Abstract
Coxsackievirus A9 (CAV9) is a member of the human enterovirus B species within the Enterovirus genus of the family Picornaviridae. It has been shown to utilize αV integrins, particularly αVβ6, as its receptors. The endocytic pathway by which CAV9 enters human cells after the initial attachment to the cell surface has so far been unknown. Here, we present a systematic study concerning the internalization mechanism of CAV9 to A549 human lung carcinoma cells. The small interfering RNA (siRNA) silencing of integrin β6 subunit inhibited virus proliferation, confirming that αVβ6 mediates the CAV9 infection. However, siRNAs against integrin-linked signaling molecules, such as Src, Fyn, RhoA, phosphatidylinositol 3-kinase, and Akt1, did not reduce CAV9 proliferation, suggesting that the internalization of the virus does not involve integrin-linked signaling events. CAV9 endocytosis was independent of clathrin or caveolin-1 but was restrained by dynasore, an inhibitor of dynamin. The RNA interference silencing of β2-microglobulin efficiently inhibited virus infection and caused CAV9 to accumulate on the cell surface. Furthermore, CAV9 infection was found to depend on Arf6 as both silencing of this molecule by siRNA and the expression of a dominant negative construct resulted in decreased virus infection. In conclusion, the internalization of CAV9 to A549 cells follows an endocytic pathway that is dependent on integrin αVβ6, β2-microglobulin, dynamin, and Arf6 but independent of clathrin and caveolin-1.
Coxsackievirus A9 (CAV9), a member of the human enterovirus B species in the family Picornaviridae, is a significant human pathogen. It causes infections of the central nervous system, myocarditis, and respiratory diseases and may occasionally cause fatal generalized infections in newborns (6, 22, 26). The CAV9 particle is about 30 nm in diameter and consists of a naked capsid with an icosahedral symmetry, surrounding a positive-sense RNA genome of approximately 7,400 nucleotides (30). The capsid is made up of 60 copies of each of the four proteins VP1 to VP4 and interacts with cell surface integrins during the early stages of infection via arginine-glycine-aspartic acid (RGD) motif that resides in the C terminus of the VP1 protein (11). While CAV9 binds to both integrin αVβ3 and αVβ6 in vitro (53, 61), our recent data show that integrin αVβ6 is the primary receptor of the virus (29).
Viruses can utilize several endocytic pathways to enter mammalian cells: macropinocytosis and clathrin-mediated, caveolin-mediated, and clathrin- and caveolin-independent routes (14, 40-41, 50). Recent studies have shown that some of these pathways differ only slightly from each other, and certain endocytic components can participate in more than just one pathway (35, 41, 55). Most of the research carried out on enterovirus endocytosis has been done with echovirus 1 (EV1), coxsackievirus B3 (CBV3), and poliovirus (PV). Recently, Karjalainen et al. showed that EV1 enters SAOS cells via tubulovesicular structures in a dynamin-independent manner that resembles fluid-phase endocytosis and macropinocytosis and that at later stages of infection is targeted to caveosomes (33). EV1 entry to CV-1 cells, on the other hand, was shown to be strictly dynamin dependent (49). PV is endocytosed to HeLa cells by a rapid clathrin- and caveolin-independent pathway, whereas in brain microvascular endothelial cells it uses slower, caveolin- and dynamin-dependent endocytosis (4, 7, 17). CBV3 enters HeLa cells by clathrin-mediated endocytosis (13) and polarized epithelial CaCo-2 cells by a process that combines features of caveolar endocytosis and macropinocytosis (16, 18). Foot-and-mouth-disease virus (FMDV), a member of the Aphthovirus genus of the family Picornaviridae, binds to several αV-integrins, including αVβ6, and is internalized through the clathrin-mediated pathway (5, 19, 31). In the light of these examples, it is evident that enterovirus internalization to human cells is a complex phenomenon wherein a virus may use different mechanisms to enter different cell types.
In the CAV9 infection cycle, the steps that follow the initial attachment of the virus to the cell surface integrins are still poorly characterized. An early electron microscopic work by Hecker et al. has shown that single CAV9 particles enter monkey kidney cells in vesicles, which then occasionally fuse and form larger structures (28). Interestingly, they found that most internalized virus particles became eventually trapped in large vacuoles, presumably lysosomes, where they were confined without proceeding to capsid uncoating and RNA release. More recently, a number of cell surface molecules have been proposed to contribute to CAV9 internalization. A subunit of major histocompatibility complex class I (MHC-I) complex, β2-microglobulin (β2M), has been shown to be essential for the infection of several picornaviruses, including CAV9, and it is supposed to have a postattachment role (12, 59, 61). In addition, heat shock 70-kDa protein 5 (HSPA5 protein, also known as glucose-regulated protein 78-kDa, or GRP78) has been suggested to function as a coreceptor for the virus and to mediate CAV9 infection by its interaction with β2M on the cell surface (57). CAV9 entry has been proposed to occur through lipid microdomains, where a number of signaling events takes place (58).
The aim of this study was to elucidate the internalization mechanism of CAV9 in A549 human lung carcinoma cells. We used chemical inhibitors, RNA interference (RNAi) silencing, and the expression of dominant negative constructs combined to virus infectivity assays and confocal imaging to examine which cellular molecules are involved in the entry process. The results indicate that CAV9 internalization is dependent on integrin αVβ6, β2M, dynamin 2, and Arf6 (ADP-ribosylation factor 6) but not clathrin or caveolin-1.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The human lung carcinoma A549 cell line was obtained from the American Type Culture Collection (ATCC), and the caveolin-1-negative human hepatocellular carcinoma cell line HuH7 (60) was from Elina Ikonen (University of Helsinki, Finland). The Phoenix Gag-Pol packaging cell line (http://www.stanford.edu/group/nolan/index.html) was obtained from Aki Manninen (University of Oulu, Finland) with authorization by Garry Nolan (School of Medicine, Stanford, University, Stanford, CA). A549 cells were maintained in Ham's F12 medium supplemented with 7% fetal calf serum (FCS) and gentamicin. HuH7 and Phoenix cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS and gentamicin.
CAV9 (Griggs strain) (11, 30) was propagated in A549 cells and purified in sucrose gradient as described previously (1). Culture medium for virus infections was supplemented with 1% FCS.
Polyclonal rabbit antiserum against CAV9 was produced as described earlier (51), and mouse monoclonal antibody ([MAb] K6) against CAV9 (9) was obtained from Lucia Fiore (Instituto Superiore di Sanita, Rome, Italy). MAb against β2-microglobulin and Arf6 and rabbit polyclonal antiserum specific to caveolin-1 were from Santa Cruz Biotechnology (catalog items sc-51509, sc-7971, and sc-894, respectively). Rabbit polyclonal hemagglutinin (HA) antiserum was from Zymed Laboratories Inc. (catalog item 71-5500), and integrin αVβ6 MAb (MAB2074Z), which does not block function, was from Chemicon. Rabbit polyclonal antiserum specific to Erk1 (sc-94) was from Santa Cruz Biotechnology. Alexa Fluor (AF) 488-, 546-, and 568-labeled anti-mouse and anti-rabbit secondary antibodies were from Molecular Probes, and horseradish peroxidase (HRP)-labeled anti-rabbit secondary antibody was from Pierce. The function-inhibiting antibodies used in the blocking assays were against integrin αV (L230; ATCC), integrin αVβ3 (MAB1976Z; Chemicon), and integrin αVβ6 (MAB2077Z; Chemicon).
Chemical inhibitors.
A549 cells were incubated at 37°C in medium supplemented with 100 μM 5-(N-ethyl-N-isopropyl)amiloride ([EIPA] catalog no. A3085; Sigma), 25 μM chlorpromazine (C8138; Sigma), 1 mM methyl-β-cyclodextrin ([MβC] C4555; Sigma), 33.2 μM nocodazole (M1404; Sigma), 5 μg/ml cytochalasin D (C8273; Sigma), 2 μM jasplakinolide (420127; Calbiochem), 1 μM latrunculin A (L5163; Sigma), 100 nM wortmannin (W1628; Sigma), or with the combination of 25 μg/ml nystatin (N3503; Sigma) and 10 μg/ml progesterone (P8783; Sigma). The inhibitors were present for 30 min prior to and throughout the CAV9 infection assay. The importance of endosome acidification was studied by incubating the cells with 0.5 mM, 2 mM, 5 mM, or 25 mM NH4Cl at 37°C for 30 min and performing the virus infection assay in the presence of the salt. Cell viability after the treatment with NH4Cl was determined by the cell viability assay (see below).
The function of chlorpromazine, MβC, and cytochalasin D was confirmed by following the uptake of specific endocytic markers. AF 488-conjugated cholera toxin B (0.2 μg/ml; Molecular Probes) and AF 546-conjugated transferrin (10 μg/ml; Molecular Probes) were used to control the function of chlorpromazine and MβC, while AF 546-conjugated dextran (Molecular Probes) was used to control the function of EIPA (at 1 μM, 50 μM, 100 μM, and 150 μM) and cytochalasin D (at 250 μg/ml). A549 cells were incubated with the inhibitor for 30 min at 37°C, after which the marker was added. The incubation was continued for 15 min, the cells were fixed and stained with Hoechst 33342 (Sigma-Aldrich), and the data were analyzed by confocal imaging.
To analyze the effect of dynasore (D7693; Sigma) on CAV9 infection, A549 cells were incubated at 37°C for 30 min with 80 μM dynasore or 0.4% dimethyl sulfoxide (DMSO) diluted in serum-free cell medium. The CAV9 infection assay was then performed in the presence of the drug. The effective inhibitory concentration of dynamin was determined by incubating the cells with variable concentrations of dynasore (10 μM, 30 μM, 50 μM, and 80 μM), followed by feeding the cells with AF 546-conjugated transferrin (10 μg/ml) for 2 min at room temperature room temperature. Unbound transferrin was removed by washing the cells with serum-free medium containing dynasore or DMSO, and the transferrin uptake was followed for 15 min at 37°C. The cell plate was then transferred onto ice, and transferrin bound to the cell surface was removed by rinsing with an acidic solution (100 mM glycine, pH 2.5, and 150 mM NaCl). The internalization of virus and transferrin was visualized by confocal microscopy.
To study the simultaneous internalization of CAV9 and dextran, A549 cells were washed with ice-cold serum-free medium, and CAV9 was allowed to attach to cell surfaces on ice for 1 h. Cells were then washed with ice-cold serum-free medium before the addition of AF 546-conjugated dextran (250 μg/ml) diluted in prewarmed serum-free medium. Cells were transferred to 37°C, where they were incubated for the desired times (5 min, 15 min, or 30 min), followed by cell fixation, permeabilization, staining, and visualization by confocal microscopy.
Plasmid constructs, transfection, and transduction with adenoviral vectors.
HA-tagged caveolin-3 and caveolin-3DGV (an amino-terminal truncation mutant) expression plasmids for wild-type (wt) and dominant negative (DN) caveolin-3, respectively (54), were obtained from Robert Parton (The University of Queensland, Brisbane, Australia). Green fluorescent protein (GFP)-dynamin 2aa and GFP-dynamin 2aaK44A (where 2aaK44A is the dynamin 2aa isoform with a K44A mutation) expression plasmids (10, 47) for wt and DN dynamin 2, respectively, were from Mark McNiven (Mayo Clinic, Rochester, MN); Eps15-GFP and Eps15EΔ95/295-GFP (where Eps15EΔ95/295 is Eps15E with a deletion of residues 95 to 295) constructs (2) for wt and DN Eps15, respectively, were from Alice Dautry-Varsat (Pasteur Institute, Paris, France); and the DN AP180C construct (23) was from Dieter Blaas (University of Vienna, Austria). Plasmids pcDNA3 HA Arf6 and pcDNA3 HA Arf6 DN T27N (25) for the expression of wt and DN Arf6, respectively, were from Addgene (10834 and 10831, respectively; www.addgene.org). Recombinant GFP-tagged Eps15-adenoviral vectors rAd:DIII and rAd:DIIIΔ2 (3, 36) for wt and DN Eps15, respectively, were provided by Yves Rouillè (Pasteur Institute, Lille, France).
For transient transfection, A549 cells were cultured up to 50% confluence and transfected with expression plasmids using FuGENE 6 or FuGENE HD (Roche), according to the protocol recommended by the manufacturer. Transfection efficiencies were approximately 5 to 10%. The adenovirus vectors were produced in HEK293T cells and used to transduce semiconfluent A549 cells. The CAV9 infection assay was done at 48 h after plasmid transfection or adenovirus transduction.
siRNA transfections.
Two individual small interfering RNA (siRNA) molecules for each gene (Qiagen) (see Table S1 in the supplemental material) were used. To transfect A549 cells in 96-well plates, 0.5 pmol of siRNA in 25 μl of H2O was mixed with 0.2 μl of siLentFect (Bio-Rad) diluted in 25 μl of serum-free medium and incubated for 30 min at room temperature. A total of 13,000 cells were then added in 150 μl of serum-supplemented medium and cultured at 37°C in 5% CO2 for 48 h. The transfection conditions were optimized by transfecting the cells with siRNA targeting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and measuring the GAPDH enzyme activity with a KDalert GAPDH assay kit (Applied Biosystems). The optimal conditions that produced an approximately 70% reduction in the GAPDH activity without being toxic to the cells were then used throughout the work. To transfect cells on 24-well plates, the method was scaled up accordingly.
Determination of silencing efficiencies.
The silencing efficiencies were measured by quantitative reverse transcription-PCR (qRT-PCR) or Western analysis. For qRT-PCR, the mRNA was isolated at 48 h after siRNA transfection with PolyATract System 1000 (Promega) according to the manufacturer's instructions. The cDNA was synthesized using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega) and oligo(dT)15 primer (Finnzymes) according to the protocol recommended for M-MLV by the manufacturer. The real-time PCR was done with Maxima SYBR green qPCR Master Mix (Fermentas) using a QuantiTect Primer Assay and primer set (Qiagen) for each studied gene. The run was performed with a RotorGene 6000 real-time PCR cycler (Corbett). The expression levels of β2M or GAPDH were used to standardize the qRT-PCR results.
For Western analysis, the cells were suspended in a lysis buffer (1% Triton X-100 in Tris-buffered saline [TBS]), supplemented with protease inhibitor cocktail and benzonase (both from Sigma-Aldrich), incubated at room temperature for 15 min, and centrifuged at 5,000 rpm for 5 min in an Eppendorf 5810 R centrifuge. The soluble fraction was recovered, and the precipitated membrane fraction was dissolved in one volume of denaturing buffer (8 M urea-10 mM β-mercaptoethanol in TBS). The total protein concentration of the soluble fraction was measured with a bicinchoninic acid (BCA) protein assay kit (Pierce). Protein samples corresponding to 30 μg of total protein were separated in a 15% SDS-PAGE gel (Bio-Rad) and transferred onto a Hybond-P membrane (Amersham). The membrane was treated with blocking buffer (5% skim milk in phosphate-buffered saline [PBS]) at room temperature for 3 h and incubated overnight at 4°C with a primary antibody diluted in blocking buffer. The membrane was washed and incubated with HRP-labeled secondary antibody at room temperature for 1.5 h. After the membrane was washed, it was soaked in enhanced chemiluminescence (ECL) reagent (Pierce) and exposed to Biomax XAR films (Kodak). The films were scanned, and band intensities were analyzed with UVP VisionWorksLS Image Acquisition and Analysis Software.
Cell viability assay.
The cell viability at 48 h after siRNA transfection and NH4Cl treatment was tested by double staining the cells with the dead-cell marker Sytox Orange nucleic acid stain (Molecular Probes) and Hoechst. Cells incubated with 10 mM MβC for 40 min at 37°C were used as negative controls, and nontransfected, mock-transfected, and scramble-transfected cells were used as positive controls. Transfected and MβC-treated cells were incubated in a solution containing Sytox Orange (1.7 μM) and Hoechst (1 μg/ml) in TBS for 30 min at room temperature. The stain was removed, and fluorescence intensities were measured with a Victor3 multilabel reader (Perkin Elmer). The proportion of Hoechst signal to Sytox Orange signal was calculated, and the value of each sample was given as a percentage of the mean value of positive controls. The experiment was repeated three times, and the mean was considered the final result. Cutoff values were calculated as the positive-control mean ± 2 standard deviations (SDs).
Generation of caveolin-1-silenced cell line by retrovirus-mediated RNAi.
A retroviral vector, RVH1-cav1-KD-puro, a derivative of pQCXIH (Clontech), bearing a short hairpin RNA (shRNA) target sequence against canine caveolin-1 coding sequence (gi:50979109) (37, 56), and plasmid pMD.G, expressing the vesicular stomatitis virus G protein (VSV-G), were obtained from Aki Manninen (University of Oulu, Finland). Recombinant viruses were generated by transfecting Phoenix cells at 70% confluence with 2 μg of pRVH1-cav1-KD-puro or the control vector pRVH1-puro, 0.2 μg of pMD.G, and 6 μl of FuGENE 6 per well of a 24-well plate according to the manufacturer's instructions. The cells were cultured at 37°C for 24 h, after which the medium was changed, and incubation was continued at 37°C. The medium was collected at 2, 3, 4, and 5 days posttransfection; centrifuged for 2 min at 1,000 × g to remove cell debris; and passed through a 0.2-μm-pore-size syringe filter. The aliquots were combined and frozen in liquid nitrogen.
To generate caveolin-1-silenced A549 cell line, the cells were seeded onto 24-well plates and cultured overnight in DMEM supplemented with 7% FCS and gentamicin. After 24 h, the medium was removed from the subconfluent (50 to 70%) cells, and 300 μl of RVH-cav1-KD-puro or RVH-puro retrovirus-containing supernatant from Phoenix cells was added. Polybrene (hexadimethrine bromide; Sigma-Aldrich) was added to virus preparations prior to use at 4 μg/ml. One hour later, 1 ml of DMEM-7% FCS was added, and incubation was continued overnight. The transduction procedure was repeated, after which the cells were trypsinized and cultured in DMEM containing 2 μg/ml of puromycin (BD Biosciences) to select retrovirus-transduced cells. Single-cell clones were selected on 96-well plates from the cell population that survived puromycin treatment. Silencing efficiencies were determined by Western analysis and confocal microscopy.
Virus infectivity assays.
In experiments where the efficiency of virus infection was analyzed by microscopy, the cells were infected with a virus dilution aiming at 60% efficiency of infection in untreated cells. After 1 h of incubation on ice, unbound virus was removed by washing with cold medium. Warm medium was added, and the cells were transferred to 37°C. In all virus infectivity assays, the addition of warm medium marked 0-min time point. The infection was allowed to proceed for different time periods, after which the cells were fixed with 4% formalin and permeabilized with 0.2% Triton X-100 (the 0-min samples were not permeabilized). The cells were then stained with the desired antibodies and Hoechst. For the inhibitor assays, the efficiency of infection was determined as the ratio of infected cells to the total cell number, and the values were calculated from at least three independent experiments. In the assays concerning transfection of expression plasmids or adenovirus transduction, the infection efficiency was determined by enumerating transfected or transduced cells in 5 to 10 images.
In the siRNA screen, transfected cells were inoculated with a CAV9 dilution that infected approximately 10% of the untreated cells. Noninfected cells were used as negative controls, and nontransfected, mock-transfected, and scramble-transfected cells were used as positive controls. After 6 h of incubation at 37°C, the cells were fixed and permeabilized as above and stained with rabbit polyclonal CAV9 antiserum combined with AF 488-labeled secondary antibody and Hoechst. Fluorescence intensities were measured with a Victor3 apparatus, and the ratio of virus-specific signal to Hoechst signal was determined and used as a measure of the infection efficiency. The assay was repeated for a total of five times, and the results of the individual experiments were standardized according to the mean value of positive controls. For each siRNA, the mean of the standardized values was considered the final result. The cutoff values were specified as the positive-control mean ± 3 SDs.
For the Arf6 silencing assay, the virus infection assay was performed as in the siRNA screen. Altogether, 33 wells for each sample were analyzed in three separate assays. The statistical analysis between treated and control cells was done by a paired-sample t test, and a P value of <0.05 was considered significant. For the NH4Cl test, the infection assay was done as above. The values were calculated from five wells for each NH4Cl concentration. The test was done two times, and the mean indicated the final result.
The antibody blocking assays were performed as described elsewhere (29).
Confocal microscopy and image analysis.
Fixed, permeabilized, and immunostained cells were mounted on microscope slides in Mowiol 4-88 (Calbiochem-Novabiochem), 25% glycerol, and 0.1 M Tris-HCl, pH 8.5, with 25 mg/ml (diazabicyclooctane) ([DABCO] Sigma-Aldrich) and examined with a Zeiss LSM510 META confocal microscope using a Plan-Apochromat objective (63× oil objective; 1.4 numerical aperture). The colocalization analysis was performed from three to four images by quantifying the proportion of colocalizing channel intensities with BioImageXD software (32). The statistical significance of the colocalization was evaluated using the algorithm of Costes et al.(15) embedded in the software. The images were converted to TIFF format for the adjustment of brightness and contrast in the ImageJ (http://rsb.info.nih.gov/ij) or Photoshop CS3 program.
RESULTS
The role of macropinocytosis in CAV9 internalization.
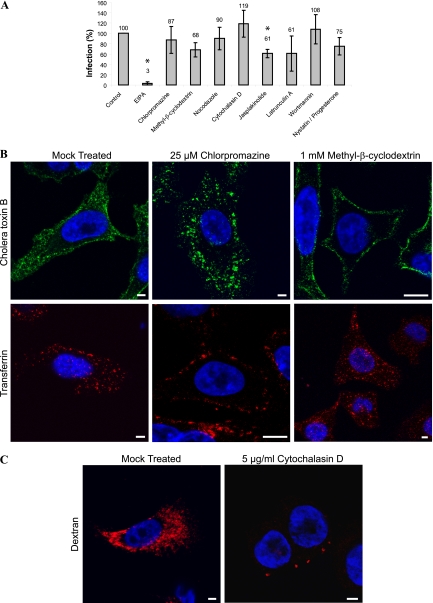
In order to elucidate the endocytic pathway utilized by CAV9 in A549 cells, the effects of chemical inhibitors of endocytosis were studied. The inhibitors used and their mode of action are described in Table 1. As shown in Fig. 1A, EIPA and jasplakinolide caused a statistically significant reduction in CAV9 proliferation (P = 1.6 × 10−5 and 0.013, respectively), whereas chlorpromazine, methyl-β-cyclodextrin (MβC), nocodazole, cytochalasin D, latrunculin A, wortmannin, and a mixture of nystatin-progesterone had no statistically significant effect. The functionality of chlorpromazine and MβC was verified by studying their effects on the internalization of transferrin and cholera toxin B, and dextran was used to monitor the effect of cytochalasin D. As expected, chlorpromazine inhibited the internalization of transferrin but had no effect on cholera toxin B, whereas MβC inhibited the internalization of cholera toxin B but did not affect transferrin (Fig. 1B). Cytochalasin D totally blocked the internalization of dextran (Fig. 1C).
TABLE 1.
Pharmacological inhibitors used in this work
| Inhibitor | Effect | Concn |
|---|---|---|
| EIPA | Inhibits Na+/H+ exchange | 100 μM |
| Chlorpromazine | Inhibits clathrin-dependent endocytosis | 25 μΜ |
| Methyl-β-cyclodextrin | Extracts cholesterol from lipid membranes | 1 mM |
| Nocodazole | Depolymerizes microtubules | 33.2 μΜ |
| Cytochalasin D | Disrupts actin polymerization | 5 μg/ml |
| Latrunculin A | Disrupts actin polymerization | 1 μM |
| Jasplakinolide | Stabilizes actin microfilaments | 2 μM |
| Wortmannin | Inhibits phosphoinositide-3-kinase | 100 nM |
| Nystatin + progesterone | Disrupts caveolae/lipid rafts | 25 μg/ml + 10 μg/ml |
FIG. 1.
Effects of endocytosis inhibitors on CAV9 infection on A549 cells. (A) A549 cells were preincubated at 37°C for 30 min with EIPA (100 μM), chlorpromazine (25 μM), methyl-β-cyclodextrin (MβC; 1 mM), nocodazole (33.2 μM), cytochalasin D (5 μg/ml), jasplakinolide (2 μM), latrunculin A (1 μM), wortmannin (100 nM), or with a combination of nystatin (25 μg/ml) and progesterone (10 μg/ml). The cells were infected with CAV9 at 60% efficiency of infection and incubated on ice for 1 h. The unbound virus was removed, and cells were transferred to 37°C and incubated for 6 h. The inhibitors were present throughout the experiment. Cells were fixed, permeabilized, and stained with Hoechst and virus-specific antibody, and the efficiency of infection was calculated from confocal images as the ratio of infected cells to the total cell number. The experiment was performed three times, and the mean is shown. Statistical significance was calculated with a paired-sample t test, in which a P of <0.05 was considered significant. Inhibitors showing a statistically significant effect are shown with an asterisk. Effects of chlorpromazine (25 μM) and MβC (1 mM) on the internalization of cholera toxin B and transferrin (B) and of cytochalasin D (5 μg/ml) on the internalization of dextran (C) are shown. A549 cells were incubated with the inhibitors for 30 min at 37°C, after which AF 488-conjugated cholera toxin B (0.2 μg/ml), AF 546-conjugated transferrin (10 μg/ml), or AF 546-conjugated dextran (250 μg/ml) was added. The incubation was continued for 15 min, the cells were fixed and stained with Hoechst, and confocal images were taken. Cholera toxin B is shown in green, transferrin and dextran are in red, and nuclei are in blue. Bar, 10 μm.
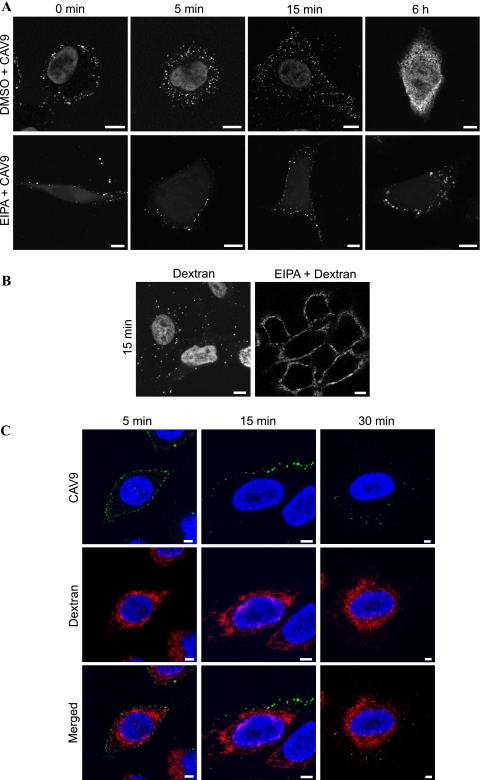
The inhibitory effect of EIPA was studied further by confocal imaging of virus infection at different time points. As shown in Fig. 2A, the virus infection was arrested in EIPA-treated cells, and virus particles were seen in clusters or in vesicle-like structures at or close to the cell periphery. This effect is similar to that found earlier with EV1 (33). To demonstrate that the concentration of EIPA was effective, it was used to inhibit the internalization of a fluid-phase marker, AF 568-conjugated dextran (Fig. 2B).
FIG. 2.
Macropinocytosis is not required for CAV9 internalization. (A) A549 cells were preincubated at 37°C for 30 min with EIPA (100 μM) or DMSO (0.4%). CAV9 was added at 60% efficiency of infection, and cells were incubated on ice for 1 h. Unbound virus was removed by washing, and cells were transferred to 37°C for incubation for different time periods (0, 5, and 15 min and 6 h). Cells were fixed, permeabilized, and stained prior to confocal imaging. In the top row, CAV9 is shown as white spots, and the nucleus is shown as a larger circle in the middle. In the lower row, only the virus is shown in white since the autofluorescence of EIPA stains the cell, making the visualization of the nucleus difficult. (B) A549 cells were preincubated with 100 μM EIPA, and dextran uptake (white) was followed for 15 min at 37°C. Cells incubated with 0.4% DMSO were used as a control. (C) The simultaneous internalization of CAV9 and dextran was studied by infecting the A549 cells with CAV9 in the presence of AF 546-conjugated dextran (250 μg/ml) and following the infection for 5, 15, and 30 min before cell fixation, permeabilization, staining, and confocal imaging. CAV9 is shown in green, dextran is in red, and nuclei are in blue. In panel C, the cells at 15 min postinfection were imaged closer to the nucleus than in panel A; thus, the virus appears near the cell surface. Bar, 10 μm.
EIPA is inhibits macropinocytosis and is sometimes considered the main diagnostic test for this pathway (42). Yet its function has been shown to be cell type dependent and in some cases unspecific (24). We found that even though actin-stabilizing jasplakinolide inhibited CAV9 infection in a statistically significant manner, the actin-disrupting agents cytochalasin D and latrunculin (Fig. 1A) did not cause statistically significant inhibition on CAV9 infection. This contradicts the involvement of macropinocytosis in CAV9 internalization. To further study the role of macropinocytosis in CAV9 entry, a cointernalization assay with CAV9 and dextran was performed. Dextran entered A549 cells clearly faster than CAV9 (Fig. 2C) as it had entered the cytoplasm by 5 min postinfection when CAV9 was still closer to the cell surface. Furthermore, siRNAs against macropinocytosis-regulatory molecules (phosphatidylinositol 3-kinase [PI(3)K], Rac1, PacI, Cdc42, and Rab5) did not reduce CAV9 proliferation (see below). In conclusion, our results suggest that the endocytosis of CAV9 to A549 cells does not depend on macropinocytosis.
siRNA screen.
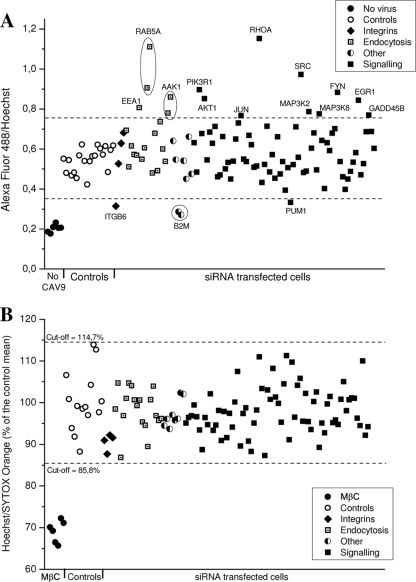
In order to obtain more information about the cell entry mechanism of CAV9 and cellular molecules involved in its infectious cycle, an siRNA panel was designed. The panel included siRNAs targeting central endocytosis effectors and regulators as well as a selected set of signaling molecules. Two individual siRNA molecules for each gene were used (see Table S1 in the supplemental material), and siRNAs against reported CAV9 receptor genes, integrin subunits β3 and β6, were chosen as controls. The effects of the siRNA panel on CAV9 infection of A549 cells are shown in Fig. 3A and Table S1 in supplemental material. The siRNAs increasing CAV9 proliferation are shown as values exceeding the cutoff (positive-control mean ± 3 SDs), and the siRNAs inhibiting the virus are shown as values below the cutoff. The strict cutoff was used to avoid the identification of false siRNAs, even at the expense of missing weak effects. The silencing efficiencies of some of the siRNAs were measured by qRT-PCR (Table S1 in supplemental material), but as exact data about protein knockdown levels are missing, the siRNA results need to be considered preliminary. To evaluate cell viability after siRNA transfections, the cells were stained with the dead-cell indicator Sytox Orange. As shown in Fig. 2B, the transfections did not cause significant cell death, indicating that the observed changes in virus proliferation were due to the silencing of the specific gene and not to the overall effects on cell viability.
FIG. 3.
The effect of siRNA transfections on CAV9 infection on A549 cells. (A) siRNA-transfected A549 cells were cultured for 48 h and infected with CAV9 at 10% efficiency of infection. Unbound virus was removed, and cells were transferred to 37°C. After 6 h, the wells were fixed, permeabilized, and stained with CAV9-specific antibody, AF 488-labeled secondary antibody, and Hoechst. Fluorescence intensities were measured with a Victor3 multilabel counter, and the ratio of AF 488 signal to Hoechst signal was considered the measure of efficiency of infection. The experiment was performed five times, and the mean was calculated. Cutoff values were defined as positive-control mean ± 3 SDs. Noninfected cells served as negative controls; positive controls included nontransfected, mock-transfected, and scramble-transfected cells. Genes whose silencing increase or decrease the virus infection outside the cutoff values are indicated. (B) The cell viability at 48 h posttransfection was monitored by staining the cells with a mixture of Hoechst and the dead-cell stain Sytox Orange. Cells treated with 10 mM MβC were used as negative controls; nontransfected, and mock-transfected, and scramble-transfected cells were used as positive controls. Fluorescence intensities were measured with a Victor3 apparatus; the ratio of Hoechst signal to Sytox Orange signal was calculated, and the value of each sample was given as a percentage of the mean of positive controls. The experiment was done three times, and the mean was calculated. Cutoff values were defined as positive-control mean ± 2 SDs.
One out of two siRNAs against integrin β6 inhibited CAV9 infection. The reason for the different outcome of the β6 siRNAs was most probably their different silencing efficiency (89% versus 57%). Neither of the β3 siRNAs suppressed CAV9 infection even though one of them had very good silencing efficiency (>99%). The other genes whose silencing inhibited virus proliferation were β2M and Pumilio homolog 1 (PUM1). For β2M, both siRNAs silenced their target efficiently (91% and 80%) and showed a similar effect on virus infection. The dependence on β2M will be discussed below. For the siRNAs targeting PUM1, the one showing higher silencing efficiency (85% versus 70%) inhibited CAV9 infection. However, as PUM1 was recently shown to be a central regulator that regulates the expression of over 10% of human genes (43), it is very difficult to deduce the reason for this suppression. To our surprise, there were more siRNAs that enhanced CAV9 infection than inhibited it. The siRNAs that promoted virus proliferation targeted genes involved in endocytosis and cell signaling and will be discussed below.
Role of integrins in CAV9 cell entry.
CAV9 has previously been shown to utilize αVβ6 and αVβ3 as its receptors (53, 61). However, our recent results demonstrate that the virus binds to integrin αVβ6 with higher affinity than to αVβ3 (29). In our siRNA screen, the silencing of integrin subunit β6 but not of β3 inhibited the virus infection (Fig. 3A), which strengthens the hypothesis that αVβ6 is the main receptor for CAV9 in A549 cells. A further support to this result came from an experiment showing that function-blocking antibody against αVβ6 could inhibit CAV9 infection by 60%, whereas αVβ3 antibody inhibited the virus by only 30%. Function-blocking αV antibody inhibited CAV9 by 80% (not shown).
To our surprise, double labeling of CAV9 and integrin αVβ6 in CAV9-infected A549 cells (Fig. 4) revealed that there is no significant colocalization or clustering between the virus and integrin at the 0-min, 5-min, or 20-min time points (2%, 3%, and 6% of the virus colocalized with αVβ6, respectively). The colocalization was calculated as the mean of four images at each time point (data not shown). This finding is in clear contrast to that with echovirus 1, which has been shown to cluster and internalize into the cells in association with its receptor integrin α2β1 (39), and with FMDV, which colocalizes with αVβ6 both on the cell surface and inside the cells at 5 min postinfection (5). These results thus indicate that even though the integrin αVβ6 is crucial for the CAV9 infection, the virus and αVβ6 are not internalized in association with each other. Interestingly, the siRNAs targeting integrin-linked signaling molecules either did not alter CAV9 infection or increased it, with the enhancing effect being seen with one out of two siRNAs against Src, Fyn, RhoA, PI(3)K, and Akt1 (Fig. 3A; see also Table S1 in the supplemental material). The siRNA results are supported by the finding that wortmannin, a PI(3)K inhibitor, did not inhibit CAV9 infection (Fig. 1A). Based on these observations, it seems that integrin-linked signaling is not required for CAV9 entry to A549 cells.
FIG. 4.
CAV9 is not internalized into A549 cells in association with αVβ6 integrin. A549 cells were grown on coverslips for 24 h and infected with CAV9 at 60% efficiency of infection. Unbound virus was removed, warm medium was added (0 min), and cells were transferred to 37°C. The cells were incubated for 5 or 20 min before they were fixed and permeabilized. Cells were stained with virus-specific polyclonal antiserum and AF 488-labeled secondary antibody (green) and with integrin αvβ6-specific monoclonal antibody and AF 568-labeled secondary antibody (red) prior to confocal imaging. Bar, 10 μm.
Collectively, the above results indicate that integrin αVβ6 and, to a lesser extent, αVβ3 mediate CAV9 infection in A549 cells. However, the virus is not internalized to the cell together with the integrin, and integrin-linked signaling does not appear to have a role in CAV9 infection.
Clathrin-mediated endocytosis is not involved in CAV9 cell entry.
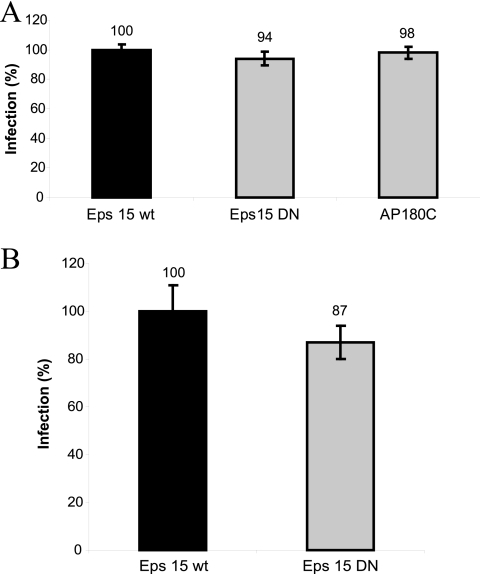
Since the internalization of CAV9 into A549 cells did not seem to follow the macropinocytic pathway, it was essential to examine whether the central endocytic molecules, clathrin, caveolin, and dynamin, participate in the cell entry of the virus. Chlorpromazine did not inhibit viral infection (Fig. 1A), suggesting that clathrin-dependent endocytosis does not play a role in CAV9 internalization. To confirm this observation, the ability of two DN constructs of clathrin route markers, Eps15EΔ95/295 and AP180C, to inhibit virus infection was studied. As shown in Fig. 5A, cells transfected with these constructs were infected with CAV9 as efficiently as control cells transfected with wt Eps15-GFP plasmid. A similar result was obtained with cells transduced with adenovirus vectors carrying wt and DN Eps15 (Fig. 5B).
FIG. 5.
Clathrin route markers Eps15 and AP180C are not involved in CAV9 infection. (A) Plasmids expressing dominant negative constructs Eps15EΔ95/295 and AP180C were transiently transfected into A549 cells. Eps15 wild-type was used as a control. (B) A549 cells were transduced with adenovirus vectors carrying wild-type (DIIIΔ2) and a dominant negative (DIII) Eps15 gene. In both assays, cells were cultured for 48 h and infected with CAV9 at 60% efficiency of infection. After 1 h of incubation on ice, unbound virus was removed, and the cells were transferred to 37°C. The cells were incubated for 6 h before they were fixed, permeabilized, and stained with virus-specific antibody. Infection efficiency was counted from microscopic images of 300 to 500 transfected or transduced cells in three separate experiments, and the infection in control cells was set to 100%. Error bars indicate SD.
Silencing of genes involved in clathrin-mediated endocytosis either increased CAV9 infection or did not have any effect on it (Fig. 3A; see also Table S1 in the supplemental material). siRNAs against EEA1 (1/2), Rab5 (2/2), and AAK1 (2/2) enhanced CAV9 infection, whereas siRNAs against clathrin heavy chain showed no effect. The silencing efficiencies of the siRNAs, however, were variable (Rab5, 98% and 52%; EEA1, 67% and 17%; and AAK1, 35% and 0%); thus, the results need to be interpreted with some caution. The silencing efficiencies of clathrin siRNAs were not measured.
Endosomal acidification is often linked to clathrin-mediated endocytosis (48), and the dependence of an acidic pH may indicate that a virus utilizes clathrin-mediated endocytosis. The incubation of cells with 0.5 mM, 2 mM, 5 mM, and 25 mM NH4Cl did not inhibit CAV9 proliferation (data not shown). This indicates that CAV9 infection is not sensitive to inhibition of endosomal acidification, giving further support to the assumption that CAV9 entry does not rely on clathrin-mediated endocytosis. Altogether, the results obtained with chlorpromazine, dominant negative Eps15 and AP180 constructs, and siRNA transfections indicate that clathrin-mediated endocytosis does not play a role in CAV9 internalization.
Caveolae-mediated endocytosis is not required for CAV9 internalization.
Enteroviruses have been shown to be internalized through caveolin-mediated endocytosis in some cell lines (16-17, 39). Thus, we wanted to find out whether this is also the case for CAV9. As shown in Fig. 1A, a mixture of nystatin-progesterone, an inhibitor of caveolar endocytosis, did not significantly affect CAV9 infection. However, the overexpression of the DN form of caveolin-3 inhibited the virus in a statistically significant way (P = 0.0026), with inhibition of approximately 50% compared to the corresponding wt construct (Fig. 6A). Caveolin-3 is considered a muscle-specific isoform, but the DN caveolin-3 construct used in this study has earlier been shown to efficiently suppress caveolar endocytosis of simian virus 40 (SV40) infection in different cell types (54).
FIG. 6.
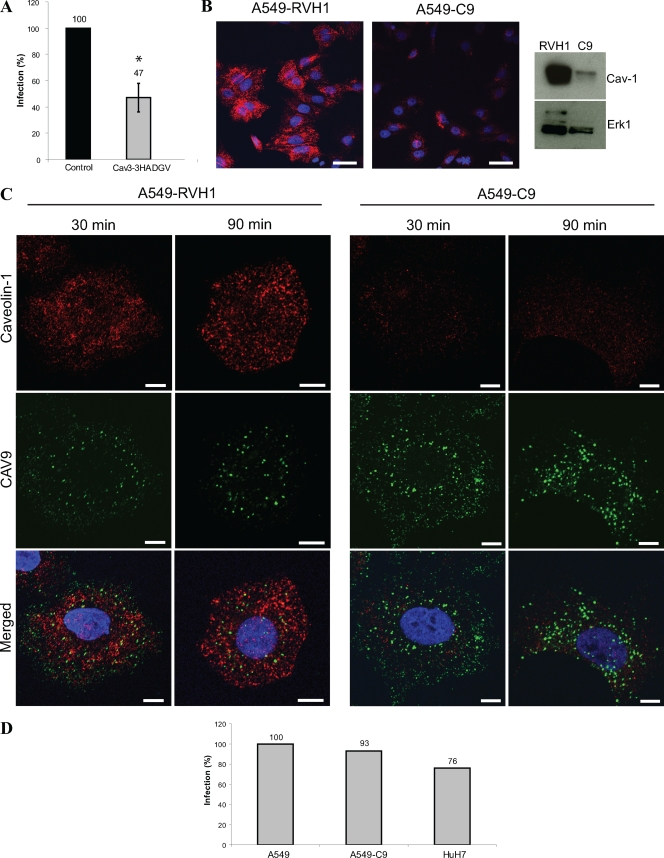
Caveolin-1 is not needed for CAV9 infection. (A) Plasmid constructs expressing wild-type and dominant negative caveolin-3 were transiently transfected to A549 cells. After 48 h, the cells were infected with CAV9 at 60% efficiency of infection, incubated on ice for 1 h, and washed. Warm medium was added, and the cells were transferred to 37°C. The cells were incubated for 6 h prior to fixation and staining. Infection efficiency was counted from microscopic images of 100 to 200 transfected cells in three separate experiments, and the infection in wild-type control cells was set to 100%. Error bar indicates SD. (B) The caveolin-1-silenced cell line A549-C9 was generated by infecting A549 cells with retrovirus vector carrying caveolin-1-silencing shRNA. The vector RVH1 was used as a control. The silencing efficiency was analyzed by confocal imaging and Western analysis. For confocal imaging, the cells were cultured for 24 h on coverslips, fixed, and permeabilized, after which caveolin-1 was stained with rabbit polyclonal antiserum and AF 546-labeled secondary antibody (red). For Western analysis, protein samples (30 μg) were separated in a 15% SDS-PAGE gel and transferred to a Hybond-P membrane. Caveolin-1-specific antibody combined to HRP-labeled secondary antibody was used for detection, and Erk1-specific antibody was used as a loading control. (C) Cell lines A549-RVH1 and A549-C9 were infected with CAV9 as above. The infection was allowed to proceed for 30 or 90 min before fixation, permeabilization, and immunostaining. CAV9 is shown in green, caveolin-1 is in red, and nuclei are in blue. Bar, 10 μm. (D) A549, A549-C9, and HuH-7 cells were infected with CAV9 as above. After 6 h of incubation at 37°C, cells were fixed, permeabilized, and stained with CAV9-specific antibody. Infection efficiency was counted from microscopic images of 100 to 200 cells, and the infection in A549 cells was set to 100%.
In the siRNA screen, caveolin-1 siRNAs did not affect CAV9 proliferation (Fig. 2A and Table S1 in the supplemental material). The silencing efficiencies of the siRNAs were 83% and 75% by Western analysis (data not shown). Since the results with the caveolin-3 DN construct and caveolin-1 siRNAs were rather contradictory, a caveolin-1-silenced cell line, A549-C9, was generated by using a retroviral RNAi vector. A caveolin-1 silencing efficiency of about 69% in A549-C9 was verified by confocal imaging and Western analysis (Fig. 6B). As shown in Fig. 6C, CAV9 was endocytosed into the caveolin-1-silenced cell line as efficiently as into the vector control cells (A549-RVH1). To confirm that this result was not due to the residual caveolin-1 expression, the ability of CAV9 to infect HuH7 cells, shown to be devoid of caveolae (60), was tested. The CAV9 infection in HuH7 cells was very efficient (Fig. 6D), demonstrating that caveolin-1 is not required for CAV9 internalization.
Dynamin 2 is needed for CAV9 endocytosis.
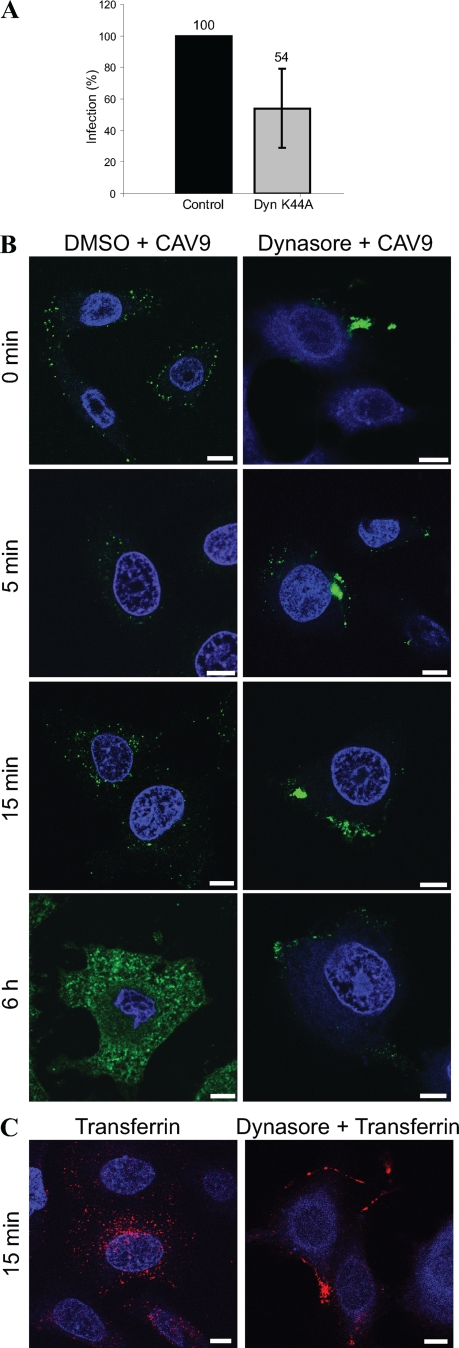
Dynamin is a large GTPase that promotes the fission of endocytic membranes. It was originally considered to participate in clathrin-dependent endocytosis only but has recently been implicated in several other endocytic pathways (20). Since dynamin 2 has been shown to be required for the internalization of numerous viruses, we analyzed its role in CAV9 infection. For this, the effect of the dominant negative form of dynamin 2 was tested. The expression of DN dynamin 2aaK44A resulted in approximately 50% inhibition compared to the corresponding wt construct (Fig. 7A), but the inhibition was not statistically significant (P = 0.082). In the siRNA panel, dynamin 2 siRNAs did not exhibit any effect on virus proliferation (Fig. 3A and Table S1 in the supplemental material). The silencing efficiencies of the siRNAs were relatively good (80% and 82%), but it is still possible that the remaining 20% dynamin 2 expression is sufficient to promote the virus entry. Therefore, the role of dynamin 2 in CAV9 infection was further studied by dynasore, a cell-permeable, noncompetitive dynamin GTPase activity inhibitor. Dynasore effectively blocked the virus infection and caused CAV9 accumulation at the cell periphery (Fig. 7B). The function of dynasore was verified by its ability to inhibit transferrin uptake (Fig. 7C). The result with dynasore thus confirmed that dynamin 2 is needed for CAV9 endocytosis.
FIG. 7.
CAV9 infection is dependent on dynamin 2. (A) A549 cells transiently transfected with plasmids expressing wild-type and dominant negative (K44A) forms of dynamin-2 were infected with CAV9 at 60% efficiency of infection and incubated on ice for 1 h. Unbound virus was removed by washing, and cells were transferred to 37°C, where they were incubated for 6 h. Cells were fixed, permeabilized, and stained with CAV9-specific antibody, and confocal images were taken. Infection efficiency was counted from microscopic images of 100 to 200 transfected cells in three separate experiments, and the infection in wild-type control cells was set to 100%. Error bar indicates SD. (B) A549 cells were preincubated with 80 μM dynasore or 0.4% DMSO for 30 min before CAV9 infection. The infection was performed as above and incubated at 37°C for 0 min, 5 min, 15 min, and 6 h before fixation, permeabilization, staining with CAV9-specific antibody, and confocal imaging. Dynasore was present throughout the assay. The virus is shown in green, and nuclei are in blue. (C) To verify the function of dynasore, A549 cells were preincubated with 80 μM dynasore, and the control cells were preincubated with DMSO. AF 546-conjugated transferrin was added and incubated for 2 min at room temperature. The cells were washed with medium containing dynasore or DMSO, and the cells were incubated for 15 min at 37°C. The cell plate was then transferred onto ice, and transferrin bound to the cell surface was removed by acidic washing. The internalization of transferrin was visualized by confocal microscopy. Bar, 10 μm.
CAV9 internalization is dependent on β2-microglobulin and Arf6.
The dependence of CAV9 on β2-microglobulin has earlier been shown by antibody blocking (59). The role of β2M in virus infection has remained speculative, with indications that it is not required for virus attachment to the cell surface but is involved in some postattachment step(s) (59). In the siRNA screen, both β2M siRNAs efficiently inhibited CAV9 infection in A549 cells (Fig. 3A and Table S1 in the supplemental material). In the antibody blocking assay, β2M antibody inhibited CAV9 proliferation by 80% (data not shown).
Confocal imaging of cells transfected with β2M and negative-control siRNAs showed that CAV9 was attached efficiently to the surface of cells lacking β2-microglobulin, but the endocytosis was arrested, and the virus still remained on the cell periphery at 20 min postinfection. In comparison, in the control siRNA-transfected cells, the virus had already begun to enter the cytoplasm after 5 min (Fig. 8). Interestingly, at 1 h postinfection, the CAV9 endocytosis also continued in the β2M-silenced cells, but this delayed entry never resulted in the production of a new virus progeny (data not shown). This might indicate that CAV9 is directed to an unproductive endocytic pathway in the absence of β2M. The proportion of the virus that was colocalized with β2M at 0 min, 5 min, and 20 min postinfection was quantified from three confocal images at each time point, and the mean colocalization values were 40%, 16%, and 18%, respectively (data not shown).
FIG. 8.
β2M is required for CAV9 infection. A549 cells were transfected with control siRNA or β2M siRNA (Hs_β2M_3) and cultured for 48 h. The cells were infected with CAV9 at 60% efficiency of infection, incubated on ice for 1 h, and washed. Warm medium was added at the 0-min time point; the cells were transferred to 37°C and incubated for 5 min, 20 min, and 6 h before fixing, permeabilization (except for the 0-min sample), staining with CAV9- and β2M-specific antibodies, and confocal imaging. CAV9 is shown in green, β2M is in red, and nuclei are in blue. A silenced cell in the 0-min images is indicated by an arrow. Bar, 10 μm.
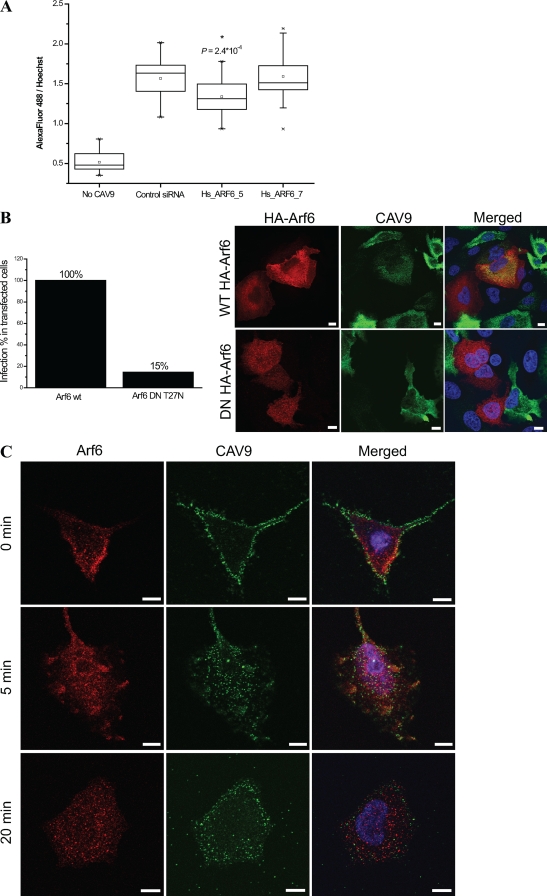
Arf6 (ADP-ribosylation factor 6) is a small GTPase involved in the regulation of endosomal membrane traffic and structural organization of the cell (21). It is known to mediate the internalization of several ligands, including major histocompatibility class I (MHC-I) proteins and β1 integrin (8, 45, 52). Arf6 was recently shown to be needed for baculovirus entry into 293 cells (34) and to direct CBV3 into an unproductive internalization pathway (38). We studied the involvement of Arf6 in CAV9 endocytosis by measuring virus proliferation in cells transfected with two individual Arf6 siRNAs. Figure 9A shows that one of the siRNAs inhibited CAV9 infection in a statistically significant manner (P = 2.4 × 10−4), whereas the other had no effect. The silencing efficiencies of the siRNAs were 89% and 91% by qRT-PCR. In order to verify the siRNA result, A549 cells were transfected with plasmid constructs overexpressing wt and DN forms of Arf6. As shown in Fig. 9B, the expression of DN Arf6 resulted in an 85% decrease in CAV9 infection efficiency compared to cells expressing wt Arf6. Confocal imaging and colocalization analysis of CAV9 and Arf6 at 0 min, 5 min, and 20 min postinfection showed a similar pattern of staining (Fig. 9C). The colocalization (calculated as the mean of four images at each time point [data not shown]) was modest, with 7%, 9%, and 9% of CAV9 colocalizing with Arf6 at the indicated time points, respectively. To conclude, the results obtained with Arf6 siRNA and overexpression of DNA Arf6 indicate that CAV9 internalization in A549 cells is dependent on Arf6.
FIG. 9.
Arf6 mediates CAV9 endocytosis. (A) A549 cells transfected with negative-control siRNA and two individual Arf6 siRNAs were infected with a CAV9 at 10% efficiency of infection, incubated on ice for 1 h, and washed. Warm medium was added, and the cells were transferred to 37°C and incubated for 6 h, after which they were fixed and permeabilized. The cells were stained with CAV9-specific antibody, AF 488-labeled secondary antibody, and Hoechst. Fluorescence intensities were measured with a Victor3 multilabel counter, and the ratio of AF 488 signal to Hoechst signal was considered the measure of efficiency of infection. For each sample, 33 wells in three separate assays were analyzed. Statistical significance between control siRNA- and Hs_ARF6_5 and Hs_ARF6_7 siRNA-silenced cells was calculated with a paired-sample t test, in which a P value of <0.05 was considered significant. In the box plot, mean, median, upper and lower quartiles, upper and lower 95% values, and maximal and minimal values are indicated by square, horizontal line, box boundaries, vertical lines, and a cross, respectively. Statistically significant difference is shown with an asterisk. (B) Plasmids expressing HA-conjugated wild-type and dominant negative Arf6 were transfected into A549 cells and cultured for 48 h. The cells were infected with CAV9 at 60% efficiency of infection by a procedure described above. Cells were stained with CAV9-specific antibody combined with AF 488-labeled secondary antibody and with HA-specific antibody combined with AF 568-labeled secondary antibody and Hoechst. The efficiency of CAV9 infection was calculated by counting the proportion of infected cells in cells transfected with wt and DN Arf6 (total values of six and seven images, respectively). HA tag is shown in red, CAV9 is in green, and nuclei are in blue. (C) A549 cells were infected with CAV9 as above and incubated at 37°C for 0 min, 5 min, and 20 min prior to fixation, permeabilization (not in the 0-min sample), and staining with antibodies specific to CAV9 and Arf6 and with Hoechst. Arf6 is shown in red, CAV9 is in green, and nuclei are in blue. Bar, 10 μm.
DISCUSSION
Enteroviruses utilize several mechanisms to enter their host cells. The early steps of CAV9 infection have earlier been reported to be dependent on αV integrins, β2-microglobulin, and lipid microdomains (29, 58-59, 61). In this work, we conducted a systematic study in order to identify the internalization mechanism of CAV9 in A549 lung carcinoma cells.
Integrins αVβ3 and αVβ6 have previously been shown to function as receptors for CAV9 (53, 61). A recent work from our group illustrated that soluble αVβ6 but not αVβ3 bound to immobilized CAV9 and that soluble αVβ6 but not αVβ3 blocked virus infectivity (29). The results presented in this study show that siRNA silencing of integrin subunit β6 but not of β3 inhibited CAV9 infection, and a similar effect was seen with antibody blocking. It thus seems that αVβ6 and, to a lesser extent, αVβ3 mediate CAV9 infection in this cell model. The role of αVβ6 as an internalizing agent was, however, complicated by the observation that no clustering or colocalization between integrin αVβ6 and the virus was seen at the early time points of infection. The situation seems thus substantially different from FMDV, where αVβ6 serves to deliver the virus to acidic endosomes via clathrin-dependent endocytosis (5). The siRNA silencing of a number of integrin-linked signaling molecules did not inhibit CAV9 infection; on the contrary, several of these siRNA molecules enhanced virus proliferation. This implies that integrin-linked signaling is not required for CAV9 infection in A549 cells. It has earlier been shown by Triantafilou and Triantafilou that CAV9 infection activates the Raf/mitogen-activated protein kinase (MAPK) pathway in GMK cells (58). However, these investigators did not examine whether this activation was essential for the infection. It is thus possible that signaling cascades become activated upon the contact of CAV9 and integrin, but this activation does not promote virus entry and proliferation. In conclusion, integrin αVβ6 is essential for CAV9 infection in A549 cells, but the virus is not internalized in association with it. In addition, integrin-linked signaling does not seem to be involved in the process. The exact role of integrin αVβ6 in CAV9 infection is not yet understood and still needs further study. One explanation might be that it mediates the primary binding and concentration of CAV9 on the cell surface. Since our protocol included an incubation of the infected cells for 1 h on ice before cells were fixed and stained, it is possible that a weak and transient association was missed.
The finding that EIPA inhibited CAV9 endocytosis suggests that CAV9 might enter A549 cells through the macropinocytic pathway. However, several lines of evidence illustrate that this is not the case: siRNAs targeting regulators of macropinocytosis [Cdc42, Rac1, Pak1, and PI(3)K] did not reduce CAV9 infection, and a similar result was obtained with wortmannin, a PI(3)K inhibitor. Chemicals inhibiting actin dynamics either had no effect on virus proliferation or caused only partial inhibition. Furthermore, the kinetics of virus internalization was clearly different from that of dextran, a generally used fluid-phase marker. The criteria for macropinocytic entry of viruses, proposed by Mercer and Helenius, include the sensitivity to actin inhibitors and to inhibition of the above-mentioned regulators (42). Thus, we conclude that CAV9 is not internalized in A549 cells through macropinocytosis. Even though EIPA is generally considered an inhibitor of macropinocytosis, it may have a number of other effects, such as the alteration of the morphology and subcellular localization of early and late endosomes and lysosomes (24). In a recent work, EIPA was shown to inhibit CBV3 replication, most probably by inhibiting the virus RNA polymerase (27). Since CAV9 and CBV3 RNA polymerases are highly similar, having 95% amino acid identity (11), it is possible that the inhibitory effect of EIPA on CAV9 proliferation is due to its effect on CAV9 replication.
In order to identify the internalization route of CAV9, we analyzed whether clathrin- and caveolin-mediated pathways are involved. We found that chlorpromazine, endosomal acidification with NH4Cl, the expression of Eps15 and AP180 dominant negative constructs, and siRNA silencing of genes participating in the clathrin route all allowed efficient CAV9 infection. This implies that clathrin-mediated endocytosis is not involved in CAV9 cell entry.
Methyl-β-cyclodextrin has earlier been shown to inhibit CAV9 infection, which has been considered to indicate the dependence of the virus on lipid microdomains (58). In addition, it has been suggested that caveolin-mediated entry is involved in the endocytosis of other enteroviruses (16-17, 39). In our work, silencing of caveolin-1 either by siRNA or retroviral RNAi vector did not prevent virus proliferation. Furthermore, CAV9 was able to efficiently infect the caveolin-1-negative cell line HuH7. On the other hand, the overexpression of DN caveolin-3 caused approximately 50% inhibition in CAV9 infection. The reason for this inhibition is not totally clear, but it might resemble the situation with EV1, where DN caveolin-3 does not prevent endocytosis but inhibits infection at later stages (33). In conclusion, our results indicate that caveolin-1 is not essential for CAV9 entry.
Dynamin is a high-molecular-weight GTPase involved in membrane fission during endocytic events. Its function is necessary for clathrin-mediated endocytosis, but it has also been implicated in other internalization pathways, such as phagocytosis and caveolin-1-, ILRβ2-, and flotillin-dependent pathways (20). Since dynamin is a central regulator of endocytosis and is needed for the internalization of numerous viruses, we wanted to analyze whether it is involved in the entry process of CAV9. The DN form of dynamin-2 inhibited virus infection by approximately 50%. Even though this inhibition was not statistically significant, it might suggest that dynamin 2 has a role in CAV9 endocytosis. The dynamin 2 siRNAs had no effect on virus proliferation, but this may be explained by the functionality of residual dynamin. To further elucidate the role of dynamin, we used dynasore, a recently introduced chemical inhibitor. Dynasore completely blocked CAV9 infection and resulted in the trapping of the virus close to the cell surface. This finding thus confirmed that dynamin-2 is required for the internalization of CAV9.
In our siRNA screen, the siRNAs targeting β2-microglobulin effectively inhibited CAV9 infection. The dependence of CAV9 infection on β2M has earlier been shown by antibody blocking (59, 61), but the role of β2M in virus infection has not been identified. Although CAV9 is not directly bound to β2M on the cell surface (59), our confocal imaging revealed that β2M and the virus are in close association at early time points of infection. In cells depleted of β2M, endocytosis was arrested, and the virus remained close to the cell surface even at 20 min postinfection. After 1 h, the virus also began to enter the β2M-silenced cells, but this delayed entry did not result in the production of new virus progeny. Thus, without β2M, CAV9 seems to be directed to an unproductive endocytic pathway The existence of such an unproductive pathway is supported by the early finding of Hecker et al., who showed that most endocytosed CAV9 particles became accumulated in lysosomes without causing an infection (28).
Arf6 is a small GTPase that has multiple roles in the regulation of endosomal membrane traffic and other cellular functions (19). The dependence of CAV9 infection on β2M and the finding that Arf6 regulates the endocytosis of MHC-I proteins (44) directed us to study whether Arf6 is involved in the entry process of the virus. One of two Arf6 siRNAs inhibited CAV9 infection in a statistically significant manner, and overexpression of DN Arf6 resulted in a clear decrease in the infection efficiency compared to the corresponding wt construct. The results thus indicate that Arf6 controls the endocytosis of CAV9 to A549 cells.
In conclusion, we propose a model for CAV9 entry into A549 cells in which the virus is first attached to the cell surface by a process requiring integrin αVβ6 and then rapidly transferred forward to form an association with β2M It is possible that other molecules are also involved. The virus is then internalized by an Arf6-controlled route, perhaps still in association with β2M. The function of dynamin 2 is needed for the pathway, but its exact role and relation to Arf6 are still unclear. Although dynamin is not generally thought to be involved in Arf6-dependent endocytosis, there is a report by Nishi and Saigo showing that herpes simplex virus structural protein VP22 is internalized in HeLa cells in an Arf6- and dynamin-dependent manner (46). Interestingly, the endocytoses of CAV9 and VP22 appear very similar in being independent of clathrin, caveolin, and the Rho family GTPases RhoA, Rac1, and Cdc42 while requiring dynamin and Arf6. We thus present the first demonstration of an infectious virus entering the cell via a dynamin/Arf6-dependent endocytosis pathway.
Supplementary Material
Acknowledgments
Elina Ikonen (University of Helsinki, Finland), Aki Manninen (University of Oulu, Finland), Lucia Fiore (Instituto Superiore di Sanita, Rome, Italy), Robert Parton (University of Queensland, Brisbane, Australia), Mark McNiven (Mayo Clinic, Rochester, MN), Alice Dautry-Varsat (Institute Pasteur, Paris, France), Dieter Blaas (University of Vienna, Austria), and Yves Rouillè (Institute Pasteur de Lille, France) are warmly thanked for providing cell lines, antibodies, and expression constructs. We acknowledge Harri Savilahti and Ari Helenius for their comments concerning the manuscript and Ritva Kajander for her assistance in the laboratory work.
This work was supported by research grants from the Academy of Finland (number 122215 to T.H., 114721 to V.M., and 128539 to P.S.), the Sigrid Juselius Foundation, and Turku University Graduate School.
Footnotes
Published ahead of print on 20 January 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abraham, G., and R. J. Colonno. 1984. Many rhinovirus serotypes share the same cellular receptor. J. Virol. 51:340-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benmerah, A., M. Bayrou, N. Cerf-Bensussan, and A. Dautry-Varsat. 1999. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J. Cell Sci. 112:1303-1311. [DOI] [PubMed] [Google Scholar]
- 3.Benmerah, A., C. Lamaze, B. Begue, S. L. Schmid, A. Dautry-Varsat, and N. Cerf-Bensussan. 1998. AP-2/Eps15 interaction is required for receptor-mediated endocytosis. J. Cell Biol. 140:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson, J. M. 2008. New (fluorescent) light on poliovirus entry. Trends Microbiol. 16:44-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berryman, S., S. Clark, P. Monaghan, and T. Jackson. 2005. Early events in integrin αvβ6-mediated cell entry of foot-and-mouth disease virus. J. Virol. 79:8519-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomqvist, S., A. Paananen, C. Savolainen-Kopra, T. Hovi, and M. Roivainen. 2008. Eight years of experience with molecular identification of human enteroviruses. J. Clin. Microbiol. 46:2410-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandenburg, B., L. Y. Lee, M. Lakadamyali, M. J. Rust, X. Zhuang, and J. M. Hogle. 2007. Imaging poliovirus entry in live cells. PLoS Biol. 5:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, F. D., A. L. Rozelle, H. L. Yin, T. Balla, and J. G. Donaldson. 2001. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154:1007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttinelli, G., V. Donati, F. M. Ruggeri, P. Joki-Korpela, T. Hyypiä, and L. Fiore. 2003. Antigenic sites of coxsackie A9 virus inducing neutralizing monoclonal antibodies protective in mice. Virology 312:74-83. [DOI] [PubMed] [Google Scholar]
- 10.Cao, H., F. Garcia, and M. A. McNiven. 1998. Differential distribution of dynamin isoforms in mammalian cells. Mol. Biol. Cell 9:2595-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, K. H., P. Auvinen, T. Hyypiä, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9; implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 12.Chevaliez, S., J. Balanant, P. Maillard, Y. C. Lone, F. A. Lemonnier, and F. Delpeyroux. 2008. Role of class I human leukocyte antigen molecules in early steps of echovirus infection of rhabdomyosarcoma cells. Virology 381:203-214. [DOI] [PubMed] [Google Scholar]
- 13.Chung, S. K., J. Y. Kim, I. B. Kim, S. I. Park, K. H. Paek, and J. H. Nam. 2005. Internalization and trafficking mechanisms of coxsackievirus B3 in HeLa cells. Virology 333:31-40. [DOI] [PubMed] [Google Scholar]
- 14.Conner, S. D., and S. L. Schmid. 2003. Regulated portals of entry into the cell. Nature 422:37-44. [DOI] [PubMed] [Google Scholar]
- 15.Costes, S. V., D. Daelemans, E. H. Cho, Z. Dobbin, G. Pavlakis, and S. Lockett. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86:3993-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyne, C. B., and J. M. Bergelson. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119-131. [DOI] [PubMed] [Google Scholar]
- 17.Coyne, C. B., K. S. Kim, and J. M. Bergelson. 2007. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 26:4016-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyne, C. B., L. Shen, J. R. Turner, and J. M. Bergelson. 2007. Coxsackievirus entry across epithelial tight junctions requires occludin and the small GTPases Rab34 and Rab5. Cell Host Microbe 2:181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dicara, D., A. Burman, S. Clark, S. Berryman, M. J. Howard, I. R. Hart, J. F. Marshall, and T. Jackson. 2008. Foot-and-mouth disease virus forms a highly stable, EDTA-resistant complex with its principal receptor, integrin αvβ6: implications for infectiousness. J. Virol. 82:1537-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doherty, G. J., and H. T. McMahon. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78:857-902. [DOI] [PubMed] [Google Scholar]
- 21.D'Souza-Schorey, C., and P. Chavrier. 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7:347-358. [DOI] [PubMed] [Google Scholar]
- 22.Eisenhut, M., B. Algawi, T. Wreghitt, J. Foweraker, T. McKee, R. Miles, and J. Challener. 2000. Fatal coxsackie A9 virus infection during an outbreak in a neonatal unit. J. Infect. 40:297-298. [DOI] [PubMed] [Google Scholar]
- 23.Ford, M. G., B. M. Pearse, M. K. Higgins, Y. Vallis, D. J. Owen, A. Gibson, C. R. Hopkins, P. R. Evans, and H. T. McMahon. 2001. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291:1051-1055. [DOI] [PubMed] [Google Scholar]
- 24.Fretz, M., J. Jin, R. Conibere, N. A. Penning, S. Al-Taei, G. Storm, S. Futaki, T. Takeuchi, I. Nakase, and A. T. Jones. 2006. Effects of Na+/H+ exchanger inhibitors on subcellular localisation of endocytic organelles and intracellular dynamics of protein transduction domains HIV-TAT peptide and octaarginine. J. Control. Release 116:247-254. [DOI] [PubMed] [Google Scholar]
- 25.Furman, C., S. M. Short, R. R. Subramanian, B. R. Zetter, and T. M. Roberts. 2002. DEF-1/ASAP1 is a GTPase-activating protein (GAP) for ARF1 that enhances cell motility through a GAP-dependent mechanism. J. Biol. Chem. 277:7962-7969. [DOI] [PubMed] [Google Scholar]
- 26.Grist, N. R., E. J. Bell, and F. Assaad. 1978. Enteroviruses in human disease. Prog. Med. Virol. 24:114-157. [PubMed] [Google Scholar]
- 27.Harrison, D. N., E. V. Gazina, D. F. Purcell, D. A. Anderson, and S. Petrou. 2008. Amiloride derivatives inhibit coxsackievirus B3 RNA replication. J. Virol. 82:1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecker, W., J. Meyer, R. Boeni, and K. Bienz. 1974. Pinocytotic uptake and intralysosomal crystal formation of coxsackievirus A9 in monkey kidney cells. An electron microscopic autoradiographic study. Arch. Gesamte Virusforsch. 46:167-174. [DOI] [PubMed] [Google Scholar]
- 29.Heikkilä, O., P. Susi, G. Stanway, and T. Hyypiä. 2009. Integrin αVβ6 is a high-affinity receptor for coxsackievirus A9. J. Gen. Virol. 90:197-204. [DOI] [PubMed] [Google Scholar]
- 30.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypia, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527-1538. [DOI] [PubMed] [Google Scholar]
- 31.Johns, H. L., S. Berryman, P. Monaghan, G. J. Belsham, and T. Jackson. 2009. A dominant-negative mutant of rab5 inhibits infection of cells by foot-and-mouth disease virus: implications for virus entry. J. Virol. 83:6247-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kankaanpää, P., K. Pahajoki, V. Marjomäki, J. Heino, D. White, and X. D. BioImage. 2006. BioImageXD. University of Jyväskylä, Jyväskylä, Finland, and University of Turku, Turku, Finland. http://www.bioimagexd.net.
- 33.Karjalainen, M., E. Kakkonen, P. Upla, H. Paloranta, P. Kankaanpää, P. Liberali, G. H. Renkema, T. Hyypiä, J. Heino, and V. Marjomäki. 2008. A Raft-derived, Pak1-regulated entry participates in α2β1 integrin-dependent sorting to caveosomes. Mol. Biol. Cell 19:2857-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laakkonen, J. P., A. R. Makela, E. Kakkonen, P. Turkki, S. Kukkonen, J. Peranen, S. Yla-Herttuala, K. J. Airenne, C. Oker-Blom, M. Vihinen-Ranta, and V. Marjomäki. 2009. Clathrin-independent entry of baculovirus triggers uptake of E. coli in non-phagocytic human cells. PLoS One 4:e5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau, A. W., and M. M. Chou. 2008. The adaptor complex AP-2 regulates post-endocytic trafficking through the non-clathrin Arf6-dependent endocytic pathway. J. Cell Sci. 121:4008-4017. [DOI] [PubMed] [Google Scholar]
- 36.Lecot, S., S. Belouzard, J. Dubuisson, and Y. Rouille. 2005. Bovine viral diarrhea virus entry is dependent on clathrin-mediated endocytosis. J. Virol. 79:10826-10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manninen, A., P. Verkade, S. Le Lay, J. Torkko, M. Kasper, J. Fullekrug, and K. Simons. 2005. Caveolin-1 is not essential for biosynthetic apical membrane transport. Mol. Cell. Biol. 25:10087-10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchant, D., A. Sall, X. Si, T. Abraham, W. Wu, Z. Luo, T. Petersen, R. G. Hegele, and B. M. McManus. 2009. ERK MAP kinase-activated Arf6 trafficking directs coxsackievirus type B3 into an unproductive compartment during virus host-cell entry. J. Gen. Virol. 90:854-862. [DOI] [PubMed] [Google Scholar]
- 39.Marjomäki, V., V. Pietiäinen, H. Matilainen, P. Upla, J. Ivaska, L. Nissinen, H. Reunanen, P. Huttunen, T. Hyypiä, and J. Heino. 2002. Internalization of echovirus 1 in caveolae. J. Virol. 76:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsh, M., and A. Helenius. 2006. Virus entry: open sesame. Cell 124:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayor, S., and R. E. Pagano. 2007. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8:603-612. [DOI] [PubMed] [Google Scholar]
- 42.Mercer, J., and A. Helenius. 2009. Virus entry by macropinocytosis. Nat. Cell Biol. 11:510-520. [DOI] [PubMed] [Google Scholar]
- 43.Morris, A. R., N. Mukherjee, and J. D. Keene. 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol. Cell. Biol. 28:4093-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naslavsky, N., R. Weigert, and J. G. Donaldson. 2004. Characterization of a nonclathrin endocytic pathway: membrane cargo and lipid requirements. Mol. Biol. Cell 15:3542-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naslavsky, N., R. Weigert, and J. G. Donaldson. 2003. Convergence of non-clathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol. Biol. Cell 14:417-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishi, K., and K. Saigo. 2007. Cellular internalization of green fluorescent protein fused with herpes simplex virus protein VP22 via a lipid raft-mediated endocytic pathway independent of caveolae and Rho family GTPases but dependent on dynamin and Arf6. J. Cell Biol. 282:27503-27517. [DOI] [PubMed] [Google Scholar]
- 47.Ochoa, G. C., V. I. Slepnev, L. Neff, N. Ringstad, K. Takei, L. Daniell, W. Kim, H. Cao, M. McNiven, R. Baron, and P. De Camilli. 2000. A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 150:377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelkmans, L., and A. Helenius. 2003. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 15:414-422. [DOI] [PubMed] [Google Scholar]
- 49.Pietiäinen, V., V. Marjomäki, P. Upla, L. Pelkmans, A. Helenius, and T. Hyypiä. 2004. Echovirus 1 endocytosis into caveosomes requires lipid rafts, dynamin II, and signaling events. Mol. Biol. Cell 15:4911-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietiäinen, V. M., V. Marjomäki, J. Heino, and T. Hyypiä. 2005. Viral entry, lipid rafts and caveosomes. Ann. Med. 37:394-403. [DOI] [PubMed] [Google Scholar]
- 51.Pulli, T., H. Lankinen, M. Roivainen, and T. Hyypiä. 1998. Antigenic sites of coxsackievirus A9. Virology 240:202-212. [DOI] [PubMed] [Google Scholar]
- 52.Radhakrishna, H., and J. G. Donaldson. 1997. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139:49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roivainen, M., L. Piirainen, T. Hovi, I. Virtanen, T. Riikonen, J. Heino, and T. Hyypiä. 1994. Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology 203:357-365. [DOI] [PubMed] [Google Scholar]
- 54.Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J. F. Hancock, and R. G. Parton. 1999. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1:98-105. [DOI] [PubMed] [Google Scholar]
- 55.Sandvig, K., M. L. Torgersen, H. A. Raa, and B. van Deurs. 2008. Clathrin-independent endocytosis: from nonexisting to an extreme degree of complexity. Histochem. Cell Biol. 129:267-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuck, S., A. Manninen, M. Honsho, J. Fullekrug, and K. Simons. 2004. Generation of single and double knockdowns in polarized epithelial cells by retrovirus-mediated RNA interference. Proc. Natl. Acad. Sci. U. S. A. 101:4912-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Triantafilou, K., D. Fradelizi, K. Wilson, and M. Triantafilou. 2002. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J. Virol. 76:633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Triantafilou, K., and M. Triantafilou. 2003. Lipid raft microdomains: key sites for coxsackievirus A9 infectious cycle. Virology 317:128-135. [DOI] [PubMed] [Google Scholar]
- 59.Triantafilou, M., K. Triantafilou, K. M. Wilson, Y. Takada, N. Fernandez, and G. Stanway. 1999. Involvement of β2-microglobulin and integrin αvβ3 molecules in the coxsackievirus A9 infectious cycle. J. Gen. Virol. 80:2591-2600. [DOI] [PubMed] [Google Scholar]
- 60.Vainio, S., S. Heino, J. E. Mansson, P. Fredman, E. Kuismanen, O. Vaarala, and E. Ikonen. 2002. Dynamic association of human insulin receptor with lipid rafts in cells lacking caveolae. EMBO Rep. 3:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, C. H., T. Kajander, T. Hyypiä, T. Jackson, D. Sheppard, and G. Stanway. 2004. Integrin αvβ6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 78:6967-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.