Abstract
Flaviviruses transmitted by arthropods represent a tremendous disease burden for humans, causing millions of infections annually. All vector-borne flaviviruses studied to date suppress host innate responses to infection by inhibiting alpha/beta interferon (IFN-α/β)-mediated JAK-STAT signal transduction. The viral nonstructural protein NS5 of some flaviviruses functions as the major IFN antagonist, associated with inhibition of IFN-dependent STAT1 phosphorylation (pY-STAT1) or with STAT2 degradation. West Nile virus (WNV) infection prevents pY-STAT1 although a role for WNV NS5 in IFN antagonism has not been fully explored. Here, we report that NS5 from the virulent NY99 strain of WNV prevented pY-STAT1 accumulation, suppressed IFN-dependent gene expression, and rescued the growth of a highly IFN-sensitive virus (Newcastle disease virus) in the presence of IFN, suggesting that this protein can function as an efficient IFN antagonist. In contrast, NS5 from Kunjin virus (KUN), a naturally attenuated subtype of WNV, was a poor suppressor of pY-STAT1. Mutation of a single residue in KUN NS5 to the analogous residue in WNV-NY99 NS5 (S653F) rendered KUN NS5 an efficient inhibitor of pY-STAT1. Incorporation of this mutation into recombinant KUN resulted in 30-fold greater inhibition of JAK-STAT signaling than with the wild-type virus and enhanced KUN replication in the presence of IFN. Thus, a naturally occurring mutation is associated with the function of NS5 in IFN antagonism and may influence virulence of WNV field isolates.
The continued emergence and reemergence of flaviviruses transmitted by mosquitoes and ticks is associated with significant human morbidity and mortality worldwide. These viruses include West Nile virus (WNV), Japanese encephalitis virus (JEV), dengue virus (DENV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV). Despite their importance as human pathogens, no specific therapies exist for treatment of infection with any of the flaviviruses. Host type I interferon (IFN-α/β) responses are critical to recovery from infection (27, 51, 54), and IFN-α2a has been tested in human clinical trials as a potential therapeutic for flavivirus infection. However, such treatment has had limited success (48, 55). One reason for ineffectiveness of IFN may be that flaviviruses can suppress IFN-mediated signal transduction and thus dampen the antiviral effects of IFN on infected cells (5). Indeed, in the case of WNV and JEV, virus virulence correlates positively with the ability to inhibit IFN-mediated signal transduction (19, 22). Therefore, determining how flaviviruses suppress this vital host response will facilitate the understanding of virus virulence. Furthermore, this work will identify targets for the development of therapeutics that, when administered with IFN, potentiate its actions as an antiviral treatment.
Following cellular recognition of virus infection, IFN-α/β is secreted and binds in an autocrine and paracrine manner to cell surface receptors, IFN-α receptor subunits 1 and 2 (IFNAR1 and IFNAR2), to activate Janus kinase-signal transducer and activator of transcription (JAK-STAT) signal transduction (47). Briefly, IFN binding ligates the receptors, which promotes trans- and auto-phosphorylation of JAKs (Jak1 and Tyk2) associated with the receptor subunits (16). The JAKs then phosphorylate the intracellular domains of the receptors, creating a docking site for STAT1 and STAT2. The STATs, in turn, are phosphorylated by the JAKs, inducing heterodimerization of STAT1 and STAT2 and binding of a third component, IFN-regulatory factor-9 (IRF-9), to form the transcription factor IFN-stimulated gene factor 3 (ISGF3) (15, 47). ISGF3 then translocates to the nucleus, where it binds to the IFN-stimulated response element (ISRE) in the promoter region of IFN-stimulated genes (ISGs) (9), such as protein kinase R, the Mx proteins, 2′,5′-oligoadenylate synthetase, and ISG15. ISG expression contributes to the cellular antiviral state and modulates cell proliferation, cell death, and, depending on the cell type, immune responses to infection (18).
All flaviviruses examined thus far, including WNV (11), JEV (24), Langat virus (LGTV; a member of the TBEV antigenic complex) (6), and DENV (14), can suppress IFN-mediated JAK-STAT signaling by inhibiting JAK phosphorylation. This block prevents downstream signaling including tyrosine phosphorylation and nuclear localization of STAT1 and STAT2 as well as ISG expression. DENV imposes an additional block to signaling by reducing the cellular levels of STAT2 expression (17). We previously identified the nonstructural protein NS5 of LGTV as a potent antagonist of STAT1 phosphorylation and downstream signaling (6). NS5 is approximately 900 amino acids in length and is highly conserved between flaviviruses owing to the fact that it encodes the viral methyltransferase (MTase) and RNA-dependent RNA polymerase (RdRp). The IFN antagonist domain of LGTV NS5 maps between amino acids 355 and 735 and thus is contained within the RdRp domain (44). Similarly, NS5 proteins from TBEV (59) and JEV (23) antagonize STAT1 phosphorylation, most likely as a result of suppression of JAK activation. Finally, NS5 from DENV has recently been shown to contribute to IFN antagonism by binding and degrading STAT2 (2, 33). Hence, the flavivirus NS5 protein appears critical to flavivirus resistance to IFN.
Other flavivirus nonstructural proteins besides NS5 can contribute to flavivirus IFN resistance. The flavivirus genome encodes one large polyprotein that is cleaved into three structural proteins (core, membrane [derived from a precursor, designated prM], and the envelope protein [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (25). Expression of the NS4B protein from DENV (serotype 2) suppresses STAT1 phosphorylation in IFN-treated cells (38). The ability of NS4B to prevent STAT1 activation was dependent on the 23-amino-acid signal peptide derived from the NS4A coding sequence (termed 2K for 2,000-molecular-weight peptide) (37); its activity was augmented by the addition of NS2A and NS4A (38). The NS4B proteins including the 2K fragment (2KNS4B) from WNV and YFV were similar to 2KNS4B of DENV-2 in their abilities to suppress JAK-STAT signaling (37). Thus, 2KNS4B is thought to be the primary antagonist of STAT1 phosphorylation encoded by these three viruses. Additional studies have been performed using Kunjin virus (KUN), an attenuated subtype of WNV endemic to Australia that only rarely causes cases of clinical disease in humans. This work demonstrated that multiple nonstructural proteins may contribute to antagonism of IFN signaling, including NS2A, NS2B, NS3, NS4A, and NS4B. A role for KUN NS5 in IFN antagonism was not detected in this study (26).
Given the ability of JEV to utilize NS5 as an IFN antagonist (23), we hypothesized that NS5 from WNV (which belongs to the same antigenic complex as JEV) may also suppress IFN responses. Furthermore, we reasoned that this activity may not have been previously recognized using KUN NS5 if the relative suppressive activity of IFN-antagonist proteins differs between virulent and attenuated virus strains. To test these questions, we used an NS5 expression construct corresponding to the virulent NY99 strain of WNV (WNV-NY99) and examined its effect on IFN-β-dependent JAK-STAT signaling. We also compared the ability to suppress STAT1 phosphorylation of 2KNS4B and NS5 proteins derived from a number of flaviviruses from the TBEV and JEV antigenic complexes with various degrees of virulence in humans. This work revealed WNV-NY99 NS5 as a potent suppressor of IFN-mediated JAK-STAT signaling while KUN NS5 was a poor inhibitor. We found that a single residue in KUN NS5 at position 653 was associated with reduced IFN antagonism during virus replication, suggesting that NS5 function in suppression of IFN responses may influence virus virulence in humans. Taken together, these studies begin to dissect potential mechanisms of flavivirus resistance to IFN and thus have direct implications for live attenuated vaccine design.
MATERIALS AND METHODS
Cells, virus, and transfection.
HEK293T, HEK293, and Vero cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. Recombinant Newcastle disease virus expressing green fluorescent protein (NDV-GFP) was grown in 10-day-old embryonated chicken eggs as previously described (45). All transfections were performed using Lipofectamine 2000 in OptiMEM (Invitrogen).
Generation of 2KNS4B and NS5 expression constructs.
For use in the NDV-GFP bioassay and ISRE activity assay, cDNA encoding DENV-2 core protein and NS5 was derived from the full-length clone pD2/IC-30P (provided by R. Kinney, National Center for Infectious Diseases, CO), and WNV NS5 was derived by reverse transcription-PCR (RT-PCR) of RNA isolated from Vero cells containing the WNV NY3356 replicon (53) (provided by P. Y. Shi, Novartis). This WNV NS5 protein sequence is derived from WNV strain NY 2000-crow3356 and is identical to the WNV-NY99 NS5 sequence. The genes were cloned into the mammalian expression vector pCAGGS in frame with a C-terminal hemagglutinin (HA) epitope tag. The pCAGGS-HA-Nipah virus (NiV) V plasmid was a kind gift from M. Shaw (Mount Sinai School of Medicine, NY).
LGTV NS5 and 2KNS4B were derived following PCR amplification using the LGTV E5 infectious cDNA clone as the template (kindly provided by A. Pletnev, NIAID, NIH). TBEV (strain Hypr) and JEV-SA14-14-2 (JEV-SA) cDNAs for NS proteins were obtained following RT-PCR of RNA isolated from virus-infected cells. This work with TBEV was performed in biosafety level 4 facilities at the University of Texas Medical Branch. KUN and WNV-NY99 NS protein cDNAs were amplified by PCR from infectious molecular cDNA clones (20; also W. Liu, V. Mokhonov, and A. Khromykh, unpublished data), whereas JEV Nakayama (JEV-N) NS proteins were PCR amplified from replicon cDNA (P. Mason, unpublished data). Primers for each amplification are detailed in Table 1.
TABLE 1.
Primer sequences used to PCR amplify viral genes
| Virus | 2KNS4B primer sequence |
NS5 primer sequence |
||
|---|---|---|---|---|
| 5′ Primera | 3′ Primer | 5′ Primera | 3′ Primer | |
| LGTV | CACCATGAGTAGTGATGACAACAAACTGGCATA | ACGCCGTGTCCCAGTGGTCCGGAGCC | CACCATGGGTGGATCCGAGGGAGAC | AAATATTGAGCTCTCCAGTTTGAGCTC |
| TBEV | CACCATGAGCAGTGACGACAACAAACTGGC | ACGCCTACCCCCAGAAGCTCGAAGCC | CACCATGGGTGGTTCTGAGGGAGAC | GATTATTGAGCTCTCCAGTCTGAGCTC |
| WNV-NY99 | CACCATGTCGCAGACAGACAACCAGCTAGC | TCTTTTTAGTCCTGGTTTTTCCATG | CACCATGGGTGGGGCAAAAGGACGC | CAGTACTGTGTCCTCAACCAAAGTTG |
| KUN | CACCATGTCGCAGACAGACAACCAGCTAGC | TCTCTTCAGCCCTGGTTTTTCCATG | CACCATGTCGCAGACAGACAACCAGCTAGC | CAATACTGTATCCTCAACCAATG |
| JEV-Nakayama | CACCATGTCACAGACAGATAACCAACTGGC | CCTTTTCAAGGAGGGCTTGTCAGCG | CACCATGGGAAGGCCTGGGGGCAGG | GATGACCCTGTCTTCCTGGATC |
| JEV-SA14142 | CACCATGTCACAGACAGATAACCAACTGGC | CCTTTTCAAGGAGGGCTTATCAGCG | CACCATGGGAAGGCCTGGGGGCAGG | GATGACCCTGTCTTCCTGGATCAAGAC |
Bases in italics were added to the 5′ primers to insert start codons and enable cloning into the TOPO Gateway system (Invitrogen).
After PCR amplification, each gene was directionally cloned into Gateway entry vectors (Invitrogen, Carlsbad, CA), followed by subcloning into pcDNA6.2DEST/V5 (Invitrogen) to generate C-terminal V5 epitope-tagged genes (Table 1). The sequence of each construct was verified by DNA sequencing.
Site-directed mutants of NS5 were made using a QuikChange Lightning site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene) with the primers detailed in Table 2. Mutations were made in pENTR/SD/D-TOPO entry vector, followed by sequencing and recombination into pcDNA6.2DEST/V5.
TABLE 2.
Primer sequences used to mutate WNV-NY99 or KUN NS5 expression constructs
| Primer name | Sense primer sequence (5′ to 3′) | Antisense primer sequence (5′ to 3′) |
|---|---|---|
| NY99 N377A | GAGTGAAGTACGTGCTCGCCGAGACCACCAACTGGT | ACCAGTTGGTGGTCTCGGCGAGCACGTACTTCACTC |
| NY99 N381A | GCTCAACGAGACCACCGCCTGGTTGTGGGCGTTT | AAACGCCCACAACCAGGCGGTGGTCTCGTTGAGC |
| NY99 E627A | CTGGTGAGGATGATGGCAGGGGAAGGAGTGATT | AATCACTCCTTCCCCTGCCATCATCCTCACCAG |
| NY99 E629A | GGATGATGGAAGGGGCAGGAGTGATTGGCCC | GGGCCAATCACTCCTGCCCCTTCCATCATCC |
| NY99 VI631/632AA | GATGGAAGGGGAAGGAGCGGCTGGCCCAGATGATGTGG | CCACATCATCTGGGCCAGCCGCTCCTTCCCCTTCCATC |
| NY99 W651A | GACCCAAAGTCAGGACCGCGCTGTTTGAGAATGGGG | CCCCATTCTCAAACAGCGCGGTCCTGACTTTGGGTC |
| NY99 F653S | CAAAGTCAGGACCTGGCTGTCTGAGAATGGGGAAG | CTTCCCCATTCTCAGACAGCCAGGTCCTGACTTTG |
| NY99 E378A | AAGTACGTGCTCAACGCGACCACCAACTGGTTG | CAACCAGTTGGTGGTCGCGTTGAGCACGTACTT |
| NY99 W382A | TCAACGAGACCACCAACGCGTTGTGGGCGTTTTTGG | CCAAAAACGCCCACAACGCGTTGGTGGTCTCGTTGA |
| KUN S653F | CCCAAGGTTAGAACCTGGCTGTTTGAGAATGGGGA | TCCCCATTCTCAAACAGCCAGGTTCTAACCTTGGG |
NDV-GFP bioassay.
Vero cells were transfected with either the empty pCAGGS plasmid or plasmids encoding various viral proteins as detailed in specific experiments. Expression of DENV-2 core protein was included as a negative control for IFN antagonism, whereas the NiV V, DENV-2 NS5, and LGTV NS5 proteins were included as positive controls. At 24 h posttransfection, cells were treated with 1,000 U/ml of human IFN-β (PBL). Following 24 h of IFN-β treatment, cells were infected with NDV-GFP as described previously (45). Fluorescence images were obtained at 14 h postinfection (hpi).
Immunofluorescence.
To examine virus protein expression and STAT1 phosphorylation in cells, Vero cells expressing each protein or infected with KUN (multiplicity of infection [MOI] of 1) were treated with human IFN-β for 15 min, fixed in ice-cold 100% methanol for 10 min, and stained using anti-phosphotyrosine 701-STAT1 (Cell Signal Technologies) and either anti-V5 (Invitrogen) antibodies as previously described (6) or a cocktail of monoclonal antibodies to WNV NS5 at a 1:20 dilution (13). Images were captured using a Zeiss Axio Scope with Axiovision software or a Zeiss LSM710 confocal microscope.
Reporter gene assays.
HEK293T cells were cotransfected with pCAGGS plasmids encoding various viral proteins, the IFN-inducible chloramphenicol acetyltransferase (CAT) reporter (ISRE54-CAT) plasmid, and a plasmid constitutively expressing the firefly luciferase protein. The V5-tagged LGTV NS5 plasmid also served as a positive control. At 24 h posttransfection, cells were treated with 1,000 U/ml IFN-β for another 24 h prior to harvest and assay for CAT activity, as previously described (46). pCAGGS-firefly luciferase and pISRE-54-CAT reporter plasmids were kind gifts from L. Martinez-Sobrido (University of Rochester, NY). Alternatively, HEK293 cells were cotransfected with plasmids encoding an IFN-inducible firefly luciferase reporter (pISRE-luc; Stratagene) and the constitutive Renilla firefly expression plasmid, pRL-TK (Promega). At 24 h posttransfection, the HEK293 cells were infected with wild-type (WT) KUN or KUN NS5 carrying the mutation S653F ([NS5:S653F] MOI of 1). At 24 hpi, cells were treated with 1,000 U/ml of human IFN-β. Following 6 to 7 h of treatment with IFN, cells were lysed and measured for luciferase activities according to the manufacturer's instructions (Promega). The reporter activity of the IFN-treated sample was normalized to the constitutively expressed luciferase value of that sample to control for transfection efficiency.
Flow cytometry.
Vero cells transfected with the empty vector or various NS5 expression constructs were treated with 1,000 U/ml IFN-β for 15 min, washed twice in cold Dulbecco's phosphate-buffered saline (DPBS), and trypsinized for 10 min at 37°C to dislodge cells. Cells were resuspended in freshly prepared 2% paraformaldehyde-DPBS and incubated for 10 min at 37°C, followed by permeabilization in 90% methanol for 10 min on ice. Cells were washed once in stain buffer (BD Pharmingen, San Diego, CA), followed by incubation with anti-pY(701)-STAT1 conjugated to Alexa Fluor 647 (BD Pharmingen) and anti-V5 conjugated to fluorescein isothiocyanate (FITC; Invitrogen) for 45 min at room temperature in the dark. Alexa Fluor 647- and FITC-conjugated mouse immunoglobulin G2a (IgG2a) were used as isotype controls. Cells were washed once in stain buffer and analyzed using a FACSCalbur or FACSAria flow cytometer (BD Biosciences) and FlowJo software (Tree Star). After V5-positive cells were gated, the percent tyrosine-phosphorylated STAT1 (pY-STAT1) inhibition was determined as the fraction of V5-positive cells that were pY-STAT1 negative.
Generation of recombinant KUN containing NS5 S653F.
The NS5 S653F mutation was generated using QuikChange PCR on an intermediate plasmid that was constructed in two steps. First, a 1,958-bp region comprising the last 1,334 nucleotides of the NS5 gene and complete 3′ untranslated region (UTR) was amplified from the FLSDX 250pro plasmid (12, 20) using the following primer pair: KUN NS5-HindIII, GGCAAGCTTGAGTTGTTGGACGGGGA (engineered to contain a HindIII restriction site, indicated in bold); and KUN XhoI-R, CCCCCTCGAGCAATTGT. The amplification product was cloned into a pcDNA3 vector using HindIII and XhoI restriction sites. The HindIII and XbaI fragment from pcDNA3-NS5 plasmid containing the area of interest was then cloned into a pUC18 vector. QuikChange PCR was carried out on the pUC18-NS5 plasmid with Pfu DNA polymerase using the 5′ to 3′ sense primer GTTAGAACCTGGCTGTTCGAGAATGGGGAGGAA and antisense primer TTCCTCCCCATTCTCGAACAGCCAGGTTCTAAC. The resulting mutated fragment was then cloned back into the full-length FLSDX 250pro plasmid using the SgrAI and XhoI restriction sites, and the S653F mutation was confirmed by sequencing.
To produce infectious virus, in vitro transcription was carried out with SP6 polymerase using 1 μg of XhoI-linearized plasmid as a template; the resulting RNA transcript was electroporated into 5 × 106 BHK-21 cells. Virus recovered following electroporation was used to infect Vero76 cells at an MOI of 0.1, and the supernatant was collected at approximately 72 h postinfection, centrifuged at low speed, and then aliquoted for storage at −80°C. Virus replication was quantified by a focus-forming assay as previously described (36) using a cocktail of monoclonal antibodies to NS5 (13) diluted 1:50.
Western blot analysis.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (1 mM EDTA, 50 mM Tris, 150 mM NaCl, 0.5% deoxycholate [DOC], 0.1% SDS, 1% NP-40) and centrifuged at 200 × g for 10 min at 4°C. Protein in the cleared lysate was quantified using a Bradford protein assay (Bio-Rad Laboratories, Hercules, CA) and subjected to SDS-PAGE, followed by transfer to nitrocellulose membranes. Membranes were blocked and probed for V5, STAT1, pY-STAT1, or actin as previously described (6, 44). Alternatively, membranes were probed with anti-HA (Sigma), a mouse anti-WNV E polyclonal (kind gift of R. Tesh, University of Texas Medical Branch, TX), or a cocktail of monoclonal antibodies to WNV NS5 at 1:500 (13).
Structural modeling.
To determine amino acids that may be important to WNV NS5 function, the NS5 sequences from LGTV, WNV-NY99, and KUN were aligned using Clustal W alignment within DNAStar Lasergene software. Residues of interest were modeled on the KUN NS5 RdRP structure (Protein Data Bank code 2HFZ) (31) using PyMol.
Statistical analysis.
Data from ISRE reporter assays and flow cytometry using IFN-β at 1,000 U/ml were analyzed by one-way analysis of variance (ANOVA) with either Dunnett's multiple comparison test or Tukey's posttest to determine significant differences (P < 0.05) between individual groups. Virus titration data were analyzed by a two-tailed t test or Mann-Whitney U test as indicated in the figure legends.
RESULTS
Identification of WNV-NY99 NS5 as an IFN antagonist.
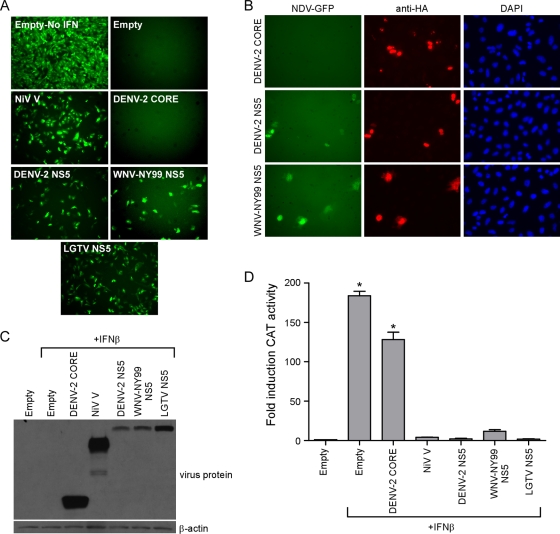
The NS5 proteins from LGTV, TBEV, JEV, and DENV disrupt IFN-mediated JAK-STAT signaling, albeit through varied mechanisms (2, 6, 23, 59). It is well established that WNV antagonizes IFN-mediated signal transduction (11, 26) although the contribution of NS5 to this is not fully resolved. To examine the contribution of WNV-NY99 NS5 to IFN antagonism, we first analyzed its impact on replication of NDV-GFP in the presence of IFN. NDV-GFP is highly sensitive to the antiviral effects of IFN. Thus, stimulation of cells with IFN prior to infection prevents NDV-GFP replication, as demonstrated by a lack of GFP expression. NDV-GFP replication can be rescued by expressing antagonists of IFN signaling such as the NiV V protein in cells prior to infection (45, 52). Vero cells were transfected with an empty plasmid or plasmids expressing DENV-2 core (negative control), NiV V, DENV-2 NS5, LGTV NS5 (as positive controls), or WNV-NY99 NS5 and treated with IFN-β. Twenty-four hours after IFN treatment, cells were infected with NDV-GFP and examined at 14 hpi for GFP expression (Fig. 1A). NDV-GFP replication was not detected in cells transfected with an empty plasmid or in those expressing the DENV-2 core protein. However, the presence of the NiV V protein, DENV-2 NS5, LGTV NS5, or WNV NS5 facilitated NDV-GFP replication (Fig. 1A). By immunofluorescence staining, NDV-GFP was present only in cells expressing the flavivirus NS5 proteins (Fig. 1B). These results indicate that NS5 from WNV-NY99 can function as a suppressor of host IFN responses.
FIG. 1.
WNV NS5 antagonizes host IFN-β responses by inhibiting JAK-STAT signaling. (A) Effect of WNV NS5 expression on NDV-GFP replication in the presence of IFN. Vero cells transfected with an empty plasmid or a plasmid expressing individual virus proteins as indicated were treated with 1,000 U/ml IFN-β for 24 h and then infected with NDV-GFP. NDV-GFP replication was monitored by GFP fluorescence at 14 hpi. (B) Examples from panel A demonstrating NDV-GFP replication (green) in cells expressing flavivirus NS5 proteins (red). Nuclei are counter stained with 4′,6′-diamidino-2-phenylindole (DAPI; blue). (C) Western blot demonstrating the relative expression of each viral protein in HEK293T cells. (D) Effect of WNV NS5 on ISRE-dependent gene expression. HEK293T cells were transfected with the ISRE-CAT reporter plasmid, a plasmid constitutively expressing firefly luciferase, and either an empty plasmid or a plasmid expressing the virus protein indicated and treated with IFN for 24 h. CAT activity in cell lysates was determined and normalized to the luciferase activity in each sample. Fold induction of CAT activity was determined relative to the normalized CAT activity value of cells transfected with the empty vector and not treated with IFN. Error bars indicate standard errors of the mean (SEM) from three individual experiments performed in triplicate; asterisks indicate significant differences from cells transfected with the empty vector and not treated with IFN (P < 0.05, by ANOVA followed by Dunnett's multiple comparison test).
We next wanted to determine if WNV NS5 specifically inhibits JAK-STAT signaling in response to IFN. Thus, we examined ISRE promoter activation in HEK293T cells expressing NS5 from WNV-NY99, DENV-2, or LGTV. Expression of DENV-2 core or NiV V proteins was again included as a negative and positive control, respectively. The expression of each protein is shown in Fig. 1C. Plasmids encoding the different virus proteins were cotransfected with the reporter plasmid pISRE-54-CAT as well as a plasmid driving the constitutive expression of firefly luciferase. After a 24-h treatment with IFN-β, cell lysates were harvested and assayed for CAT and luciferase activities (Fig. 1D). IFN treatment of cells transfected with the empty vector or expressing DENV-2 core protein resulted in a significant increase in CAT activity (P < 0.05), demonstrating activation of JAK-STAT signaling. However, CAT activity in IFN-treated cells expressing NiV V, DENV-2 NS5, WNV-NY99 NS5, or LGTV NS5 was not statistically different from activity in cells transfected with an empty plasmid and not treated with IFN (Fig. 1D), suggesting that JAK-STAT signaling was not active in these cultures. Thus, WNV-NY99 NS5 suppresses IFN responses specifically by interfering with JAK-STAT signaling, similar to NS5 from LGTV or DENV-2.
Comparison of NS5 and 2KNS4B function in inhibition of pY-STAT1.
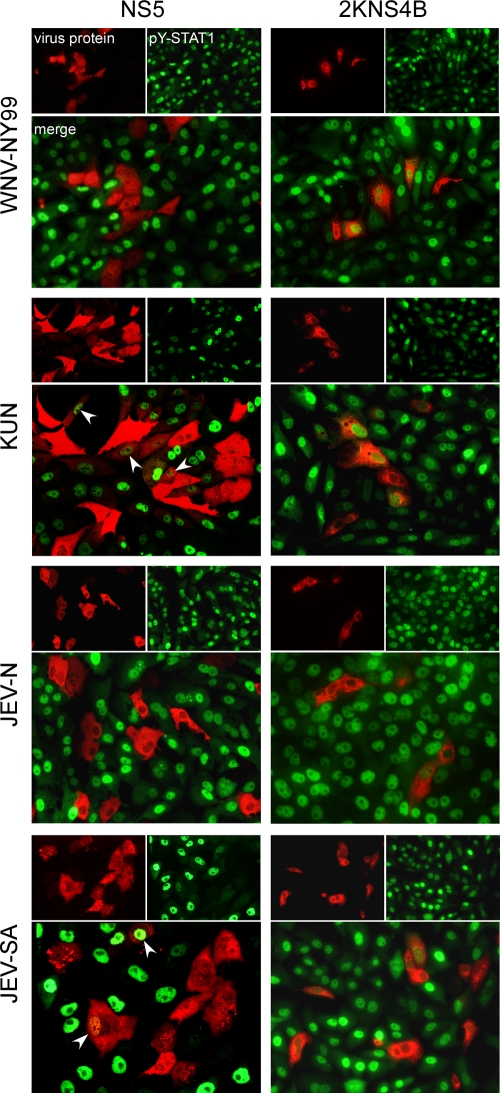
In cells infected with WNV, JEV, or LGTV, suppression of signaling is associated with the failure of both STAT1 and STAT2 to be phosphorylated on tyrosine residues (6, 11, 24, 26). In turn, this prevents STAT nuclear translocation and ISRE-driven gene expression. The 2KNS4B protein from WNV has been demonstrated to prevent STAT1 phosphorylation in IFN-treated cells (26, 37). To compare the impact of NS5 and 2KNS4B from virulent and attenuated strains of these viruses on STAT1 activation, we examined phosphorylation and nuclear localization of STAT1 (pY-STAT1) by immunofluorescence assay (IFA) in IFN-treated cells expressing NS5 or 2KNS4B derived from WNV-NY99 and KUN or the virulent JEV Nakayama strain (JEV-N) (8, 39) and the live attenuated vaccine strain, JEV-SA14-14-2 (JEV-SA) (40, 42) (Fig. 2).
FIG. 2.
Immunofluorescence of STAT1 phosphorylation and nuclear localization in cells expressing flavivirus NS5 and 2KNS4B proteins. Vero cells were transfected with plasmids encoding the NS5 or 2KNS4B proteins derived from the indicated viruses, with each fused to a C-terminal V5 epitope tag. At 24 h posttransfection, cells were treated with IFN-β for 15 min, fixed in methanol, and stained for anti-V5 (red) and anti-pY-STAT1 (green). Arrowheads indicate examples of NS5-positive cells that are also positive for pY-STAT1.
In Vero cells transfected with the empty expression plasmid (pcDNA6.2/V5) and treated with IFN-β, pY-STAT1 was readily detected in the nucleus of the vast majority of cells (data not shown). However, the majority of cells expressing NS5 from WNV-NY99 or JEV-N and treated with IFN-β were negative for pY-STAT1 (Fig. 2). This was similar to results obtained with LGTV or TBEV NS5 (6; also data not shown). In contrast, nuclear pY-STAT1 was detectable in numerous cells expressing low levels of NS5 from KUN or in JEV-SA NS5-expressing cells (Fig. 2, arrowheads). Phosphorylated STAT1 was observed in the nucleus of cells expressing 2KNS4B from all viruses tested (Fig. 2). These observations suggest that NS5 from WNV-NY99 prevents the phosphorylation and nuclear translocation of STAT1 in response to IFN-β and, hence, support results obtained using the NDV complementation and ISRE activity assays. As expected (23), NS5 derived from virulent JEV-N also efficiently prevented pY-STAT1 accumulation.
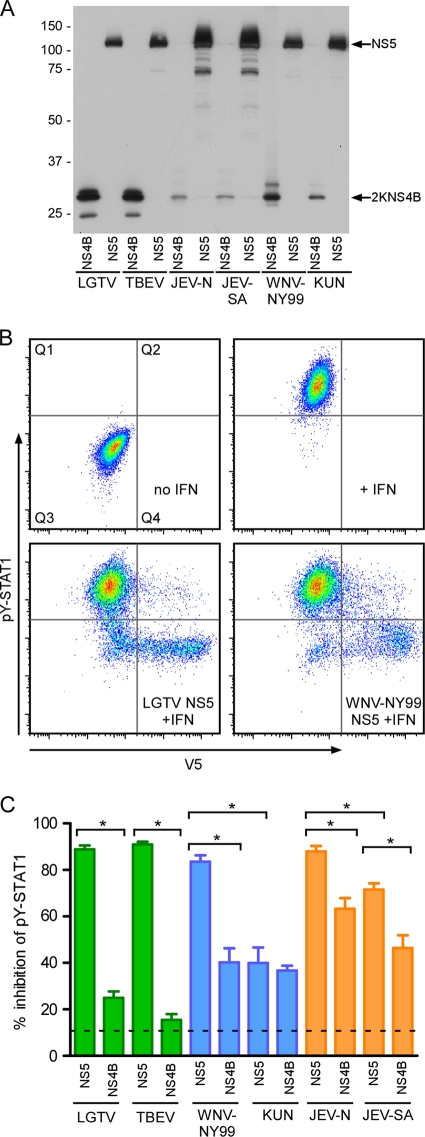
To quantify the intrinsic ability of each 2KNS4B and NS5 protein to impede JAK-STAT signaling, we used flow cytometry to measure pY-STAT1 in cells expressing V5 epitope-tagged 2KNS4B or NS5. This quantitative technique to measure pY-STAT1 provides advantages over other measurements because the transfection efficiency between samples can be directly normalized by gating V5-positive cells (44). Vero cells transiently expressing each V5 fusion protein (expression levels are shown in Fig. 3A) were stimulated with IFN-β, fixed, permeabilized, and incubated with pY-STAT1- and V5-specific antibodies. During analysis, the V5-positive cell population was gated, and the percent inhibition of pY-STAT1 for each protein was defined as the proportion of V5-expressing cells that were pY-STAT1 negative (Fig. 3B). NS5 and 2KNS4B from LGTV were used as positive and negative controls for pY-STAT1 inhibition, respectively.
FIG. 3.
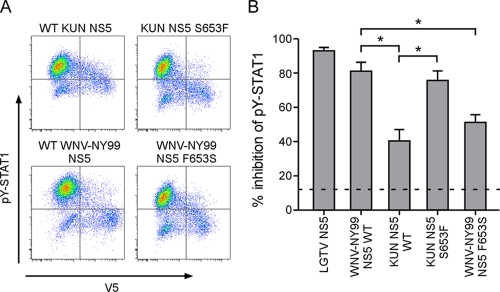
Quantification of inhibition of STAT1 phosphorylation by flavivirus NS5 and 2KNS4B proteins. (A) Western blot demonstrating the expression of NS5 and 2KNS4B proteins, each in frame with a C-terminal V5 epitope tag, in Vero cells. (B) Flow cytometry analysis to determine the percent inhibition of pY-STAT1 in Vero cells expressing flavivirus nonstructural proteins. Vero cells were fixed, permeabilized, and stained using antibodies that recognize pY(701)-STAT1 and V5. The examples shown are cells transfected with an empty plasmid and either untreated or treated with IFN-β (top panels) or cells expressing LGTV NS5 or WNV-NY99 NS5 and treated with IFN-β (lower panels). (C) Quantification of pY-STAT1 inhibition by each protein analyzed by flow cytometry. V5-positive cells were gated and examined for pY-STAT1. The percent pY-STAT1 inhibition is the percentage of V5-positive cells (contained in quadrant 2 [Q2] and Q4 in B) that were pY-STAT1 negative (in Q4). The average level of V5-negative cells that are also pY-STAT1 negative in IFN-β-treated cultures (cells present in Q3) is illustrated by the dotted line as an indication of the background levels in this assay. Error bars indicate SEM from four independent experiments; asterisks indicate significant differences (P < 0.05, by ANOVA followed by Tukey's test).
NS5 from WNV-NY99 was an efficient antagonist of signaling, with approximately 85% of NS5-positive cells negative for pY-STAT1 (Fig. 3C). This level of inhibition was significantly greater than that of the WNV-NY99 2KNS4B protein. In contrast, KUN NS5 suppressed pY-STAT1 in significantly fewer cells (in approximately 42% of NS5-positive cells) than WNV-NY99 NS5 (Fig. 3C). This level of inhibition by KUN NS5 was similar to that produced by the KUN 2KNS4B protein. Taken together, these results suggest that NS5 derived from the virulent WNV-NY99 is the most potent antagonist of IFN-mediated JAK-STAT signaling encoded by this virus. Furthermore, the results suggest that KUN NS5 is an inefficient IFN antagonist.
As also shown in Fig. 3C, NS5 derived from the virulent JEV-N strain was an efficient suppressor of signal transduction, with approximately 90% of IFN-β-treated cells negative for pY-STAT1. Expression of JEV-N 2KNS4B also resulted in a pronounced level of suppression, at about 65%. Interestingly, suppression of pY-STAT1 by JEV-SA NS5 was significantly lower than that by JEV-N NS5 and not different from that by JEV-N 2KNS4B. There was no significant difference between the relative abilities of the 2KNS4B proteins from the two JEV strains to inhibit signaling. Consistent with previously published work (23), these results suggest that NS5 derived from JEV is a more efficient antagonist of IFN-mediated JAK-STAT signaling than 2KNS4B but that JEV 2KNS4B likely contributes to suppression of this signaling pathway in infected cells. These results also indicate that NS5 from the live attenuated vaccine strain is a less efficient antagonist than NS5 from virulent JEV strains.
Finally, expression of NS5 and 2KNS4B from TBEV-Hypr resulted in approximately 90% and 15% inhibition of pY-STAT1, respectively (Fig. 3C). These levels of inhibition were not statistically different from their LGTV-derived counterparts. The finding that TBEV NS5 is an efficient antagonist of IFN-mediated signaling is consistent with the recent findings of Werme et al. (59).
Identification of residues important for WNV NS5 function as an IFN antagonist.
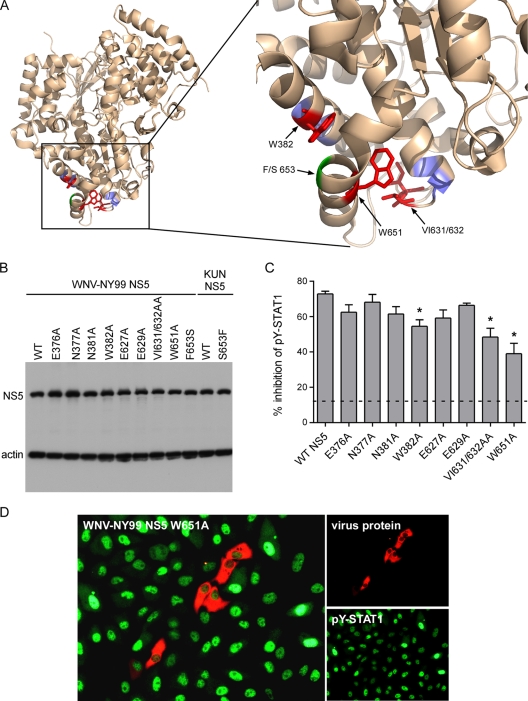
We previously identified a number of amino acids within LGTV NS5 required for its IFN antagonist function (44). The residues identified were positioned in two noncontiguous areas of the protein, between amino acids 374 to 380 and 624 to 647, that mapped proximal to each other when modeled onto the KUN RdRp crystal structure (31, 44). To determine if the specific residues identified for LGTV NS5 were also important for WNV-NY99 NS5 function, we initially made site-to-alanine mutations at the analogous residues in WNV-NY99 NS5 (N377, N381, E627, E629, VI631/632, and W651) (Fig. 4A) and examined the resulting degree of suppression using flow cytometry. The mutations did not appear to affect NS5 expression levels (Fig. 4B). Mutation at VI631/632AA and W651A significantly decreased the ability of WNV-NY99 NS5 to suppress IFN signaling, with W651A reducing the activity of NS5 by approximately 45% (Fig. 4C). By IFA, cells expressing NY99 NS5:W651A showed predominantly nuclear accumulation of pY-STAT1, suggesting that this protein had reduced capacity to inhibit JAK-STAT signaling (Fig. 4D). The mutations E627A and E629A did not affect WNV-NY99 NS5 antagonist function (Fig. 4C). Furthermore, the mutations N377A and N381A did not affect NS5 function, but unlike their counterparts in LGTV NS5, these WT residues have no charge. We reasoned that the two residues adjacent to these may have a more pronounced role due to their charge (E376) or aromatic side chain (W382). Mutation at W382A had a modest but significant effect on NY99 NS5-mediated suppression of IFN signaling, while E376A had no effect (Fig. 4C). Thus, WNV NS5 residues W382, VI631/632, and W651 are important to its function as an IFN antagonist.
FIG. 4.
Identification of residues important for WNV NS5 function as an IFN antagonist. (A) Position of residues selected for mutation on the WNV KUN RdRp structure (31). The entire RdRp is shown on the left; the region of interest in IFN antagonism is shown on the right. The residues demonstrated in this study to have a role in WNV-NY99 NS5 function in IFN antagonism are depicted in red (W382, VI631/632, and W651) or green (residue 653), whereas those that were mutated to alanine with no effect on NS5 function are depicted in blue. (B) Western blot demonstrating the relative expression of each WNV-NY99 or KUN NS5 mutant used in these studies. (C) Quantification of pY-STAT1 inhibition by WNV-NY99 NS5 site mutants analyzed by flow cytometry. Error bars indicate SEM; asterisks indicate significant differences from WT NY99 NS5 (P < 0.05, by ANOVA followed by Dunnett's test). (D) IFA of WNV-NY99 NS5 containing a W651A mutation in IFN-β-treated Vero cells. NS5 is shown in red, and pY-STAT1 is shown in green.
As demonstrated in the experiment shown in Fig. 3C, NS5 derived from WNV-NY99 suppressed pY-STAT1 accumulation better than KUN NS5. There are 10 amino acid differences between these two NS5 proteins, of which 9 represent relatively conserved substitutions (Table 3). However, the mutation at residue 653 from Phe (NY99) to Ser (KUN) represents a change in hydrophobicity and maps within the IFN-antagonist domain identified for LGTV NS5 (Fig. 4A). To determine if this residue is responsible for the different levels of inhibition, we made an S653F mutation in KUN NS5 as well as the converse mutation (F653S) in WNV-NY99 NS5 and tested the ability of the mutant NS5 proteins to suppress pY-STAT1 by flow cytometry. KUN NS5:S653F yielded a flow cytometry profile that was more similar to that of WT NY99 NS5 (Fig. 5A), suppressing pY-STAT1 in approximately 76% of cells, a result not significantly different from WT NY99 NS5 (Fig. 5B). The reverse mutation, F653S in WNV-NY99 NS5, reduced the ability of this molecule to inhibit signaling to levels similar to inhibition by WT KUN NS5 (Fig. 5B). Thus, the residue at position 653 is a critical determinant of WNV NS5 antagonist function.
TABLE 3.
Differences in NS5 amino acid sequences between WNV strain NY99 and WNV subtype KUN
| Virus | NS5 amino acid residue at position: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 33 | 162 | 177 | 247 | 312 | 531 | 653 | 731 | 877 | |
| NY99 | T | I | I | R | R | D | K | F | V | A |
| KUN | I | T | L | K | K | E | R | S | T | S |
FIG. 5.
Residue 653 is largely responsible for the differences between NY99 and KUN NS5 function in IFN antagonism. (A) Flow cytometry to determine the percent inhibition of pY-STAT1 in IFN-β-treated Vero cells expressing KUN and WNV-NY99 mutants. The examples shown are cells expressing WT KUN NS5, KUN NS5:S653F, WT WNV-NY99, and NY99 NS5:F653S. (B) Quantification of pY-STAT1 inhibition by WNV-NY99 and KUN NS5 mutants by flow cytometry. LGTV NS5 was included as a positive control for pY-STAT1 inhibition. Error bars indicate SEM from four independent experiments; asterisks indicate significant differences (P < 0.05, by ANOVA followed by Tukey's test).
WNV NS5 residue S653F has an important role in IFN antagonism during virus replication.
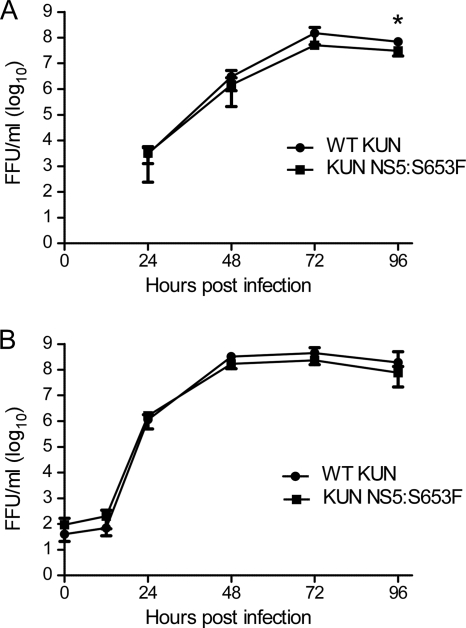
To determine if the NS5 residue at position 653 has relevance to IFN antagonism in the context of virus replication, the NS5:S653F mutation was introduced into KUN using reverse genetics. WT KUN had a small replication advantage in Vero cells but only at 96 hpi (Fig. 6A). WT and NS5:S653F KUN viruses replicated equally well in HEK293 cells (Fig. 6B). Taken together, these results suggest that mutation at NS5:S653F did not drastically compromise the ability of KUN to replicate, despite the fact that this mutation resides in the RdRP domain.
FIG. 6.
Replication of WT KUN and KUN NS5:S653F in cell culture. Supernatants from Vero cells infected at an MOI of 0.001 (A) or HEK293 cells infected at an MOI of 0.1 (B) were assayed for infectious virus by focus-forming assay at the times indicated. Error bars indicate SEM from three independent experiments performed in triplicate; the asterisk indicates significant difference (P < 0.05, Student's t test). FFU, focus-forming units.
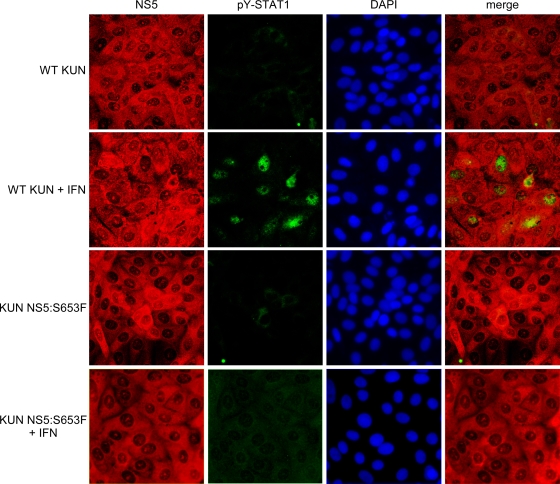
We first assessed the effect of the S653F mutation on IFN antagonism using IFA. Vero cells were infected with WT and mutant KUN for 48 h and then left untreated or treated with 1,000 U/ml IFN-β for 15 min. The cells were then stained for NS5 and pY-STAT1. Although the majority of cells infected with WT KUN and treated with IFN were negative for pY-STAT1, a substantial number of infected cells contained nuclear pY-STAT1 (Fig. 7). In contrast, pY-STAT1 was not observed in IFN-treated cells infected with KUN NS5:S653F (Fig. 7).
FIG. 7.
Immunofluorescence of pY-STAT1 in WT and NS5:S653F KUN-infected cells. Vero cells were infected with WT or NS5:S653F KUN and treated with 1,000 U/ml IFN-β at 48 hpi, followed by staining for NS5 (red) and anti-pY-STAT1 (green). Nuclei were counterstained with DAPI.
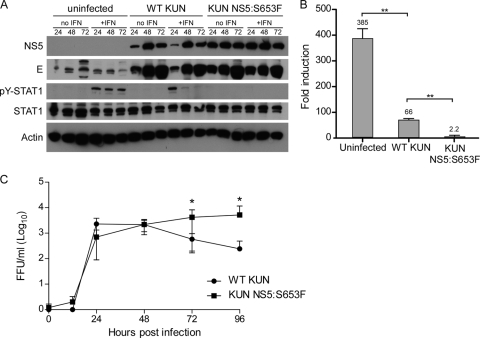
The ability of WT and mutant viruses to suppress pY-STAT1 was also compared by Western blot analysis (Fig. 8A). Phosphorylated STAT1 was readily detected in uninfected HEK293 cells treated with 1,000 U/ml IFN-β. Suppression of pY-STAT1 in WT KUN-infected cells was evident at 48 hpi. In contrast, KUN NS5:S653F replication was associated with an almost complete lack of pY-STAT1 in IFN-treated cells at 24 hpi. Even though the two viruses grew equally well in HEK293 cells (Fig. 6B), the expression of NS5 and E proteins in KUN NS5:S653F-infected cells was higher at 24 hpi, and NS5 expression tended to be higher at 72 hpi. We also observed higher NS5 expression at 12 and 24 hpi in KUN NS5:S653F-infected Vero cells (data not shown). These results support the IFA results and demonstrate that the presence of S653F results in more robust suppression of IFN-mediated JAK-STAT signaling.
FIG. 8.
Replication of NS5:S653F KUN is associated with greater IFN resistance than WT KUN. (A) Western blot demonstrating increased efficiency of IFN antagonism by KUN NS5:S653F. HEK293 cells were left uninfected or were infected with WT or NS5:S653F KUN at an MOI of 1. At the times indicated, cells were treated with 1,000 U/ml of IFN-β for 15 min, and cell lysates were analyzed for virus protein expression (NS5 and E) and STAT1 phosphorylation. (B) Effect of S653F mutation on ISRE-dependent gene expression during KUN replication. HEK293 cells were transfected with the pISRE-luc reporter plasmid and a plasmid constitutively expressing Renilla luciferase. Cells were infected 24 h later with WT or NS5:S653F KUN at an MOI of 1 and incubated for a further 24 h. Cells were treated with 1,000 U of IFN-β for 7 to 8 h prior to assaying for dual luciferase activities. Fold induction of firefly luciferase activity was determined relative to the normalized luciferase activity value of virus-infected cells not treated with IFN. Error bars indicate SEM from three individual experiments; asterisks indicate significant differences from cells transfected with the empty vector and not treated with IFN (P < 0.005, by Student's t test). (C) KUN replication in the presence of IFN. Vero cells were infected with WT or NS5:S653F KUN at an MOI of 0.001 and treated with high-dose IFN-β at 12 hpi. Supernatants were assayed for infectious virus by focus-forming assay at the times indicated. Error bars indicate SEM from five independent experiments performed in triplicate; an asterisk indicates significant difference (P = 0.0079, Mann-Whitney test). FFU, focus forming units.
To quantify inhibition of signaling by WT and KUN NS5:S653F viruses, we examined ISRE promoter activation in HEK293 cells treated with IFN at 24 hpi. WT KUN replication resulted in a 5.8-fold reduction in ISRE activity compared to uninfected cells, whereas infection with KUN NS5:S653F resulted in a 175-fold reduction. Thus, the presence of the 653F mutation in NS5 resulted in a 30-fold greater inhibition of IFN-dependent signaling than the presence of WT residue in the context of virus replication.
Finally, we examined virus replication in the presence of IFN. Vero cells were infected at an MOI of 0.001 and treated with high-dose IFN-β (100 to 1,000 U/ml) at 12 hpi. Infectious virus in supernatants was measured at the times indicated in the legend to Fig. 8C by focus-forming assay. In the presence of IFN, replication of KUN NS5:S653F was significantly greater than that of WT KUN at 72 and 96 hpi (Fig. 8C). Thus, the S653F mutation in NS5 confers greater resistance to the antiviral effects of IFN.
DISCUSSION
A major mechanism by which WNV evades the host antiviral response is to suppress IFN-stimulated JAK-STAT signaling (11, 19). In this report, we have demonstrated that NS5 from the virulent NY99 strain of WNV is a potent inhibitor of IFN-mediated signal transduction. WNV NS5 expression prevented the development of the cellular antiviral state, as demonstrated by its ability to augment NDV-GFP replication in IFN-treated cells (Fig. 1A). As observed during infection (11, 19), IFN antagonism mediated by WNV-NY99 NS5 was associated with failure of STAT1 to be phosphorylated (Fig. 3), translocate to the nucleus (Fig. 2), and participate in the expression of ISRE-dependent genes (Fig. 1D). This work adds WNV-NY99 to the number of highly pathogenic flaviviruses that utilize NS5 as an efficient IFN antagonist (including TBEV, DENV, and JEV), suggesting that this function of NS5 is essential to the success of flaviviruses as emerging and reemerging pathogens (28).
Effective host IFN responses are critical to recovery from flavivirus infection. Thus, the relative ability of these viruses to subvert the IFN response may be a decisive factor in their virulence. In support of this concept, we found that NS5 from WNV-NY99 was a potent suppressor of IFN responses, whereas NS5 from the closely related but attenuated KUN was not (Fig. 3). These results are consistent with previous work that examined the ability of individual KUN proteins to suppress ISRE-dependent responses and did not find a role for NS5 (26). A single residue at position 653 is largely responsible for this difference since its mutation in KUN NS5 to the corresponding NY99 residue (S653F) conferred an ability to antagonize signaling similar to that of WT NY99 NS5 (Fig. 5B). Furthermore, introduction of F653S to NY99 NS5 compromised the ability of this protein to prevent pY-STAT1 accumulation, suggesting that this residue is more generally important for WNV NS5 function in IFN antagonism. Incorporation of the NS5 mutation S653F into a recombinant KUN increased the virus's ability to suppress IFN-β-mediated STAT1 phosphorylation and ISRE-dependent gene expression. Strikingly, KUN NS5 bearing the S653F mutation during transient expression demonstrated only a 2-fold increase in its ability to inhibit pY-STAT1, yet replication of a recombinant KUN bearing this mutation resulted in a 30-fold increase in inhibition of signaling compared to WT virus (Fig. 8B). This more potent antagonism was associated with greater resistance to the antiviral effects of IFN during WNV replication (Fig. 8C). The importance of S653F during virus replication provides definitive evidence for the biological relevance of NS5 and, specifically, the residue at position 653, in IFN antagonism.
Interestingly, we found that viral proteins accumulated to higher levels at 24 hpi in KUN NS5:S653F-infected cells than in cells infected with WT virus (Fig. 8A) without an increase in infectious virus (Fig. 6). Because E and NS5 protein levels were greater in both IFN-competent and -incompetent cells infected with KUN NS5:S653F at 24 hpi, it is possible that the S653F mutation not only increases resistance to IFN (Fig. 5B) but also stabilizes NS5 expression. This may also result in a general acceleration of protein expression (and perhaps RNA replication) by stabilizing the replication complex. Incorporation of S653F into KUN NS5 expressed ectopically did not alter its expression level (Fig. 4B). However, NS5 turnover is likely to be more complex during virus replication, as exemplified by the fact that DEN NS5-mediated degradation of STAT2 was observed only when NS5 was expressed as part of a cleavable polyprotein (2). Thus, the mutation may affect NS5 stability only after cleavage. Alternatively, NS5 may be stabilized through increased binding to a cellular target induced during virus replication. Future experiments will more precisely address the mechanism of IFN antagonism and its relationship to WNV NS5 turnover.
Residue 653 lies within not only the IFN antagonism domain previously identified for LGTV NS5 but also the three-dimensional pocket we previously proposed to mediate much of LGTV NS5's function in IFN resistance (44). In addition, mutagenesis studies demonstrated that at least three WNV NS5 residues located in this site, W382, VI631/632, and W651, were important for IFN antagonism. Hence, this site appears more broadly important to NS5 function, suggesting that the mechanism of STAT1 inhibition, at least in part, may be common to NS5 proteins from both TBEV and JEV serogroups. NS5 proteins from JEV-N and JEV-SA also demonstrated significantly different abilities to prevent pY-STAT1 accumulation (Fig. 3) and differ from each other at eight amino acids. Based on the experiments presented here, we predict that residue 640 within JEV NS5, located within the same site of NS5 and divergent between JEV-N and JEV-SA strains, is responsible for these differences. However, although LGTV NS5 residues 355 to 735 are sufficient to inhibit IFN signaling equally as well as the full-length protein, the analogous truncation of WNV-NY99 or TBEV NS5 did not function efficiently as antagonists (data not shown). While we did not finely map the antagonism domains in these two proteins, only expression constructs corresponding to residues 1 to 735 retained resistance to IFN in both cases. This is consistent with previous mapping studies of JEV NS5 (23) and with the requirement for sequences in the MTase domain of TBEV NS5 for optimal inhibition (59). Thus, additional features of some NS5 molecules may also contribute to suppression.
The relationship between NS5 function and virulence of the corresponding virus was not observed for the tick-borne flaviviruses. NS5 from attenuated LGTV and pathogenic TBEV both exhibited the same high degree of pY-STAT1 suppression (Fig. 3C). Of course, flaviviruses encode factors other than NS5 that contribute to pathogenicity. The E protein, for example, is particularly important in flavivirus virulence since it mediates virus binding to cellular receptors and entry to the host cell (reviewed in reference 32). The presence of specific glycosylation sites in E is associated with WNV virulence (3, 4), and the WNV E protein can suppress innate immune responses to double-stranded RNA, a phenomenon dependent on E glycosylation status (1). The E protein has recently been demonstrated to affect sensitivity of JEV to host IFN responses since a mutation in E that reduced replication efficiency also reduced the capacity to antagonize IFN-mediated JAK-STAT signaling (22). Hence, while NS5 function in IFN resistance is likely required for virus replication and pathogenesis, it is not the only candidate for defining flavivirus virulence.
The accumulated data presented here and previously (2, 6, 23, 44, 59) suggest that NS5 is the most potent of the flavivirus-encoded IFN antagonists in mammalian cells. However, NS4B also antagonizes responses, a function that is dependent on the 2K signal sequence derived from NS4A, and is enhanced in the presence of the other small hydrophobic NS proteins, NS2A and NS4A (37, 38). During flavivirus replication, these three proteins are involved in endoplasmic reticulum membrane proliferation, membrane anchoring of the viral replication complex, and RNA replication (28, 29, 34, 35, 50, 58). In the case of WNV and probably all flaviviruses, membrane rearrangement is concomitant with redistribution of cellular cholesterol to sites of viral replication (30). The resulting loss of cholesterol-rich lipid rafts in the plasma membrane is associated with reduced IFN-mediated JAK-STAT signal transduction (30). Thus, it is highly possible that the functions of NS4A, NS4B, and the intervening 2K signal sequence in membrane rearrangement contribute to their IFN antagonism. However, this does not readily explain why 2KNS4B from JEV can suppress STAT1 phosphorylation at levels far greater than other 2KNS4B molecules, for example, from TBEV (Fig. 3C), unless their roles differ in membrane alteration and potentially cholesterol metabolism, which seems unlikely. Thus, a more specific mechanism of NS4B-mediated IFN antagonism may exist.
The use of multiple proteins to suppress IFN-mediated JAK-STAT signaling, as well as using one relatively conserved protein to target this pathway using different mechanisms, is not unique to the flaviviruses. The best described examples of this are the paramyxoviruses, a large family of negative-stranded RNA viruses that includes several important human pathogens such as measles virus, mumps virus, and NiV. The V protein from mumps virus targets both STAT1 and STAT3 for proteasomal degradation (41, 57) whereas the simian virus 5 V protein degrades only STAT1 (10, 56), and the type II human parainfluenza virus V protein degrades only STAT2 (43, 56). The NiV P gene encodes four proteins, P, V, W, and C, all capable of functioning in IFN antagonism (21, 45). NiV V and P proteins sequester STAT1 and STAT2 in the cytoplasm in high-molecular-weight complexes (49, 52), whereas the W protein, which shares a common N terminus with P and V, sequesters unphosphorylated STAT1 in the nucleus (52). As has been speculated for NiV (52), encoding multiple IFN antagonists may be associated with the high virulence of some flaviviruses or contribute to their broad host range (both invertebrate and vertebrate) by overcoming IFN responses from multiple species.
The most outstanding question raised by the current study, given the clear effect of the S653F mutation on NS5-mediated IFN antagonism, is what is its role in WNV virulence? We are currently addressing this question in the mouse model. Interestingly, in a comparison of sequences from WNV strains of high and low virulence in humans, the virulent SPU116/89 strain (WNV lineage II, isolated from a fatal infection in South Africa) had a number of variable residues in NS5 (7). Four out of five of these (residues 623, 635, 641, and 643) map within the same pocket on NS5 as residue 653 (data not shown). Thus, we speculate that this virus may have an increased capacity to suppress IFN responses compared to its closely related but less virulent South African strains. A greater understanding of the precise roles of specific residues required for IFN antagonism by WNV NS5 will shed light on their role in virulence and could be exploited in the development of live attenuated vaccines or antiviral therapeutics.
Acknowledgments
We thank Anita Mora for graphical expertise and members of the laboratory for critical reviews of the manuscript. We thank Richard Kinney, Pei Y. Shi, Megan Shaw, Luis Martinez-Sobrido, Alexander Pletnev, and Bob Tesh for kindly providing reagents and Richard Cádagan and Roselyn Hallett for excellent technical assistance.
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID) and by grants 5UO1AI66321 and U54 AI057158 from NIAID, NIH, to A.A.K. and A.G.-S., respectively. M.L.-R. is the recipient of an NIH fellowship (FAI077333A).
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Arjona, A., M. Ledizet, K. Anthony, N. Bonafe, Y. Modis, T. Town, and E. Fikrig. 2007. West Nile virus envelope protein inhibits dsRNA-induced innate immune responses. J. Immunol. 179:8403-8409. [DOI] [PubMed] [Google Scholar]
- 2.Ashour, J., M. Laurent-Rolle, P. Y. Shi, and A. Garcia-Sastre. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beasley, D. W., C. T. Davis, J. Estrada-Franco, R. Navarro-Lopez, A. Campomanes-Cortes, R. B. Tesh, S. C. Weaver, and A. D. Barrett. 2004. Genome sequence and attenuating mutations in West Nile virus isolate from Mexico. Emerg. Infect. Dis. 10:2221-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beasley, D. W., M. C. Whiteman, S. Zhang, C. Y. Huang, B. S. Schneider, D. R. Smith, G. D. Gromowski, S. Higgs, R. M. Kinney, and A. D. Barrett. 2005. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J. Virol. 79:8339-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S. M., D. N. Mitzel, and M. E. Bloom. 2006. Action and reaction: the arthropod-borne flaviviruses and host interferon responses. Future Virol. 1:447-459. [Google Scholar]
- 6.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botha, E. M., W. Markotter, M. Wolfaardt, J. T. Paweska, R. Swanepoel, G. Palacios, L. H. Nel, and M. Venter. 2008. Genetic determinants of virulence in pathogenic lineage 2 West Nile virus strains. Emerg. Infect. Dis. 14:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, J. X., H. Ni, M. R. Wills, G. A. Campbell, B. K. Sil, K. D. Ryman, I. Kitchen, and A. D. Barrett. 1995. Passage of Japanese encephalitis virus in HeLa cells results in attenuation of virulence in mice. J. Gen. Virol. 76:2757-2764. [DOI] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, R. A., A. A. Khromykh, J. M. Mackenzie, J. H. Scherret, T. I. Khromykh, and J. S. Mackenzie. 1999. Loss of dimerisation of the nonstructural protein NS1 of Kunjin virus delays viral replication and reduces virulence in mice, but still allows secretion of NS1. Virology 264:66-75. [DOI] [PubMed] [Google Scholar]
- 13.Hall, R. A., S. E. Tan, B. Selisko, R. Slade, J. Hobson-Peters, B. Canard, M. Hughes, J. Y. Leung, E. Balmori-Melian, S. Hall-Mendelin, K. B. Pham, D. C. Clark, N. A. Prow, and A. A. Khromykh. 2009. Monoclonal antibodies to the West Nile virus NS5 protein map to linear and conformational epitopes in the methyltransferase and polymerase domains. J. Gen. Virol. 90:2912-2922. [DOI] [PubMed] [Google Scholar]
- 14.Ho, L. J., L. F. Hung, C. Y. Weng, W. L. Wu, P. Chou, Y. L. Lin, D. M. Chang, T. Y. Tai, and J. H. Lai. 2005. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J. Immunol. 174:8163-8172. [DOI] [PubMed] [Google Scholar]
- 15.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ihle, J. N. 1995. The Janus protein tyrosine kinase family and its role in cytokine signaling. Adv. Immunol. 60:1-35. [DOI] [PubMed] [Google Scholar]
- 17.Jones, M., A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79:5414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadowaki, N. 2009. The divergence and interplay between pDC and mDC in humans. Front Biosci. 14:808-817. [DOI] [PubMed] [Google Scholar]
- 19.Keller, B. C., B. L. Fredericksen, M. A. Samuel, R. E. Mock, P. W. Mason, M. S. Diamond, and M. Gale, Jr. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 80:9424-9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni, S., V. Volchkova, C. F. Basler, P. Palese, V. E. Volchkov, and M. L. Shaw. 2009. Nipah virus edits its P gene at high frequency to express the V and W proteins. J. Virol. 83:3982-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, J. J., C. L. Liao, J. T. Liao, Y. L. Lee, and Y. L. Lin. 2009. A Japanese encephalitis virus vaccine candidate strain is attenuated by decreasing its interferon antagonistic ability. Vaccine 27:2746-2754. [DOI] [PubMed] [Google Scholar]
- 23.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbach, B. D., and C. M. Rice. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23-61. [DOI] [PubMed] [Google Scholar]
- 26.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of Interferon Signaling by the New York 99 Strain and Kunjin Subtype of West Nile Virus Involves Blockage of STAT1 and STAT2 Activation by Nonstructural Proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobigs, M., A. Mullbacher, Y. Wang, M. Pavy, and E. Lee. 2003. Role of type I and type II interferon responses in recovery from infection with an encephalitic flavivirus. J. Gen. Virol. 84:567-572. [DOI] [PubMed] [Google Scholar]
- 28.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 30.Mackenzie, J. M., A. A. Khromykh, and R. G. Parton. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host. Microbe 2:229-239. [DOI] [PubMed] [Google Scholar]
- 31.Malet, H., M. P. Egloff, B. Selisko, R. E. Butcher, P. J. Wright, M. Roberts, A. Gruez, G. Sulzenbacher, C. Vonrhein, G. Bricogne, J. M. Mackenzie, A. A. Khromykh, A. D. Davidson, and B. Canard. 2007. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 282:10678-10689. [DOI] [PubMed] [Google Scholar]
- 32.Mandl, C. W. 2005. Steps of the tick-borne encephalitis virus replication cycle that affect neuropathogenesis. Virus Res. 111:161-174. [DOI] [PubMed] [Google Scholar]
- 33.Mazzon, M., M. Jones, A. Davidson, B. Chain, and M. Jacobs. 2009. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 200:1261-1270. [DOI] [PubMed] [Google Scholar]
- 34.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873-8882. [DOI] [PubMed] [Google Scholar]
- 35.Miller, S., S. Sparacio, and R. Bartenschlager. 2006. Subcellular localization and membrane topology of the Dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854-8863. [DOI] [PubMed] [Google Scholar]
- 36.Mitzel, D. N., S. M. Best, M. F. Masnick, S. F. Porcella, J. B. Wolfinbarger, and M. E. Bloom. 2008. Identification of genetic determinants of a tick-borne flavivirus associated with host-specific adaptation and pathogenicity. Virology 381:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni, H., and A. D. Barrett. 1996. Molecular differences between wild-type Japanese encephalitis virus strains of high and low mouse neuroinvasiveness. J. Gen. Virol. 77:1449-1455. [DOI] [PubMed] [Google Scholar]
- 40.Ni, H., G. J. Chang, H. Xie, D. W. Trent, and A. D. Barrett. 1995. Molecular basis of attenuation of neurovirulence of wild-type Japanese encephalitis virus strain SA14. J. Gen. Virol. 76:409-413. [DOI] [PubMed] [Google Scholar]
- 41.Nishio, M., D. Garcin, V. Simonet, and D. Kolakofsky. 2002. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology 300:92-99. [DOI] [PubMed] [Google Scholar]
- 42.Nitayaphan, S., J. A. Grant, G. J. Chang, and D. W. Trent. 1990. Nucleotide sequence of the virulent SA-14 strain of Japanese encephalitis virus and its attenuated vaccine derivative, SA-14-14-2. Virology 177:541-552. [DOI] [PubMed] [Google Scholar]
- 43.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 44.Park, G. S., K. L. Morris, R. G. Hallett, M. E. Bloom, and S. M. Best. 2007. Identification of residues critical for the interferon antagonist function of Langat virus NS5 reveals a role for the RNA-dependent RNA polymerase domain. J. Virol. 81:6936-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Percy, N., W. S. Barclay, A. Garcia-Sastre, and P. Palese. 1994. Expression of a foreign protein by influenza A virus. J. Virol. 68:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 48.Rahal, J. J., J. Anderson, C. Rosenberg, T. Reagan, and L. L. Thompson. 2004. Effect of interferon-α2b therapy on St. Louis viral meningoencephalitis: clinical and laboratory results of a pilot study. J. Infect. Dis. 190:1084-1087. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roosendaal, J., E. G. Westaway, A. Khromykh, and J. M. Mackenzie. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samuel, M. A., and M. S. Diamond. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 79:13350-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw, M. L., A. Garcia-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 54.Shresta, S., J. L. Kyle, H. M. Snider, M. Basavapatna, P. R. Beatty, and E. Harris. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 78:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solomon, T., N. M. Dung, B. Wills, R. Kneen, M. Gainsborough, T. V. Diet, T. T. Thuy, H. T. Loan, V. C. Khanh, D. W. Vaughn, N. J. White, and J. J. Farrar. 2003. Interferon alfa-2a in Japanese encephalitis: a randomised double-blind placebo-controlled trial. Lancet 361:821-826. [DOI] [PubMed] [Google Scholar]
- 56.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 57.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Werme, K., M. Wigerius, and M. Johansson. 2008. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell Microbiol. 10:696-712. [DOI] [PubMed] [Google Scholar]