Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) accessory protein 6 (p6) is a 63-amino-acid multifunctional Golgi-endoplasmic reticulum (ER) membrane-associated protein, with roles in enhancing virus replication and in evading the innate immune response to infection by inhibiting STAT1 (signal transducer and activator of transcription factor 1) translocation to the nucleus. Here, we demonstrate that p6 has an N-terminal region-cytoplasm-C-terminal region-cytoplasm configuration with residues 2 to 37 likely membrane embedded. Expression of p6, or of its N-terminal 41-amino-acid region, in the absence of other viral proteins, induced the formation of membranous structures, some of which were similar to double membrane vesicles involved in virus replication. Consistent with a role in virus replication, p6 partially colocalized with nonstructural protein 3 (nsp3), a marker for virus replication complexes. Further, while the C-terminal region is required for preventing STAT1 translocation to the nucleus, our results also indicated that the N-terminal 18 amino acids were necessary for maximal inhibition. Collectively, these results support the notion that p6 is a two-domain protein, although the function of each is not completely independent of the other.
The severe acute respiratory syndrome coronavirus (SARS-CoV) was the causative agent for a severe respiratory disease outbreak that affected approximately 8,000 patients in 2002 to 2003. Ten percent of infected patients died, with the highest mortality occurring in those over 60 years of age (35). SARS-CoV, a group 2 coronavirus, shares many characteristics with other coronaviruses, including a large positive-strand RNA genome and a novel mechanism of discontinuous transcription (27, 37). All coronaviruses encode at least four structural proteins (spike [S], nucleocapsid [N], transmembrane [M], and envelope [E]) and several accessory proteins of largely unknown function that are interspersed between and contained within the structural proteins at the 3′ end of the genome (26, 42). SARS-CoV encodes eight accessory proteins (reviewed in reference 30). Four of these proteins appear to be virion associated, although this is controversial (13, 14, 16, 32, 39). These accessory proteins are not required for virus replication in tissue culture cells when cells are infected at high multiplicities of infection (MOIs) (54). Previous studies have suggested that these proteins have anti- and proapoptotic activities, inhibit interferon (IFN) signaling, and enhance virus virulence (5, 9, 17, 20, 22, 24, 30, 38, 39, 48, 53).
One accessory protein, protein 6 (p6), encoded by open reading frame 6 (ORF6), is a 63-amino-acid membrane-associated protein with a hydrophobic N-terminal region of about 41 amino acids and a hydrophilic C-terminal tail (see Fig. 1A). p6 inhibits IFN signaling by binding via its C-terminal region to karyopherin α2, a nuclear import protein, thereby sequestering it in the cytoplasm and indirectly inhibiting translocation of STAT1 (signal transducer and activator of transcription factor 1) to the nucleus (9). p6 inhibits nuclear import not only of STAT1 but of any cellular protein that contains a classical nuclear localization signal (15).
FIG. 1.
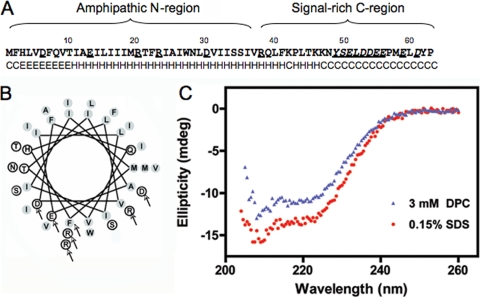
p6 is predicted to contain an α-helix. (A) p6 sequence. Secondary structure predictions are indicated as helical (H), extended (E), or random coil (C). (B) Helical wheel projection of residues 1 to 38 of p6. Shaded circles represent hydrophobic residues while open circles represent hydrophilic residues. Arrows point to each amino acid that was replaced with a cysteine in the cysteine modification assay. (C) Circular dichroism. Far UV CD spectra were recorded as described in Materials and Methods using a protein concentration of 0.285 mg/ml (34.3 μM in 5.2 mM SDS) or 0.415 mg/ml (50 μM in 3 mM DPC). Ellipticity is shown in millidegrees (mdeg).
The N-terminal region of p6 is predicted to form an amphipathic helix. By analogy with other α-helical viral membrane proteins, such as the hepatitis C virus (HCV) nonstructural protein 5A (NS5A) (36), this region might have a role in membrane rearrangement. SARS-CoV replication takes place on membrane scaffolds, largely originating from the endoplasmic reticulum (ER) (19). Recent work suggests that nsp3, nsp4, and nsp6 are involved in the induction of double membrane vesicles (DMV) and convoluted membranes (CM), both involved in coronavirus replication (6, 33, 34). p6 is associated with Golgi membranes and the endoplasmic reticulum and colocalizes with nsp3 and nsp8, involved in virus replication, and with newly synthesized viral RNA (49). Deletion of p6 from the SARS-CoV genome results in a delay in the kinetics of virus replication at a low MOI (55). Furthermore, p6 insertion into the genome of an attenuated strain of mouse hepatitis virus (MHV) enhances virus replication in tissue culture cells and virulence in mice (31, 38). In the presence of p6, MHV RNA and protein synthesis is detected at an earlier time postinfection (p.i.), resulting in enhanced production of infectious virus (15, 31, 38). Thus, the N-terminal region of p6 is predicted to have two functions: inducing membrane rearrangement with enhancement of virus replication and stabilization of the C-terminal domain, enabling its binding to karyopherin α2.
MATERIALS AND METHODS
Cells and viruses.
293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) (GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS). Murine 17Cl-1 cells were grown in DMEM supplemented with 5% tryptose phosphate and 5% FBS. Vero E6 cells were grown in DMEM supplemented with 10% FBS, 25 mM HEPES, 2% l-glutamine, and 1% nonessential amino acids. For electron microscopy studies, 293T cells were transfected with 8 μg DNA by calcium phosphate transfection. For immunofluorescence studies or complementation assays, 17Cl-1 or Vero E6 cells, respectively, were transfected with DNA encoding each mutant using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). MHV-A59 was grown in 17Cl-1 cells, and the titers of the virus were determined in HeLa cells expressing the MHV receptor, CEACAM1 (carcinoembryonic antigen-related cell adhesion molecule 1) (51). Recombinant SARS-CoV (rSARS-CoV) or rSARS-CoV in which ORF6 had been deleted (rSARS-CoV-Δ6) was generated and propagated, and the titers of the virus were determined as previously described (55).
Constructions of plasmids expressing p6 mutants.
p6-HA was cloned as described previously and inserted into a pCAGGS vector using standard PCR techniques (38). pCAGGS-based plasmids expressing hemagglutinin-tagged p6 (HA-p6), p6, and mutants Δ3-10 (mutant in which residues 3 to 10 had been deleted), Δ3-18, Δ11-18, Δ17-24, Δ23-30, Δ29-36, Δ35-42, and 7a-HA, were generated by PCR. p6-GFP and its mutants 1-41-GFP, 9-41-GFP, and 40-63-GFP were generated by PCR and insertion into a pEGFP-N1 vector (BD Biosciences Clontech, San Jose, CA). All plasmids were sequenced prior to use in experiments.
Bacterial expression of p6.
A construct expressing p6 flanked by N-terminal T7 and C-terminal His tags was codon optimized for growth in bacteria and produced synthetically (Geneart, Burlingame, CA). The construct was subcloned into a pET21a vector and expressed in Escherichia coli BL21(DE3) cells (Invitrogen, Eugene, OR). Protein was expressed at low levels after autoinduction in high-density cultures (45). T7-p6-His was purified by affinity chromatography using a cobalt-based metal affinity chromatography resin (Talon; Clontech Laboratories Inc., Mountain View, CA). Routinely, 0.1 to 0.5 mg/liter of protein was obtained.
Circular dichroism.
Spectra were recorded using an Aviv 62DS instrument with a 0.1-cm-path-length quartz cuvette and by scanning from 260 to 205 nm in 0.5-nm increments. Five scans were averaged for each sample. The spectra were determined in 50 mM Na2HPO4 (pH.8.0), 100 mM NaCl, and either sodium dodecyl sulfate (SDS) (5.2 mM) or dodecylphosphocholine (DPC) (3 mM).
Cysteine modification assays.
The cysteine modification assay was performed as previously described (50). Briefly, 293T cells were transfected with 4 μg pCAGGS vector expressing either unmodified p6 or a mutant with one of the following residues substituted with a cysteine: F2, D6, E13, R20, R23, D30, R38, or E59. Mutants were generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All proteins were hemagglutinin (HA) tagged. At 20 h posttransfection, the cells were harvested and resuspended in HCN buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 2 mM CaCl2). The cells were treated with either 0.01% digitonin or 1% Triton X-100 and incubated for 20 min at 4°C. Maleimide polyethylene glycol (maleimide PEG) (5 kDa) was added to each reaction mixture at a final concentration of 1 mM and allowed to react for 30 min at 4°C. Dithiothreitol (DTT) (20 mM) was added to each reaction mixture, and samples were incubated for 10 min at 4°C. Triton X-100 (1%) was added to each reaction mixture. Proteins were separated on a 15% SDS-polyacrylamide gel and detected using mouse anti-HA antibody, followed by streptavidin-horseradish peroxidase (HRP)-conjugated anti-mouse antibody (Jackson Immunoresearch).
Immunofluorescence assays.
17Cl-1, 293T, and Vero E6 cells were used in these studies. 17Cl-1 cells were used in studies of localization, since these cells have a well demarcated Golgi apparatus, 293T cells were used in instances where electron microscopy studies were performed in parallel to facilitate high levels of transfection, and Vero E6 cells were used in STAT1 translocation studies to eliminate the effects of endogenously expressed IFN. The cells were cultured on poly-l-lysine-treated coverslips (BD Biosciences), fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) or 0.005% digitonin in 300 mM sucrose, 100 mM KCl, 2.5 mM MgCl2, 1 mM EDTA, and 10 mM HEPES (pH 6.9). HA-tagged p6 was detected using mouse anti-HA antibody (Covance, Berkeley, CA), followed by fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse antibody (Jackson Immunoresearch, West Grove, PA). In some experiments, Golgi membranes were labeled with rabbit antigiantin (Covance) followed by Cy3-conjugated donkey anti-rabbit antibody (Jackson Immunoresearch). The ER was labeled with mouse anti-KDEL (KDEL stands for Lys-Asp-Glu-Leu) (Stressgen, Victoria, British Columbia, Canada) or mouse anti-PDI (PDI stands for protein disulfide isomerase) (Calbiochem, La Jolla, CA), followed by Cy3-conjugated donkey anti-mouse antibody. In STAT1 localization assays, cells were fixed with cold methanol and then detected using polyclonal rabbit anti-phospho-STAT1 (Tyr701) (P-STAT1) (Cell Signaling, Danvers, MA) antibody, followed by Cy3-conjugated anti-rabbit antibody. MHV nsp3 was detected with rabbit anti-nsp3 antibody (kindly provided by Susan Baker, Loyola University), followed by Cy3-conjugated anti-rabbit antibody. SARS-CoV nsp8 was detected with rabbit anti-nsp8 antibody (kindly provided by Mark Denison, Vanderbilt University). Nuclei were stained with ToPro-3 (Molecular Probes, Carlsbad, CA). The cells were examined using a Zeiss confocal microscope. To obtain quantitative information about the levels of expression of the various constructs, the cells were transfected and examined by confocal microscopy at a fixed gain. The intensity of immunofluorescence was determined by visual inspection. Constructs were compared this way in 5 to 10 independent experiments.
SARS-CoV complementation assays.
Vero E6 cells were transfected with 4.0 μg DNA for 20 h, followed by infection with rSARS-CoVΔ6 or rSARS-CoV at an MOI of 0.01 PFU/cell. Infected cells were harvested at 8 h p.i., and viral titers were determined as described previously (55). For controls, cells were transfected with the same amount of empty vector. Transfection efficiency was determined by green fluorescent protein (GFP) expression. This experiment was performed in a biosafety level 3 (BSL3) laboratory.
Electron microscopy.
The cells were fixed with 2.5% glutaraldehyde (in 0.1 M sodium cocadylate buffer [pH 7.4]) overnight at 4°C and then postfixed with 1% osmium tetroxide for 1 h. Following serial alcohol dehydration (50%, 75%, 95%, and 100%), the samples were embedded in Epon 12 (Ted Pella, Redding, CA). Ultramicrotomy was performed, and ultrathin sections (70 nm) were poststained with uranyl acetate and lead citrate. Samples were examined with a JEOL 1230 transmission electron microscope (TEM) (Tokyo, Japan).
Cryoultramicrotomy and immuno-TEM.
The cells were fixed in 4% paraformaldehyde with 0.2% glutaraldehyde in PBS, embedded in 10% gelatin, and frozen by direct immersion in liquid nitrogen. Ultrathin frozen sections (90 to 95 nm) were prepared using a Leica EM UC6 ultramicrotome (Bannockburn, IL) and then loaded onto a Formvar plus carbon-coated 300-mesh Ni grid. p6-GFP was labeled with rabbit anti-GFP antibody (Invitrogen, Eugene, OR), followed by 10-nm gold-conjugated goat anti-rabbit antibody (EMS, Hatfield, PA). In dual-labeling experiments, samples were stained sequentially with anti-GFP antibody and 20-nm gold-conjugated anti-rabbit antibody (Ted Pella), extensively washed, and then blocked with normal goat serum prior to treatment with rabbit anti-nsp3 antibody and 10-nm gold-conjugated anti-rabbit antibody. The grids were treated with methylcellulose and uranyl acetate, and the samples were examined using a JEOL 1230 TEM.
RESULTS
Residues 2 to 37 form an α-helix and are membrane embedded.
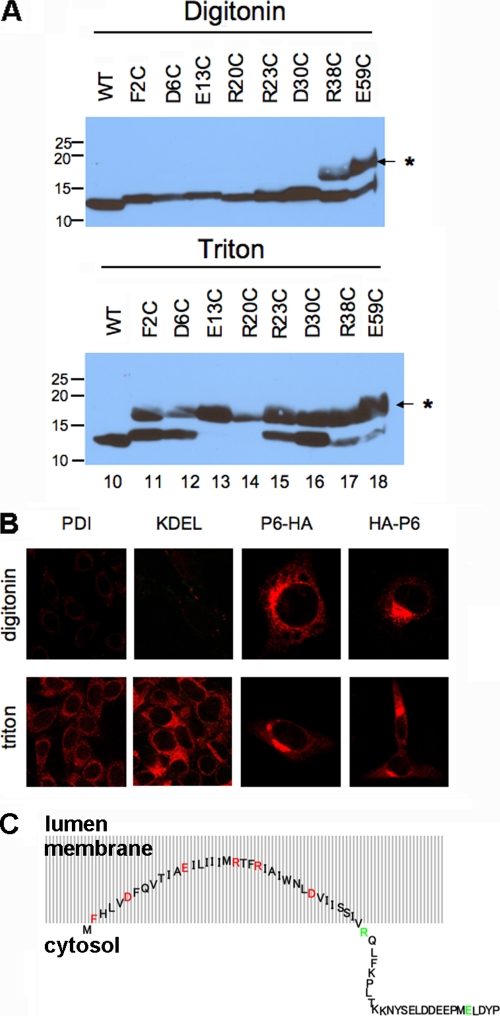
The predicted structure of p6 is an amphipathic α-helix (Fig. 1A and B) (31). To test this prediction, we initially expressed p6 with N- and C-terminal T7 and His tags, respectively, in bacteria and solubilized the resulting protein in membrane mimetics dodecylphosphocholine (DPC) and sodium dodecyl sulfate (SDS). Analysis by circular dichroism suggested that the protein was partially α-helical when solubilized in either detergent (Fig. 1C). The α-helix may form a hairpin or may be positioned parallel to the membrane. If the latter were true, the hydrophilic charged residues located on one side of the helix would likely be accessible to the cytosol. To determine whether these residues were accessible at the cytosol-membrane interface, we individually replaced each charged residue within the N-terminal hydrophobic domain of p6-HA with a cysteine (Fig. 1B). This approach was feasible because unmodified p6-HA contains no cysteines. We then assessed whether the sulfhydryls could be modified by the membrane-impermeable reagent maleimide PEG (5 kDa) after selective membrane permeabilization with digitonin. We also mutated the phenylalanine (F) at position 2 to cysteine to determine how much of the N terminus was exposed to the cytosol. The p6-HA mutants were expressed in 293T cells, and cysteine modification of the proteins was examined by changes in electrophoretic mobility. We found that the cysteines at position R38 and E59 were readily modified after digitonin permeabilization, but E13, R20, R23, and D30 were not accessible to the modifying agent (Fig. 2A). F2 was also not membrane accessible. For a control, all proteins were modified, albeit to different degrees, when the ER membrane was permeabilized with Triton X-100 (Fig. 2A).
FIG. 2.
Residues 2 to 37 of p6 are membrane embedded with both termini in the cytoplasm. (A) Each charged residue within the N-terminal hydrophobic domain of p6-HA, as well as the Phe at position 2, was replaced singly with a cysteine and expressed in 293T cells. The cells were permeabilized with digitonin (top) or Triton X-100 (bottom) and then treated with maleimide PEG (MP) (5 kDa). SDS-PAGE was performed, followed by immunoblotting with anti-HA antibody. The positions of molecular mass standards (in kilodaltons) are indicated to the left of the gels. The position of the MP-modified band is indicated by the asterisk and small arrow to the right of the gels. WT, wild type. (B) 17Cl-1 cells were transfected with p6-HA or HA-p6 for 18 h and then fixed with 4% paraformaldehyde. The cells were permeabilized with either digitonin or Triton X-100 as indicated and then stained with anti-HA antibody (red). For both constructs, the HA tag could be detected with either method of permeabilization. Control experiments were performed by staining cells for two proteins in the lumen, PDI and KDEL. (C) Schematic diagram showing the putative conformation of p6 relative to intracellular membranes. Residues 2 to 37 are buried in the membrane, although the N-terminal amino acids are likely to be near the membrane/cytosol interface. Although drawn as an extended structure, the precise structure is not known, and p6 may actually form a hairpin. Mutated residues were located in the membrane (inaccessible to maleimide PEG [red]) or in the cytosol (accessible [green]).
We previously showed that both the N and C termini were present on the cytoplasmic side of the membrane, using GFP-appended constructs (31). Given the inaccessibility of cysteine at position 2 to maleimide PEG, we reexamined this issue using p6 containing an N-terminal HA tag, since GFP may be large enough to modify p6 orientation. We assayed the accessibility of the HA tag on HA-p6 and p6-HA after selective permeabilization of the outer cell membrane with digitonin. We stained cells with either anti-PDI or anti-KDEL antibodies as controls, as these antibodies detect proteins that are known to localize in the ER lumen (8). PDI and KDEL were readily detected after treatment with Triton X-100, which permeabilizes all cellular membranes, but not after digitonin treatment (Fig. 2B). In contrast, we found that the HA tags of both p6-HA and HA-p6 were accessible after permeabilization with either digitonin or Triton X-100 (Fig. 2B), indicating that both the N and C termini of p6 were localized to the cytoplasmic side of intracellular membranes, in accordance with our previous observations (31).
From the results described above, residue 38 is exposed to the cytoplasm while residues 2 and 30 are membrane embedded. Since the residues from positions 31 to 37 are hydrophobic, they are also likely to be embedded in the membrane (Fig. 2C). The N terminus is probably located at the cytoplasm/membrane interface, since residue 2 was not exposed to the cytoplasm but N-terminal GFP and HA tags were. Further, since the average length of a transmembrane α-helix is about 21 residues (1), the N-terminal anchor region of p6 is too short to cross the membrane twice, making it unlikely that p6 contains a transmembrane domain.
p6 induces membrane rearrangements.
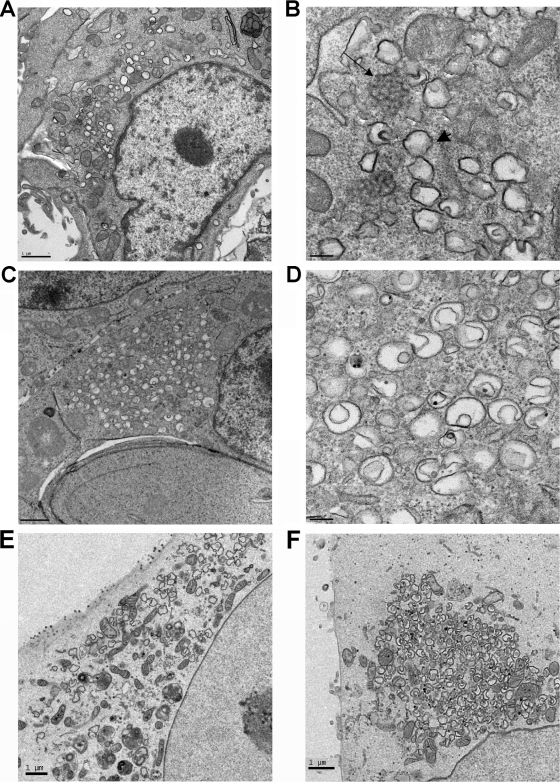
Infection with coronaviruses induces formation of DMV and CM (10, 19, 43) (Fig. 3A and B). These membrane structures are associated with viral RNA synthesis and are required for virus replication. p6 enhances coronavirus replication at early times p.i. and contains an α-helix that may form a hairpin, characteristics found in cellular proteins, such as reticulons which are able to modify membrane curvature (40). Therefore, we reasoned that p6 might function by inducing membrane changes that could serve as sites for virus replication. To examine this possibility, we used transmission electron microscopy (TEM) to visualize any membrane modifications induced by p6. 293T cells were transfected with p6-HA for 24 h and then processed for TEM. Expression of p6-HA induced formation of multiple small vesicles of about 200 to 400 nm in the perinuclear region (Fig. 3C and D). Many of these small vesicles were in various stages of engulfing cytoplasmic material, and in some instances, “pinched-off” membrane vesicles were present within larger vesicles. The size and morphology of these vesicles were very similar to those of DMV formed during viral infection (Fig. 3B and D). These vesicular structures were not detected in cells transfected with empty vector or 7a-HA (data not shown). 7a is a membrane-associated SARS-CoV protein with multiple functions, but it is not believed to be involved in virus replication (30). However, p6 was not required for DMV formation in SARS-CoV-infected cells, because similar numbers of structures were detected in cells infected with SARS-CoV or SARS-CoV-Δ6, lacking p6 expression (Fig. 3E and F).
FIG. 3.
p6-HA-induced formation of small double membrane vesicles. (A and B) 17Cl-1 cells were infected with MHV-A59 for 20 h and prepared for TEM. DMV (arrowhead) and CM (arrow) were readily detected, in agreement with previous results (10, 19, 43). (C and D) 293T cells were transfected with p6-HA for 24 h and then processed for TEM. p6 expression induced formation of vesicles (C and D) that were similar to DMV detected in MHV-infected cells (A and B). (E and F) Vero E6 cells were infected with rSARS-CoV (E) or rSARS-CoV-Δ6 (F). Similar numbers of DMV were detected in cells infected with either virus. Bars, 1 μm (A, C, E, and F) and 100 nm (B and D).
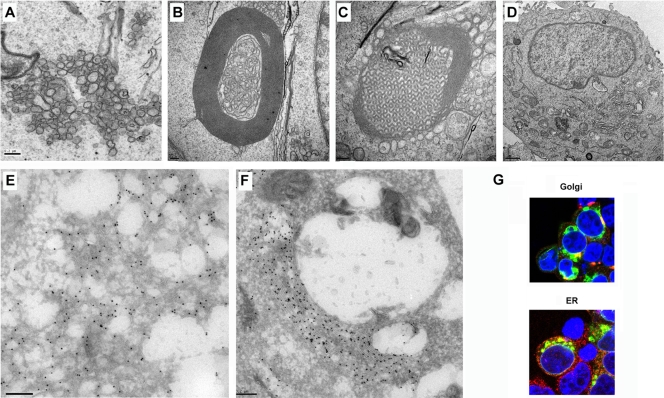
We next examined cells transfected with p6-GFP. p6-GFP was chosen for these studies because GFP molecules are known to weakly dimerize (52). Overexpression of integral ER membrane proteins appended to GFP is sufficient to induce the formation of organized structures, such as stacked cisternae, whorls, and sinusoidal arrays (2, 41). Thus, we reasoned that this property would increase the visibility of any membrane rearrangements induced by p6. In addition, GFP expression provides a stronger signal than antibody detection in immunofluorescence studies, and anti-GFP antibodies are more sensitive and specific than anti-HA antibodies for studies using immunoelectron microscopy (immuno-EM). Transfection of p6-GFP induced extensive membrane rearrangement, including membranous vesicles that resembled DMV (Fig. 4A and E) as well as membranous whorls (Fig. 4B), sinusoidal arrays (Fig. 4C), and stacked cisternae (karmellae and lamellae) (Fig. 4F). These structures were detected in cells transfected with p6-GFP but not in GFP-transfected cells (Fig. 4D). We showed that p6 was associated with these membranous structures by immuno-EM using an anti-GFP antibody (Fig. 4E and F). These membranes were partly derived from the endoplasmic reticulum (ER) and partially derived from Golgi membrane, since p6-GFP colocalized with KDEL (an ER marker) or giantin (a Golgi membrane marker) (Fig. 4G). Of note, only membranous vesicles were detected in p6-HA-transfected cells, suggesting that the GFP appendage was required for the other structures detected in cells transfected with p6-GFP.
FIG. 4.
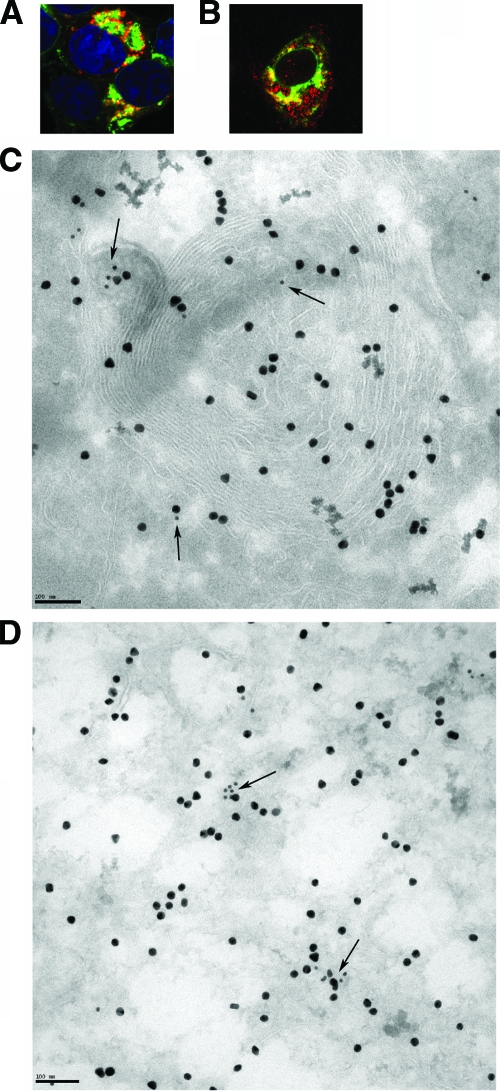
Expression of p6-GFP induced intracellular membrane rearrangement. 293T cells were transfected with p6-GFP (A to C and E to G) or GFP (D) for 24 h and then prepared for TEM (A to D), immuno-EM (E and F), or confocal microscopy (G). Ten-nanometer gold particles indicate the presence of p6-GFP (E and F). p6-GFP induced and is localized with membranous vesicles (A and E), whorls (B), sinusoidal arrays (C), and lamellae (F). Bars, 200 nm (A to C and E and F) and 1 μm (D). (G) p6-GFP induced membrane structures were derived from ER and Golgi membranes. Green shows p6-GFP, red shows giantin (Golgi membrane marker) or KDEL (ER marker), and blue shows nuclei stained with ToPro-3 (blue).
These data suggest that p6 has the ability to induce intracellular membrane rearrangement when expressed from DNA plasmids. To determine the relationship of these membrane structures induced by p6 to virus replication, we transfected cells with p6-GFP and then infected them with the A59 strain of MHV (MHV-A59). We previously showed that p6 enhances growth of an attenuated strain of MHV-JHM (38), but MHV-A59 grows to higher titers than any MHV-JHM strain and does not induce extensive syncytium formation, thus facilitating EM studies. We then examined the colocalization of p6-GFP and nsp3 in infected cells. nsp3 is commonly used as a marker for DMV and for sites of virus replication, although it should be noted that nsp3 is detected in CM and not in the interior of DMV, where the bulk of viral RNA is found (19). By confocal microscopy, we detected a moderate amount of overlap between p6-GFP and nsp3 (Fig. 5A). While DMV and CM were detected in infected cells expressing p6-GFP or GFP, the large membranous structures described above were present only in p6-GFP-transfected cells. p6-GFP, but not nsp3, was detected in these structures (Fig. 5C). p6-GFP and nsp3 were detected in close proximity at other sites in the perinuclear areas in infected cells, most likely on membranes of small vesicles (Fig. 5D). We also showed that p6-GFP colocalized with nsp8, another protein involved in viral replication, in p6-GFP expressing Vero E6 cells that were infected with rSARS-CoV-Δ6 (Fig. 5B).
FIG. 5.
p6 partially overlapped with nsp3. (A, C, and D) 293T cells were transfected with p6-GFP and MHV receptor CEACAM1 for 24 h and then were infected with MHV-A59 for 8 h. (B) Vero E6 cells were transfected with p6-GFP for 24 h and then infected with rSARS-CoV-Δ6 for 8 h. Samples were then processed for confocal microscopy (A and B) or immuno-EM (C and D). Green shows p6-GFP, red shows nsp3 (A) or nsp8 (B), and blue shows nuclei. Original magnification of panels A and B, ×63. (C) p6-GFP was labeled with 20-nm gold, and nsp3 was labeled with 10-nm gold. Arrows point to nsp3. Bar, 100 nm. Note the lack of overlap in areas with p6-GFP-induced whorls (C) but the overlap in regions adjacent to whorls, associated with vesicles (C and D).
Expression of the N-terminal part of p6 was sufficient to complement growth of p6-deleted rSARS-CoV and to induce membrane rearrangement.
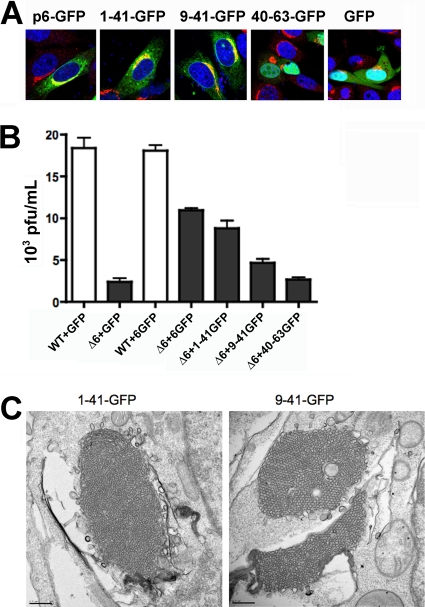
The N-terminal region of p6 is hydrophobic and is predicted to be responsible for the appearance of the membranous vesicles, stacked cisternae, membranous whorls, and sinusoidal arrays described above and for the enhancement of viral replication. To determine whether the N-terminal region could, in fact, perform these functions in the absence of C-terminal sequences, we constructed a set of p6 deletions, including 1-41-GFP, 9-41-GFP, and 40-63-GFP, and used them to complement growth of rSARS-CoV-Δ6, as described previously (55) and in electron microscopy studies. 1-41-GFP and 9-41-GFP showed localization patterns similar to that of p6-GFP, overlapping with the Golgi membrane marker, giantin (Fig. 6A). As expected, deletion of the N-anchor region (40-63-GFP) disrupted its association with membranes so that its localization was indistinguishable from that of GFP (Fig. 6A).
FIG. 6.
Expression of the N-terminal part of p6 was sufficient to complement growth of p6-deleted rSARS-CoV. (A) Localization of p6-GFP, 1-41-GFP, 9-41-GFP, 40-63-GFP, and GFP in 17Cl-1 cells. Green shows GFP constructs, red shows giantin, and blue shows the nuclei. Original magnification, ×63. (B) Vero E6 cells were transfected in triplicate with p6-GFP, 1-41-GFP, 9-41-GFP, 40-63-GFP, or control GFP vector and then infected with rSARS-CoVΔ6 (black bars) for 8 h. GFP- or 6-GFP-transfected cells were also infected with wild-type (WT) rSARS-CoV as controls (white bars). Virus titers were determined as described in Materials and Methods. The titers in 6-GFP-, 1-41-GFP-, and 9-41-GFP-transfected cells were significantly higher than those in GFP-transfected cells (P < 0.05). The titers in 40-63-GFP- and GFP-transfected cells were not significantly different (P > 0.5). Titers in 1-41-GFP- and p6-GFP-transfected cells were not significantly different (ND) (P > 0.5) but were different from those in 9-41-GFP-transfected cells (P < 0.05). The data shown are representative of 2 or 3 individual experiments. Statistical significance was calculated by Student t tests. (C). Expression of 1-41-GFP or 9-41-GFP induced membrane rearrangement in 293T cells. Bars, 0.5 μm.
We previously showed that rSARS-CoV-Δ6 grew to titers lower than those of rSARS-CoV in Vero E6 cells and that this defect was complemented if p6 was provided in trans by expression of p6 tagged with HA (55). GFP-tagged constructs were used in the assays shown in Fig. 6B to facilitate identification of transfected cells prior to SARS-CoV infection. The transfection efficiency was approximately 70% based on GFP expression, and the mutant proteins were expressed at levels similar to that of the wild-type protein (data not shown). Similar to our previous results, we observed that p6-GFP complemented rSARS-CoV-Δ6 growth. We found that p6 expressing only amino acids 1 to 41 complemented rSARS-CoV-Δ6 growth to a level similar to that of full-length p6, and even a construct expressing only residues 9 to 41 showed some enhancing activity (Fig. 6B). As expected, a construct deleted in N-terminal residues 1 to 39 (40-63-GFP) was unable to complement rSARS-CoV-Δ6 at all (Fig. 6B). Consistent with these results, 1-41-GFP and 9-41-GFP both induced extensive membrane rearrangement in transfected cells (Fig. 6C). 40-63-GFP did not enhance rSARS-CoV-Δ6 replication or induce membrane rearrangement (data not shown).
An intact N-terminal region was required for maximal inhibition of STAT1 translocation to the nucleus.
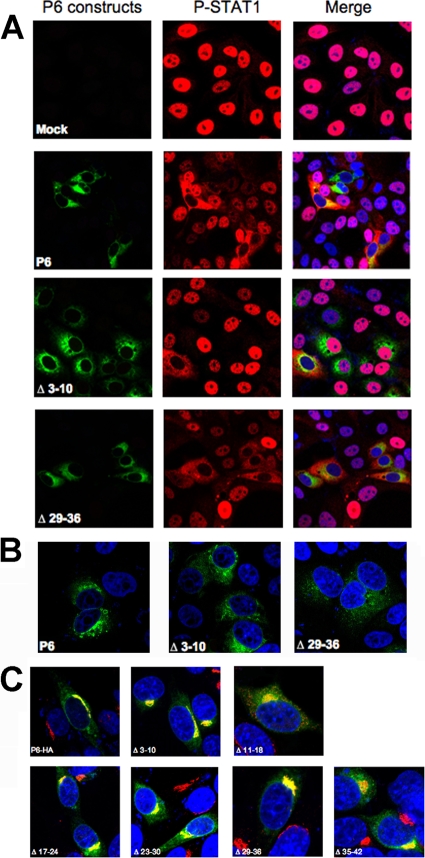
While the C-terminal region of p6 is required for karyopherin α2 binding and inhibition of STAT1 translocation to the nucleus (9), the N-terminal region serves as a membrane anchor. To begin to define the structural requirements for this aspect of p6 function, we constructed a series of staggered 8 amino acid deletion mutants beginning at residue 17 and extending to residue 42. We also included two previously described deletion mutants, encompassing residues 3 to 10 and 11 to 18 in these analyses. We then transfected Vero E6 cells with these p6-HA variants, treated them with 100 units/ml human gamma interferon (hIFN-γ), and then examined the localization of STAT1 using an anti-phosphorylated-STAT1 (P-STAT1) antibody. Vero E6 cells were used in these assays because they lack endogenous IFN expression. The inhibitory effect was incomplete in the case of Δ3-10 and Δ11-18, in which STAT1 remained in the cytoplasm in less than 20% of the cells (Fig. 7A, shown for Δ3-10). All the other mutants were able to inhibit STAT1 translocation equivalently to native p6 (Fig. 7A, only Δ29-36 is shown). Differences in the ability to inhibit STAT1 translocation did not reflect differences in expression levels because Δ17-24, Δ23-30, Δ29-36, and Δ35-42, which were expressed at lower levels than p6-HA, were able to inhibit STAT1 translocation (Fig. 7A and B, only Δ29-36 is shown), whereas Δ3-10, which was expressed at a level similar to that of wild-type p6, did not (Fig. 7A and B). A simple explanation for these results is that the deletion mutants localized differently in cells than native p6. However, we found that deletion of amino acids 3 to 10, 11 to 18, 17 to 24, 23 to 30, 29 to 36, or 35 to 42 did not significantly change the localization pattern, with substantial costaining with the Golgi marker giantin (Fig. 7C). These latter assays were performed in 17Cl-1 cells, because Golgi membrane morphology is less distinct in Vero E6 cells.
FIG. 7.
An intact N-terminal region is required for maximal inhibition of STAT1 translocation to the nucleus. (A) Vero E6 cells were transfected with wild-type or mutant p6-HA for 18 h and then treated with 100 units/ml human IFN-γ for 30 min. (B) Comparison of expression levels of p6, Δ3-10, and Δ29-36 showing that Δ29-36 is expressed at a level lower than that of either p6 or Δ3-10. (C) Localization of p6 mutants in 17Cl-1 cells. Green shows wild-type or mutant p6-HA, red shows P-STAT1 (A) or giantin (C), and blue shows nuclei. Original magnifications, ×40 (A and B) and ×63 (C).
DISCUSSION
Here, we show that SARS-CoV accessory protein p6 is able to distort membrane curvature, providing a possible explanation for its ability to enhance virus replication. p6 expressed with an HA tag induced vesicle formation, but its effects were most striking when expressed as a chimeric protein with GFP. GFP is known to weakly dimerize. When expressed as the cytoplasmic tail of an integral membrane protein, these weak interactions are sufficient to induce extensive membrane rearrangements (2). Expression of p6-GFP most likely anchors GFP molecules in close proximity on membranes, thereby resulting in the formation of the membranous structures shown in Fig. 4. Such structures have been described previously and are believed to be smooth ER generated from non-ribosome-containing parts of the ER (41). They were not detected in cells infected with SARS-CoV or with MHV engineered to express p6, only in cells transfected with p6-GFP prior to infection. These structures were not sites of virus replication, since they did not colocalize with nsp3, a marker of virus replication (Fig. 5C). Rather, nsp3 appeared to overlap partly with p6 in the perinuclear areas where DMV are located (Fig. 5D). Consistent with these findings, p6 overlaps with SARS-CoV nsp8, a putative primase involved in RNA replication (Fig. 5B) (21). Although the most dramatic membrane rearrangement occurs only when p6 is overexpressed, it illustrates the ability of p6 to induce membrane curvature, which may facilitate DMV formation during a natural coronavirus infection.
Since p6 enhances virus replication at early times p.i. and at low MOI (55), this leads us to postulate that p6 is most effective during early stages of infection when a small degree of membrane rearrangement might result in an increase in the number of replication complexes, and as a consequence, a detectable augmentation in viral RNA and protein synthesis. We could not detect a difference in the number and appearance of DMV by TEM when cells infected with rSARS-CoV or rSARS-CoV-Δ6 (Fig. 3E and F) were compared or when MHV expressing p6 or not expressing p6 was compared (data not shown). This likely occurred because the extensive membrane changes that occurred during infection made inapparent the subtle changes induced by the relatively small amounts of p6 present in these cells. It should be noted that while p6 overlap with replication-associated proteins is detected in all studies (Fig. 5) (21, 49), the extent of overlap is variable. This variability may reflect differences in cell strains, detergents used to permeabilize cells, or p6 expression levels. In the presence of higher levels of p6, protein may localize at additional membrane sites within infected cells. Of note, p6 overlaps with the M protein in MHV-infected cells (38). Whether p6 also has a role in virus assembly requires further investigation. An alternative possibility, given its overlap with the M protein, is that virion-incorporated p6 enhances the initiation of viral replication. Although p6 has been detected in SARS-CoV virions (14), we could not detect it in virions released from cells infected with recombinant MHV expressing p6 (49). This suggests that virion-associated p6 is not responsible for the observed augmentation of RNA replication, at least during MHV infection.
Membrane rearrangement is critical for both positive-strand RNA virus replication and the development of host cell structures, including the ER. Another member of the nidovirus family, equine arteritis virus (EAV), uses a replication strategy similar to that of coronaviruses and also induces DMV and CM. nsp2 and nsp3 of EAV, when expressed in transfection assays, are able to induce membrane rearrangements nearly identical to those observed in infected cells (44). Membrane rearrangement as a critical component in RNA virus replication is not limited to nidoviruses, as this phenomenon is also detected in cells infected with picornaviruses and other positive-strand RNA virus viruses (reviewed in reference 28). Expression of poliovirus transmembrane proteins 2BC and 3A, in isolation, is sufficient to induce membrane rearrangements similar to those detected in infected cells (46).
Unlike these viral proteins, p6 does not contain a transmembrane domain. Residues 2 to 37 in the N-terminal region form an α-helix and are buried in the cytoplasmic membrane. It should be noted that the cysteine modification assay (Fig. 2A), used as the basis for this conclusion, has limitations that temper conclusions concerning the geometry of polar residues in the stretch from positions 2 to 37. Amino acids that contain a long alkyl side chain, including arginine, can “snorkel” from deep within membranes to present their polar head groups into the aqueous environment (29). What can be inferred is that the alpha-carbon core of the p6 sequence from positions 2 to 37 is largely occluded from the cytoplasmic environment (Fig. 2C). p6 most likely forms a hairpin, although because of this caveat, it remains possible that p6 lies parallel to the lipid bilayer.
This predicted structure of p6 is closer to that of host cell membrane proteins, such as members of the DP1/reticulon family, and viral proteins, such as HCV nonstructural proteins 4B and 5A and brome mosaic virus (BMV) protein 1a (11, 12, 25, 36, 40). DP1 and reticulon proteins are monotopic and form hairpins, which are postulated to cause sufficient membrane distortion to result in curvature (40). HCV NS4B, NS5A, and BMV protein 1a have also been shown to contain an α-helix and induce membrane curvature involved in formation of viral replication complexes. Among them, HCV NS5A, also a monotopic protein, is postulated to lie in the cytoplasmic leaflet of the intracellular membrane (3, 36). The localization patterns of NS5A and p6 are quite similar, with both overlapping Golgi-ER markers (3) (Fig. 4G). There are also functional similarities, since both proteins facilitate viral replication and antagonize the IFN response, although through different mechanisms. HCV NS5A antagonizes the IFN response by suppressing STAT1 phosphorylation in hepatocyte-derived cell lines and inhibiting IFN-induced protein kinase PKR activation (23, 47).
In addition to enhancing virus replication, p6 interferes with IFN signaling by binding karyopherin α2 via its C-terminal hydrophilic residues, thereby indirectly inhibiting STAT1 translocation to the nucleus (9). Our results show that the N-terminal membrane-associated region is also required for this function, but no region of 8 amino acids in the N terminus is absolutely required for this function (Fig. 7A). However, deletion of residues 3 to 10 or 11 to 18 resulted in less inhibition of STAT1 translocation compared to the other deletions (Fig. 7A). An α-helix of variable length is predicted to be present in all of the 8 amino acid deletions that we analyzed, including Δ3-10 and Δ11-18 (NNPREDICT and PSIPRED) (4, 7, 18). Further, our results showed that none of these deletions significantly changed the localization of p6. It is possible that deletions at the N-terminal region affect an additional function of p6, such as interacting with other host or viral proteins, thereby decreasing p6's ability to inhibit STAT1 nuclear translocation. Further, the fact that 9-41-GFP localized similarly to 1-41-GFP but only partially complemented the growth of SARS CoV-Δ6 (Fig. 6) is consistent with an additional role for the N-terminal region.
In conclusion, our results show that the N-terminal region of p6 is sufficient to induce membrane rearrangement and enhance virus replication. The effects of p6 on SARS-CoV replication are relatively modest in tissue culture cells and in mice. However, this protein has been conserved as the virus jumped species from bats to infect palm civets and humans, suggesting that this protein has an important role in viral infection in its natural host.
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health (PO1 AI060699). D.F. was supported by an NIH training grant (T32 HL07638).
We thank Lei Shi and Dwight Look for providing human IFN-γ.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Arkin, I. T., and A. T. Brunger. 1998. Statistical analysis of predicted transmembrane alpha-helices. Biochim. Biophys. Acta 1429:113-128. [DOI] [PubMed] [Google Scholar]
- 2.Borgese, N., M. Francolini, and E. Snapp. 2006. Endoplasmic reticulum architecture: structures in flux. Curr. Opin. Cell Biol. 18:358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 4.Bryson, K., L. J. McGuffin, R. L. Marsden, J. J. Ward, J. S. Sodhi, and D. T. Jones. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36-W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. Y., Y. H. Ping, H. C. Lee, K. H. Chen, Y. M. Lee, Y. J. Chan, T. C. Lien, T. S. Jap, C. H. Lin, L. S. Kao, and Y. M. Chen. 2007. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 196:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clementz, M. A., A. Kanjanahaluethai, T. E. O'Brien, and S. C. Baker. 2008. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375:118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davoust, N., J. Jones, P. F. Stahel, R. S. Ames, and S. R. Barnum. 1999. Receptor for the C3a anaphylatoxin is expressed by neurons and glial cells. Glia 26:201-211. [DOI] [PubMed] [Google Scholar]
- 8.Ellgaard, L., M. Molinari, and A. Helenius. 1999. Setting the standards: quality control in the secretory pathway. Science 286:1882-1888. [DOI] [PubMed] [Google Scholar]
- 9.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81:9812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouttenoire, J., V. Castet, R. Montserret, N. Arora, V. Raussens, J. M. Ruysschaert, E. Diesis, H. E. Blum, F. Penin, and D. Moradpour. 2009. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J. Virol. 83:6257-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouttenoire, J., R. Montserret, A. Kennel, F. Penin, and D. Moradpour. 2009. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J. Virol. 83:11378-11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, C., N. Ito, C. T. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 80:7287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, C., C. J. Peters, and S. Makino. 2007. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 81:5423-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain, S., S. Perlman, and T. M. Gallagher. 2008. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J. Virol. 82:7212-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 79:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan, S., B. C. Fielding, T. H. Tan, C. F. Chou, S. Shen, S. G. Lim, W. Hong, and Y. J. Tan. 2006. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 122:20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kneller, D. G., F. E. Cohen, and R. Langridge. 1990. Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 214:171-182. [DOI] [PubMed] [Google Scholar]
- 19.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, P., V. Gunalan, B. Liu, V. T. Chow, J. Druce, C. Birch, M. Catton, B. C. Fielding, Y. J. Tan, and S. K. Lal. 2007. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology 366:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, C. W., Z. R. Chan, D. G. Yang, W. H. Lo, Y. K. Lai, M. D. Chang, and Y. C. Hu. 2006. Accelerated induction of apoptosis in insect cells by baculovirus-expressed SARS-CoV membrane protein. FEBS Lett. 580:3829-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan, K. H., K. L. Lan, W. P. Lee, M. L. Sheu, M. Y. Chen, Y. L. Lee, S. H. Yen, F. Y. Chang, and S. D. Lee. 2007. HCV NS5A inhibits interferon-alpha signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. J. Hepatol. 46:759-767. [DOI] [PubMed] [Google Scholar]
- 24.Law, P. T., C. H. Wong, T. C. Au, C. P. Chuck, S. K. Kong, P. K. Chan, K. F. To, A. W. Lo, J. Y. Chan, Y. K. Suen, H. Y. Chan, K. P. Fung, M. M. Waye, J. J. Sung, Y. M. Lo, and S. K. Tsui. 2005. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 86:1921-1930. [DOI] [PubMed] [Google Scholar]
- 25.Liu, L., W. M. Westler, J. A. den Boon, X. Wang, A. Diaz, H. A. Steinberg, and P. Ahlquist. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5:e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 27.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra, V. K., M. N. Palgunachari, J. P. Segrest, and G. M. Anantharamaiah. 1994. Interactions of synthetic peptide analogs of the class A amphipathic helix with lipids. Evidence for the snorkel hypothesis. J. Biol. Chem. 269:7185-7191. [PubMed] [Google Scholar]
- 30.Narayanan, K., C. Huang, and S. Makino. 2008. SARS coronavirus accessory proteins. Virus Res. 133:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Netland, J., D. Ferraro, L. Pewe, H. Olivares, T. Gallagher, and S. Perlman. 2007. Enhancement of murine coronavirus replication by severe acute respiratory syndrome coronavirus protein 6 requires the N-terminal hydrophobic region but not C-terminal sorting motifs. J. Virol. 81:11520-11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuman, B. W., J. S. Joseph, K. S. Saikatendu, P. Serrano, A. Chatterjee, M. A. Johnson, L. Liao, J. P. Klaus, J. R. Yates III, K. Wuthrich, R. C. Stevens, M. J. Buchmeier, and P. Kuhn. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82:5279-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oostra, M., M. C. Hagemeijer, M. van Gent, C. P. Bekker, E. G. Te Lintelo, P. J. Rottier, and C. A. de Haan. 2008. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 82:12392-12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oostra, M., E. G. te Lintelo, M. Deijs, M. H. Verheije, P. J. Rottier, and C. A. de Haan. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 81:12323-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peiris, J. S., Y. Guan, and K. Y. Yuen. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88-S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279:40835-40843. [DOI] [PubMed] [Google Scholar]
- 37.Perlman, S., and J. Netland. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pewe, L., H. Zhou, J. Netland, C. Tangadu, H. Olivares, L. Shi, D. Look, T. M. Gallagher, and S. Perlman. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaecher, S. R., J. M. Mackenzie, and A. Pekosz. 2007. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 81:718-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata, Y., G. K. Voeltz, and T. A. Rapoport. 2006. Rough sheets and smooth tubules. Cell 126:435-439. [DOI] [PubMed] [Google Scholar]
- 41.Snapp, E. L., R. S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 45.Studier, F. W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207-234. [DOI] [PubMed] [Google Scholar]
- 46.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 48.Tan, Y. X., T. H. Tan, M. J. Lee, P. Y. Tham, V. Gunalan, J. Druce, C. Birch, M. Catton, N. Y. Fu, V. C. Yu, and Y. J. Tan. 2007. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 81:6346-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tangudu, C., H. Olivares, J. Netland, S. Perlman, and T. Gallagher. 2007. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 81:1220-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voeltz, G. K., W. A. Prinz, Y. Shibata, J. M. Rist, and T. A. Rapoport. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573-586. [DOI] [PubMed] [Google Scholar]
- 51.Williams, R. K., G. Jiang, and K. V. Holmes. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 88:5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, F., L. G. Moss, and G. N. Phillips, Jr. 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14:1246-1251. [DOI] [PubMed] [Google Scholar]
- 53.Ye, Z., C. K. Wong, P. Li, and Y. Xie. 2008. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochim. Biophys. Acta 1780:1383-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, J., A. Falcon, H. Zhou, J. Netland, L. Enjuanes, P. Perez Brena, and S. Perlman. 2009. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 83:2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]