endoplasmic reticulum (ER) transport (38, 39, 55). Microtubule-dependent transport is an energy-dependent active transport system that includes both positive-end (directed away from the MTOC) and negative-end (directed toward the MTOC) transport. The direction of transport depends on cargo interactions with the molecular motors directing this transport, with dynein being associated with negative-end transport and kinesin with positive-end transport. BicD1 colocalizes with Rab6a in the trans-Golgi network and on cytoplasmic vesicles that associate with Golgi membranes in a Rab6-dependent manner secondary to a Rab6 binding domain at the C terminus of BicD1, suggesting an important role for BicD1 as an adaptor for dynein-dependent transport in the cell (55). In addition to having a role in the Golgi-ER trafficking, BicD1 has been shown to regulate anchoring of microtubules to the centrosome, as BICD1/2 knockdown induced microtubule unfocusing, with microtubules no longer appearing to radiate from the centrosome (26). BicD1 binds to its cargo via its C-terminal domain and to the dynein motor via its N-terminal domain (38). In this study we demonstrated that pp150 and BicD1 interact and that this interaction was required for localization of pp150 to the AC in virus-infected cells. In addition, we demonstrated that inhibition of BicD1 expression by short hairpin RNA (shRNA) led to a reduction in the yield of infectious virus. Finally, we demonstrated that formation of the AC and the assembly of infectious virions were dynein dependent, suggesting a critical role in microtubules in the production of infectious HCMV. Together, these results argue that HCMV replication is dependent on efficient localization of pp150 to the AC through its interaction with BicD1 and that pp150 localization to the AC is dynein dependent.
MATERIALS AND METHODS
Cells, virus and antibodies.
HCMV strain AD169 was propagated in primary human foreskin fibroblast (HFF) cells grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum (NCS) and penicillin-streptomycin. Infectious virus stocks were prepared from supernatants of infected HFF cells which exhibited a 100% cytopathic effect and stored at −80°C until use, and titers were determined as described previously (1). COS-7 cells were grown in DMEM supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin. HEK 293 cells (Grip Tite cells; Invitrogen, Carlsbad, CA) were cultured in DMEM supplemented with 10% FCS, 0.1% nonessential amino acids, and 50 μg/ml G-418. HCMV proteins were detected with monoclonal antibodies (MAbs) that have been previously described (48, 67-69, 74). MAbs used in the present study include those specific for pp150 (MAb 36-14), pp28 (MAb 41-18), gM/gN (MAb 14-16A), IE1 (MAb P63-27), and gM (MAb IMP). Bicaudal D1 was detected using a previously described rabbit polyclonal antibody (55). β-Tubulin was detected using a previously described mouse monoclonal antibody (68). Commercially available antibodies were used to detect actin (Chemicon, Temecula, CA), TGN 46 (AbD Serotec, Oxford, United Kingdom), and green fluorescent protein (GFP) (Invitrogen, Carlsbad, CA). The myc epitope was detected with mouse monoclonal antibody 9E10 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA).
Plasmids.
The Bicaudal D1-GFP plasmid has been previously described (55) and was a gift from Anna Akhmanova (Erasmus Medical Center, Rotterdam, The Netherlands). Using this plasmid as a template, we PCR amplified and cloned BicD1 into pcDNA 4/TO/myc-His C (Invitrogen, Carlsbad, CA) to construct a C-terminally myc-tagged BicD1 (forward primer, 5′GTGTGGATCCGCCACCATGGCCGCAGAAGAGGTATTGCAG3′; reverse primer, 5′GTGTGCGGCCGCGTTGTGAGCACTGTGGCGGTACGG3′). The bacterial artificial chromosome (BAC) encoding HCMV strain AD169 (HB-5), provided by M. Messerle and U. Koszinowski (University of Munich, Germany), was used as a template to PCR amplify the UL32 ORF encoding pp150, which was cloned into pcDNA 4/TO/myc-His C (Invitrogen) and myc tagged on its C terminus (forward primer, 5′GTGTGGATCCGCCACCATGAGTTTGCAGTTTATCGGTCTAC3′; reverse primer, 5′GTGTGCGGCCGCTTCCTCCGTGTTCTTAATCTTCTCG3′), into pEGFPN1 (Clontech, Mountain View, CA) to make a C-terminally GFP-tagged construct, into pmCherry N1 to make a C-terminally mCherry-tagged construct (forward primer, 5′GTGTAGATCTGCCACCATGAGTTTGCAGTTTATCGGTCTAC3′; reverse primer, 5′GTGTAAGCTTTTCCTCCGTGTTCTTAATCTTCTCG3′), and into pcDNA 3.1(+) (Invitrogen) to make an untagged full-length pp150 (forward primer, 5′GTGTGGATCCGCCACCATGAGTTTGCAGTTTATCGGTCTAC3′; reverse primer, 5′GTGTGCGGCCGCCTATTCCTCCGTGTTCTTAATCTTCTCG3′). To generate a GFP-mCherry fusion protein for use as a positive control in fluorescence resonance energy transfer (FRET) experiments, the GFP ORF was PCR amplified from pEGFPN1 vector (Clontech) and cloned into pmCherryN1 vector (Clontech) (forward primer, 5′GTGTAAGCTTCGCCACCATGGTGAGCAAGGGCGAG3′; reverse primer, 5′GTGTGGATCCGCCTTGTACAGCTCGTCCATGCCGAG3′). The pp28GFP plasmid has been described previously (74) and was subcloned into pmCherryN1 vector (Clontech). The myc-tagged pp28 plasmid has been described previously (75). Using the Bicaudal D1-GFP plasmid as template, we PCR amplified into the BglII/SalI sites of pEGFP-C2 vector (Clontech) the following amino acid regions of BicD1: 1 to 276 (forward primer, 5′GTGTGGATCCATATGGCCGCAGAAGAGGTATTGC3′; reverse primer, 5′GTGTGTCGACCTACCCATCCTCGGCAAATTTGAGTCC3′), 1 to 482 (forward primer, 5′GTGTGGATCCATATGGCCGCAGAAGAGGTATTGC3′; reverse primer, 5′GTGTGTCGACCTAGGCCATCTTCTCACCACTCTCCTTG3′), 343 to 482 (forward primer, 5′GTGTGGATCCATCTTTTGGCCAACCTACAGGAGTCAC3′; reverse primer, 5′GTGTGTCGACCTAGGCCATCTTCTCACCACTCTCCTTG3′), 343 to 820 (forward primer, 5′GTGTGGATCCATCTTTTGGCCAACCTACAGGAGTCAC3′; reverse primer, 5′GTGTGTCGACCTATTTAGGGCTGCCGATCTTGCTCTTTCC3′), and 630 to 820 (forward primer, 5′GTGTGGATCCATGCCATAATCCGGGACCAAATCAAGC3′; reverse primer, 5′GTGTGTCGACCTATTTAGGGCTGCCGATCTTGCTCTTTCC3′).
Yeast two-hybrid assay.
The ProQuest yeast two-hybrid (Y2H) system with Gateway technology (Invitrogen) was used to identify proteins interacting with pp150, according to the manufacturer's instructions. The pp150 protein sequence was analyzed using the PHYRE (Protein Homology/AnalogY Recognition Engine) protein fold recognition server (Imperial College, London) (46) to identify potential secondary structures. A putative alpha-helical domain was identified between amino acids (aa) 620 and 823 of pp150. The region of pp150 spanning amino acids proline 614 to serine 842 was PCR amplified using primers containing 5′ attB1 and 3′ attB2 recombination sites and cloned into pDONR222.1 vector (Invitrogen) to make the entry clones, followed by recombinatorial cloning in frame with the gal4 DNA binding domain in the pDEST32 yeast expression vector (Invitrogen). The pp150 bait clones were transformed into the MaV203 strain of yeast, and colonies that grew on selective SC-Leu medium were screened by PCR for the pp150/pDEST32 plasmid. Yeasts containing the pp150/pDEST32 were transformed with a Proquest human fetal brain cDNA library which contains cDNAs fused to the gal4 activation domain of pEXP-AD502 (Invitrogen). Interactions between fusion proteins in this system activate three reporter genes, those for β-galactosidase, HIS3, and URA3. Screening of ∼20 million clones resulted in the isolation of colonies that fulfilled the selection criteria of His prototropy on yeast dropout medium lacking Trp, Leu, and His in the presence of 3-aminotriazole. Positive colonies were confirmed by expression of β-galactosidase by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) filter assays and growth on yeast dropout medium lacking Leu, Trp, and Ura. Selected yeast colonies were grown, plasmids isolated, and the insert cDNA PCR amplified with primers specific for pEXP-AD502 (Invitrogen). The PCR products were sequenced using pEXP-AD502 sequencing primers and analyzed using the NCBI BLAST software, and interacting proteins were identified.
FRET by acceptor photobleaching. (i) FRET in transfected COS-7 cells.
Plasmids expressing mCherry-tagged pp150 and GFP-tagged BicD1 were cotransfected into COS-7 cells grown to 60% confluence on 13-mm glass coverslips. At 48 h posttransfection the cells were fixed with 3% paraformaldehyde for 15 min, washed with phosphate-buffered saline (PBS)-Tween 20 (0.05%), and mounted on glass slides, using n-propylgallate (0.2%) in 9:1 (vol/vol) glycerol-PBS as the mounting medium. GFP-mCherry fusion protein was used as a positive control. Fluorescence resonance energy transfer (FRET) by acceptor photobleaching was measured using a donor-dequenching and acceptor-photobleaching protocol on a confocal laser scanning microscope (Leica SP2; Leica, Bannockburn, IL) as described previously (74). An excitation wavelength of 488 nm and an emission range of 500 to 550 nm were used to acquire images of GFP-tagged proteins, and an excitation wavelength of 561 nm and an emission range of 570 to 700 nm were used to acquire images of mCherry-tagged proteins. The acceptor, mCherry, was photobleached to 30% of the original fluorescence signal by using full power of the 561-nm line. An image of GFP fluorescence and mCherry fluorescence after photobleaching was obtained. Similar data were collected from multiple cells. Multiple regions of interest (ROI) in the photobleached area per cell were selected, and the mean GFP fluorescences before and after photobleaching were obtained by using Leica confocal software. FRET efficiency (%) was calculated as [(Dpost − Dpre)/Dpost] ×100. Here, Dpost is the fluorescence intensity of the GFP (donor) after photobleaching, and Dpre is the fluorescence intensity of the GFP (donor cells) before photobleaching. A nonbleached region in the cell was selected as an internal control for FRET analysis. Negative controls included COS-7 cells cotransfected with GFP-tagged pp150 and mCherry-tagged pp28, with GFP-tagged BicD1 and mCherry-tagged pp28, and with pEGFPN1 and pmCherry N1 vectors.
(ii) FRET in HCMV-infected HFF cells.
HFF cells cultured to confluence on 13-mm glass coverslips were infected with HCMV AD169 at a multiplicity of infection (MOI) of 1. At 5 days postinfection, the cells were fixed with 3% paraformaldehyde for 15 min. The cells were permeabilized with 0.1% Triton X-100 and blocked with 2.5% goat serum in PBS. The cells were reacted with anti-pp150 monoclonal antibody (XPA 36-14), anti-BicD1 rabbit antiserum (number 2296; Anna Akhmanova, Erasmus Medical Center, Rotterdam, The Netherlands), and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse IgG2b and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibodies. The coverslips were processed for FRET. FRET was measured in infected cells in a fashion similar to that for COS-7 cells. In this case, an excitation wavelength of 488 nm and an emission range of 500 to 545 nm were used to acquire images of FITC-labeled proteins, and an excitation wavelength of 561 nm and an emission range of 575 to 630 nm were used to acquire images of TRITC-labeled proteins. FRET was measured in the assembly compartment (AC) as well as in the cytoplasmic vesicles in infected cells. As a negative control, FRET was measured between pp28 and pp150 using specific antibodies. A nonbleached region in the cell was selected as an internal control for FRET analysis.
Depletion of Bicaudal D1 by RNA interference (RNAi).
SureSilencing shRNA plasmids (SuperArray Bioscience Corp., Frederick, MD) were used to deplete BicD1 expression. This system has four plasmids expressing shRNA sequences targeting different regions of the BicD1 ORF and a negative-control plasmid expressing a scrambled shRNA sequence cloned into a humanized GFP (hMGFP) vector that allows identification of shRNA-expressing cells by GFP expression. HFF cells were electroporated using the Amaxa normal human dermal fibroblast (NHDF) basic nucleofection kit (Lonza, Walkersville, MD) with 4 μg of shRNA plasmid mix or a negative-control plasmid. The efficiency of electroporation was determined by an immunofluorescence assay. After electroporation, HFF cells were plated on 13-mm coverslips, and at 2, 4, and 6 days postelectroporation, cells were washed in Dulbecco's phosphate-buffered saline and fixed in 3% paraformaldehyde. The cells were permeabilized with 0.5% Triton X-100, and the nuclei were stained with Hoechst 33342 (Invitrogen) to visualize the total number of cells on the coverslips. We then calculated the number of cells expressing GFP per 100 cells stained with Hoechst 33342.
To assess the efficiency of depletion of BicD1, HEK 293 cells were cotransfected with a myc-tagged BicD1 and BicD1 shRNA mix or shRNA control plasmid using Trans-LT1 reagent (Mirus Bio Corporation, WI). At 2, 4, and 6 days posttransfection, cell pellets were harvested, lysed (50 mM HEPES [pH 7.2], 250 mM NaCl, 10% glycerol, 2 mM EDTA, 0.5% NP-40, and protease inhibitors), and stored at −80°C until used. The lysates were mixed with 2× SDS-PAGE loading buffer (20% SDS, 500 mM Tris [pH 7.6], 1% bromophenol blue, 50% glycerol, 1 M dithiothreitol [DTT]), boiled, and resolved by SDS-PAGE. After transfer to nitrocellulose membrane, immunoblots were blocked in 2.5% nonfat dry milk in PBS with 0.2% Tween 20, probed with anti-myc MAb 9E10, and developed with goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody, followed by incubation with ECL reagents (Pierce, Rockford, IL). Actin was used as a loading control and detected using mouse monoclonal antiactin antibody (Millipore, Billerica, MA). To confirm that the shRNA targeting BicD1 was able to deplete BicD1 in HFFs, HFFs were transfected with BicD1 shRNA mix or control shRNA, and 4 days later, the cells were washed once with PBS and lysed (50 mM HEPES [pH 7.2], 250 mM NaCl, 10% glycerol, 2 mM EDTA, 0.5% NP-40, and protease inhibitors). The lysates were resolved by SDS-PAGE, and the level of expression of endogenous BicD1 was checked by Western blotting, as described above, using a specific rabbit anti-BicD1 antibody (number 2296; Anna Akhmanova, Erasmus Medical Center, Rotterdam, The Netherlands). Nontransfected HFF lysates were used as a positive control. Actin was used as a loading control.
HFF cells were electroporated with BicD1 shRNA mix or scrambled shRNA as described above and grown on 13-mm glass coverslips; 24 h later the cells were infected with HCMV AD169 at a multiplicity of infection of 1.0. At 5 days postinfection, the cells were fixed with 3% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100, and blocked with 2.5% goat serum. The cells were reacted with specific MAbs to pp150, pp28, and gM/gN and rabbit polyclonal anti-BicD1 anti-serum and with the respective secondary antibodies as required. Nuclei were stained with TO-PRO-3 iodide (Invitrogen). Confocal images were taken using an Olympus Fluoview laser confocal microscope.
Immunofluorescence assays.
COS-7 cells were grown on 13-mm glass coverslips in 24-well BD-Falcon cell culture plates (BD Biosciences, CA) and transfected at 60% of confluence with 0.5 μg of each plasmid/coverslip using Mirus LT1 reagent (Mirus Bio, Madison, WI). Cells were processed for immunofluorescence assay at 24 h posttransfection. The HFF cells were electroporated with 4 μg of plasmid DNA using Amaxa as described above and grown on 13-mm glass coverslips in 24-well plates. At 24 h postelectroporation, HFF cells were infected with HCMV, and immunostaining was done at 5 days postinfection. The coverslips were washed with PBS, fixed in 3% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 2.5% goat serum in PBS. Following incubation with primary antibody and washing with PBS containing 0.1% Tween 20, antibody binding was detected with the appropriate secondary antibody conjugated with either FITC or TRITC (Southern Biotech, Birmingham, AL). The nuclei were stained with TO-PRO-3 iodide.
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate (SDS)-polyacrylamide electrophoresis (PAGE) under reducing conditions and immunoblotting were carried out as described previously (74). Solubilized proteins were subjected to SDS-PAGE and transferred onto nitrocellulose membranes. After reacting with the appropriate primary antibodies, the blot was developed with appropriate HRP-conjugated antibodies, followed by chemiluminescence detection as described above.
Coimmunoprecipitation.
HEK 293 GripTite cells (Invitrogen) grown to 60% confluence in 10-cm tissue culture dishes were transfected with appropriate expression plasmids such as pcDNA pp150myc and BicD1-GFP as described above, and 48 h later, the growth medium was aspirated, and the cells washed with PBS and lysed (50 mM HEPES [pH 7.2], 250 mM NaCl, 10% glycerol, 2 mM EDTA, 0.5% NP-40, and protease inhibitors) at 4°C. The lysate was sonicated and centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was kept at −80°C or used immediately. The myc-tagged proteins in the cell lysates were immunoprecipitated using magnetic beads conjugated with anti-myc antibodies (Miltenyi Biotec, Auburn, CA). The eluate was subjected to SDS-PAGE followed by Western blotting, treated with appropriate primary (e.g., rabbit anti-GFP antiserum) and secondary (e.g., goat anti-rabbit IgG-HRP) antibodies, and developed as described above. As a negative control we used magnetic beads conjugated with antihemagglutinin (anti-HA) antibodies (Miltenyi Biotec).
RESULTS
Formation and maintenance of the assembly compartment is microtubule and dynein dependent.
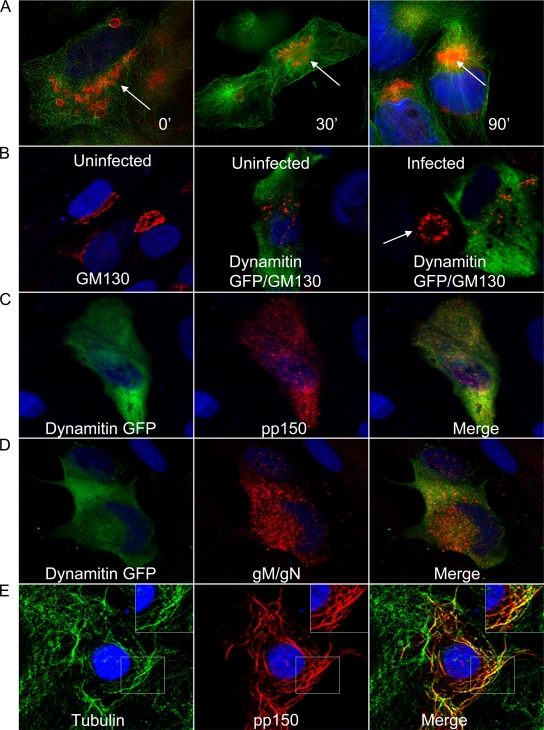
Previously it has been shown that the microtubule network is required for trafficking of the HCMV viral capsid to the nucleus during viral entry, a finding consistent with more extensive studies of herpes simplex virus (HSV) (21, 52, 54, 60). In addition, our earlier studies demonstrated that the viral AC colocalized with the MTOC and was dispersed into smaller vesicles when infected cells were treated with the microtubule-depolymerizing agent nocodazole (68). Initially we investigated the potential role of the microtubule network during the later stages of viral infection. HCMV-infected HFF cells, at 5 days postinfection, were treated with 2 μM nocodazole for 1.5 h, washed, and allowed to recover in growth medium. Nocodazole inhibition of microtubule polymerization is reversible, and its removal allowed repolymerization of the microtubules. The infected cells were fixed at various time points after nocodazole washout and reacted with an anti-pp150 MAb to visualize the AC. Nocodazole treatment caused dispersion of the AC (Fig. 1A). After nocodazole washout, pp150-positive cytoplasmic vesicles presumably derived from the assembly compartment were localized within a perinuclear site, and the morphology of the AC (Fig. 1A, arrows) reformed within 90 min (Fig. 1A). These data suggested that an intact microtubule network was essential for formation and maintenance of the morphological integrity of the AC. The role of microtubule transport in the morphogenesis of the AC was next examined using a dominant-negative dynamitin (p50), a component of the dynactin complex of proteins which, together with dynein, is required for microtubule transport. Overexpression of dynamitin can disrupt dynein-dependent microtubule transport, including formation of membrane organelles such as the Golgi apparatus secondary to the disruption of the dynactin-dynein complex (14). HFFs were transfected with a dynamitin-GFP expression construct and 24 h later infected with HCMV. At 5 days postinfection, dynamitin overexpression disrupted the viral AC, as evidenced by dispersed vesicles positive for the tegument protein pp150 and the abundant glycoprotein complex gM/gN (Fig. 1C and D). The Golgi apparatus was also disrupted by dynamitin overexpression in both uninfected and infected cells (Fig. 1B). Cells which failed to express dynamitin but were infected with HCMV revealed the previously described GM130-positive structures (Golgi apparatus) arranged around the periphery of the viral AC (Fig. 1B, arrow) (68). These data provided additional evidence for the role of microtubules in the maintenance of the AC and a potential role for dynein-mediated transport in the morphogenesis of the AC. In the course of these studies, we observed that in most of the transfected COS-7 cells, pp150 formed cytoplasmic tubules which colocalized with β-tubulin, an observation that suggested that this protein trafficked to the AC by a microtubule- and dynein-dependent pathway (Fig. 1E).
FIG. 1.
Requirement for microtubule network- and dynein-dependent trafficking in the structural integrity of the viral assembly compartment (AC). (A) HCMV-infected HFFs were treated with nocodazole at day 5 postinfection and harvested at 30 and 90 min after nocodazole washout as described in Materials and Methods. The AC was detected with anti-pp150 MAb (red), β-tubulin with a mouse MAb (green), and nuclei with TOPRO 3 (blue). White arrows show the AC. (B) Overexpression of dynamitin-GFP inhibits dynein-dependent trafficking and causes dispersal of the Golgi apparatus (GM 130) in uninfected (red; middle panel) and in HCMV-infected (red; right panel) HFFs. The left panel shows the Golgi apparatus (GM130; red) in nontransfected, uninfected HFFs. The white arrow in the right panel shows the AC in an infected HFF showing altered Golgi apparatus morphology. (C and D) Overexpression of dynamitin-GFP causes dispersion of the AC as evidenced by dispersion of pp150 (red in panel C) and gM/gN (red in panel D). (E) COS-7 cells transfected with a pp150-expressing plasmid were fixed and stained with anti-pp150 MAb (red), β-tubulin with a mouse MAb (green), and nuclei with TOPRO 3 (blue). The inset shows an enlarged view of a portion of the cell imaged. All images are at a magnification of ×600 and were collected on a confocal microscope.
HCMV tegument protein pp150 interacts with Bicaudal D1.
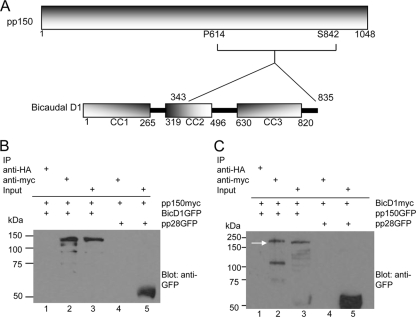
We next carried out yeast two-hybrid (Y2H) screening experiments to identify proteins that interacted with pp150. Initially, putative secondary structures of pp150 were identified using the PHYRE (Protein Homology/AnalogY Recognition Engine) protein fold recognition server (Imperial College, London) in order to perform the screen with structured domains of this large protein (46). A putative alpha-helical domain was identified between amino acids 620 and 823 of pp150. We used this region of pp150, encompassing proline 614 to serine 842, as bait and a human fetal brain cDNA library containing cDNAs fused to the gal4 activation domain of pEXP-AD502 (Invitrogen, Carlsbad, CA) as prey in multiple yeast two-hybrid screens. Although several interactors were identified, the finding that a fragment of Bicaudal D1 corresponding to amino acids 343 to 835 interacted with pp150 was of particular interest because BicD1 has been shown to be a dynein adaptor protein (Table 1). BicD1 has three putative coiled-coil domains (CC1, CC2, and CC3), consisting of amino acids 1 to 265, 319 to 496, and 630 to 820, respectively (Fig. 2A) (38, 39, 57). The amino-terminal coiled-coil domain (CC1) has been shown to interact with dynein, while the carboxyl-terminal coiled-coil domain (CC3) has been shown to interact with the GTPase Rab6 (38, 39, 55, 57, 76). To confirm the findings from the yeast two hybrid assays, we determined that myc-tagged pp150 could pull down GFP-tagged BicD1 when both constructs were cotransfected into HEK293 cells (Fig. 2B, lane2). The negative control HA-tagged magnetic beads failed to pull down myc-tagged pp150 or GFP-tagged BicD1 from the cell lysates (Fig. 2B, lane 1). The pulldown was specific to pp150-BicD1 interaction, since myc-tagged pp150 was unable to pull down GFP-tagged HCMV tegument protein pp28 (Fig. 2B, lane 4). The expression of myc-tagged pp150 in the cotransfected cells was confirmed by probing with anti-myc antibodies (data not shown). As further confirmation of the pp150-BicD1 interaction, we performed a reciprocal set of coimmunoprecipitation experiments in which myc-tagged BicD1 pulled down GFP-tagged pp150 in cotransfected HEK293 cells (Fig. 2C, lane 2, arrow). HA-tagged magnetic beads failed to pull down either BicD1 or pp150 from these cell lysates (Fig. 2C, lane 1). Myc-tagged BicD1 did not pull down GFP-tagged pp28 (Fig. 2C, lane 4). Similar to the results discussed above, probing the blot with anti-myc antibody confirmed the expression of myc-tagged BicD1 in cotransfected cells (data not shown). Together, these data confirmed our findings from the Y2H screens and provided evidence of the interaction between pp150 and the cellular protein BicD1 in the absence of other HCMV-encoded proteins.
TABLE 1.
Results of yeast two-hybrid screening
| Yeast transformant | Growth phenotype in auxotrophic medium |
β-Galactosidase assay result | |||
|---|---|---|---|---|---|
| −Leu, −Trp | −Leu, −Trp, −His, +25 mM 3-aminotriazole | −Leu, −Trp, −Ura | −Leu, −Trp, +0.2% 5-fluoroorotic acid | ||
| pDEST22 + pDEST32 | Growth | No growth | No growth | Growth | White |
| pDEST22 + pp150614-842pDEST32 | Growth | No growth | No growth | Growth | White |
| pDEST32 + BICD1pEXP-AD502a | Growth | No growth | No growth | Growth | White |
| pp150614-842pDEST32 + BICD1pEXP-AD502a | Growth | Growth | Growth | No growth | Blue |
Clone from the prey cDNA library pulled out as interactor.
FIG. 2.
HCMV tegument protein pp150 interacts with Bicaudal D1. (A) Schematic of the pp150 fragment (proline 614 to serine 842) used as bait in Y2H assays and of Bicaudal D1 (BicD1). BicD1 has three coiled-coil domains (CC1, CC2, and CC3) consisting of amino acids 1 to 265, 319 to 496, and 630 to 820, respectively. The Y2H results indicated that pp150P614-S842 interacts with a region of BicD1 from amino acid 343 to 835. (B) Coimmunoprecipitation of pp150 and BicD1. Protein complexes from lysates of HEK 293 cells cotransfected with pp150-myc and BicD1-GFP or pp28-GFP for 48 h were pulled down using anti-myc magnetic beads. Lane 1, negative control in which cell lysates were treated with anti-HA magnetic beads. Lane 2, pp150-myc pulldown of BicD1-GFP. Lane 3, input lysate of BicD1-GFP. Lane 4, lack of pulldown of pp28-GFP by pp150-myc. Lane 5, input lysate control for pp28-GFP. The blot was probed with anti-GFP antibody. (C) Coimmunoprecipitation of BicD1 and pp150. Protein complexes from lysates of HEK 293 cells cotransfected with BicD1-myc and pp150-GFP were pulled down using anti-myc magnetic beads. Lane 1, negative control in which cell lysates were treated with anti-HA magnetic beads. Lane 2, BicD1-myc pulling down pp150-GFP (the arrow points to pp150-GFP). Lane 3, input lysate of pp150-GFP. Lane 4, lack of pulldown of nonspecific control pp28-GFP. Lane 5, input lysate of pp28-GFP. The blot was probed with anti-GFP antibody.
HCMV tegument protein pp150 colocalizes with Bicaudal D1.
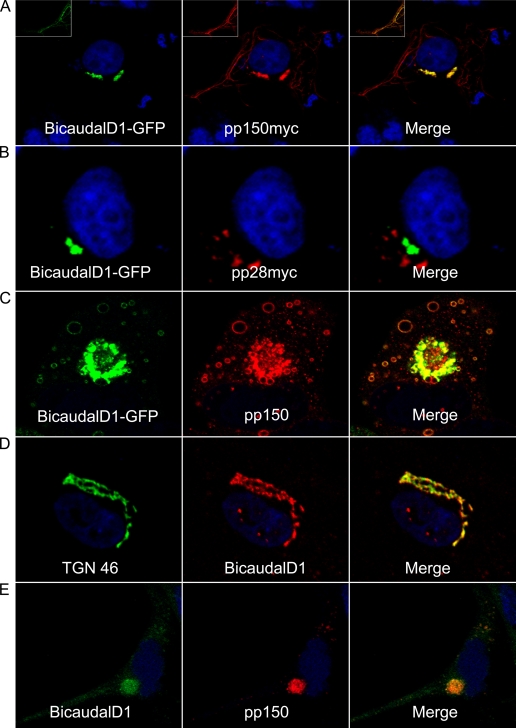
For a protein-protein interaction to be biologically relevant, the interacting proteins presumably must localize within the same cellular compartments at some point during HCMV replication. We found that pp150 colocalized with BicD1 in a perinuclear site and on cytoplasmic tubules in cotransfected COS-7 cells (Fig. 3A), while no colocalization was observed between BicD1-GFP and myc-tagged pp28 (Fig. 3B). We then transfected human foreskin fibroblasts (HFFs) with BicD1-GFP and 24 h later infected them with HCMV strain AD169. At 5 days postinfection, pp150 was visualized using a pp150-specific MAb. In infected cells, transfected BicD1-GFP colocalized with virus-expressed pp150 in cytoplasmic vesicles and in the assembly compartment, indicating that the two proteins localized to the same cellular compartments (Fig. 3C). To further document the colocalization of these proteins in virus-infected cells, a rabbit anti-BicD1 antibody (kindly provided by Anna Akhmanova, Erasmus Medical Center, Rotterdam, The Netherlands) was used to detect BicD1 in HCMV-infected HFFs. Bicaudal D1 staining was observed in a distinct perinuclear region in uninfected cells and, as previously shown, could be colocalized with markers of the trans-Golgi network (Fig. 3D) (21). In infected cells, BicD1 staining coincided with pp150 staining in the AC and in cytoplasmic vesicles (Fig. 3E). These results indicated that pp150 and BicD1 localized to the same cellular compartments in transfected COS-7 cells and in transfected and subsequently infected HFFs. Importantly, endogenous BicD1 was also detected at the AC in HCMV infected HFFs.
FIG. 3.
HCMV tegument protein pp150 colocalizes with BicD1. (A) COS-7 cells cotransfected with BicD1-GFP and pp150-myc plasmids were fixed and stained with anti-myc antibody as described in Materials and Methods. (A) BicD1-GFP (green) colocalized with pp150 (red) in perinuclear vesicular structures and in the cytoplasm. The inset shows a region of cytoplasm of the cell imaged at a higher laser intensity to visualize cytoplasmic tubules. (B) Lack of colocalization between BicD1-GFP and pp28-myc (red) expressed in COS-7 cells. Cells were stained with anti-myc antibody. (C) HFFs transfected with BicD1-GFP, infected with HCMV as described in Materials and Methods, fixed 5 days later, and stained with anti-pp150 MAb. BicD1-GFP (green) and pp150 (red) colocalized in the assembly compartment and cytoplasmic vesicles in transfected-infected cells. (D) HFFs showing endogenous BicD1 localization. The left panel shows the trans-Golgi network stained with sheep anti-TGN 46 antibody (green), the middle panel shows BicD1 (red) stained with a specific rabbit antiserum, and the right panel shows the merge image showing BicD1 trafficking to the trans-Golgi network. (E) HFFs infected with HCMV showing endogenous BicD1. BicD1 was stained using a specific rabbit antiserum, and pp150 was stained with mouse MAb. BicD1 colocalized with pp150 in the perinuclear assembly compartment and in cytoplasmic vesicles. All images are at a magnification of ×600.
FRET between HCMV tegument protein pp150 and Bicaudal D1.
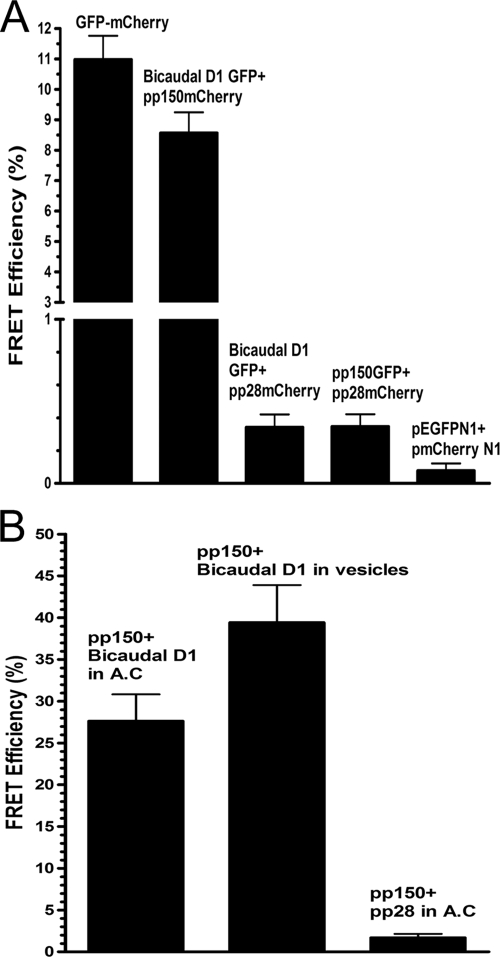
To determine if pp150-BicD1 interactions were binary, as suggested from our Y2H studies, we investigated interactions between these proteins utilizing fluorescence resonance energy transfer (FRET) assays, a technique commonly used to investigate molecular interactions and conformational changes in proteins (72). Our FRET assay is based on the ability of a higher-energy fluorophore (donor; GFP) to directly transfer energy to a lower-energy fluorophore (acceptor; mCherry), leading to a quenching of the donor fluorescence. FRET can be demonstrated as an increase in donor fluorescence after photobleaching the acceptor (donor dequenching and acceptor photobleaching) (7, 18). BicD1-GFP was coexpressed in COS-7 cells with mCherry-tagged pp150, and FRET was carried out. A GFP-mCherry fusion protein was used as the positive control, as this construct provided FRET secondary to an intramolecular interaction. Negative controls included COS-7 cells cotransfected with BicD1-GFP and pp28-mCherry, with pp150-GFP and pp28-mCherry, and with pEGFPN1 and pmCherry N1 vectors. A region of interest (ROI) in the cell was selected, and the acceptor fluorophore (mCherry-tagged protein) was bleached to 30% of its prebleach intensity. We selected multiple smaller ROIs within the bleached region, and the FRET efficiency at these ROIs was recorded. FRET efficiency was also measured in ROIs outside the bleached region, as internal negative controls. The mean FRET efficiency (Fig. 4A) for the pp150-mCherry-BicD1GFP pair was 8.57%, similar to that of the positive control, GFP-mCherry fusion (11.0%). The negative controls resulted in only background levels of FRET, i.e., 0.34% for the Bicaudal D1 GFP-pp28-mCherry pair, 0.35% for the pp150-GFP-pp28-mCherry pair, and 0.08% for the pEGFPN1-pmCherryN1 pair. The internal negative controls did not give any FRET. The results from these FRET experiments in transfected COS-7 cells indicated that pp150 and BicD1 interacted directly with each other and in the absence of other HCMV proteins. To confirm that a similar interaction occurred in HFFs infected with HCMV, we performed similar FRET experiments with infected HFFs at 5 days postinfection (Fig. 4B). BicD1 and pp150 were immunostained with specific antibodies, and FRET was performed between the secondary fluorophores in the juxtanuclear viral assembly compartment and within cytoplasmic vesicles. As a negative control, we measured FRET between pp150 and pp28, two tegument proteins previously shown to colocalize in the AC (68). A mean FRET efficiency of 27.6% was observed for the pp150-BicD1 pair in the viral assembly compartment, and a mean FRET efficiency of 39.4% was observed for the same protein pair in cytoplasmic vesicles that could be resolved from the AC (Fig. 4B). The mean FRET efficiency for the pp150-pp28 pair in viral assembly compartment was 1.69%. These results strongly argued that pp150 and BicD1 interacted directly and that within HCMV-infected HFF cells, these proteins localized within the cytoplasmic sites of viral assembly.
FIG. 4.
FRET analysis of pp150-BicD1 interaction. (A) COS-7 cells were transfected with plasmids expressing GFP-tagged Bicaudal D1 and mCherry-tagged pp150, GFP-tagged Bicaudal D1 and mCherry-tagged pp28 (negative control), GFP-tagged pp150 and mCherry-tagged pp28 (negative control), or pEGFPN1 and pmCherryN1 vectors (negative control) or with a plasmid expressing a GFP-mCherry fusion protein (positive control). The cells were harvested 48 h later, fixed with 3% paraformaldehyde, and analyzed by FRET assays as described in Materials and Methods. A region of interest (ROI) in the cell was selected, and the mCherry was bleached to 30% of its intensity at the ROI. The increase in the intensity of GFP after photobleaching of mCherry was determined, and the FRET efficiency between the donor (GFP) and the acceptor (mCherry) was calculated as described in Materials and Methods. FRET was observed only between the pp150-mCherry and BicD1-GFP pair and compared with the FRET seen in the positive-control GFP-mCherry fusion protein. Error bars indicate standard deviations. (B) FRET between pp150 and Bicaudal D1 was measured in HCMV-infected HFFs as described in Materials and Methods. Briefly, HCMV-infected HFF cells were fixed at 5 days postinfection with 3% paraformaldehyde. The cells were reacted with anti-pp150 MAb, anti-BicD1 rabbit antiserum, and TRITC-conjugated goat anti-mouse IgG2b and FITC-conjugated goat anti-rabbit secondary antibodies, respectively. FRET was measured between FITC and TRITC-labeled proteins in the assembly compartment (AC) as well as in the cytoplasmic vesicles in infected cells. As a negative control, FRET was measured between pp28 and BicD1 using anti-pp28 MAb and anti-BicD1 rabbit antibody and using TRITC-conjugated goat anti-mouse IgG2a and FITC-conjugated goat anti-rabbit secondary antibodies, respectively.
Bicaudal D1 interacts with pp150 through a domain in its carboxyl terminus.
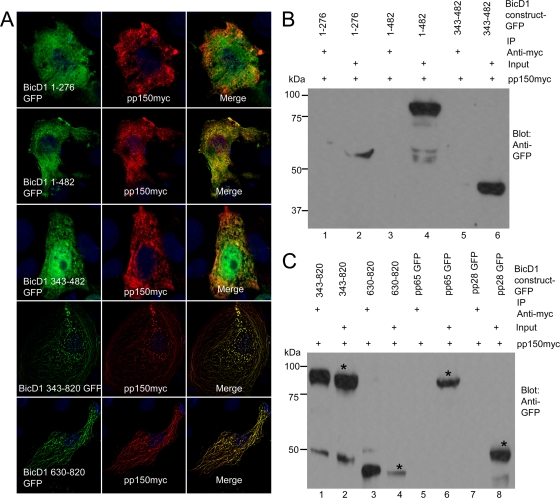
To map the region of BicD1 interacting with pp150, GFP fusion proteins containing amino acids 1 to 276, 1 to 482, 343 to 482, 343 to 820, and 630 to 820 of BicD1 were constructed and were cotransfected with myc-tagged pp150 in COS-7 cells (Fig. 5A). In transfected COS-7 cells, BicD1 amino acids 1 to 276 and 343 to 482 appeared to partition in both the cytoplasm and nucleus, BicD1 amino acids 1 to 482 were expressed diffusely in the cytoplasm, and BicD1 amino acids 343 to 820 and 630 to 820 appeared to localize to perinuclear vesicular structures (data not shown). Upon cotransfection, we observed that pp150 did not colocalize with the constructs containing BicD1 amino acids 1 to 276, 1 to 482, and 343 to 482 (Fig. 5A) whereas it colocalized with those containing BicD1 amino acids 343 to 820 and 630 to 820 constructs (Fig. 5A) in perinuclear vesicular structures as well as in cytoplasmic tubules, much like the pattern seen when pp150 colocalizes with full-length BicD1 in COS-7 cells (Fig. 3A). These observations suggested that BicD1 amino acids 630 to 820 were responsible for the interaction with pp150, a finding consistent with results from our Y2H screening. To confirm these observations, we performed coimmunoprecipitation experiments with HEK 293 cells cotransfected with plasmids expressing myc-tagged pp150 and GFP-tagged BicD1 constructs (Fig. 5B and C). Myc-tagged pp150 failed to immunoprecipitate BicD1 constructs containing amino acids 1 to 276 (Fig. 5B, lane 1), 1 to 482 (Fig. 5B, lane 3) and 343 to 482 (Fig. 5B, lane 5), whereas it did pull down BicD1 amino acids 343 to 820 (Fig. 5C, lane 1) and 630 to 820 (Fig. 5B, lane3). Figure 5B, lanes 2, 4, and 6, shows the input cell lysates of BicD1 amino acids 1 to 276, 1 to 482, and 343 to 482, respectively, before pulldown with myc-tagged magnetic beads, while Fig. 5C, lanes 2 and 4, shows the input cell lysates of BicD1 amino acids 343 to 820 and 630 to 820, respectively. myc-tagged pp150 did not pull down other HCMV tegument proteins, pp65 and pp28, that were included as negative controls (Fig. 5C, lanes 5 and 7; lanes 6 and 8 show the respective input lysates). These observations together with results from our imaging studies argued that a domain within the C terminus of BicD1 between amino acids 630 and 820 was responsible for the interaction with pp150.
FIG. 5.
BicD1 amino acids 630 to 820 interact with pp150. (A) Colocalization of BicD1-GFP (green) constructs and pp150-myc (red) in COS-7 cells. COS-7 cells cotransfected with the indicated plasmids were fixed and stained with anti-myc antibody as described in Materials and Methods. (B) Lysates of HEK 293 cells cotransfected with pp150-myc and BicD1-GFP constructs for 48 h and precipitated using anti-myc magnetic beads. Constructs were BicD1 aa 1 to 276-GFP (lane 1), BicD1 aa 1 to 482-GFP (lane 3), and BicD1 aa 343 to 482-GFP (lane 5). Cell lysate input is in lanes 2, 4, and 6. (C) Lysates of HEK 293 cells cotransfected with pp150myc and BicD1-GFP constructs for 48 h and precipitated using anti-myc magnetic beads. Constructs were BicD1 aa 343 to 820-GFP (lane 1), BicD1 aa 630 to 820-GFP (lane 3), pp65-GFP (lane 5), and pp28-GFP (lane 7); asterisks indicate GFP-tagged BicD1 constructs. Cell lysate input is in lanes 2, 4, 6, and 8. The blots were probed with a rabbit anti-GFP antibody. All images are at a magnification of ×1,000.
Recruitment of Bicaudal D1 to the viral assembly compartment depends on its C-terminal domain.
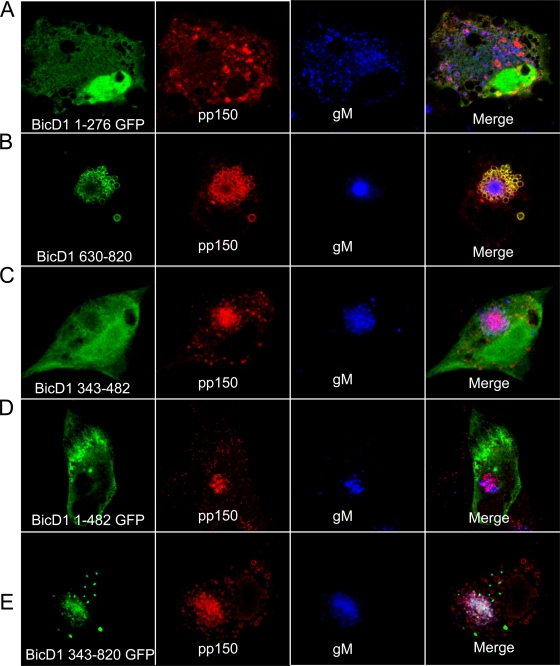
To further characterize the recruitment of BicD1 to the viral assembly compartment, GFP fusion proteins containing amino acids 1 to 276, 1 to 482, 343 to 482, 343 to 820, and 630 to 820 of BicD1 were transiently expressed in HFFs which were then infected with HCMV. We used specific MAbs to detect pp150 and the viral glycoprotein gM. Expression of GFP-tagged BicD1 1 to 276 in HFFs resulted in dispersion of pp150- and gM-positive vesicles (Fig. 6A). This finding was consistent with previous studies that indicated that a BicD1 construct containing these amino acids had a dominant-negative effect on dynein-dependent transport and provided further evidence for a role of dynein in pp150 intracellular trafficking (38, 39, 55). GFP-tagged BicD1 amino acids 630 to 820 colocalized with pp150-positive vesicles that were localized to the AC (Fig. 6B). This region of BicD1 has been reported to interact with Rab6, and interaction with Rab6 has been shown to be important for localizing BicD1 to Golgi membranes (55). Interestingly, this region of BicD1 has been shown to be required for BicD1 multimerization through its interactions with the coiled-coil region in the amino terminus, suggesting that the GFP-tagged BicD1 amino acids 630 to 820 could dimerize with endogenous BicD1 (55). GFP-tagged BicD1 amino acids 343 to 482 lack both the dynein binding and the Rab6 binding domains. This construct was observed to be dispersed in the cytoplasm and the nucleus, it did not colocalize with pp150, and its expression did not inhibit formation of AC or trafficking of viral proteins to the AC as evidenced by the intracellular expression of pp150 and gM (Fig. 6C). A GFP-tagged BicD1 construct containing amino acids 1 to 482 that lacks the Rab6 binding domain but expresses the dynein binding domain (amino acids 1 to 276) was dispersed in the cytoplasm, did not colocalize with pp150, and did not inhibit formation of AC or trafficking of the viral proteins pp150 and gM to the AC (Fig. 6D). The GFP-tagged construct containing BicD1 amino acids 343 to 820 lacked the dynein binding domain and was observed to colocalize with pp150 in the AC (Fig. 6E). Together, this imaging data indicated that recruitment of BicD1 to the AC and its colocalization with pp150 in infected cells was dependent on domains in its C terminus that included the previously described Rab6 binding domain of BicD1 (55).
FIG. 6.
The C-terminal domain of BicD1 is involved in its trafficking to the viral assembly compartment. HFFs were transfected with truncated BicD1-GFP constructs and 24 h later were infected with HCMV. Five days later they were fixed and stained with anti-pp150 (red)- or anti-gM (blue)-specific MAbs and TRITC-conjugated goat anti-mouse IgG2b and Alexafluor-647 conjugated goat anti-mouse IgG1, respectively. (A) BicD1 aa 1 to 276-GFP (green), pp150 (red) and gM (blue). (B) BicD1 aa 630 to 820-GFP (green), pp150 (red), and gM (blue). (C) BicD1 aa 343 to 482-GFP (green), pp150 (red), and gM (blue). (D) BicD1 aa 1 to 482-GFP (green), pp150 (red), and gM (blue). (E) BicD1 aa 343 to 820-GFP (green), pp150 (red), and gM (blue). All images are at a magnification of ×600.
BicD1 expression and its interactions with pp150 are essential for localization of pp150 to the AC and assembly of infectious virus.
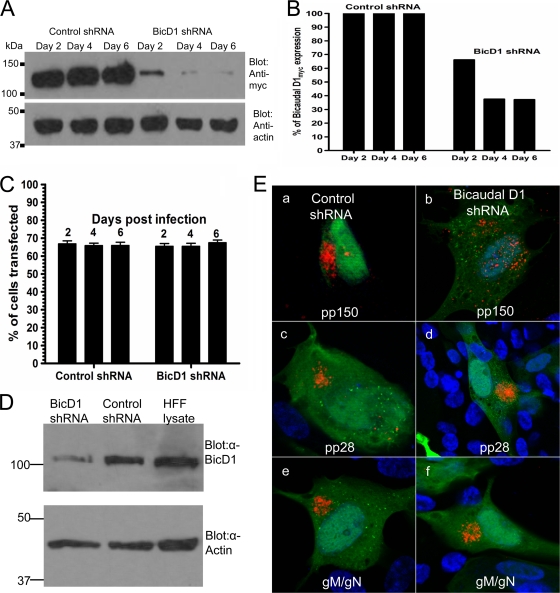
The interactions between an essential virion tegument protein, pp150, and BicD1 that lead to localization of these proteins in the cytoplasmic AC suggested that inhibition of this interaction could limit virus assembly. To determine the importance of pp150-BicD1 interaction in HCMV assembly, we used shRNA-expressing plasmids to deplete endogenous BicD1 in HFFs followed by infection with HCMV. A panel of four plasmids expressing shRNAs specific to four distinct regions of the BicD1 ORF were used. A plasmid expressing a scrambled shRNA sequence was used as a negative control. Each of these plasmids also expresses GFP, which permitted the identification of shRNA-expressing cells. We chose to pool all four shRNA plasmids to ensure maximum levels of long-term depletion of BicD1. The efficiency of BicD1 depletion was monitored temporally in HEK 293 cells transfected with a myc-tagged BicD1 expression vector and the shRNA expression vectors. By day 4 posttransfection, BicD1 expression was reduced by 66.67% in cells transfected with the depleting shRNA mix, whereas it remained unchanged in cells transfected with control scrambled shRNA (Fig. 7A and B). For studies directed at inhibiting virus assembly, it was important to obtain a high level of transfection efficiency in HFFs and to determine that HCMV infection did not downregulate shRNA expression. To assay the efficiency of transfection in HFFs, we counted the number of GFP-expressing cells versus total number of cells at various time points posttransfection. The transfection efficiency ranged from 61.8% to 71.8% (Fig. 7C) through 6 days after infection. Thus, HFFs could be efficiently transfected, and the expression of transfected constructs remained detectable for at least until 6 days postinfection (7 days posttransfection). To confirm that the shRNA targeting BicD1 was able to deplete BicD1 in HFFs, HFFs were transfected with BicD1 shRNA mix or control shRNA, and 4 days later, the cells were harvested and the level of expression of endogenous BicD1 was checked by Western blotting using a specific rabbit anti-BicD1 antibody (number 2296). It was found that the BicD1-depleting shRNA was able to deplete endogenous BicD1 in HFFs by 73% (Fig. 7D). To study the effects of BicD1 depletion on HCMV assembly, HFF cells were transfected with BicD1-depleting shRNA or scrambled shRNA and then infected with HCMV. We examined the phenotype of the assembly compartment as well as infectious virus yield after 5 days of infection. In HFFs transfected with the control scrambled shRNA, the viral AC phenotype appeared as previously described (Fig. 7E). In HFFs depleted of BicD1, we found that at 5 days postinfection, a compact, pp150-expressing juxtanuclear assembly compartment failed to form and pp150-containing vesicles were dispersed throughout the cytoplasm (Fig. 7E). The altered intracellular localization of pp150 in cells expressing the BicD1 knockdown shRNA could either result from a defect in the intracellular trafficking of pp150 to a cytoplasmic site(s) of virus assembly or be secondary to disruption of the AC in cells depleted of BicD1. As an off-target control and a control for nonspecific disruption of the cellular architecture, we determined the cellular localization of the tegument protein pp28 and the envelope glycoprotein complex gM/gN. We found that infected HFFs expressing scrambled shRNA showed a normal-appearing juxtanuclear AC when assayed for pp150 (Fig. 7E), pp28 (Fig. 7E), and gM/gN (Fig. 7E), whereas 92.62% of HFFs depleted of BicD1 showed defective pp150 localization (Fig. 7E). The effect of BicD1 depletion on trafficking of HCMV proteins was specific for pp150, because in infected HFFs depleted of BicD1, both pp28 and the gM/gN complex trafficked normally to the assembly compartment (Fig. 7E). One hundred percent of both HFFs depleted of BicD1 and HFFs expressing scrambled shRNA showed a normal assembly compartment phenotype and normal trafficking of pp28 and gM/gN (Fig. 7E). These results indicated that BicD1 directed trafficking of pp150 to the HCMV assembly compartment.
FIG. 7.
Depletion of BicD1 inhibits trafficking of pp150 to the assembly compartment. (A) Evidence of depletion of BicD1 using shRNA-expressing plasmids. HEK 293 cells were cotransfected with BicD1-myc plasmid and either with shRNA plasmids to deplete BicD1 expression or with scrambled control shRNA plasmid. Cells were lysed in equal volumes of lysis buffer on days 2, 4, and 6 posttransfection and subjected to SDS-PAGE followed by Western blotting. Western blots were probed with anti-myc antibody to detect expression of BicD1-myc (top panel) and with antiactin antibody as a loading control (bottom panel). (B) Histogram of densitometry signals from depletion of BicD1, illustrating efficiency of Bicaudal D1 depletion. (C) Efficiency of transfection of HFFs following electroporation with shRNA plasmids to deplete BicD1 expression or with scrambled control shRNA. The cells were infected by HCMV at 24 h postelectroporation, and on days 2, 4 and 6 postinfection cells were fixed and stained with Hoechst 33342 to visualize total number of cells on the coverslips and GFP expressing cells counted. The efficiency of transfection was estimated by the number of GFP-expressing cells per 100 cells. Error bars indicate standard deviations. (D) Evidence of depletion of endogenous Bicaudal D1 in HFFs using shRNA. HFF cells were transfected either with shRNA plasmids to deplete BicD1 expression or with scrambled control shRNA plasmid. Cells were lysed in equal volumes of lysis buffer at 4 days posttransfection and subjected to SDS-PAGE followed by Western blotting. Western blots were probed with anti-BicD1 antibody to detect expression of BicD1 (top panel) and with anti-actin antibody as a loading control (bottom panel). An HFF lysate prepared from nontransfected cells was used as a positive control. (E) HFF cells were electroporated with shRNA plasmids to deplete BicD1 expression (b, d, and f) or with scrambled control shRNA plasmid (a, c, and e) and then infected on the following day. At 5 days postinfection, cells were fixed and stained with MAbs to pp150, pp28, or gM/gN followed by TRITC-conjugated secondary antibodies. Plasmids that encode either scrambled control shRNA or BicD1 shRNA express GFP (green) in transfected cells. All images are at a magnification of ×600.
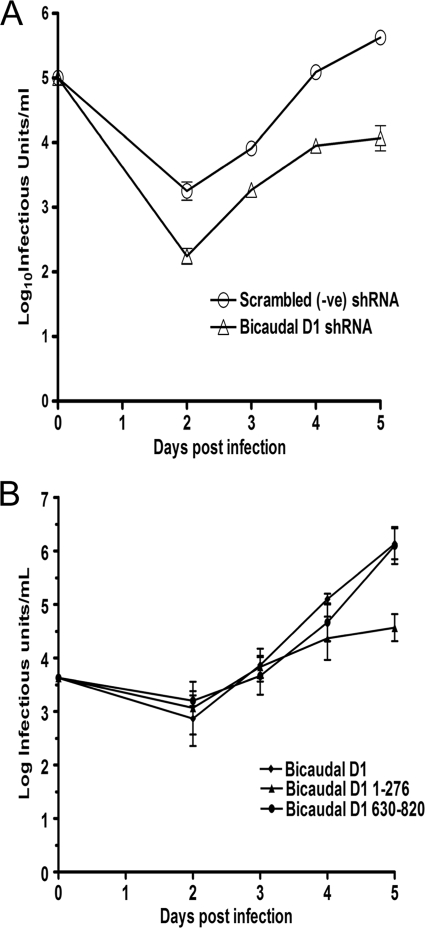
To determine the effect of BicD1 depletion on virus replication, we infected HFFs depleted of BicD1; harvested them at days 2, 4, and 6 postinfection; and assayed the total yield (cellular and supernatant) of infectious virus. As shown in Fig. 8A, cells depleted of BicD1 produced about 1.5 log10 less infectious virus than cells expressing scrambled shRNA. Taken together, these data suggest that BicD1 depletion specifically affects the trafficking of the essential tegument protein, pp150, to the sites of viral assembly, consequently reducing the yield of infectious virus.
FIG. 8.
Decreased viral replication associated with shRNA depletion of BicD1 and overexpression of dominant-negative BicD1 aa 1 to 276. (A) HFFs expressing shRNA vectors to deplete BicD1 or control scrambled shRNA were infected with AD169 at an MOI of 0.1, and total infectious virus from supernatant and cells was assayed at the indicated time points postinfection and expressed as log10 infectious units/ml of sample. (B) HFFs were transfected with the indicated GFP-tagged Bicaudal D1 (aa 1 to 276) constructs and 24 h later were infected with HCMV AD169 at an MOI of 0.1. Virus from supernatant and cells was harvested at the indicated times and assayed for the amount of infectious particles. Results are expressed as log10 infectious units/ml of sample. Error bars indicate standard deviations.
Overexpression of the dynein-interacting domain of BicD1 has been reported to function in a dominant-negative manner by sequestering dynein, thus blocking dynein-dependent transport (26, 38, 39, 55). To determine the effect of overexpression of the BicD1 dominant-negative amino acids 1 to 276 on pp150 localization to the AC and virus replication, we overexpressed GFP-tagged BicD1 amino acids 1 to 276 in HFF cells and infected them with HCMV 24 h later. Similar to findings using the dominant-negative dynamitin, the AC was disrupted and pp150 was not localized within a recognizable AC (Fig. 6A). Virus yields were decreased in HFF cells expressing the GFP-tagged construct containing BicD1 amino acids 1 to 276 by about 1.5 log10, confirming our previous findings that formation of the AC and assembly of infectious virus were dynein dependent (Fig. 8B). Together, these studies argued that the observed BicD1 dependence of pp150 localization to the AC was dynein dependent as a result of its interaction with this dynein adaptor protein.
DISCUSSION
The assembly of an infectious HCMV virion is an exceedingly complex process requiring the localization of a large number of proteins to sites of particle assembly. Layering of the nucleocapsid with tegument proteins (tegumentation) of the virion presumably requires orderly incorporation of tegument proteins, perhaps governed by protein-protein interactions that organize individual proteins into subunits that are incorporated into the assembling particle. The HCMV pp150 is an example of an essential tegument protein whose incorporation into the virion tegument is required for the assembly of an infectious particle. It has not been reported to have enzymatic or transcriptional activating functions, and thus it likely serves an essential structural function in the assembly of the infectious virion. Under steady-state conditions, the vast majority of pp150 can be found in the cytoplasm, but rapid shuttling between the nucleus and cytoplasm cannot be ruled out. More importantly, it remains unclear how pp150 localizes to the AC in virus-infected cells during the assembly of infectious virions, because it has no recognizable trafficking signals that could direct it to this membranous compartment within the infected cell. Extensive Y2H screening has failed to identify interactions of pp150 with essential viral proteins that could direct it to sites of virus assembly (data not shown). Thus, we favor the possibility that pp150 interacts with a cellular protein that directs its intracellular localization to the AC in virus-infected cells. Consistent with this hypothesis, we have identified an interaction between pp150 and a dynein binding protein, BicD1, which could serve to link pp150 to microtubule transport through its interaction with dynein (55). This interaction between BicD1 and pp150 could therefore target pp150 and potentially other tegument proteins that interact with pp150 to the AC during virus infection. Moreover, the AC has been localized in close proximity to the MTOC, an intracellular compartment that is the endpoint of dynein-mediated (negative-end) microtubule transport (68). Several lines of evidence presented in this report are consistent with this postulated mechanism of pp150 localization to the AC, including (i) the colocalization of pp150 with microtubules and colocalization of pp150 with the dynein binding protein BicD1, (ii) the disruption of pp150 localization to the AC by treatment of cells with the microtubule-depolymerizing agent nocodazole and following overexpression of dynamitin, (iii) disruption of pp150 localization to the AC by depletion of BicD1 using shRNA, and (iv) altered pp150 localization to the AC in infected cells and a reduction in the production of infectious virions following disruption of dynein-dependent trafficking by overexpression of the dominant-negative N-terminal domain of BicD1 (39, 55).
Our findings from the Y2H screen and our analysis using FRET strongly argued that the interaction between pp150 and BicD1 was direct and occurred in the absence of other HCMV proteins. In order to assess the functional significance of this interaction, we used shRNA to decrease BicD1 expression in HFFs followed by analysis of virus replication. Knockdown of BicD1 expression decreased production of infectious virus by approximately 1.5 log10, demonstrating a significant role of this protein in the production of infectious virus. In this same experiment, BicD1 depletion was shown to inhibit trafficking of pp150 to the assembly compartment, whereas other viral proteins, including pp28, a tegument protein, and the viral glycoprotein complex gM/gN, appeared to traffic normally to the viral AC. These results further argued that BicD1-pp150 interactions were specific and were responsible for the intracellular trafficking of pp150 to the AC in infected cells. Interestingly, it has been previously reported that a viral mutant lacking the aspartic acid residue at position 719 in pp150 was replication defective (92). This amino acid lies within the region of pp150 that we have identified as the BicD1-interacting domain.
BicD1 is one of two human homologues of the Drosophila melanogaster protein Bicaudal D (Bic-D), which, along with the microtubule network and the negative-end-directed microtubule motor dynein, is involved in establishing asymmetric cytoplasm in the developing oocyte, germ line cyst development, oocyte specification, asymmetric mRNA localization within the oocyte, and egg chamber formation (3, 61, 62, 82, 83, 89). The N-terminal domains of both BicD1 and D2 interact with dynein intermediate chain and with the dynactin subunits dynamitin and p150Glued, whereas the C-terminal domain interacts with isoforms of the small GTPase Rab6, thus linking dynein-dependent motility to vesicular trafficking (5, 25, 38, 39, 55, 88). BicD2 also weakly interacts with kinesin-1 isoforms KIF5A and KIF5B (31). The C terminus of BicD2 also binds to dynamitin (38). The N and C termini of BicD1 and -D2 self-interact, leading to their autoinhibition, a mechanism that appears to regulate their function (38, 39). Binding of BicD1 with dynein intermediate chain, but not with dynamitin (p50) or p150Glued, is dependent on its phosphorylation by the serine/threonine kinase glycogen synthase kinase 3β (GSK-3β) (26). Transgenic mice having neuron-specific expression of the BicD2 N terminus were observed to have chronic impairment of dynein/dynactin function and developed amyotrophic lateral sclerosis (ALS)-like features in their motor neurons (85). BicD2 has also been reported to be a substrate of Nek8, a mammalian NIMA-related kinase (36). BicD1 is also known to associate with Chlamydia trachomatis inclusions in a biovar-specific manner; however, the functional significance of such interactions is still unknown, and the Chlamydia proteins that interact with BicD1 have not been identified (57). It is of considerable interest that during its intracellular replication Chlamydia forms an inclusion body in immediate proximity to the MTOC in HeLa cells and that its localization to this site is dynein dependent although not inhibited by overexpression of dynamitin, an observation suggesting a dynein-dependent but dynactin-independent mechanism of microtubule transport (30). Recently, it has been shown that BicD1 localized to the Chlamydia inclusion in the absence of Rab6 activity, suggesting that BicD1 dynein binding activity and interactions with as-yet-undefined Chlamydia proteins were the only requirements for the association of BicD1 with the Chlamydia inclusion (57). However, in contrast to our studies of the AC in HCMV-infected cells, those authors demonstrated dispersion of membranous compartments in the cell, i.e., the Golgi apparatus, by expression of both BicD1 amino acids 1 to 276 and dynamitin but no effect on the Chlamydia inclusion body (30, 57). Thus, although it is clear that BicD1 can function as a dynein adaptor protein and through its Rab6 binding domains provides a potential membrane interaction function, it is unclear if each of these functions is utilized in a similar fashion by intracellular microbes that use dynein-mediated transport during their replication.
Several viruses are known to utilize the cellular cytoskeletal network and the associated motor complexes in their life cycle (64). Viruses, including HIV, Ebola virus, adenovirus, simian virus 40 (SV40), and HSV, utilize the actin network and associated myosin motors for entry into cells (58, 63, 80, 81, 90). A number of viruses, including adeno-associated virus, murine polyomavirus, influenza virus, canine parvovirus, and Kaposi's sarcoma-associated herpesvirus (KSHV), utilize microtubules and dynein motors for efficient nuclear targeting (49, 59, 70, 71, 78). Proteins of several viruses, including adenovirus, African swine fever virus, canine parvovirus, foamy virus, HSV, Lyssa virus, pseudorabies virus, and rabies virus, may also directly interact with components of the dynein machinery (29, 50, 51) and with kinesins (47, 79). Cytoskeletal components, including α-actin, β-actin, α-tubulin, β-tubulin, cofilin, keratin, and vimentin, form constituents of the HCMV proteome (4, 87). In the current study we have shown that pp150, an essential tegument protein of HCMV, could interact with dynein indirectly through its interactions with BicD1 and that this interaction is required for trafficking of this essential tegument protein to cytoplasmic sites of viral assembly. This mechanism of pp150 intracellular trafficking is attractive because it would allow pp150 and potentially any viral protein(s) that interacts with pp150 to localize to the AC through BicD1 interactions with dynein. Furthermore, the presence of a Rab6 binding domain on BicD1 could also provide a membrane interaction function to address pp150 and associated viral proteins to intracellular membranes that serve as sites of virion envelopment and assembly. Such a mechanism would suggest a potential involvement of Rab6 GTPases in HCMV assembly. In instances in which the role of Rab6 has been studied, the redistribution of intracellular membranous compartments appears to be microtubule dependent, suggesting a role for dynein and kinensin, a finding that is consistent with the binding of Rab6 to p150Glued, a component of the dynactin protein complex (76), and also with Rabkinesin (23, 24). As noted above, Rab6 has been proposed to have a role in the formation of the Chlamydia inclusion body, although the precise function has not been described (57). Similarly, its potential role in the assembly of HCMV is not understood. Currently our laboratory has studies under way to dissect the potential role of this small GTPase in the formation of the AC and in the assembly of the infectious HCMV virion.
Acknowledgments
We thank Anna Akhmanova (Erasmus Medical Center, Rotterdam, The Netherlands) for her kind gift of enhanced-GFP-tagged Bicaudal D1 plasmid and Bicaudal D1-specific rabbit antisera, Albert Tousson of the UAB High Resolution Imaging Facility for help with the FRET experiments, and UAB neuroscience core C.
UAB neuroscience core C is supported by grant P30-NS47466. This work was supported by grants AI035602-13 and AI050189-07 from the NIH to W.J.B.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. [DOI] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., G. B. Smith, C. D. Meiering, and E. S. Mocarski. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baens, M., and P. Marynen. 1997. A human homologue (BICD1) of the Drosophila bicaudal-D gene. Genomics 45:601-606. [DOI] [PubMed] [Google Scholar]
- 4.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, F. A., and B. Short. 2003. Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 15:405-413. [DOI] [PubMed] [Google Scholar]
- 6.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berdiev, B. K., E. Cormet-Boyaka, A. Tousson, Y. J. Qadri, H. M. Oosterveld-Hut, J. S. Hong, P. A. Gonzales, C. M. Fuller, E. J. Sorscher, G. L. Lukacs, and D. J. Benos. 2007. Molecular proximity of cystic fibrosis transmembrane conductance regulator and epithelial sodium channel assessed by fluorescence resonance energy transfer. J. Biol. Chem. 282:36481-36488. [DOI] [PubMed] [Google Scholar]
- 8.Blum, A., M. Giladi, M. Weinberg, G. Kaplan, H. Pasternack, S. Laniado, and H. Miller. 1998. High anti-cytomegalovirus (CMV) IgG antibody titer is associated with coronary artery disease and may predict post-coronary balloon angioplasty restenosis. Am. J. Cardiol. 81:866-868. [DOI] [PubMed] [Google Scholar]
- 9.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 10.Britt, W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417-470. [DOI] [PubMed] [Google Scholar]
- 11.Britt, W. J., and S. Boppana. 2004. Human cytomegalovirus virion proteins. Hum. Immunol. 65:395-402. [DOI] [PubMed] [Google Scholar]
- 12.Britt, W. J., M. Jarvis, J. Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Britt, W. J., M. A. Jarvis, D. D. Drummond, and M. Mach. 2005. Antigenic domain 1 is required for oligomerization of human cytomegalovirus glycoprotein B. J. Virol. 79:4066-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkhardt, J. K., C. J. Echeverri, T. Nilsson, and R. B. Vallee. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 17.Cobbs, C. S., L. Harkins, M. Samanta, G. Y. Gillespie, S. Bharara, P. H. King, L. B. Nabors, C. G. Cobbs, and W. J. Britt. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62:3347-3350. [PubMed] [Google Scholar]
- 18.Daelemans, D., S. V. Costes, S. Lockett, and G. N. Pavlakis. 2005. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol. Cell. Biol. 25:728-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demmler, G. J. 1991. Infectious Diseases Society of America and Centers for Disease Control. Summary of a workshop on surveillance for congenital cytomegalovirus disease. Rev. Infect. Dis. 13:315-329. [DOI] [PubMed] [Google Scholar]
- 20.Doerr, H. W., R. Braun, and K. Munk. 1985. Human cytomegalovirus infection: recent developments in diagnosis and epidemiology. Klin. Wochenschr. 63:241-251. [DOI] [PubMed] [Google Scholar]
- 21.Dohner, K., K. Radtke, S. Schmidt, and B. Sodeik. 2006. Eclipse phase of herpes simplex virus type 1 infection: Efficient Dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Echard, A., A. el Marjou, and B. Goud. 2001. Expression, purification, and biochemical properties of rabkinesin-6 domains and their interactions with Rab6A. Methods Enzymol. 329:157-165. [DOI] [PubMed] [Google Scholar]
- 24.Echard, A., F. Jollivet, O. Martinez, J. J. Lacapere, A. Rousselet, I. Janoueix-Lerosey, and B. Goud. 1998. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science 279:580-585. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs, E., B. Short, and F. A. Barr. 2005. Assay and properties of rab6 interaction with dynein-dynactin complexes. Methods Enzymol. 403:607-618. [DOI] [PubMed] [Google Scholar]
- 26.Fumoto, K., C. C. Hoogenraad, and A. Kikuchi. 2006. GSK-3beta-regulated interaction of BICD with dynein is involved in microtubule anchorage at centrosome. EMBO J. 25:5670-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson, W. 1981. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology 111:516-537. [DOI] [PubMed] [Google Scholar]
- 28.Gibson, W., and A. Irmiere. 1984. Selection of particles and proteins for use as human cytomegalovirus subunit vaccines. Birth Defects Orig. Artic. Ser. 20:305-324. [PubMed] [Google Scholar]
- 29.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124:741-754. [DOI] [PubMed] [Google Scholar]
- 30.Grieshaber, S. S., N. A. Grieshaber, and T. Hackstadt. 2003. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell Sci. 116:3793-3802. [DOI] [PubMed] [Google Scholar]
- 31.Grigoriev, I., D. Splinter, N. Keijzer, P. S. Wulf, J. Demmers, T. Ohtsuka, M. Modesti, I. V. Maly, F. Grosveld, C. C. Hoogenraad, and A. Akhmanova. 2007. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell 13:305-314. [DOI] [PubMed] [Google Scholar]
- 32.Grosse, S. D., D. S. Ross, and S. C. Dollard. 2008. Congenital cytomegalovirus (CMV) infection as a cause of permanent bilateral hearing loss: a quantitative assessment. J. Clin. Virol. 41:57-62. [DOI] [PubMed] [Google Scholar]
- 33.Harkins, L., A. L. Volk, M. Samanta, I. Mikolaenko, W. J. Britt, K. I. Bland, and C. S. Cobbs. 2002. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet 360:1557-1563. [DOI] [PubMed] [Google Scholar]
- 34.Hensel, G., H. Meyer, S. Gartner, G. Brand, and H. F. Kern. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591-1601. [DOI] [PubMed] [Google Scholar]
- 35.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holland, P. M., A. Milne, K. Garka, R. S. Johnson, C. Willis, J. E. Sims, C. T. Rauch, T. A. Bird, and G. D. Virca. 2002. Purification, cloning, and characterization of Nek8, a novel NIMA-related kinase, and its candidate substrate Bicd2. J. Biol. Chem. 277:16229-16240. [DOI] [PubMed] [Google Scholar]
- 37.Homman-Loudiyi, M., K. Hultenby, W. Britt, and C. Soderberg-Naucler. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoogenraad, C. C., A. Akhmanova, S. A. Howell, B. R. Dortland, C. I. De Zeeuw, R. Willemsen, P. Visser, F. Grosveld, and N. Galjart. 2001. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 20:4041-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoogenraad, C. C., P. Wulf, N. Schiefermeier, T. Stepanova, N. Galjart, J. V. Small, F. Grosveld, C. I. de Zeeuw, and A. Akhmanova. 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 22:6004-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 41.Jahn, G., T. Kouzarides, M. Mach, B. C. Scholl, B. Plachter, B. Traupe, E. Preddie, S. C. Satchwell, B. Fleckenstein, and B. G. Barrell. 1987. Map position and nucleotide sequence of the gene for the large structural phosphoprotein of human cytomegalovirus. J. Virol. 61:1358-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jahn, G., and M. Mach. 1990. Human cytomegalovirus phosphoproteins and glycoproteins and their coding regions. Curr. Top. Microbiol. Immunol. 154:171-185. [DOI] [PubMed] [Google Scholar]
- 43.Jones, T. R., and S. W. Lee. 2004. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 78:1488-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalejta, R. F. 2008. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72:249-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalejta, R. F., J. T. Bechtel, and T. Shenk. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 47.Kim, W., Y. Tang, Y. Okada, T. A. Torrey, S. K. Chattopadhyay, M. Pfleiderer, F. G. Falkner, F. Dorner, W. Choi, N. Hirokawa, and H. C. Morse III. 1998. Binding of murine leukemia virus Gag polyproteins to KIF4, a microtubule-based motor protein. J. Virol. 72:6898-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krzyzaniak, M., M. Mach, and W. J. Britt. 2007. The cytoplasmic tail of glycoprotein M (gpUL100) expresses trafficking signals required for human cytomegalovirus assembly and replication. J. Virol. 81:10316-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakadamyali, M., M. J. Rust, H. P. Babcock, and X. Zhuang. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 100:9280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leopold, P. L., and K. K. Pfister. 2006. Viral strategies for intracellular trafficking: motors and microtubules. Traffic 7:516-523. [DOI] [PubMed] [Google Scholar]
- 51.Lyman, M. G., and L. W. Enquist. 2009. Herpesvirus interactions with the host cytoskeleton. J. Virol. 83:2058-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mabit, H., M. Y. Nakano, U. Prank, B. Saam, K. Dohner, B. Sodeik, and U. F. Greber. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 76:9962-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mach, M., B. Kropff, M. Kryzaniak, and W. Britt. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marozin, S., U. Prank, and B. Sodeik. 2004. Herpes simplex virus type 1 infection of polarized epithelial cells requires microtubules and access to receptors present at cell-cell contact sites. J. Gen. Virol. 85:775-786. [DOI] [PubMed] [Google Scholar]
- 55.Matanis, T., A. Akhmanova, P. Wulf, E. Del Nery, T. Weide, T. Stepanova, N. Galjart, F. Grosveld, B. Goud, C. I. De Zeeuw, A. Barnekow, and C. C. Hoogenraad. 2002. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 4:986-992. [DOI] [PubMed] [Google Scholar]
- 56.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moorhead, A. R., K. A. Rzomp, and M. A. Scidmore. 2007. The Rab6 effector Bicaudal D1 associates with Chlamydia trachomatis inclusions in a biovar-specific manner. Infect. Immun. 75:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakano, M. Y., K. Boucke, M. Suomalainen, R. P. Stidwill, and U. F. Greber. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74:7085-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naranatt, P. P., H. H. Krishnan, M. S. Smith, and B. Chandran. 2005. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J. Virol. 79:1191-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogawa-Goto, K., K. Tanaka, W. Gibson, E. Moriishi, Y. Miura, T. Kurata, S. Irie, and T. Sata. 2003. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J. Virol. 77:8541-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh, J., K. Baksa, and R. Steward. 2000. Functional domains of the Drosophila bicaudal-D protein. Genetics 154:713-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh, J., and R. Steward. 2001. Bicaudal-D is essential for egg chamber formation and cytoskeletal organization in Drosophila oogenesis. Dev. Biol. 232:91-104. [DOI] [PubMed] [Google Scholar]
- 63.Ojala, P. M., B. Sodeik, M. W. Ebersold, U. Kutay, and A. Helenius. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radtke, K., K. Dohner, and B. Sodeik. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell. Microbiol. 8:387-400. [DOI] [PubMed] [Google Scholar]
- 65.Roby, C., and W. Gibson. 1986. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 59:714-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samanta, M., L. Harkins, K. Klemm, W. J. Britt, and C. S. Cobbs. 2003. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J. Urol. 170:998-1002. [DOI] [PubMed] [Google Scholar]
- 67.Sanchez, V., P. C. Angeletti, J. A. Engler, and W. J. Britt. 1998. Localization of human cytomegalovirus structural proteins to the nuclear matrix of infected human fibroblasts. J. Virol. 72:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanjuan, N., A. Porras, and J. Otero. 2003. Microtubule-dependent intracellular transport of murine polyomavirus. Virology 313:105-116. [DOI] [PubMed] [Google Scholar]
- 71.Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Brauchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 294:1929-1932. [DOI] [PubMed] [Google Scholar]
- 72.Selvin, P. R. 2000. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7:730-734. [DOI] [PubMed] [Google Scholar]
- 73.Seo, J. Y., and W. J. Britt. 2007. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J. Virol. 81:6536-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seo, J. Y., and W. J. Britt. 2008. Multimerization of tegument protein pp28 within the assembly compartment is required for cytoplasmic envelopment of human cytomegalovirus. J. Virol. 82:6272-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seo, J. Y., and W. J. Britt. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Short, B., C. Preisinger, J. Schaletzky, R. Kopajtich, and F. A. Barr. 2002. The Rab6 GTPase regulates recruitment of the dynactin complex to Golgi membranes. Curr. Biol. 12:1792-1795. [DOI] [PubMed] [Google Scholar]
- 77.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith, G. A., and L. W. Enquist. 2002. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18:135-161. [DOI] [PubMed] [Google Scholar]
- 79.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. U. S. A. 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 81.Sodeik, B. 2002. Unchain my heart, baby let me go—the entry and intracellular transport of HIV. J. Cell Biol. 159:393-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suter, B., L. M. Romberg, and R. Steward. 1989. Bicaudal-D, a Drosophila gene involved in developmental asymmetry: localized transcript accumulation in ovaries and sequence similarity to myosin heavy chain tail domains. Genes Dev. 3:1957-1968. [DOI] [PubMed] [Google Scholar]
- 83.Swan, A., and B. Suter. 1996. Role of Bicaudal-D in patterning the Drosophila egg chamber in mid-oogenesis. Development 122:3577-3586. [DOI] [PubMed] [Google Scholar]
- 84.Tandon, R., and E. S. Mocarski. 2008. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J. Virol. 82:9433-9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teuling, E., V. van Dis, P. S. Wulf, E. D. Haasdijk, A. Akhmanova, C. C. Hoogenraad, and D. Jaarsma. 2008. A novel mouse model with impaired dynein/dynactin function develops amyotrophic lateral sclerosis (ALS)-like features in motor neurons and improves lifespan in SOD1-ALS mice. Hum. Mol. Genet. 17:2849-2862. [DOI] [PubMed] [Google Scholar]
- 86.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wanschers, B. F., R. van de Vorstenbosch, M. A. Schlager, D. Splinter, A. Akhmanova, C. C. Hoogenraad, B. Wieringa, and J. A. Fransen. 2007. A role for the Rab6B Bicaudal-D1 interaction in retrograde transport in neuronal cells. Exp. Cell Res. 313:3408-3420. [DOI] [PubMed] [Google Scholar]
- 89.Wharton, R. P., and G. Struhl. 1989. Structure of the Drosophila BicaudalD protein and its role in localizing the the posterior determinant nanos. Cell 59:881-892. [DOI] [PubMed] [Google Scholar]
- 90.Yonezawa, A., M. Cavrois, and W. C. Greene. 2005. Studies of Ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J. Virol. 79:918-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zipeto, D., F. Baldanti, E. Percivalle, G. Gerna, and G. Milanesi. 1993. Identification of a human cytomegalovirus mutant in the pp150 matrix phosphoprotein gene with a growth-defective phenotype. J. Gen. Virol. 74:1645-1648. [DOI] [PubMed] [Google Scholar]