Abstract
Hepatitis E virus (HEV) is a causative agent of acute hepatitis, and it is the sole member of the genus Hepevirus in the family Hepeviridae. The open reading frame 1 (ORF1) protein of HEV encodes nonstructural polyprotein with putative domains for methyltransferase, cysteine protease, helicase and RNA-dependent RNA polymerase. It is not yet known whether ORF1 functions as a single protein with multiple domains or is processed to form separate functional units. On the basis of amino acid conserved motifs, HEV helicase has been grouped into helicase superfamily 1 (SF-1). In order to examine the RNA helicase activity of the NTPase/helicase domain of HEV, the region (amino acids 960 to 1204) was cloned and expressed as histidine-tagged protein in Escherichia coli (HEV Hel) and purified. HEV Hel exhibited NTPase and RNA unwinding activities. Enzyme hydrolyzed all rNTPs efficiently, dATP and dCTP with moderate efficiency, while it showed less hydrolysis of dGTP and dTTP. Enzyme showed unwinding of only RNA duplexes with 5′ overhangs showing 5′-to-3′ polarity. We also expressed and purified two HEV Hel mutants. Helicase mutant I, with substitution in the nucleotide-binding motif I (GKS to GAS), showed 30% ATPase activity. Helicase mutant II, with substitutions in the Mg2+ binding motif II (DEAP to AAAP), showed 50% ATPase activity. Both mutants completely lost ability to unwind RNA duplexes with 5′ overhangs. These findings represent the first report demonstrating NTPase/RNA helicase activity of the helicase domain of HEV ORF1.
Viruses with single-strand positive-sense RNA genomes represent the largest class of viruses, which includes numerous pathogens of humans, plants, and animals. In these viruses, RNA replication occurs through negative-strand RNA intermediate, which may also act as the template for synthesis of subgenomic RNAs in some viruses. During replication, various nonstructural proteins remain associated with the viral polymerase in a small compartmentalized replisome. Most of the other accessory proteins are obtained from the cellular machinery.
Helicase seems to be essential for RNA replication by many positive-sense RNA viruses (19). Many positive-strand RNA viruses encode their own RNA helicases and besides RNA-dependent RNA polymerase, helicase is the most conserved viral sequence in these viruses. It has been shown by direct mutagenesis studies in poliovirus (26, 39), alphaviruses (31), brome mosaic virus (2, 41), nidoviruses (40), and flaviviruses (15) that helicase functions are essential for viral replication. In addition, it may be involved in RNA translocation, genome packaging, protection of RNA at the replication center, modulating RNA-protein interactions, etc.
Helicases are classified into six superfamilies, SF-1 to SF-6 (11, 35), and can be classified further into subfamilies, A (3′→5′) or B (5′→3′) depending on their unwinding directionality. Classic helicases (exhibiting both NTPase and unwinding activities) are referred to as subtype α, while translocases (with no unwinding activity) are referred to as subtype β (35). SF-1 and SF-2 constitute largest of these superfamilies with seven signature motifs (I, Ia, II, III, IV, V, and VI), which form core of the enzyme. Although these motifs are not comparable between SF-1 and SF-2, universal features of core domains include (i) conserved residues involved in binding and hydrolysis of the NTP and (ii) an arginine finger that plays a key role in energy coupling.
Hepatitis E virus (HEV) is a nonenveloped virus in the genus Hepevirus of the family Hepeviridae. Hepatitis E is an important public health disease in many developing countries and is also endemic in some industrialized countries (8). Infection by HEV has a known association with increased mortality during pregnancy (22, 23). HEV has a positive-sense RNA genome of ∼7.2 kb, consisting of a 5′ noncoding region (5′NCR) of 27 to 35 nucleotides (nt), followed by three open reading frames (ORFs)—ORF1, ORF2, and ORF3—and a 3′NCR of 65 to 74 nt, ending with a poly(A) tail of variable length (37). The 5′ end has m7G cap (18). ORF1 is known to encode for the viral nonstructural polyprotein with a proposed molecular mass of ∼186 kDa (3). Based on protein sequence homology, the ORF1 polyprotein is proposed to contain four putative domains indicative of methyltransferase, papain-like cysteine protease, RNA helicase (Hel), and RNA-dependent RNA polymerase (RdRp) (24). ORF2 encodes the major structural protein (capsid protein), which has N-terminal signal peptide and three glycosylation sites and is translocated across the endoplasmic reticulum (ER). ORF2 protein associates with the 5′ end of the viral RNA, suggesting its regulatory role in the virus replication (36, 37, 44, 45). ORF3 encodes a protein which gets phosphorylated by the cellular mitogen activated protein kinase and is associated with cellular membranes and cytoskeleton fractions (43).
HEV belongs to an “alpha-like” supergroup of positive-sense single-stranded RNA (+ssRNA) viruses with conserved motifs of replication-related proteins in the ORF1, with typical signature sequences homologous with the other members of the family (11, 12, 13). ORF1 of HEV encodes additional domains such as the Y domain, papainlike protease, “proline-rich hinge,” and the X domain. Methyltransferase (25), RdRp (1), and X domain (binding to poly-ADP-ribose) (9) in ORF1 have been characterized, whereas the functions of the other domains are yet to be identified. Intracellularly expressed RdRp localizes itself in the ER membranes (30), suggesting that HEV replicates probably in ER in the cytosolic compartment of the cells. It is still unknown whether ORF1 polyprotein undergoes cleavages to form separate functional units of the replication machinery or functions as a single protein with multiple functional domains.
The putative RNA helicase of HEV contains all of the seven conserved segments typical of the SF-1 helicase (12, 13). Putative SF-1 helicases are extremely widespread among +ssRNA viruses. Based on sequence comparisons, such helicases have been identified in a variety of plant virus families, as well as in animal viruses such as alphavirus, rubivirus, hepatitis E virus, and coronavirus (11). When compared to other +ssRNA viral helicases belonging to SF-1, HEV helicase showed the highest overall similarity with the helicase of beet necrotic yellow vein virus, a plant furovirus. HEV helicase was speculated to have N-terminal NTPase and C-terminal RNA-binding domains (24). A major obstacle in studying HEV replication has been lack of cell culture system. We report here experimental verification of the helicase activity of the recombinant helicase domain protein of HEV.
MATERIALS AND METHODS
Cloning, expression, and purification of putative HEV NTPase/helicase domain.
The putative HEV Helicase domain (amino acids 960 to 1204 of ORF1) was amplified by PCR from the plasmid pGEM-TEASY-HEVT1FG (a plasmid encoding complete genome of genotype 1 HEV; GenBank accession no. DQ459342). Start codon/NdeI and stop codon/BamHI sites were incorporated in the forward and reverse primers, respectively. The PCR product was cloned in vector pET15b in frame with N-terminal polyhistidine tag (pET15b.HEV Hel). After sequence confirmation, the recombinant vector was transformed into BL21(DE3)/pLysS host bacterial cells for protein expression. Protein induction was done with 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 33°C.
The fusion protein was purified from bacterial culture pellets by using a ProBond nickel chelating resin column (Invitrogen, Carlsbad, CA). Briefly, cell pellet was lysed by using buffer containing 6 M guanidinium-HCl, 20 mM sodium phosphate (pH 7.8), and 0.5 M NaCl. Cell lysate was spun at 10,000 × g for 30 min, filtered through a 0.45-μm-pore-size syringe filter (Millipore), and loaded on to the column preequilibrated with binding buffer composed of 8 M urea, 20 mM sodium phosphate (pH 7.8), and 0.5 M NaCl. Elution of the protein was performed using elution buffer (20 mM sodium phosphate [pH 7.8], 0.5 M NaCl, 250 mM imidazole). Collected fractions were analyzed by SDS-15% PAGE. Fractions containing protein of the expected size were combined and concentrated by using Amicon membrane columns (cutoff, 10 kDa; Millipore). The protein was further purified by gel filtration chromatography (Sephacryl HR100, CV-120 ml; Amersham Biosciences) by using an Akta Basic 100 HPLC system (Amersham Pharmacia). Fractions were analyzed by SDS-15% PAGE. Fractions containing purified protein were combined and then concentrated by using an Amicon membrane column, and the buffer was exchanged to 50 mM HEPES (pH 7.0). Glycerol and dithiothreitol (DTT) were added to final concentrations of 20% and 2 mM, respectively, and stored at −70°C in aliquots. Western blot analysis was done using anti-His monoclonal antibodies (Sigma Chemicals, St. Louis, MO). The protein concentration was determined by the Lowry method.
Generation of mutants by site-directed mutagenesis.
To modify underlined amino acid residues from the Walker A motif (motif I), AGVPGSGKS to AGVPGSGAS, and the Walker B motif (motif II), VIDEAP to VIAAAP, of HEV helicase, site-directed mutagenesis was carried out with the pET15b-HEV Hel clone as a template and the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions (the primer sequences are given in Table 1). Clones were confirmed by sequencing and transformed into Escherichia coli BL21(DE3)/pLysS cells. Protein induction and purification were performed as described above.
TABLE 1.
PCR primers/oligonucleotides used in this study
| Primer or oligonucleotide | Sequence (5′-3′) | mer |
|---|---|---|
| Primers | ||
| Helicase forward | CATCCATATGGGCTGTCGGGTTACCCCCGG | 30 |
| Helicase reverse | CATCGGATCCTCAGAAAAAGTTATTAACGATTG | 33 |
| SM GKS forward | GGT GTG CCC GGG TCC GGC GCG TCC CGC TCT ATC ACC | 36 |
| SM GKS reverse | GGT GAT AGA GCG GGA CGC GCC GGA CCC GGG CAC ACC | 36 |
| SM DE forward | GGG CGC CGG GTT GTC ATT GCT GCG GCT CCA TCC CTC | 36 |
| SM DE reverse | GAG GGA TGG AGC CGC AGC AAT GAC AAC CCG GCG CCC | |
| RNA oligonucleotides | ||
| RNA1 | UUU UUU UUU UUU CGC UGA UGU CGC CUG G | 28 |
| RNA2 | CCA GGC GAC AUC AGC G | 16 |
| RNA3 | CGC UGA UGU CGC CUG GUU UUU UUU UUU U | 28 |
| RNA4 | CCA GGC GAC AUC AGC GAA AAA AAA AAA A | 28 |
| Poly(A) | AAA AAA AAA AAA AAA AAA A | 19 |
| Poly(G) | GGG GGG GGG GGG GGG GGG G | 19 |
| Poly(C) | CCC CCC CCC CCC CCC CCC C | 19 |
| Poly(U) | UUU UUU UUU UUU UUU UUU U | 19 |
| DNA oligonucleotides | ||
| DNA1 | TTT TTT TTT TTT CGC TGA TGT CGC CTG G | 28 |
| DNA2 | CCA GGC GAC ATC AGC G | 16 |
| DNA3 | CGC TGA TGT CGC CTG GTT TTT TTT TTT T | 28 |
| DNA4 | CCA GGC GAC ATC AGC GAA AAA AAA AAA A | 28 |
| Poly(A) | AAA AAA AAA AAA AAA AAA A | 19 |
| Poly(G) | GGG GGG GGG GGG GGG GGG G | 19 |
| Poly(C) | CCC CCC CCC CCC CCC CCC C | 19 |
| Poly(T) | TTT TTT TTT TTT TTT TTT T | 19 |
NTP-binding analysis.
Nucleoside triphosphate (NTP) and deoxynucleoside triphosphate (dNTP) binding analysis was carried out essentially as described previously (7). All radiochemicals were purchased from BRIT (Hyderabad, India). Briefly, assays were done separately for different NTPs in a 20-μl total volume, each containing 2 μg of protein, 50 mM HEPES (pH 7.0), 2 mM MgCl2, 10 mM KCl, 5 mM DTT, and 1 μCi of each of the following α-32P-labeled NTPs: ATP (3,000 Ci/mmol), GTP (3,200 Ci/mmol), CTP (3,200 Ci/mmol), UTP (3,200 Ci/mmol), dATP (3,200 Ci/mmol), dCTP (3,200Ci/mmol), and dGTP (2,500 Ci/mmol). The reaction mixture was incubated in a microtiter plate (96 well) on ice for 15 min, followed by cross-linking by UV irradiation (254 nm) for 45 min on ice. The reaction was terminated by adding 0.5 μl of cold 100 mM ATP. Proteins were separated by SDS-12.5% PAGE, and the gel was stained with Coomassie brilliant blue and processed further for autoradiography.
NTPase assay.
NTPase assays were performed using two methods. The first was a thin-layer chromatography assay as described earlier (10). Briefly, reaction was carried out in a 20 μl-volume containing 1 to 100 pmol of protein, 50 mM HEPES (pH 7.0), 2 mM MgCl2, 10 mM KCl, 0.05 mg of bovine serum albumin (BSA)/ml, 2 mM DTT, 0.1 or 0.5 mM ATP, 1 μCi of [γ-32P] ATP, and (3,000 Ci/mmol). Reactions were incubated at 37°C for 40 min and then terminated by the addition of EDTA to a final concentration of 20 mM. Analysis of the products of hydrolysis was done by spotting a 1-μl reaction mixture onto a polyethyleneimine-cellulose thin-layer chromatography plate (Merck), and separation was carried out with 0.375 M potassium phosphate (pH 3.5) as the mobile phase. Plate was air dried and exposed to X-ray film. ATPase assays were carried out with different protein concentrations, cold ATP concentrations, and poly(U) oligonucleotide (19 nt; 25 μg/ml, final concentration).
The second assay was a colorimetric method performed in a 96-well plate using malachite green reagent. In a 50-μl reaction, 100 pmol of enzyme, 50 mM HEPES (pH 7.0), 2 mM MgCl2, 10 mM KCl, 0.05 mg of BSA/ml, 2 mM DTT, and 0.1 mM NTP were added, followed by incubation at 37°C for 40 min. When indicated, various 19-nt oligonucleotides [either poly(U), poly(A), poly(C), or poly(G) RNA or poly(T), poly(A), poly(G), or poly(C) DNA] were included in the reaction mixture at a concentration of 25 μg/ml. The reaction was terminated by the addition of EDTA to a final concentration of 20 mM, and color was developed by the addition of an equal volume of malachite green-molybdate reagent (to prepare malachite green-molybdate reagent, solution 1 [0.045% malachite green] and solution 2 [4.2% ammonium molybdate in 4 N HCl] were freshly mixed in a 3:1 ratio, respectively, and filtered through a 0.22-μm-pore-size filter, and Tween 20 was added to a final concentration of 0.01%), followed by incubation at room temperature for 5 min. The absorbance was measured at 630 nm by using μQuant (Bio-Tek) 96-well plate reader. All of the reactions were carried out in duplicates, and average values were obtained to calculate the NTPase activity in the form of the amount of inorganic phosphate produced through hydrolysis of NTP using a standard curve (made by using known concentrations of potassium dihydrogen phosphate in the assay). All of the results given with this quantitative assay are averages of two repeat experiments.
Preparation of helicase substrates.
The oligonucleotides DNA1 (28 bases) and DNA2 (16 bases) (all sequences are listed in Table 1) (28) were used to generate DNA duplexes with a 5′ overhang. For duplexes with a 3′ overhang, DNA3 (28 bases) and DNA2 were used, and for blunt-end duplexes, DNA4 (28 bases) and DNA1 were used. To generate RNA substrates, oligonucleotides with the same sequences were used (by replacing T with U). All DNA and RNA oligonucleotides were obtained commercially (IDT).
To generate RNA duplex substrates with a 5′ overhang, 0.2 nmol of RNA2 (16 bases) was 32P radiolabeled at the 5′ end using T4 polynucleotide kinase (NEB) and [γ-32P]ATP (3,000 Ci/mmol) according to the manufacturer's protocol. The radiolabeled oligonucleotide was mixed with 0.2 nmol of RNA1 (28 bases) in annealing buffer (10 mM Tris-Cl [pH 7.4], 100 mM NaCl, 1 mM EDTA), heated at 94°C for 5 min, and allowed to cool slowly to room temperature. Annealed products were resolved on 20% nondenaturing PAGE, and a band corresponding to double-stranded nucleic acid was cut from the gel, minced, and soaked in buffer containing 0.5 M ammonium acetate, 10 mM EDTA, and 1% SDS at room temperature for 5 to 6 h for elution. An equal volume of chloroform was added and mixed in, and the mixture was allowed to stand for 30 min. The aqueous phase was collected, precipitated with an equal volume of isopropanol, left overnight at −20°C, and spun, and the pellet was dissolved in nuclease-free water.
All duplexes were generated using the same strategy.
Strand displacement assays.
The 20-μl unwinding reaction containing 50 mM HEPES (pH 7.0), 2 mM MgCl2, 10 mM KCl, 0.05 mg of BSA/ml, 2 mM DTT, 1 mM ATP, 1 pmol of labeled RNA-DNA duplex, and 100 pmol of protein was incubated at 37°C for 2 h. The reaction was terminated by using 0.2% SDS and 20 mM EDTA (final concentration), followed by proteinase K treatment for 10 min at 37°C. The products were resolved by 20% PAGE, and the gels were processed for autoradiography.
RESULTS
Expression and purification of a putative HEV NTPase/helicase domain and its mutants.
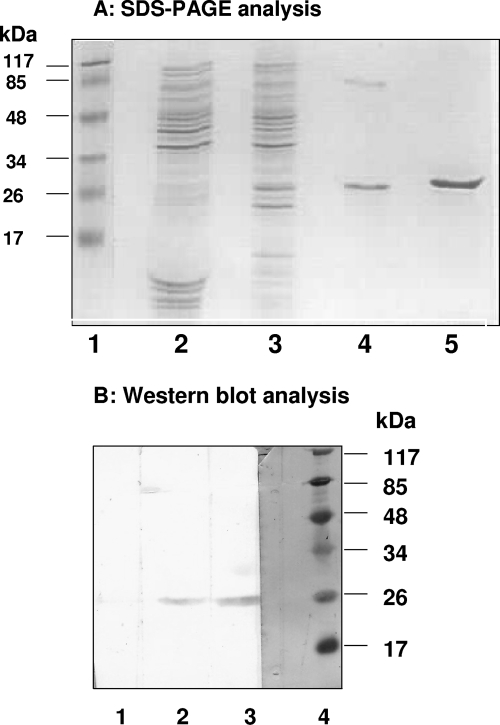
Full-length HEV NTPase/helicase (amino acids 960 to 1204 of the ORF1, 245 amino acids long) (24) was expressed as an N-terminal histidine tagged fusion protein in a bacterial expression system and purified from inclusion bodies by using nickel affinity chromatography with a hybrid buffer system, i.e., loading of the sample in denaturing conditions and elution in a native buffer condition. Analysis of the protein by SDS-PAGE revealed that purity of the protein was ca. 90% and needed further purification. Nickel resin-purified protein was purified by gel filtration chromatography using high-pressure liquid chromatography (HPLC) in the next step. The purified protein was had a mass of ∼26 kDa when analyzed by SDS-PAGE and Western blotting (Fig. 1). The HPLC gel filtration peak corresponding to HEV helicase protein was in the range of 25 to 30 kDa (data not shown), indicating that the protein existed in monomeric form at physiological conditions (1× phosphate-buffered saline).
FIG. 1.
SDS-PAGE and Western blot analysis. (A) SDS-15% PAGE analysis of HEV Hel protein purification at each stage of purification. Lanes: 1, prestained protein molecular mass marker; 2, uninduced cell extract; 3, induced cell extract; 4, protein purified with a Ni-chelating resin; 5, HPLC-purified protein. (B) Western blot analysis of HEV Hel protein using anti-histidine monoclonal antibodies for detection. Lanes: 1, induced cell lysate; 2, protein purified using Ni-chelating resin; 3, HPLC-purified protein; 4, amido black stained molecular mass marker.
Two mutants of the HEV helicase (Hel mut I and Hel mut II) were generated by site-directed mutagenesis. These proteins were also expressed as histidine-tagged proteins in E. coli and purified by using the protocol used for the wild-type enzyme. In Hel mut I, a conserved lysine residue in motif I (AGVPGSGKS), which is known to have crucial role in NTP binding was mutated to an alanine (K to A). In Hel mut II, conserved aspartic acid and glutamic acid residues from motif II (VIDEAP), known to be involved in Mg2+ binding, were modified to alanines (AA). Both proteins showed same sized bands on SDS-PAGE and Western blotting as those for the wild-type protein (data not shown).
NTP binding and NTPase activities of HEV Hel.
NTP hydrolysis is known to provide the energy required for translocation of RNA helicase, along with a single-stranded nucleic acid and for duplex unwinding. To analyze the ability and specificity of HEV Hel in binding and hydrolyzing different nucleotide substrates, assays were done using both rNTPs and dNTPs.
Binding of HEV Hel to NTPs.
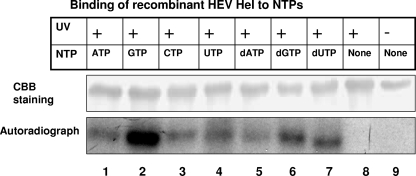
Reaction mixtures containing HEV Hel protein and different radiolabeled rNTPs and dNTPs were allowed to interact for 15 min, followed by UV cross-linking, and the products were analyzed by SDS-12.5% PAGE. Recombinant HEV helicase showed binding with rATP, rCTP, rGTP, and rUTP, as well as all of the three tested dNTPs (Fig. 2), indicating no marked specificity of the protein in NTP binding.
FIG. 2.
Binding of recombinant HEV Hel to NTPs. For NTP binding analysis, purified protein was incubated with [α-32P]NTP for 15 min, UV irradiated for 45 min, and subjected to SDS-12.5% PAGE. The gel was stained with Coomassie brilliant blue, followed by autoradiography. Lanes: 1, protein cross-linked with [α-32P]ATP; 2, [α-32P]GTP; 3, [α-32P]CTP; 4, [α-32P]UTP; 5, [α-32P]dATP; 6, [α-32P]dGTP; 7, [α-32P]dCTP; 8, UV-treated protein without NTP; 9, protein without UV treatment.
Optimization of the ATPase activity of HEV Hel.
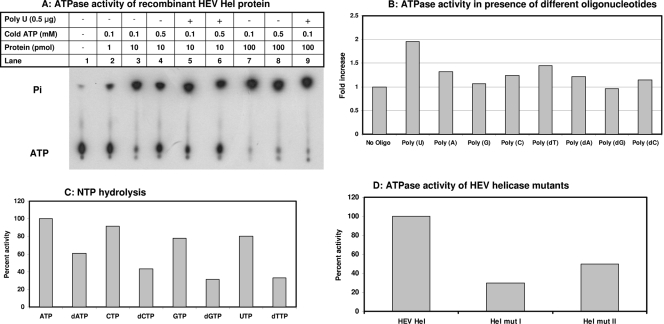
The purified HEV Hel protein was tested for ATP hydrolysis activity by using [γ-32P]ATP and thin-layer chromatography analysis and was found to be positive. The activity steadily increased with an increase in the HEV Hel concentration from 1 to 100 pmol (Fig. 3A). A quantitative colorimetric NTPase assay was used to measure the NTPase activity of HEV Hel using ATP as a substrate. Optimum conditions were determined to be 2 mM MgCl2, 0.1 mM ATP, and 25 μg of poly(U)/ml and pH 7.0 for this assay. These conditions were used for further analysis of the activities.
FIG. 3.
NTPase activity of recombinant HEV proteins. (A) The ATPase activity of recombinant HEV Hel protein was tested at different protein and cold ATP concentrations. Each 20-μl reaction contained 1 μCi of [γ-32P]ATP. After incubation at 37°C for 40 min, 1 μl of the mixture was analyzed by thin-layer chromatography and processed for autoradiography. (B) The ATPase activity of recombinant HEV Hel protein was tested in quantitative assay in the presence of different oligonucleotides. ATPase assays were carried out in the presence of RNA/DNA oligonucleotides in a 50-μl reaction containing 100 pmol of protein and 0.1 mM ATP. (C) The NTPase activity of recombinant HEV Hel protein was tested with both rNTPs and dNTPs in a quantitative assay. Each 50-μl reaction contained 0.1 mM NTP and 100 pmol of protein. (D) The ATPase activity of HEV Hel mut I and mut II proteins was tested in 50-μl reactions containing 100 pmol protein, and 0.1 mM ATP.
Polynucleotide stimulation and NTPase activity.
To see the stimulatory effects of different polynucleotides on the ATPase activity of HEV Hel, a quantitative colorimetric assay was carried out in the presence of different RNA-DNA homopolymers. To calculate the fold increase in the enzymatic activity, the activity without any polynucleotide was taken as 1. Poly(U) showed an ∼2-fold increase in the enzymatic activity. Poly(A), poly(C), poly(dT), poly(dA), and poly(dC) showed <1.5-fold stimulation. Poly(G) and poly(dG) did not show any effect on the enzymatic activity (Fig. 3B).
To know whether HEV Hel was able to hydrolyze all of the bound NTPs, different NTPs were tested as substrates in the presence of poly(U). ATP was found to be the best substrate for HEV Hel. To compare hydrolysis of other NTPs, ATP hydrolysis was taken as the 100% enzymatic activity and values were calculated for different NTPs. Activity of the enzyme with ATP, CTP, GTP, and UTP ranged from 100 to 80%. Although it decreased in the order of dATP > dCTP > dTTP > dGTP with dNTPs as substrates (activity range, 60 to 30%) (Fig. 3C).
ATPase activity of helicase mutants.
Hel mut I and Hel mut II proteins were assayed for their ATPase activities in a quantitative assay (Fig. 3D). Hel mut I showed a 70% loss in the ATPase activity, while Hel mut II showed a 50% loss compared to wild-type HEV Hel. This also demonstrates that results with proteins purified from E. coli extracts were authentic activities due to HEV helicase and not due to E. coli-derived contaminant.
RNA/DNA strand displacement activities.
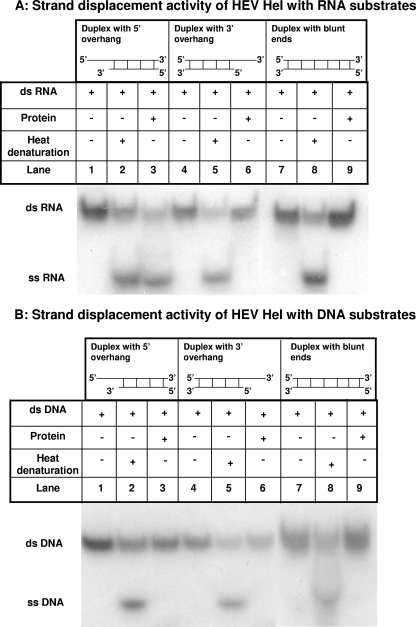
To characterize the unwinding activity of HEV Hel, RNA and DNA duplexes with either 3′ or 5′ single-stranded overhangs or duplexes with blunt ends were used. The blunt end duplexes were generated by annealing 28-nt oligonucleotides, while for duplexes with 5′ and 3′ overhangs, 28 nt and 16 nt long oligonucleotides were used, and both had 12-nt single-stranded stretches at their respective ends. One strand in each of the three RNA or DNA duplexes was 5′ end labeled. In duplexes with 5′ and 3′ overhangs, labeling of the 16-mer was done to facilitate the visualization on PAGE upon unwinding. HEV Hel showed binding with all duplexes (with blunt ends and with a 5′ overhang or a 3′ overhang) of both RNA and DNA irrespective of their overhangs, indicating no specificity for nucleic acid binding (data not shown).
Duplex unwinding assays were done in the presence of ATP and Mg2+. HEV Hel showed unwinding of only RNA duplexes with 5′ single-stranded overhang that was visualized by release of the labeled 16-mer (Fig. 4A). Enzyme did not show unwinding of DNA duplex with 5′ overhang. Similarly, there was no unwinding of RNA-DNA duplexes with blunt ends or with 3′ overhangs (Fig. 4).
FIG. 4.
Strand displacement activity of recombinant HEV helicase protein. (A) Strand displacement activity of recombinant HEV Hel was checked using different RNA substrates. The HEV Hel protein was incubated with RNA duplexes with a 5′ overhang (lanes 1, 2, and 3), with a 3′ overhang (lanes 4, 5, and 6), and with blunt ends (lanes 7, 8, and 9). (B) Strand displacement activity of recombinant HEV Hel was checked by using different DNA substrates. Wild-type HEV Hel protein was incubated with DNA duplexes with a 5′ overhang (lanes 1, 2, and 3), with a 3′ overhang (lanes 4, 5, and 6), and with blunt ends (lanes 7, 8, and 9).
Hel mut I and HEV Hel mut II proteins were also checked for their unwinding ability at a 100-pmol protein concentration using RNA duplex with a 5′ overhang. Both mutants completely lost the unwinding ability, although Hel mut II had shown 50% ATPase activity (data not shown).
DISCUSSION
ORF1 of HEV codes for a nonstructural polyprotein with a length of 1693 amino acids with multiple enzyme domains. On the basis of sequence motif analysis, HEV Hel was classified into SF-1 group of helicases. The lack of a suitable cell culture system and small animal model has been the main obstacle in understanding replication of the virus. However, several labs have attempted to evaluate the role of different viral proteins in replication, pathogenesis, and host defense by cloning individual viral genes (6, 27, 42). Putative domains of the polyprotein, such as methyl transferase, the X domain (macrodomain), and RdRp, have been expressed as individual recombinant proteins, and their activities have been successfully demonstrated in vitro (9, 25, 30). In the present study, we cloned the putative NTPase/helicase domain of genotype 1 HEV in a prokaryotic system and tested the protein for the signature activities of the enzyme.
Helicases unwind nucleic acid duplexes using NTP as an energy source, and ATPase activity is intrinsic to both viral and cellular helicases. HEV Hel exhibited efficient hydrolysis of all rNTPs. It also exhibited hydrolysis of all dNTPs, but with relatively low efficiency. It was observed in severe acute respiratory syndrome (SARS) coronavirus helicase (member of the SF-1) that, although the enzyme preferred ATP, dATP, and dCTP as substrates, it also hydrolyzed all NTPs (38).
The ATPase and helicase activities of many +ssRNA viruses have been reported, and most of the helicases were found to be stimulated in the presence of RNA—either homonucleotides or nonspecific RNA oligonucleotides. Stimulation by RNA cofactor was seen in enhancing the basal ATPase activity of the enzyme by 15- to 20-fold in SF-1 members such as the SARS coronavirus (38), human coronavirus 229E (HCo-229E) (17, 33), and equine arteritis virus (34) and in SF-2 members such as dengue virus (5) and bovine diarrhea virus (15). We observed twofold stimulation in the ATPase activity of HEV Hel in the presence of poly(U) and no stimulation with poly(G) and poly(dG), whereas other tested oligonucleotides showed <1.5-fold enhancement. SF-1 helicases of turnip yellow mosaic virus (20), rubellavirus (14), and Semliki forest virus (32) showed a <2-fold increase in the ATPase activity in the presence of ssRNA. HEV Hel belongs to this second group of viruses.
Walker A motif (motif I) has a consensus of XGX-AGXGKT in SF-1 helicases. The lysine residue is responsible for binding to the beta- and gammaphosphates of NTP-Mg2+ complexes. Mutation of this lysine abolishes the ATPase activity (16). Hel mut I in the present study showed a 70% loss in the ATPase activity due to the replacement of lysine with alanine. The conserved D residue in the Walker B motif (motif II, XXDEXD/H) has been shown to interact with catalytic Mg2+ and is important for NTPase activity, and mutation in this domain is known to affect both NTPase and helicase functions (29). Hel mut II (DE to AA) showed a 50% loss in the ATPase activity, indicating that both motifs I and II are equally important for ATP hydrolysis. Hel mut I and mut II completely lost unwinding activity when tested at a higher protein concentration (100 pmol), indicating the dependence of helicase activity on the NTPase activity of the enzyme.
Most helicases require a stretch of ssRNA of specific polarity adjacent to the duplex region to initiate strand separation. HEV Hel showed strand displacement only from RNA duplexes with 5′ overhangs exhibiting 5′-to-3′ polarity. SF-1 helicases from animal viruses such as HCo-229E (33) and SARS coronavirus (both of which are coronaviruses), equine arteritis virus (34), and porcine reproductive and respiratory syndrome virus (4) (both of which are arteriviruses), possess 5′-to-3′ unwinding polarity. It was previously suggested that all SF-1 helicases have 5′-to-3′ unwinding polarity, but further studies showed that plant viruses such as hordeivirus and potexvirus (both +ssRNA viruses) encode helicases with bidirectional unwinding activity (21). Thus, HEV Hel belongs to a distinct class of 5′-to-3′ RNA viral helicases containing the animal +ssRNA group of viruses in the SF-1 family with other members such as coronaviruses, arteriviruses, alphaviruses, and rubiviruses.
In conclusion, we report the expression, purification, and biochemical characterization of the NTPase/helicase domain of ORF1 from hepatitis E virus. We report here that the computer-predicted helicase domain of HEV is a functional enzyme exhibiting both NTPase and RNA unwinding activities. The enzyme showed a 5′-to-3′ unwinding polarity with specificity for RNA duplexes. Motif I mutant (Walker A motif) with replacement of critical lysine residue resulted in a 70% loss in ATPase activity, whereas motif II mutant (Walker B motif) with replacement of DE with AA resulted in a 50% reduction in ATPase activity. Further, the lack of unwinding activities in both mutant enzymes confirmed the dependence of helicase activity on the efficient generation of energy during unwinding. Considering their vital role during virus replication, helicases are being considered antiviral targets. We can explore this possibility for HEV. Further, characterization of other motifs in the enzyme may also help elucidate the function of SF-1 helicases and prove useful in the design of broad-spectrum antivirals.
Acknowledgments
Yogesh Karpe acknowledges Indian Council of Medical Research for a senior research fellowship.
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Agarwal, S., D. Gupta, and S. K. Panda. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA polymerase (RdRp). Virology 282:87-101. [DOI] [PubMed] [Google Scholar]
- 2.Ahola, T., J. A. den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari, I. H., S. K. Nanda, H. Durgapal, S. Agrawal, S. K. Mohanty, D. Gupta, S. Jameel, and S. K. Panda. 2000. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1). J. Med. Virol. 60:275-283. [PubMed] [Google Scholar]
- 4.Bautista, E. M., K. S. Faaberg, D. Mickelson, and E. D. McGruder. 2002. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology 298:258-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benarroch, D., B. Selisko, G. A. Locatelli, G. Maga, J. L. Romette, and B. Canard. 2004. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of dengue virus protein NS3 are Mg2+ dependent and require a functional Walker B motif in the Helicase catalytic core. Virology 328:208-218. [DOI] [PubMed] [Google Scholar]
- 6.Chandra, V., S. Taneja, M. Kalia, and S. Jameel. 2008. Molecular biology and pathogenesis of hepatitis E virus. J. Biosci. 33:451-464. [DOI] [PubMed] [Google Scholar]
- 7.Choudhury, N. R., P. S. Malik, D. K. Singh, M. N. Islam, K. Kaliappan, and S. K. Mukherjee. 2006. The oligomeric Rep protein of mungbean yellow mosaic India virus (MYMIV) is a likely replicative helicase. Nucleic Acids Res. 34:6362-6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemente-Casares, P., S. Pina, M. Buti, M. Martin, S. Bofill-Mas, and R. Girones. 2003. Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9:448-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egloff, M.-P., H. Malet, A. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80:8493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fakuda, K., H. Morioka, S. Imajou, S. Ikeda, E. Ohtsuka, and T. Tsurimoto. 1995. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J. Biol. Chem. 270:22527-22534. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., and E. V. Koonin. 1993. Comparative analysis of amino acid sequences of key enzymes of replication and expression of positive strand RNA viruses: validity of approach and functional and evolutionary implications. Sov. Sci. Rev. D Physicochem. Biol. 11:1-84. [Google Scholar]
- 12.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair, and expression of DNA and RNA genome. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1988. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 235:16-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gros, C., and G. Wengler. 1996. Identification of an RNA-stimulated NTPase in the predicted helicase sequence of the rubella virus nonstructural polyprotein. Virology 217:367-372. [DOI] [PubMed] [Google Scholar]
- 15.Gu, B., L. Changbao, J. Lin-Goerke, D. R. Maley, L. L. Gutshall, C. A. Feltenberger, and A. M. D. Vecchio. 2000. The RNA helicase and nucleotide triphosphatase activities of the bovine viral diarrhea virus NS3 protein are essential for viral replication. J. Virol. 74:1794-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 17.Ivanov, K. A., and J. Ziebuhr. 2004. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 78:7833-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabrane-Lazizi, Y., M. Zhang, R. H. Purcell, K. D. Miller, R. T. Davey, and S. U. Emerson. 2001. Acute hepatitis caused by a novel strain of hepatitis E virus most closely related to United States strains. J. Gen. Virol. 82:1687-1693. [DOI] [PubMed] [Google Scholar]
- 19.Kadaré, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadaré, G., C. David, and A. L. Haenni. 1996. ATPase, GTPase, and RNA binding activities associated with the 206-kilodalton protein of turnip yellow mosaic virus. J. Virol. 70:8169-8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalinina, N. O., D. V. Rakitina, A. G. Solovyev, J. Schiemann, and S. Y. Morozov. 2002. RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296:321-329. [DOI] [PubMed] [Google Scholar]
- 22.Khuroo, M., and S. Kamili. 2003. Aetiology, clinical course and outcome of sporadic acute viral hepatitis in pregnancy. J. Viral Hepat. 10:61-69. [DOI] [PubMed] [Google Scholar]
- 23.Khuroo, M. S., M. R. Teli, S. Skidmore, M. A. Sofi, and M. I. Khuroo. 1981. Incidence and severity of viral hepatitis in pregnancy. Am. J. Med. 70:252-255. [DOI] [PubMed] [Google Scholar]
- 24.Koonin, E. V., A. E. Gorbalenya, M. A. Purdy, M. N. Rozanov, G. R. Reyes, and D. W. Bradley. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 89:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magden, J., N. Takeda, T. Li, P. Auvinen, T. Ahola, T. Miyamura, A. Merits, and L. Kääriäinen. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 75:6249-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzayan, C., and E. Wimmer. 1992. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology 189:547-555. [DOI] [PubMed] [Google Scholar]
- 27.Moin, S. M., V. Chandra, R. Arya, and S. Jameel. 2009. The hepatitis E virus ORF3 protein stabilizes HIF-1alpha and enhances HIF-1-mediated transcriptional activity through p300/CBP. Cell Microbiol. 11:1409-1421. [DOI] [PubMed] [Google Scholar]
- 28.Nanduri, B., A. K. Byrd, R. L. Eoff, A. J. Tackett, and D. Kevin. 2002. Raney Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc. Natl. Acad. Sci. U. S. A. 99:14722-14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO. J. 11:2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehman, S., N. Kapur, H. Durgapal, and S. K. Panda. 2008. Subcellular localization of hepatitis E virus (HEV) replicase. Virology 370:77-92. [DOI] [PubMed] [Google Scholar]
- 31.Rikkonen, M. 1996. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology 218:352-361. [DOI] [PubMed] [Google Scholar]
- 32.Rikkonen, M., J. Peranen, and L. Kääriäinen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2, J. Virol. 68:5804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seybert, A., L. C. van Dinten, E. J. Snijder, and J. Ziebuhr. 2000. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 74:9586-9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singleton, M. R., M. S. Dillingham, and D. B. Wigley. 2007. Structure and mechanism of helicase and nucleic acid translocases Annu. Rev. Biochem. 76:23-50. [DOI] [PubMed] [Google Scholar]
- 36.Surjit, M., S. Jameel, and S. K. Lal. 2004. The ORF2 protein of hepatitis E virus binds the 50 region of viral RNA. J. Virol. 78:320-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam, A. W., M. M. Smith, M. E. Guerra, C. C. Huang, D. W. Bradley, K. E. Fry, and G. R. Reyes. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanner, J. N., R.M. Watt, Y. Chai, L. Lu, M. C. Lin, J. S. M. Peiris, L. L. M. Poon, H. F. Kung, and J. D. Huang. 2003. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 278:39578-39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teterina, N. L., K. M. Kean, A. E. Gorbalenya, V. I. Agol, and M. Girard. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 73:1977-1986. [DOI] [PubMed] [Google Scholar]
- 40.van Dinten, L. C., H. van Tol, A. E. Gorbalenya, and E. J. Snijder. 2000. The predicted metal-binding region of the Arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 74:5213-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, X., W. M. Lee, T. Watanabe, M. Schwartz, M. Janda, and P. Ahlquist. 2005. Brome mosaic virus 1a nucleoside triphosphatase/ helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747-13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita, T., Y. Mori, N. Miyazaki, R. H. Cheng, M. Yoshimura, H. Unno, R. Shima, K. Moriishi, T. Tsukihara, T. C. Li, N. Takeda, T. Miyamura, and Y. Matsuura. 2009. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. U. S. A. 106:12986-12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zafrullah, M., M. H. Ozdener, S. K. Panda, and S. Jameel. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 71:9045-9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zafrullah, M., M. H. Ozdener, R. Kumar, S. K. Panda, and S. Jameel. 1999. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 73:4074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zafrullah, M., Z. Khursheed, S. Yadav, D. Sahgal, S. Jameel, and F. Ahmad. 2004. Acidic pH enhances structure and structural stability of the capsid protein of hepatitis E virus. Biochem. Biophys. Res. Commun. 313:67-73. [DOI] [PubMed] [Google Scholar]