Abstract
Poxvirus host tropism at the cellular level is regulated by virus-encoded host range proteins acting downstream of virus entry. The functioning mechanisms of most host range proteins are unclear, but many contain multiple ankyrin (ANK) repeats, a motif that is known for ligand interaction through a concave surface. We report here the crystal structure of one of the ANK repeat-containing host range proteins, the vaccinia virus K1 protein. The structure, at a resolution of 2.3 Å, showed that K1 consists entirely of ANK repeats, including seven complete ones and two incomplete ones, one each at the N and C terminus. Interestingly, Phe82 and Ser83, which were previously shown to be critical for K1's function, are solvent exposed and located on a convex surface, opposite the consensus ANK interaction surface. The importance of this convex surface was further supported by our additional mutagenesis studies. We found that K1's host range function was negatively affected by substitution of either Asn51 or Cys47 and completely abolished by substitution of both residues. Cys47 and Asn51 are also exposed on the convex surface, spatially adjacent to Phe82 and Ser83. Altogether, our data showed that K1 residues on a continuous convex ANK repeat surface are critical for the host range function, suggesting that K1 functions through ligand interaction and does so with a novel ANK interaction surface.
Poxviruses are large DNA viruses that dedicate a significant portion of their coding capacity for modulating the host antiviral responses, thereby creating a suitable environment for viral replication (9). They encode so-called host range proteins, which are required for their replication in some but not all cell types, presumably because these proteins modulate some cell type-specific antiviral responses (14, 34). K1 and C7 are two critical host range proteins that are encoded by vaccinia virus (VACV), the prototypical orthopoxvirus (22). The deletion of both K1L and C7L genes from VACV results in abortive replication of the mutant in many mammalian cells and severe attenuation in mice (6, 8, 15, 24). In most mammalian cells, K1, C7, and the cowpox virus CP77 proteins function as equivalent host range factors, since any one of the three could rescue the replication defect of ΔK1LΔC7L VACV mutant (24). A notable exception is rabbit kidney RK13 cells, where either K1 or CP77 could rescue the mutant but C7 could not (24). The mechanism by which K1/C7/CP77 facilitate VACV replication in mammalian cells is unclear. Both K1 and CP77 were found to inhibit the activation of NF-κB in response to VACV infection (3, 27). However, inhibition of host NF-κB activation is neither necessary nor sufficient for the function of CP77 as the host range factor (3). More recently, K1 and C7 were found to antagonize antiviral activities induced by type I interferons, and this function of K1 appears to correlate with its function as host range factor (16).
Many poxvirus host range proteins, including K1 and CP77, contain multiple ankyrin (ANK) repeats, a 33-residue motif that is only known for a function in protein-protein interactions (26). Recently, a 68-kDa protein that is highly conserved in all orthopoxviruses was found to be a host range factor for VACV strain MVA, and it is the only ANK repeat protein that is preserved in MVA (29, 30). Besides orthopoxvirses, many other vertebrate poxviruses also encode multiple ANK repeat proteins (18), some of which are known to be host range factors. For example, the rabbit specific myxoma virus encodes four ANK repeat proteins, and one of them, M-T5, is a well-characterized myxoma host range factor (33). A hallmark for the majority of the poxvirus ANK repeat proteins is the presence of a unique C-terminal F-box-like motif, which interacts with Skp1 and Cullin-1 of the host SCF ubiquitin ligase complex (28).
ANK motif is rarely encountered in viral proteins except the poxvirus proteins, but it is quite common in eukaryotic proteins (21, 26). The 33-residue ANK motif forms a very conserved secondary and tertiary structure, which consists of a canonical helix-loop-helix-β-hairpin/loop fold. The two helices are arranged in an antiparallel mode and followed by a loop region that points outward at a nearly 90° angle. The loop forms a β-hairpin in some cases. Multiple repeats are packed together to form an elongated L-shape structure with a large solvent-accessible surface. The ANK repeat is a very versatile scaffold for creating protein domains displaying specific binding surfaces. Different ANK proteins stack various numbers of the repeats and contain variable surface residues at the repeats, performing diverse biological functions by interacting specifically with their targets (13, 21, 26).
To date, in all determined protein complex structures containing ANK repeat proteins, the interaction occurs through a concave surface formed by the β turn and the first α-helix (1, 13, 21, 26). These residues are positioned at the tip of the β turn and along the exposed surface of the first α helix. However, it is unclear whether poxvirus ANK repeat proteins function similarly. Previously, we performed a mutagenesis study of K1 and identified several residues at the second α-helix of a predicted ANK repeat to be critical for its host range function (17). In the present study, we resolved the crystal structure of K1 at 2.3 Å, which represents the first structure of a viral ANK repeat protein. We found that K1 consists entirely of ANK repeats, lacking any F-box. Furthermore, we performed additional mutagenesis study of K1 and identified residues at the second α-helix of another K1 ANK repeat to be critical for K1's function. In contrast to all previously characterized ANK repeat proteins, the critical K1 residues are all exposed on the convex surface of ANK repeats, suggesting that K1 uses a novel protein-binding mode for its function.
MATERIALS AND METHODS
Protein purification, crystallization, and structure determination.
VACV K1 was cloned into a modified pET28 vector with an N-terminal His6 tag and a Tobacco etch virus protease cleavage site. The recombinant K1 and the selenomethionine-substituted protein were expressed in Escherichia coli and purified by using Ni-NTA as previously described (11). The protein was concentrated to 11 mg ml−1 and crystallized under conditions containing 0.1 M sodium acetate (pH 5.0), 0.2 M sodium tartrate, and 15% (wt/vol) PEG 3350. A total of 20% (vol/vol) glycerol was added to the crystallization condition for cryoprotection. A set of data was collected to 2.3-Å resolution at low temperature (100 K) using synchrotron radiation source at wavelength of selenium K-absorption edge (Advanced Photon Source, beam line 19-ID; Argonne National Laboratory). The crystal belongs to space group P21212 with two molecules in the asymmetric unit. The data were processed at the beam line by using the program HKL3000 (19). Using the same program, 18 of a total of 20 selenium sites were correctly located, and the initial phasing was carried out at the beam line, which yielded clearly interpretable density maps. Subsequently, after iterative noncrystallographic symmetry averaging and density modification by the programs DM (4) and RESOLVE (31), a near-complete model was obtained by using the program Arp/Warp (25). Additional manual modeling was done by using program COOT (7). The structure was refined with the program REFMAC5 (23), and the program PROCHECK (12) was used for the final model analysis. The current model is of excellent geometry and refinement statistics (Table 1). All molecular graphic figures are generated by program PyMOL (5).
TABLE 1.
Crystallographic data and statistics
| Type of dataa | Value(s)b |
|---|---|
| Crystal data | |
| Beamline | 19-ID APS |
| Wavelength (Å) | 0.979345 |
| Space group | P21212 |
| Cell constants | a = 95.1 Å, b = 110.3 Å, c = 86.3 Å, α = 90.0°, β = 90.0°, γ = 90.0° |
| Resolution range (Å) | 50-2.3 |
| Total no. of reflections | 326,470 |
| No. of unique reflections | 37,189 |
| Completeness (%) | 89.3 (67.2) |
| I/σ | 12.4 (3.2) |
| Rsym (%) | 10.5 (48.6) |
| Refinement statistics | |
| Reflection range used (Å) | 39.3-2.3 |
| No. of reflections used | 35,204 |
| Rwork/Rfree (%) | 21.0/25.4 |
| Rmsd bond (Å) | 0.014 |
| Rmsd angle (°) | 1.436 |
| No. of atoms | |
| Protein | 4,533 |
| Waters | 226 |
| Ramachandran plot (preferred/allowed, %) | 96.80/3.20 |
Rsym = Σ | Iobs − Iavg |/Σ Iavg. Rwork = Σ ‖ Fobs | − | Fcalc ‖/Σ | Fobs |. Rfree was calculated using 5% data. I/σ was calculated as averaged data. Rmsd, root mean square deviation.
Values in parentheses are for the highest-resolution shell.
Cells and viruses.
Vero (ATCC CCL-81) and RK-13 (ATCC CCL-37) cells were cultured in minimum essential medium with Earle's balanced salts (Invitrogen) supplemented with 10% fetal bovine serum (FBS). HeLa 229 (ATCC CCL-2.1) cells were cultured in Dulbecco modified Eagle medium (Invitrogen) with 10% FBS. All recombinant viruses were propagated on Vero cells.
Plasmid construction.
The plasmids for making K1L mutants were all derived from pK1L-V5-GFP (17) with recombinant PCR techniques as previously described (17). For each K1L mutation, a pair of mutagenesis primers with complementary sequence were used, together with either primer K1L#3 or K1L#4 (17), in two separate PCRs. The PCR products were then assembled together by recombinant PCR and substituted into the K1L coding sequence of pK1L-V5-GFP using the SspI and MfeI sites. The sequences of all constructed plasmids were confirmed by DNA sequencing with an ABI 3100 Genetic Analyzer.
Recombinant virus construction.
Various K1 mutant viruses were constructed by homologous recombination of vK1L− C7L− with derivatives of pK1L-V5-GFP as described previously (17). Briefly, pK1L-V5-GFP derivatives were transfected into Vero cells that were infected with vK1L− C7L−. Recombinant viruses encoding GFP and V5-tagged K1 were picked under the fluorescence microscope and purified by four rounds of plaque purification on Vero cells.
Growth curve analysis of K1 mutants.
RK13 or HeLa cells in 12-well plates were incubated with 0.01 or 5 PFU of different K1 mutant viruses per cell for 2 h at room temperature. After adsorption, the cells were washed twice with phosphate-buffered saline and moved to a 37°C incubator to initiate viral entry and replication. The cells were harvested at 0 and 48 hpi. The virus titers in the cell lysates were determined by duplicate plaque assays on Vero cells. Infected cells expressing green fluorescent protein (GFP) were also visualized under an inverted fluorescence microscopy using a 10× objective lens and photographed with a digital camera. The exposure time was kept constant when photographing different samples.
Western blot analysis.
Infected cells were harvested 8 h postinfection (hpi) and subjected to Western blot analysis as previously described (17). The detection antibodies were mouse monoclonal antibodies (MAbs) to V5 (Sigma-Aldrich; clone V5-10), MAbs to β-actin (Sigma-Aldrich), and rabbit anti-E3L polyclonal antiserum (kindly provided by Bertram Jacobs).
Data deposition.
The atomic coordinates and structure factors have been deposited with the Protein Data Bank (www.rcsb.org) under PDB accession code 3KEA.
RESULTS
Overall structure of K1.
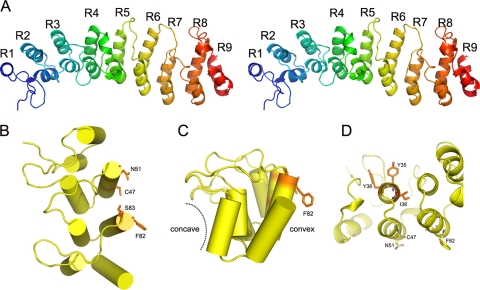
K1 adopts an extended, slightly curved structure with an overall dimension of ca. 80 by 30 by 25 Å (Fig. 1A). It consists entirely of nine ANK repeats, only six of which were predicted by previous primary sequence analysis (17). The internal seven repeats, ANK2 to ANK8, are canonical ANK repeats, each containing approximately 33 amino acid residues that form a helix-loop-helix followed by a loop. The N- and C-terminal ANK repeats (ANK1 and ANK9) are partially truncated. ANK1 has a very short leading helix, whereas ANK9 lacks the loop region. The antiparallel α-helices in each repeat stack on each other side by side, forming a highly packed helical structure. The stack was kinked ∼20° between two segments, ANK1-4 and ANK5-8. The loop regions in the first segment, ANK1-4, are extended at ∼90° to the helices, as observed in the canonical ankyrin repeats. In contrast, the loops in the second segment, ANK5-8, are oriented in the same direction as the α-helices. Therefore, the loop regions in the two segments are rotated ∼90° with respect to each other (Fig. 1A).
FIG. 1.
K1 structure. (A) Stereo view of the overall structure of K1. The secondary structures are shown in rainbow color with N termini in blue and C termini in red, respectively. K1 displays a curved structure containing nine ANK repeats. K1 uses a novel functional surface for ANK repeat proteins. In contrast to the canonical concave surface of ANK repeats, functionally important K1 residues that were identified from this and previous studies (C47, N51, F82, and S83, shown as sticks) are located on the convex surface of ANK2 and ANK3. Notice the four residues are spatially clustered, forming a contiguous interface. (B) Top view; (C) side view. (D) The concave surface on ANK2 is not involved in K1 function. Y35, Y36, and I38 are located on the concave side. Single mutations at either Y35 or Y36 had no effect on K1 function. I38 is completely buried in between the helices of ANK2 and ANK3, playing a structural role.
Previously, we showed that mutations of Phe82 and Ser83 (Phe82Lys and Ser83His) completely abolished K1's function as the host range factor in HeLa and RK13 cells (17). The K1 structure showed clearly that the side chains of both residues are solvent exposed and unlikely to play any structural role in folding the molecule (Fig. 1B, C, and D).
Identification of additional functional residues on K1: a novel functional surface for ANK repeat proteins.
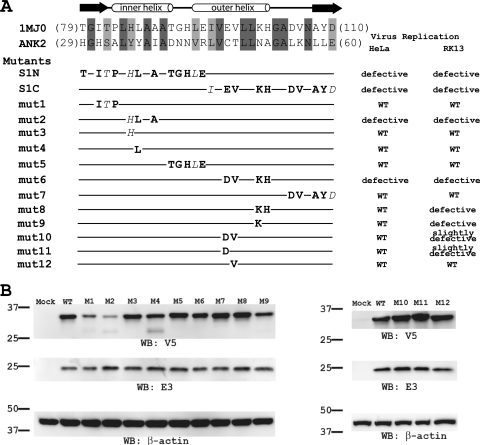
Phe82 and Ser83 locate at the tip of the second helix of ANK3, opposite the concave surface (Fig. 1B and C), which is the consensus ligand interaction surface in all solved protein complex structures containing ANK repeat proteins. This suggests that K1 may instead use the convex back surface for the host range function, which is novel for ANK repeat proteins. To test this idea, we performed a mutagenesis study to define the residues that are critical for K1's function on an ANK repeat that is adjacent to ANK3. This was done by introducing mutations into K1L of a C7L deletion mutant VACV and then assessing the effect of K1L mutation on viral replication in both HeLa and RK13 cells. By using this method, we previously showed that the substitution of ANK2 of K1 with an unrelated synthetic ANK abolished VACV replication in HeLa and RK13 cells (17). Furthermore, the substitutions at either the N- or C-terminal half of ANK2 (referred to as S1N and S1C) also abolished the host range function (17) (Fig. 2A). ANK2 was previously referred to as ANK1, since it was predicted to be the N-terminal K1 ANK repeat (17). The synthetic ANK that was used for the substitution was based on the consensus sequence of many cellular ANKs and is known to fold into the typical ANK structure (10) (PDB accession number 1MJ0). This substitution presumably altered the surface residues of K1 ANK without changing the overall structure, since different ANKs fold into an identical structure with only the surface residues that are unique (13, 26).
FIG. 2.
Mutagenesis study of K1 ANK2. (A) The amino acid sequence of K1 ANK2 is shown in an alignment with the synthetic ANK sequence from 1MJ0, with the secondary structure of the synthetic ANK labeled. The amino acids that were substituted into each K1 mutant are shown below the alignment. The nonconservative and conservative amino acid changes are shown in regular type and italics, respectively. The phenotypes of the mutants in HeLa and RK13 cells are summarized on the right. The mutants were constructed by homologous recombination of vK1L− C7L− and a plasmid encoding C-terminal V5-taged K1 and the GFP. (B) Steady-state level of K1 proteins in Vero cells that had been infected with K1L mutants. Vero cells were infected at an MOI of 5 PFU/cell. The level of K1 proteins at 8 hpi were determined by Western blotting with a monoclonal V5 antibody. On the same blot, the levels of VACV early protein E3 and host cell protein β-actin were also determined. β-Actin serves as a control for gel loading, whereas E3 serves as an indicator of VACV early protein expression. The molecular mass marker is indicated on the left in kilodaltons.
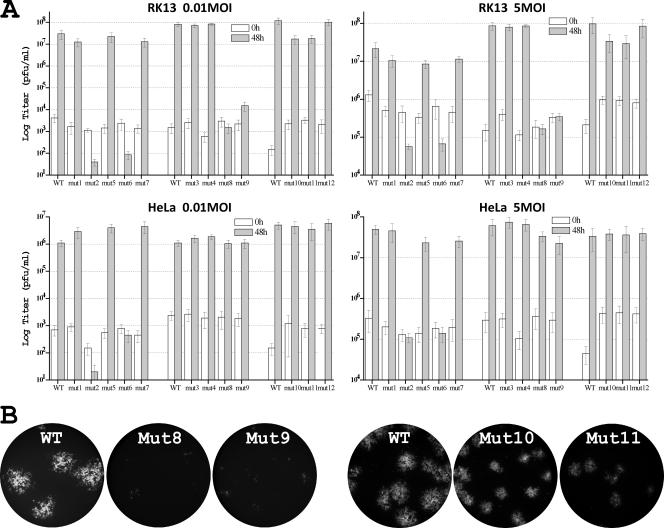
To map the exact residues of ANK2 that mediate the host range function, we introduced increasingly smaller substitutions into ANK2 (Fig. 2A). Since substituting 12 residues at the N-terminal half of ANK2 (S1N) abolished viral replication in HeLa and RK13 cells, we initially constructed three mutant viruses (mut1, mut2, and mut5), each of which contained substitutions of three or five amino acids. All of the mutants replicated efficiently in Vero cells (data not shown), where neither K1L nor C7L was required for VACV replication. mut1 and mut5 also replicated in HeLa and RK13 cells as efficiently as a virus expressing wild-type (WT) K1 (referred to as WT virus here), as measured by a similar increase in virus titers at 48 hpi at both low and high multiplicities of infection (MOIs) (Fig. 3A). However, mut2 failed to replicate in HeLa and RK13 cells, and its virus titer at 48 hpi decreased (Fig. 3A).
FIG. 3.
Phenotypes of K1 mutant VACV in RK13 and HeLa cells. (A) Growth phenotypes of K1 mutants in HeLa and RK13 cells at high (5 PFU/cell) and low (0.01 PFU/cell) MOIs. Virus yields at 0 and 48 hpi were determined by duplicate plaque assay on the permissive Vero cells. The average titers are shown with standard deviations. The mutants were assayed in three separate groups, each of which using a virus expressing WT K1 (WT) as the control. (B) Plaque morphology of K1 substitution mutants in RK13 cells. RK13 cells were infected at an MOI of 0.01 PFU/cell. At ∼36 hpi, GFP-expressing cells were visualized with a fluorescence microscope. The same exposure time was used to photograph different samples. The obtained color images were converted to grayscale, and GFP is shown as white. The mutants were assayed in two separate groups, each of which has the WT control.
In the permissive Vero cells, mut2 was found to have a reduced steady-state K1 protein level (Fig. 2B), indicating that the three amino acid substitutions (Tyr35His, Tyr36Leu, and Ile38Ala) in mut2 may reduce K1 protein stability. Indeed, in K1 structure, Ile38 is completely buried between the first α-helices of ANK1 and ANK2 and could play a critical role in stabilizing the overall integrity of the protein folding by extensive hydrophobic interactions at the helix interface (Fig. 1D). In contrast, Tyr35 and Tyr36 are located on the leading helix of ANK2 and are part of the concave surface of K1 (Fig. 1D). To assess whether the substitution of Tyr35 or Tyr36 could have contributed to the defect of mut2, we constructed two additional mutants (mut3 and mut4) by introducing Tyr35His or Tyr36Leu substitution individually into K1L. Both mutants expressed a WT level of K1 proteins and replicated efficiently in both HeLa and RK13 cells (Fig. 2B and 3A), suggesting that the defect of mut2 was not caused by changes in concave surface residues of K1 but by a reduction in K1 protein stability brought by the triple mutations.
Substituting 10 residues at the C-terminal half of ANK2 (S1C) abolished viral replication in HeLa and RK13 cells, so we initially constructed two mutant viruses (mut6 and mut7) containing four or five amino acid substitutions, respectively (Fig. 2A). Although mut7 replicated similarly to WT virus, mut6 failed to replicate in HeLa and RK13 cells (Fig. 3A). mut6 expressed a WT level of K1 proteins in Vero cells (Fig. 2B), indicating that the mutation did not reduce K1 protein stability. The four K1 residues that were substituted in mut6 (Cys47Asp, Thr48Val, Asn51Lys, and Ala52His) are clustered on the convex surface and completely solvent exposed in the K1 structure (Fig. 1B). The substitution of the first two residues (Cys47Asp and Thr48Val) slightly reduced the ability of the mutant (mut10) to replicate in RK13 cells, whereas the substitution of the latter two residues (Asn51Lys and Ala52His) severely reduced the ability of the mutant (mut8) to replicate in RK13 cells (Fig. 3). The single amino acid substitution Asn51Lys was found to be mainly responsible for the phenotype of mut8, since a mutant with only this substitution (mut9) also had a severe replication defect in RK13 cells. Similarly, the single amino acid substitution Cys47Asp was found to be responsible for the phenotype of mut10, since this substitution in mut11 resulted in a slight replication defect in RK13 cells, whereas a Thr48Val substitution in mut12 had no effect on viral replication (Fig. 3).
Although substituting Cys47 (mut11 or mut10) or Asn51 (mut9 or mut8) individually caused defect in VACV replication in RK13 cells, substituting both residues together in mut6 caused a bigger defect in VACV replication. mut6 failed to express GFP under the control of a late promoter in HeLa and RK13 cells (data not shown), indicating that its replication was blocked before the onset of viral late protein expression. In contrast, mut8, mut9, mut10, and mut11 were all able to express GFP in RK13 cells. mut8 and mut9 were able to form small foci consisted of a few GFP-expressing cells, whereas mut10 and mut11 formed GFP-positive plaques that were smaller in size than the WT (Fig. 3B), indicating that their late protein synthesis and cell-to-cell spread were reduced by different degrees. Furthermore, whereas mut6 failed to replicate in HeLa cells, mut8, mut9, mut10, and mut11 did not have any apparent replication defect in HeLa cells (Fig. 3A). This is consistent with our previous observation that HeLa cells were more tolerant to K1 mutations than RK13 cells (17). Altogether, these data suggest that Cys47 and Asn51 individually as well as synergistically contribute to K1's host range function. In the K1 structure, although Cys47 and Asn51 are on a separate repeat than Phe82 and Ser83, all four residues are spatially adjacent to each other, forming a continuous surface (Fig. 2B).
DISCUSSION
Successful replication of poxviruses in many cell types depends on virus-encoded host range proteins functioning downstream of viral entry (14, 34). Since the host range proteins are dispensable for viral replication in some cell types, they presumably play no direct role in virion biogenesis but either provide an essential function that is lacking in a particular cell type or play an essential role in downmodulating certain cell type-specific antiviral responses. Studying the functions of the host range proteins thus provides an avenue for discovering critical innate antiviral responses as well as viral strategies of evading these responses. However, the host range proteins are among the least understood in the poxvirus proteome (14, 34). In the present study, we solved the crystal structure of VACV K1, representing the first structure for any poxvirus host range proteins. The K1 structure provides a structural perspective for our previous and current mutagenesis studies of K1 (17). Collectively, the structure function studies of K1 offer insights into the molecular mechanisms of the host range proteins.
The structure of K1 shows that K1 consists entirely of ANK repeats (Fig. 1A), suggesting that K1 functions solely through ligand interaction, since ANK repeats are only known for a role in ligand interaction (13, 21, 26). The structure also confirms that K1 has no F box, which is present in the majority of poxvirus ANK repeat proteins but was predicted not to be present in K1 (18). ANK repeat proteins form the largest group of proteins encoded by the poxviruses (28). Many poxvirus ANK repeat proteins are involved in regulating the host range or modulating host signaling pathways (3, 20, 30, 32, 33). A unique feature for many poxvirus ANK repeat proteins is the presence of a C-terminal F-box, which binds to core elements of the ubiquitin ligase complex (28). It was proposed that poxvirus ANK-F-box proteins modulate diverse cellular pathways by targeting different cellular proteins for ubiquitination (18, 28). The ANK repeats presumably bind to specific cellular targets, which are ubiquitinated by the ubiquitin ligase complex that are brought in by the F-box. This, however, is unlikely to be the mechanism of action for K1, since there is no F-box in K1 structure. Similarly, although a functional F-box is present in the host range proteins CP77 and 68k-ank, the F-box is not required for their host range function (3, 30).
The K1 structure provides a much-needed structural perspective on previous and current mutagenesis study of K1. Prior to the elucidation of the atomic-level structure of K1, we performed structure function studies of K1 (17). To avoid introducing mutation that affects structure, we substituted the K1 ANK repeats with a well-defined artificial ANK repeat. The substitution presumably changed the surface residues of K1 ANK repeats without altering the overall structure, since different ANK repeats fold into almost identical structure with only the surface residues that are unique (13, 26). With this approach, we were able to identify Phe82 and Ser83 as residues that are important for K1's function (17). However, it remained possible that the substitution of these two residues might impair K1's host range function through indirect effect on protein structure. Now with the determination of K1 structure, we are able to exclude this possibility, since these two residues are on the surface of the molecule (Fig. 1B), playing no structural role. Similarly, once we identified Cys47 and Asn51 as critical residues for K1's function in the present study, we could see from the structure that these residues are also solvent exposed and unlikely to play any structural role (Fig. 1B). On the other hand, when the substitution of Tyr35, Tyr36, and Ile38 were found to disrupt K1's function and reduced the K1 protein level, we could see from the structure that these residues are involved in inter-repeats interaction and conclude that the substitutions of these residues disrupt K1's function by destabilizing the protein (Fig. 1D).
The K1 residues that were found to be essential for the host range function center on a contiguous surface (Fig. 1B), suggesting that they are part of a binding surface for some ligand. Cys47 and Asn51 are separated in primary sequence from Phe82 and Ser83, but they are spatially adjacent to each other, forming a contiguous surface. We believe that this surface is the binding surface for some protein factor and that the binding free energy is distributed over these four residues. This is probably why substitution of one of these four residues only cause partial defect in the host range function, whereas substitution of more than one residues causes a more severe defect. Previously, we found that substituting Phe82 or Ser83 caused partial defect in the replication of VACV in HeLa cells but that substituting both residues completely abolished viral replication in HeLa cells (17). In the present study, we found that substituting Cys47 only caused minor defect in replication of VACV in RK13 cells, whereas substituting Asn51 caused a more severe defect (Fig. 2). Substituting both residues, along with two other nonessential residues, caused a much more severe replication defect in RK13 and HeLa cells.
All four K1 residues that are critical for the host range function located on the convex back surface of the molecule (Fig. 1C), suggesting that K1 uses a novel model for ligand interaction. In previously studied protein complexes involving ANK repeats, the concave surface is found to bind the target molecules (13, 21, 26). The concave surface consists of stacked α-helices and β turns, at nearly 90° angle to each other (Fig. 1C). The L-shape concave surface could enclose the ligand with two perpendicular surfaces, therefore providing a much tighter binding than the flat back surface could. It is possible that K1 may only need to interact with its cellular target transiently to exert its effect, so the convex back surface of K1 may provide sufficient binding affinity. This may be why we have not been able to isolate any biologically relevant binding partners for K1 by using the affinity purification method. ACAP2, a GTPase-activating protein for ARF6, is the only cellular protein found to interact with K1 (2, 17). However, we showed previously that substitution of both Phe82 and Ser83 abolished the host range function but had no impact on ACAP2 binding (17). Similarly, we found that substituting both Asn51 and Cys47 (in mut6) impaired the host range function but had no effect on ACAP2 binding (data not shown), further supporting that ACAP2 binding is not sufficient for K1's host range function. In addition, we showed previously that the substitutions at ANK5 disrupted ACAP2 binding with no adverse effect on K1's host range function, indicating that ACAP2 binding is also not necessary for the host range function (17). K1 may interact with ACAP2 to affect some cellular processes that are not directly related to the host range restriction, and it may do so with the canonical concave surface.
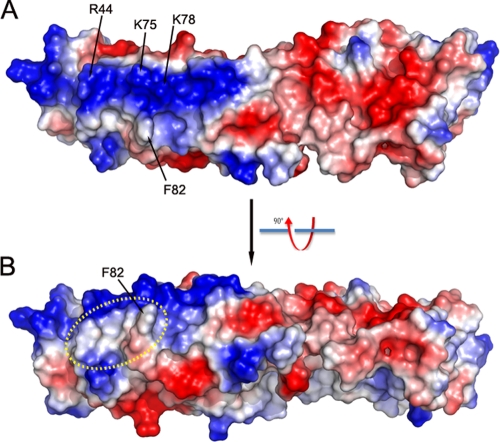
On the convex surface adjacent to the four critical K1 residues, we noticed a large positively charged surface patch comprised of at least six residues (Arg44, Lys75, Lys78, Lys111, Lys115, and Lys116) (Fig. 4). This surface patch, however, does not appear to be involved in specific charge-charge interactions, since reversing the charge of some of the residues did not affect K1's host range function. The substitution of both Lys75 and Lys78 with Glu was previously shown to have no effect on K1's host range function (17). In the present study, we showed that Arg44Glu substitution (in mut5) also had no effect on K1's function (Fig. 2). The four critical K1 residues are clustered in a nearby relatively hydrophobic region (Fig. 4), suggesting that K1-ligand recognition could be rather hydrophobic in nature rather than through charge-charge interactions.
FIG. 4.
Electro-potential of K1 surface. (A) Top view; (B) side view. The positively and negatively charged surfaces are colored blue and red, respectively. Notice the large positively (blue) charged patch on the surface of ANK1-4. The relatively hydrophobic area in dashed circle is, however, involved in K1's host range function.
Some of our K1 mutations showed different effects on VACV replication in RK13 cells than in HeLa cells. mut8 and mut9, which have substitution in Asn51 of K1, failed to replicate in RK13 cells but replicate efficiently in HeLa cells (Fig. 2). mut9 and mut10, which have substitution in Cys47 of K1, replicated with a decreased efficiency in RK13 cells but replicated normally in HeLa cells. Only when both Asn51 and Cys47 were substituted did we observe a defect in replication in both HeLa and RK13 cells. The reason for this discrepant effect of K1 mutations in different cells is unknown, but this is consistently with our previous mutagenesis study of K1 (17). For example, while individual substitution of Phe82 or Ser83 completely abolished viral replication in RK13 cells, it only reduced VACV replication in HeLa cells. RK13 cell is known to be a unique cellular host for VACV. Although both K1 and C7 can support viral replication in many human and murine cell lines, only K1 can do so in RK13 cells (24). We suspect that the host factor targeted by K1 may exist at a higher concentration or with a higher potency in RK13 cells than in HeLa cells, so mutations that slightly decrease the affinity of K1 to this host factor may be able to affect VACV replication in RK13 cells but may not be sufficient to affect VACV replication in HeLa cells. Therefore, RK13 cells serve as a more sensitive host for assessing K1 mutations, allowing us to detect mutations that individually have a minor effect but collectively have a major effect on K1's function.
Both K1 and CP77 were previously reported to inhibit the activation of NF-κB in response to VACV infection (3, 27). However, this function of the host range proteins is not responsible for alleviating the host range restriction of a VACV host range mutant with no K1L and C7L (VV-hr). Chang et al. reported previously that a mutated CP77, which failed to inhibit NF-κB, was still able to rescue the growth restriction of VV-hr in HeLa cells (3). Conversely, knocking down NF-κB protein expression in HeLa cells failed to relieve the growth restriction of VV-hr (3), suggesting that inhibition of host NF-κB activation is neither necessary nor sufficient for rescuing host range restriction of VV-hr. Our unpublished data indicate that, if C7L is absent, K1L is necessary for VACV to inhibit host NF-κB activation in HeLa cells. However, which K1 residues are critical for NF-κB inhibitory function remain to be determined. More recently, we found that K1 and C7 antagonize antiviral activities induced by type I interferons (16). K1 residues that are critical for its host range function were found to be also essential for its ability to antagonize type I interferons (16), suggesting that the interferon-induced antiviral factor that is targeted by K1 is the same factor that restricts the growth of ΔK1LΔC7L VACV mutant in nonpermissive cells.
Acknowledgments
We gratefully acknowledge the staff of beam line 19-ID at the Advanced Photon Source for their support.
This study was supported by NIH grants AI065731 (Y.X.), AI079217 (Y.X.), and AI081928 (J.D.) and by Oklahoma Agricultural Experiment Station at Oklahoma State University under Project OKL02618 (J.D.).
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Binz, H. K., P. Amstutz, A. Kohl, M. T. Stumpp, C. Briand, P. Forrer, M. G. Grutter, and A. Pluckthun. 2004. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 22:575-582. [DOI] [PubMed] [Google Scholar]
- 2.Bradley, R. R., and M. Terajima. 2005. Vaccinia virus K1L protein mediates host-range function in RK-13 cells via ankyrin repeat and may interact with a cellular GTPase-activating protein. Virus Res. 114:104-112. [DOI] [PubMed] [Google Scholar]
- 3.Chang, S. J., J. C. Hsiao, S. Sonnberg, C. T. Chiang, M. H. Yang, D. L. Tzou, A. A. Mercer, and W. Chang. 2009. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-κB activation by tumor necrosis factor alpha but is independent of its host range function. J. Virol. 83:4140-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.DeLano, W. L. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- 6.Drillien, R., F. Koehren, and A. Kirn. 1981. Host range deletion mutant of vaccinia virus defective in human cells. Virology 111:488-499. [DOI] [PubMed] [Google Scholar]
- 7.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 8.Gillard, S., D. Spehner, R. Drillien, and A. Kirn. 1986. Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc. Natl. Acad. Sci. U. S. A. 83:5573-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston, J. B., and G. McFadden. 2003. Poxvirus immunomodulatory strategies: current perspectives. J. Virol. 77:6093-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohl, A., H. K. Binz, P. Forrer, M. T. Stumpp, A. Pluckthun, and M. G. Grutter. 2003. Designed to be stable: crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. U. S. A. 100:1700-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumm, B., X. Meng, Y. Li, Y. Xiang, and J. Deng. 2008. Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein. Proc. Natl. Acad. Sci. U. S. A. 105:20711-20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 13.Li, J., A. Mahajan, and M. D. Tsai. 2006. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45:15168-15178. [DOI] [PubMed] [Google Scholar]
- 14.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng, X., J. Chao, and Y. Xiang. 2008. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 372:372-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng, X., C. Jiang, J. Arsenio, K. Dick, J. Cao, and Y. Xiang. 2009. Vaccinia virus K1L and C7L inhibit antiviral activities induced by type I interferons. J. Virol. 83:10627-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng, X., and Y. Xiang. 2006. Vaccinia virus K1L protein supports viral replication in human and rabbit cells through a cell-type-specific set of its ankyrin repeat residues that are distinct from its binding site for ACAP2. Virology 353:220-233. [DOI] [PubMed] [Google Scholar]
- 18.Mercer, A. A., S. B. Fleming, and N. Ueda. 2005. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 31:127-133. [DOI] [PubMed] [Google Scholar]
- 19.Minor, W., M. Cymborowski, Z. Otwinowski, and M. Chruszcz. 2006. HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62:859-866. [DOI] [PubMed] [Google Scholar]
- 20.Mohamed, M. R., M. M. Rahman, A. Rice, R. W. Moyer, S. J. Werden, and G. McFadden. 2009. Cowpox virus expresses a novel ankyrin repeat NF-κB inhibitor that controls inflammatory cell influx into virus-infected tissues and is critical for virus pathogenesis. J. Virol. 83:9223-9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosavi, L. K., T. J. Cammett, D. C. Desrosiers, and Z. Y. Peng. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moss, B. 2007. Poxviridae: the viruses and their replication, p. 2905-2946. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 23.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 24.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 25.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 26.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 27.Shisler, J. L., and X. L. Jin. 2004. The vaccinia virus K1L gene product inhibits host NF-κB activation by preventing IκBα degradation. J. Virol. 78:3553-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnberg, S., B. T. Seet, T. Pawson, S. B. Fleming, and A. A. Mercer. 2008. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc. Natl. Acad. Sci. U. S. A. 105:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sperling, K. M., A. Schwantes, B. S. Schnierle, and G. Sutter. 2008. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology 374:234-239. [DOI] [PubMed] [Google Scholar]
- 30.Sperling, K. M., A. Schwantes, C. Staib, B. S. Schnierle, and G. Sutter. 2009. The orthopoxvirus 68-kilodalton ankyrin-like protein is essential for DNA replication and complete gene expression of modified vaccinia virus Ankara in nonpermissive human and murine cells. J. Virol. 83:6029-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terwilliger, T. C. 2003. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374:22-37. [DOI] [PubMed] [Google Scholar]
- 32.van Buuren, N., B. Couturier, Y. Xiong, and M. Barry. 2008. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J. Virol. 82:9917-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werden, S. J., J. Lanchbury, D. Shattuck, C. Neff, M. Dufford, and G. McFadden. 2009. Myxoma virus M-T5 ankyrin-repeat host range protein is a novel adaptor that coordinately links the cellular signaling pathways mediated by Akt and Skp1 in virus-infected cells. J. Virol. 83:12068-12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werden, S. J., M. M. Rahman, and G. McFadden. 2008. Poxvirus host range genes. Adv. Virus Res. 71:135-171. [DOI] [PubMed] [Google Scholar]