Abstract
During the search for haloarchaeal viruses, we isolated and characterized a new pleomorphic lipid-containing virus, Haloarcula hispanica pleomorphic virus 1 (HHPV-1), that infects the halophilic archaeon Haloarcula hispanica. The virus contains a circular double-stranded DNA genome of 8,082 bp in size. The organization of the genome shows remarkable synteny and amino acid sequence similarity to the genome and predicted proteins of the halovirus HRPV-1, a pleomorphic single-stranded DNA virus that infects a halophilic archaeon Halorubrum sp. Analysis of the two halovirus sequences, as well as the entire nucleotide sequence of the 10.8-kb pHK2-plasmid and a 12.6-kb chromosomal region in Haloferax volcanii, allows us to suggest a new group of closely related viruses with genomes of either single-stranded or double-stranded DNA. Currently, closely related viruses are considered to have the same genome type. Our observation clearly contradicts this categorization and indicates that we should reconsider the way we classify viruses. Our results also provide a new example of related viruses where the viral structural proteins have not diverged as much as the proteins associated with genome replication. This result further strengthens the proposal for higher-order classification to be based on virion architecture rather than on genome type or replication mechanism.
Metagenomic studies have increased the amount of information on the nucleotide sequence space in our environment. It has also increased our awareness of the viral abundance and diversity not recognized before (16, 24, 26). Along with this new information, we have learned to acknowledge the significance of viruses in the evolution and behavior of other organisms (55). To reveal the dynamics and molecular interactions in the interplay between a particular virus and its host, however, isolation of single viruses and their hosts is needed. Even though a number of viruses pathogenic to humans, domestic animals, and plants, as well as some bacteriophages, have been studied in great detail, much of the diversity of the archaeal viruses has remained unknown. By the year 2007 only 44 archaeal viruses had been described (2). That embraces less than 1% of all reported viruses. Although the diversity among these few isolated archaeal viruses is considerable, a head-and-tail morphology is prevalent among isolated viruses infecting euryarchaeal cells. In contrast, viruses of Crenarchaeota are diverse and often unusual with no viruses having a head-tail morphology (53).
Archaeal haloviruses infect euryarchaeal hosts living in environments up to saturated salt. This makes them an interesting group of viruses that reside in a very restricted habitat. In samples taken from high salt environments, the Dead Sea and Spanish solar salterns, viral morphotypes most often observed were spindle-shaped, head-and-tail or tailless icosahedral particles (25, 31, 47). Isolated haloviruses, however, do not seem to reflect the proportions of different morphotypes found in the nature as nearly all of the isolates possess a head-and-tail morphology (2). Molecular level studies on only two spindle-shaped (10, 11) and one tailless icosahedral particle have been carried out (37, 51). Virus-like particles of other morphologies have also been observed in high-salt environments (47), but only one additional morphotype has been described in detail (50). This recently isolated lipid containing halovirus, HRPV-1, is the first archaeal virus containing a single-stranded DNA (ssDNA) genome (50). It infects Halorubrum sp. and has a pleomorphic appearance with glycosylated spike structures protruding from its external membrane (49, 50).
The evolution of prokaryotic viral genome sequences is very fast (18), and the assessment of viral relationships using homology of the genome sequences applies only to closely related viruses (17, 19). Current higher-order classification of viruses is based on the host organism, the nature of the genome (RNA/DNA, single stranded versus double stranded) and the virion morphology. Recently, a higher-order clustering of virus families has been proposed based on common principles of virion architectures as well as on the fold of the major capsid protein (1, 6, 12, 13, 42). Consequently, major capsid proteins most probably belong to the vertically inherited viral “self” (4), whereas proteins involved in replication of the viral genome can be swapped by horizontal exchange (21, 63). The proposal is based on observations that structurally related viruses have been found to infect organisms that reside in all three domains of life.
We have isolated a new pleomorphic virus infecting Haloarcula hispanica (Har. hispanica pleomorphic virus 1 [HHPV-1]). Here, we determine the molecular constituents of HHPV-1 and its genetic relatedness to other archaeal viruses and putative proviruses. Sequence homology and gene order (synteny) shows distinct genomic regions shared between four genetic elements separating replication, virus assembly, and integration functions. Surprisingly, in spite of the close relatedness of HRPV-1 and HHPV-1, the genome types of these two viruses differ (ssDNA and dsDNA, respectively).
MATERIALS AND METHODS
Media and growth conditions.
Modified growth medium (MGM) containing 18% (top-layer), 20% (solid), or 23% (broth) (wt/vol) artificial salt water (SW) was used (52). To prepare top-layer and solid media, 4 and 14 g of Bacto agar (Difco Laboratories) were added per liter, respectively. When samples were drawn for thin-section electron microscopy, cultivation was performed in MGM, in which 1 M Tris-HCl was replaced with 1 M 4-morpholineethanesulfonic acid (MES; pH 6.7). Halophilic archaea and bacteria used in the present study are listed in Table S1 in the supplemental material. All strains were grown aerobically at 37°C.
Isolation of halovirus HHPV-1.
Virus was isolated from a water sample acquired from a solar saltern in Margherita di Savoia, Italy. Isolation was performed by enrichment culture method in which 2 ml of the water sample was incubated with 10 ml of MGM overnight at 37°C with aeration. Enrichment culture was centrifuged (Heraeus Biofuge, 16,060 × g, 5 min, 22°C) to remove cells and debris. The water samples were then plated on 12 indicator strains (Table S1 in the supplemental material), available beforehand, using 18% MGM. After incubation at 37°C for 3 days a plaque that had appeared on a lawn of Har. hispanica was picked and purified by two consecutive single plaque purification steps.
HHPV-1 host range.
Haloarchaeal strains, indicated in Table S1 in the supplemental material, were tested for virus susceptibility. Dilutions of virus lysate were plated with different strains using 18% MGM. After incubation for 4 days, the plates were checked for plaques.
Growth and purification of HHPV-1.
Virus stock was prepared from top-layer agar plates, incubated to show semiconfluent amount of plaques by HHPV-1. Soft agar from each plate was collected into 2 ml of 23% MGM and pooled. Culture was incubated at 37°C for 1.5 h with aeration. Cells and debris were removed by centrifugation (Sorvall SLA1500, 9,700 × g, 20 min, 4°C), and the titer (PFU/ml) was determined by plaque assay. Infectivity of HHPV-1 in a stock was sustained for at least 1 month when stored at 4°C.
Viruses were precipitated from the supernatant with 10% (wt/vol) polyethylene glycol (PEG) 6000 MW, collected by centrifugation (Sorvall SLA1500, 15,200 × g, 40 min, 4°C), and resuspended in 18% SW. Titer was determined by plaque assay. Purification of virus preparation was performed by rate zonal centrifugation in a linear 5 to 20% (wt/vol) sucrose gradient in 18% SW (Sorvall AH629, 103,600 × g, 4 h, 20°C). After centrifugation, the light scattering virus zone was collected and designated as “1×-purified” virus. The A260 was measured, and the titer was determined by plaque assay. To produce “2×-purified virus,” 1×-purified virus was layered either on the top of a CsCl solution (0.3 g/ml in 18% SW) or on a linear 10 to 45% (wt/vol) sucrose gradient in 18% SW and centrifuged to equilibrium (Sorvall AH629, 79,300 × g, 16 h, 20°C). The light-scattering virus zone was either collected, or CsCl tubes were fractionated into 2-ml fractions which were analyzed by measuring the A260 and density, as well as determining the titer by plaque assay. Samples were also taken for protein gel and lipid analyses. The collected virus zone was diluted (in volume ratio of 2:1:3 of virus, 18% SW, and 18% SW without NaCl), virus was collected by differential centrifugation (Sorvall T647.5, 177,500 × g, 3 h, 20°C), and the pellet resuspended in 18% SW. For this 2×-purified virus material, the A260 was measured, the titer was determined by plaque assay, and the protein concentration was measured by the Coomassie blue method (15) with bovine serum albumin as a standard. The specific infectivity for the virus was 4.4 × 1012 PFU/mg of protein.
HHPV-1 growth curve.
A total of 30 ml of exponential-phase Har. hispanica culture (1.5 × 109 CFU/ml) was infected with HHPV-1 stock (multiplicity of infection [MOI] of 15). After an adequate adsorption period of 30 min (5 min at 22°C with no aeration, followed by 25 min at 37°C with aeration [43]), the cells were washed with MGM broth to remove the unbound viruses and resuspended into 30 ml of MGM broth. The culture was incubated at 37°C with aeration. The A550 was measured in hourly intervals of the infected and similarly treated uninfected culture. Samples were drawn concurrently with the absorbance measurements, and the number of free viruses and infective centers was determined by plaque assay. The number of viable cells (CFU/ml) was monitored in both infected and uninfected cultures. After incubation for 12 days, different-sized colonies (small, medium, and large) were picked from viable count plates of the infected culture and inoculated into MGM broth. After incubation at 37°C for 8 days with aeration the titer of the culture supernatants was determined by plaque assay.
To determine when virus production begins, a culture was prepared as described above except that an MOI of 1 was used. Samples were drawn at half-hour intervals from 1 h postinfection (hpi) onward for determination of the number of free viruses by plaque assay.
Electron microscopy.
For negative stain transmission electron microscopy, viruses were allowed to adsorb to grids for 1 min and were then stained with 1% (wt/vol) potassium phosphotungstate (pH 6.5) for ∼10 s, uranyl acetate (pH 4.5) for ∼20 s, 3.32% ammonium molybdate (pH 7.4) for ∼15 s, or 1% ammonium molybdate (pH 7.0) for ∼15 s.
For thin-section electron microscopy cultures were prepared as described for the HHPV-1 growth curve, except that MOI 40 was used. Samples were drawn 5 or 25 min postinfection or 1, 3, 5, 7, or 23 hpi from the infected culture and 5 hpi from the uninfected culture. Thin-section electron microscopy was carried out as previously described (7).
All micrographs were taken with a JEOL 1200EX electron microscope operating at 60 or 80 kV (Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki).
HHPV-1 stability.
Chloroform sensitivity was tested by mixing chloroform and virus stock in a proportion of 1:20, followed by incubation at 4°C overnight before titer determination by plaque assay.
Protein analyses.
The protein composition was analyzed by using a modified Tricine-SDS-PAGE system (devoid of spacer gel [59]) with 14% acrylamide concentration (wt/vol). The N-terminal sequences of the two structural proteins were determined in the Protein Chemistry Core Laboratory, Institute of Biotechnology, University of Helsinki, as described previously (8).
Extraction and analysis of the lipids.
Lipids from cell suspensions and 2×-purified virus preparations were extracted according to the method of Folch et al. (29) essentially as described previously (8) and stored in chloroform-methanol (9:1 [vol/vol]) at −20°C until analyzed. Total lipid extracts were subjected to thin-layer chromatography (TLC) on silica gel 60 plates (Merck), and phospholipids were separated by using chloroform-methanol-90% acetic acid (65:4:35 [vol/vol/vol]) as the solvent (8, 23). After visualizing the lipids by iodine vapor, the plates were sprayed with molybdate (27) and 0.1% orcinol-sulfuric acid reagents (60) to detect phospholipids and glycolipids, respectively. The phosphorus content of lipid spots was determined as described by Bartlett and Lewis (9). The lipids in each spot of the TLC were identified by performing a mass-spectrometric analysis and comparing their m/z values, as well as product and precursor ion profiles, to those of known lipids from halophilic archaea (8).
Genome sequencing and sequence analyses.
The genome was purified from the 2×-purified viral material by dilution of the virus 1:5 in 50 mM Tris-HCl (pH 7.2). SDS was added to a final concentration of 1.5%, EDTA was added to 1 mM, and proteinase K was added to 0.8 mg ml−1, and the preparation was incubated at 56°C for 20 min. Proteins were removed by phenol extraction and ether. Nucleic acids were precipitated with 0.3 M sodium chloride and 2 volumes of ethanol at −20°C overnight. Approximately 1.5 to 2 μg of the genome was used for each digest of DNase I (1 U of RQ1 RNase-Free DNase; Promega), RNase A (4 μg; Promega), or mung bean nuclease (0.5 U). As controls, the single-stranded and double-stranded (RFII [RF for replicative form]) viral DNAs of φX174 (New England Biolabs) were used.
To determine the preliminary genomic sequence of HHPV-1, a six-base-tagged (barcoded) library was done for Genome Sequencer FLX pyrosequencing (Roche/454 Life Science). A total of 3,000 sequences were obtained with a mean read length of 206 bp. The reads were assembled by using the gsAssembler, and the obtained assembly was verified with PCR products that covered the whole genome. The final sequence data were obtained by sequencing these PCR products. The primers used for PCR and sequencing are listed in Table S6 in the supplemental material. All sequencing was carried out by using BigDye version 3.1 and analyzed on an ABI 3130 (Applied Biosystems). Sequencing reactions were purified with CleanSeq (Agencourt) using a Beckman NXp robot (Beckman Coulter). Sequences were assembled and edited using GAP4 (61). The pHK2 sequence was determined by primer walking using pMDS1 (33) as the template. The primers used for sequence determination are listed in the Table S7 in the supplemental material. The preliminarily annotated sequence of Haloferax volcanii (Hfx. volcanii) genome was obtained from the HaloLex website (48).
Genome sequence was analyzed for the open reading frames (ORFs) by using the programs Artemis (56), GenMark (14), and pDRAW32 (AcaClone software). The basic properties of the genes and encoded proteins were studied by using the programs of the EMBOSS program package (54) at the IT Center for Science, Ltd. (www.csc.fi), in Finland and the Expasy proteomics server (30). Transmembrane helices in the polypeptides were predicted by using TMHMM server version 2.0 (41) and coiled coils were predicted using the Coils-Server (45). The global pairwise identity between two proteins was determined by using LALIGN (a global alignment method without end-gap penalty [35] at ch.EMBnet.org) using the default values and the BLOSUM50 matrix. Homologous proteins in the databases were searched by using the BLASTP search engine (3) at the National Center for Biotechnology Information (NCBI).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in the present study were deposited in GenBank with the following accession numbers: HHPV-1 genome, GU321093; and pHK2 sequence, GU321094.
RESULTS
HHPV-1 is a pleomorphic archaeal halovirus.
HHPV-1 was isolated from a solar saltern in Margherita di Savoia, Italy. HHPV-1 produced plaques on the lawn of a Har. hispanica host but was unable to produce plaques on any other strains tested (see Table S1 in the supplemental material).
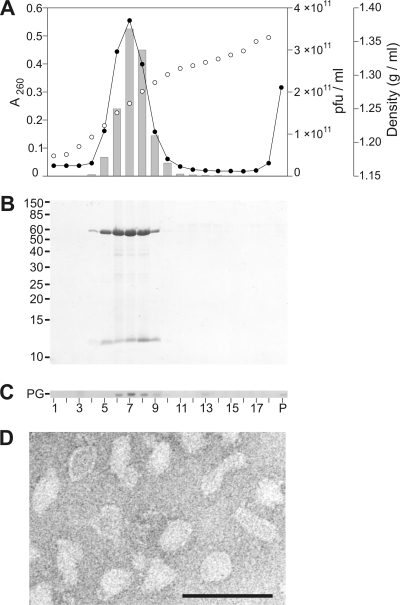
Halovirus HHPV-1 production and purification methods were established (see Materials and Methods). The virion has densities of 1.27 and 1.23 g/ml in CsCl and sucrose, respectively. Recovery and specific infectivity (PFU/A260) were monitored during the purification process (see Table S2 in the supplemental material). Virions were extremely sensitive to chloroform (data not shown). Chloroform sensitivity and low buoyant density indicated the presence of lipids as virion constituents. Gradient analysis (Fig. 1) confirmed this as A260, virus titer, proteins, and lipids peaked in the same fractions. Negative-stain transmission electron microscopy (TEM) of HHPV-1 particles purified by equilibrium centrifugation (2×-purified virus) showed pleomorphic particles (Fig. 1D).
FIG. 1.
Characterization of highly purified HHPV-1 virions. (A) Analyses of the purification in equilibrium centrifugation by A260 (•), CsCl gradient density (○), and infectivity (░⃞) of the gradient fractions. (B) Protein profiles of the gradient fractions in 14% Tricine-SDS-PAGE. Molecular masses of the standard proteins (in kilodaltons) are shown on the left. (C) TLC of the fractions showing phosphatidylglycerol (PG). The numbers of the gradient fractions for panels A to C are shown in panel C. P, pellet. (D) Negative-stained electron micrograph of 2×-purified HHPV-1. Sample was stained with 1% (wt/vol) ammonium molybdate (pH 7.0). Bar, 100 nm.
HHPV-1 is released without cell lysis.
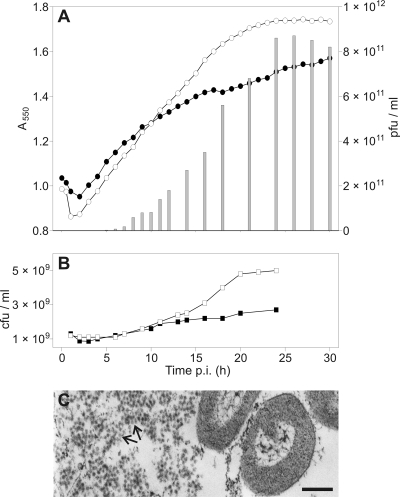
HHPV-1 did not lyse the host Har. hispanica cells (Fig. 2A). Increase in turbidity was similar for the infected and uninfected cultures until 7 hpi, after which the infected culture showed a slightly retarded growth rate. When the number of CFU was monitored, the infected culture resulted in reduced colony formation after 10 hpi compared to the uninfected one (Fig. 2B).
FIG. 2.
Production of HHPV-1 in a batch culture. (A) The turbidity (A550) of infected (•) and uninfected (○) cultures, as well as the number of free viruses (░⃞) were monitored. The time of infection was at 0 hpi. (B) The number of CFU of the infected (▪) and uninfected (□) culture. Panels A and B are shown in the same time scale. (C) Thin-section electron micrographs of the infected culture. The number of extracellular viruses grew in the course of time resulting in large clusters of viruses (arrows) at 23 hpi. Bar, 200 nm.
Virus release started 3 to 3.5 hpi, reaching its maximum, ∼8.6 × 1011 PFU/ml in the medium, around 25 hpi (Fig. 2A). The number of infective centers (ICs) started to increase after 3 hpi, indicating that not all of the cells had been infected before the onset of the release of the first viral progeny. Increases of ICs were observed up to ∼10 hpi. It should be noted that the amount of ICs could not be detected accurately because viruses seemed to be released continuously from the cells and the detected numbers of ICs were slightly higher than the CFU. Virus production seemed to influence the size of colonies because in the early stages of the life cycle (∼1 hpi) colonies formed were mostly large, and cells taken from such colonies did not produce virus. Smaller colonies dominant in the late stages, on the other hand, produced virus.
The life cycle of HHPV-1 was also studied by taking samples for thin-section electron microscopy at different time points during the life cycle. The amount of extracellular viruses was observed to increase in the course of time, and most of the viruses were detected in clusters outside the cells (Fig. 2C). However, we were not able to detect intracellular viruses or viruses in the process of budding.
There are four phospholipid classes and two major protein species in the HHPV-1 virion.
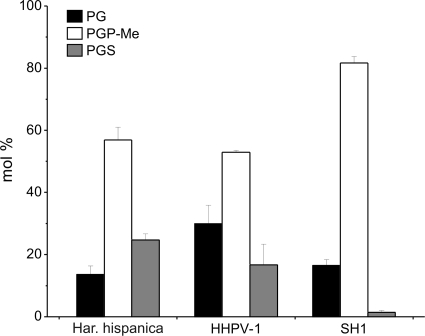
Analysis of the lipid extracts by TLC revealed that highly purified HHPV-1 virions contain phospholipids but no significant amounts of glycolipids (data not shown). The four phospholipid classes detected were cardiolipins (CL), phosphatidylglycerols (PG), phosphatidylglycerophosphate methyl esters (PGP-Me), and phosphatidylglycerosulfates (PGS). Comparison of the quantity of the three main classes of phospholipids (i.e., PG, PGP-Me, and PGS) of Har. hispanica, HHPV-1, and the icosahedral lipid-containing halovirus SH1 (8) shows that the composition of HHPV-1 is quite similar to that of the host, except that the PG is slightly enriched in the virion (Fig. 3), whereas the lipid composition of SH1 deviates significantly from that of the host.
FIG. 3.
Comparison of the phospholipid compositions of Har. hispanica, HHPV-1, and SH1. Concentrations are expressed as the mol% of total phospholipids, and only phospholipids representing >1% of the total are shown. Error bars represent standard deviations of data from at least three independent experiments. PG, phosphatidylglycerol; PGP-Me, phosphatidylglycerophosphate methyl ester; PGS, phosphatidylglycerosulfate.
Analysis of the virion in Tricine-SDS-PAGE revealed two major protein species with apparent molecular masses of approximately 60 and 12 kDa (Fig. 4A). The N-terminal sequence analysis of these two proteins gave sequences of AYSNNARGLV for the 12-kDa and VVPVVLPGVA for the 60-kDa protein. These two peptide sequences were identified as the protein products of the ORFs 3 and 4, respectively (see below). The 12-kDa protein is designated VP3 (VP for viral protein), and the 60-kDa protein is designated VP4. The protein sequence analysis also showed that VP4 contains a 50-amino-acid signal sequence that is processed.
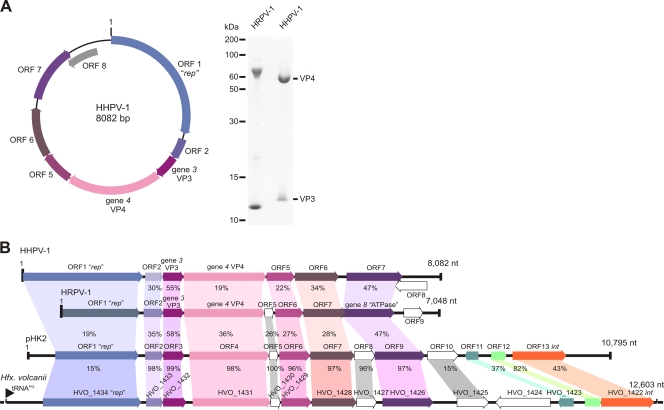
FIG. 4.
Schematic representation of the HHPV-1 genome and analysis of HHPV-1 and HRPV-1 structural proteins. (A) A schematic representation of the HHPV-1 genome (left). Identified ORFs as well as the genes encoding the two major structural proteins are indicated. On the right side of the figure is an analysis of HHPV-1 structural proteins in Tricine-SDS-PAGE, together with the related virus HRPV-1. The sizes of the protein molecular mass standards (in kilodaltons) are shown on the left. The two major structural proteins in the viral protein pattern (right) are indicated. Same amounts of the two viruses (in micrograms of protein) were loaded. (B) Alignment of the genomes of HHPV-1, HRPV-1, as well as the putative proviral elements pHK2 and the chromosomal region of Hfx. volcanii. The identities (in percentages) of the homologous translated ORFs or identified gene products are shown.
HHPV-1 genome analyses revealed relatedness to the pleomorphic virus HRPV-1.
Enzymatic digestion of the HHPV-1 genome with DNase I and mung bean nuclease showed that the genome is a dsDNA molecule (see Fig. S1 in the supplemental material). The preliminary sequence of the genome was determined by pyrosequencing, and the whole genome sequence was further confirmed by sequencing overlapping PCR-products that covered the whole genome. The ability to generate overlapping PCR-products revealed the circular nature of the genome. The HHPV-1 genome is 8,082 bp in size, and the GC percentage is 55.8%.
The HHPV-1 genome has eight putative ORFs (Fig. 4A). The properties of the ORFs and the encoded polypeptides are shown in Table S3 in the supplemental material. ORFs 3 and 4 encoding the two major structural proteins (Fig. 4A) were designated genes 3 and 4, respectively. The genome organization and protein homology showed that HHPV-1 is closely related to the recently isolated and characterized pleomorphic ssDNA virus HRPV-1 that infects Halorubrum sp. (50). The structural protein patterns are also similar (Fig. 4A). HHPV-1 ORF2, and five consecutive translated ORFs show significant identity to the translated ORFs and identified genes of HRPV-1 (Fig. 4). The highest identity (55%) was found between the HHPV-1 gene 3 encoding the small major structural protein VP3 (HHPV-1 VP3) and the VP3 protein of HRPV-1. Although both proteins are putative integral membrane proteins, they differ in the number of predicted transmembrane regions (two in HHPV-1 VP3 and four in HRPV-1 VP3). VP4, the other major structural protein of HHPV-1, is similar in size but shows relatively low identity to the HRPV-1 VP4-protein. Both of these two proteins, however, contain a signal peptide that is processed and a predicted C-terminal transmembrane domain that is preceded by a predicted coiled-coil region.
The remaining putative gene products of HHPV-1 that show homology with the translated ORFs of HRPV-1 are >20% identical (Fig. 4B). The highest identity among these (47%) was found between HHPV-1 ORF7 and the HRPV-1 ORF8. The latter has been shown to encode the minor structural protein VP8 that contains predicted ATPase motifs (50). A search using HHPV-1 ORF7 for conserved domains at the NCBI recognized domains belonging to the P-loop NTPase superfamily proteins (cl09099, Expect [E] value, 0.22). The polypeptide of the predicted ORF1 did not show significant homology to the HRPV-1 ORF1 (9% identity). A BLASTP search of the database at NCBI, however, gives best matches to the putative Rep protein of Halomicrobium mukohataei (GenBank accession no. EEJ07478.1, E-value 9e-38) and to ORF1 of plasmid pHK2 (GenBank accession no. AAB09607.1, E-value 2e-25, see below). Since ORF1 of HRPV-1 is also predicted to be a replication initiation protein (50), the two putative ORF1 polypeptides most probably serve the same function as replication initiation proteins.
Completion of the entire sequence of the pHK2-haloplasmid suggests that it is a virus.
Preliminary BLAST searches using the HHPV-1 ORFs gave hits to the ORFs of the plasmid pHK2 isolated from haloarchaeal species Haloferax sp. (34). Since similar observation was made for the HRPV-1 polypeptides (50), we completed the sequence of the pHK2-plasmid using its derivative pMDS1 (33). The previously published sequence of the 4.1-kb fragment of pHK2 (GenBank accession no. L29110.1) (34) was found to diverge from the new sequence in 12 positions. The biggest change was the addition of a G after the nucleotide (nt) 3885 in the previously determined sequence. This change removes the stop codon for ORF4 at nt 3935 and the ORF continues to ORF5. In addition, in the beginning of the previous 4.1-kb region, differences in sequence result in 94% identity between the ORF1 polypeptides predicted to encode replication initiation proteins (data not shown). The sequence obtained from the rest of the pHK2 plasmid showed homologous ORFs in the same order as in both HHPV-1 and HRPV-1 (Fig. 4B; see also Tables S4 and S5 in the supplemental material). The last annotated ORFs, ORF8 of HHPV-1 and ORF9 of HRPV-1, do not show homology to each other or to ORF10 of pHK2 (Fig. 4B). Although the HHPV-1 VP4 shows only 19% identity to the VP4 of HRPV-1, the identity between the pHK2 ORF4 encoded polypeptide and the HRPV-1 VP4 is remarkably higher, i.e., 36%. Also, HRPV-1 VP3 shows higher identity to pHK2 ORF3 (58%) than to HHPV-1 VP3 (Fig. 4B; see Table S6 in the supplemental material). The 10,795-bp pHK2 plasmid has four putative ORFs after the ORF encoding the putative ATPase (HHPV-1 ORF7, HRPV-1 VP8, and pHK2 ORF9). The encoded polypeptides of these ORFs do not show identity to either HHPV-1 or HRPV-1. ORF11, however, encodes a putative polypeptide which shows homology to φH repressorlike proteins and ORF13 encodes a putative integrase (see the supplemental material). These features strongly suggest that the pHK2 plasmid actually is a temperate virus related to both HHPV-1 and HRPV-1.
A provirus in the chromosome of Hfx. volcanii is the fourth member of the group.
A search through preliminarily annotated Hfx. volcanii genome using pHK2 ORFs as queries resulted in identification of a 12.6-kb gene region containing ORFs highly identical and in the same order as the ORFs in the pHK2 plasmid (Fig. 4B; see also the supplemental material). The region fulfills the typical features of a provirus integrated into the genome. There are ∼450 nt between tRNAArg and the ORF1 encoding a putative replication initiation protein. ORFs 2 to 9 are 96 to 98% identical to the corresponding ORFs in pHK2; the predicted ATPase encoding ORF9 is the last ORF showing high identity. After that there is no detectable similarity except for ORFs encoding the putative φH repressor-like protein (HVO_1423) and a putative integrase (HVO_1422) that are in similar positions between pHK2 and the chromosomal region of Hfx. volcanii in the end of these genetic elements (Fig. 4B). Between the preliminarily annotated ORFs HVO1423 and HVO_1422 there is a short ORF that has not been annotated in the preliminary annotation of the Hfx. volcanii genome. The predicted polypeptide, however, shows high homology to the putative ORF12 polypeptide of pHK2.
Apart from this chromosomal region in Hfx. volcanii there are several regions encoding hypothetical proteins in the genomes of halophilic archaea that show homology to the viral genomic region starting from VP4 encoding gene and ending after the putative ORF encoding an ATPase. As reported previously (50), the same region also shows homology to the similar region in another halophilic virus, His2 (10).
DISCUSSION
During a search for new haloviruses, a pleomorphic halovirus was discovered from a solar saltern on the eastern cost of Italy. This Har. hispanica pleomorphic virus 1, HHPV-1, has a pleomorphic appearance similar to the recently reported HRPV-1 virus (Fig. 1D) (50). HHPV-1 production and purification methods were set up to gain highly pure viral material. HHPV-1 virion contains four distinct phospholipid classes and two major structural proteins with predicted transmembrane regions (Fig. 4A). The other described pleomorphic halovirus HRPV-1 has also been shown to contain lipids and two major structural proteins (50). The almost identical phospholipid compositions of HHPV-1 and Har. hispanica suggest that lipids during the assembly of HHPV-1 are acquired from the host almost without selection. Interestingly, the composition of phospholipids of SH1, an icosahedral virus that also infects the Har. hispanica host, is significantly different from that of HHPV-1 (Fig. 3). This observation indicates that the lipids of SH1 are selectively incorporated into the virion most likely due to the geometrical constraints within the icosahedral protein coat and/or specific lipid-protein interactions (8, 37). Thus, the assembly pathway of HHPV-1 must differ considerably from that of SH1 and related viruses.
HHPV-1 exits the host without causing cell lysis (Fig. 2). Exit without lysis has been observed for several viruses of Crenarchaeota (58, 64) and has also been observed for the haloviruses HRPV-1, His1, and His2 (10, 50). Some of these viruses seem to exit in an as-yet-undescribed way. Interestingly, the two viruses homologous to HHPV-1, also exit the host without cell disruption (10, 50). Such a mode of exit suggests budding.
Genome synteny reveals a close relationship between HHPV-1 and the HRPV-1. In addition, the entire 10.8-kb genome of pHK2 completed in the present study as well as the genomic 12.6-kb region of Hfx. volcanii, show considerable homology to HHPV-1 and HRPV-1 viruses (Fig. 4B). pHK2 has earlier been reported to be a plasmid (32, 34). Although the viral nature of pHK2 has not been confirmed, the highly homologous genomic region of Hfx. volcanii shows the typical features of a virus integrated into tRNAArg. In addition, both pHK2 and the Hfx. volcanii chromosomal region contain a putative integrase (46), as well as genes encoding φH repressorlike proteins. We predict that these two genetic elements can be enclosed into pleomorphic viral particles and consequently will be designated as proviruses in the discussion that follows.
Comparison of the four genomes enables us to detect the most conserved putative and identified proteins of the proposed new group of pleomorphic viruses. Of the two major structural proteins, VP3 and VP4, VP3 shows the highest identity. VP4, on the other hand shows quite high diversity. The VP4 of HRPV-1 was predicted to function as the receptor binding and fusion protein (50). Viral receptor-binding proteins are often the most variable ones among the viral structural proteins (17, 44, 57). Since all of these viruses infect different hosts, variability of VP4 is in accordance with its function as a receptor interacting protein. Apart from these two proteins, ORF5, ORF6, and ORF7 of HHPV-1 show relatively high identity in all four genomes. The region containing identical and almost identical predicted proteins in the pHK2 and Hfx. volcanii elements starts from ORF2 and ends after ORF9. In most of the pairwise alignments (see the supplemental material), the predicted and identified proteins of HRPV-1 are more closely related to corresponding ORF-encoded proteins of either pHK2 or Hfx. volcanii than to predicted and identified proteins of HHPV-1. HRPV-1 genome also contains ORF5, which shows homology to the corresponding ORFs in pHK2 and Hfx. volcanii proviruses. This suggests that HRPV-1 is a closer relative to pHK2 and Hfx. volcanii proviruses than to HHPV-1.
Keeping in mind the high identity of the central region of the pHK2 and Hfx. volcanii provirus genomes, it is surprising that ORF1 and the region starting from pHK2 ORF10 and Hfx. volcanii HVO_1425 varies considerably. HRPV-1 and HHPV-1 genomes do not contain the region starting from the pHK2 ORF10 and including the φH repressor and the integrase. This shows that an integrase is not required for the life cycle of these pleomorphic viruses even though under certain conditions an integrase may confer an advantage. Similar observation have also been made for other types of viruses such as the crenarchaeal virus SSV1 (22) and an icosahedral phage P23-77 that infects Thermus thermophilus (38).
The ORF1s of the four genomes shows low identity in the pairwise alignments (the highest being 20% between HRPV-1 and Hfx. volcanii ORF1). They all are, however, predicted to function as replication initiation proteins. HHPV-1 and pHK2 ORF1 polypeptides contain the conserved domain DUF1424 (E value of < 0.01). This domain can be found in the family of haloarchaeal proteins predicted to be involved in replication. ORF1-encoded polypeptides of HRPV-1, pHK2, and Hfx. volcanii provirus also contain the three conserved motifs suggested to be essential for initiation proteins of rolling-circle replication (36). The HHPV-1 ORF1 contains domains 2 and 3 of these conserved motifs. Since at least HHPV-1, HRPV-1 and pHK2 genomes are circular, it is highly likely that also the ORF1 encodes a rolling-circle replication initiation protein. Replication and packaging of a circular ssDNA genome of bacteriophage φX174 has been described in detail (40, 63). Replication of the circular ssDNA genome of HRPV-1 and circular dsDNA genome of HHPV-1 may occur in a similar fashion. Packaging of different types of genomes just occurs in different stages of the replication process.
During our studies of new halophilic viruses we have found a new group of pleomorphic viruses including HHPV-1 and HRPV-1 and two putative proviral elements. Based on these results, two major observations can be made that both have consequences as to how viruses are classified. First, we observed that the genome of HHPV-1 is a circular dsDNA molecule, whereas the genome of HRPV-1 is a circular ssDNA molecule (50). In terms of current viral classification, this is unusual, since according to the nucleotide and deduced amino acid sequence data these viruses are closely related and should be grouped together despite the different genome types. This shows that in the era of genome sequencing, higher-order classification according to the genome type may not be justified.
Second, although associated with different hosts, these four viruses show conserved regions, including the two major structural proteins determined for HHPV-1 and HRPV-1 (50). This conserved region containing ORFs 2 to 9 in pHK2 most probably constitutes the core proteins for virion assembly. In contrast, the putative replication initiation proteins show lower identity. In addition, unlike the core proteins, multiple alignment of the ORF1 polypeptides arranges the relationships between the viruses and the proviral elements in a different order than the alignment using the core proteins. Thus, comparisons of the four genomes suggest that the viral world is indeed “… a dynamic network of relationships in which genes have diverse, variously intertwined histories …” (39) obtained both vertically and horizontally. The comparisons of the core proteins of this new pleomorphic virus group show, however, that the structural components are more conserved and most probably obtained by vertical inheritance in order to keep the viral self intact and functional for continuation of the viral lineage (1, 4-6, 12, 13, 42). Similar observations have been made for tailed phages using even more abundant sequence information (20, 21, 28, 62). As a consequence, the mode of the genome replication and the nature of the genome (RNA/DNA, ss or ds) may not be adequate determinants for classifying distant, but related viruses to higher-order taxons. The determinants for the virion architecture instead may better serve this function.
Supplementary Material
Acknowledgments
We thank Paula Collin-Olkkonen, Sari Korhonen, Päivi Laamanen, Kirsi Lipponen, Soile Storman, Eeva-Marja Turkki, and Suvi Varjonen for excellent technical assistance. Nisse Kalkkinen and Gunilla Rönnholm are acknowledged for the protein N-terminal sequencing.
This study was supported by Academy of Finland Centre of Excellence Program in Virus Research grant 1129684 (2006-2011) and Academy of Finland grant 1210253 (to D.H.B.).
Footnotes
Published ahead of print on 20 January 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abrescia, N. G. A., J. M. Grimes, E. E. Fry, J. J. Ravantti, D. H. Bamford, and D. I. Stuart. 2009. What does it take to make a virus: the concept of the viral “self.” In P. G. Stockley and R. Twarock (ed.), Emerging topics in physical virology, in press. Imperial College Press, London, England.
- 2.Ackermann, H. W. 2007. 5500 Phages examined in the electron microscope. Arch. Virol. 152:227-243. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford, D. H. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231-236. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, D. H., R. M. Burnett, and D. I. Stuart. 2002. Evolution of viral structure. Theor. Popul. Biol. 61:461-470. [DOI] [PubMed] [Google Scholar]
- 6.Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage φ6. Virology 107:222-228. [DOI] [PubMed] [Google Scholar]
- 8.Bamford, D. H., J. J. Ravantti, G. Rönnholm, S. Laurinavičius, P. Kukkaro, M. Dyall-Smith, P. Somerharju, N. Kalkkinen, and J. K. Bamford. 2005. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 79:9097-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlett, E. M., and D. H. Lewis. 1970. Spectrophotometric determination of phosphate esters in the presence or absence of orthophosphate. Anal. Biochem. 36:159-167. [DOI] [PubMed] [Google Scholar]
- 10.Bath, C., T. Cukalac, K. Porter, and M. L. Dyall-Smith. 2006. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, salterprovirus. Virology 350:228-239. [DOI] [PubMed] [Google Scholar]
- 11.Bath, C., and M. L. Dyall-Smith. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol. 72:9392-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 13.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 14.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 16.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 18.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 19.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 20.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned thus far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 21.Casjens, S. R. 2008. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res. Microbiol. 159:340-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clore, A. J., and K. M. Stedman. 2007. The SSV1 viral integrase is not essential. Virology 361:103-111. [DOI] [PubMed] [Google Scholar]
- 23.Corcelli, A., M. Colella, G. Mascolo, F. P. Fanizzi, and M. Kates. 2000. A novel glycolipid and phospholipid in the purple membrane. Biochemistry 39:3318-3326. [DOI] [PubMed] [Google Scholar]
- 24.Desnues, C., B. Rodriguez-Brito, S. Rayhawk, S. Kelley, T. Tran, M. Haynes, H. Liu, M. Furlan, L. Wegley, B. Chau, Y. Ruan, D. Hall, F. E. Angly, R. A. Edwards, L. Li, R. V. Thurber, R. P. Reid, J. Siefert, V. Souza, D. L. Valentine, B. K. Swan, M. Breitbart, and F. Rohwer. 2008. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452:340-343. [DOI] [PubMed] [Google Scholar]
- 25.Diez, B., J. Anton, N. Guixa-Boixereu, C. Pedros-Alio, and F. Rodriguez-Valera. 2000. Pulsed-field gel electrophoresis analysis of virus assemblages present in a hypersaline environment. Int. Microbiol. 3:159-164. [PubMed] [Google Scholar]
- 26.Dinsdale, E. A., R. A. Edwards, D. Hall, F. Angly, M. Breitbart, J. M. Brulc, M. Furlan, C. Desnues, M. Haynes, L. Li, L. McDaniel, M. A. Moran, K. E. Nelson, C. Nilsson, R. Olson, J. Paul, B. R. Brito, Y. Ruan, B. K. Swan, R. Stevens, D. L. Valentine, R. V. Thurber, L. Wegley, B. A. White, and F. Rohwer. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629-632. [DOI] [PubMed] [Google Scholar]
- 27.Dittmer, J. C., and R. L. Lester. 1964. A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J. Lipid Res. 15:126-127. [PubMed] [Google Scholar]
- 28.Fokine, A., P. G. Leiman, M. M. Shneider, B. Ahvazi, K. M. Boeshans, A. C. Steven, L. W. Black, V. V. Mesyanzhinov, and M. G. Rossmann. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U. S. A. 102:7163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folch, J., M. Lees, and G. H. Sloane-Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 30.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guixa-Boixereu, N., K. Lysnes, and C. Pedros-Alio. 1999. Viral lysis and bacterivory during a phytoplankton bloom in a coastal water microcosm. Appl. Environ. Microbiol. 65:1949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes, M. L., and M. L. Dyall-Smith. 1990. A plasmid vector with a selectable marker for halophilic archaebacteria. J. Bacteriol. 172:756-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes, M. L., S. D. Nuttall, and M. L. Dyall-Smith. 1991. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J. Bacteriol. 173:3807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes, M. L., F. Pfeifer, and M. L. Dyall-Smith. 1995. Analysis of the halobacterial plasmid pHK2 minimal replicon. Gene 153:117-121. [DOI] [PubMed] [Google Scholar]
- 35.Huang, X., and W. Miller. 1991. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12:337-357. [Google Scholar]
- 36.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes, and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jäälinoja, H. T., E. Roine, P. Laurinmäki, H. M. Kivelä, D. H. Bamford, and S. J. Butcher. 2008. Structure and host-cell interaction of SH1, a membrane-containing, halophilic euryarchaeal virus. Proc. Natl. Acad. Sci. U. S. A. 105:8008-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jalasvuori, M., S. T. Jaatinen, S. Laurinavičius, E. Ahola-Iivarinen, N. Kalkkinen, D. H. Bamford, and J. K. Bamford. 2009. The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J. Virol. 83:9388-9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koonin, E. V., Y. I. Wolf, K. Nagasaki, and V. V. Dolja. 2009. The complexity of the virus world. Nat. Rev. Microbiol. 7:250. [Google Scholar]
- 40.Koths, K., and D. Dressler. 1980. The rolling circle-capsid complex as an intermediate in φX DNA replication and viral assembly. J. Biol. Chem. 255:4328-4338. [PubMed] [Google Scholar]
- 41.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 42.Krupovič, M., and D. H. Bamford. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941-948. [DOI] [PubMed] [Google Scholar]
- 43.Kukkaro, P., and D. H. Bamford. 2009. Virus-host interactions in environments with a wide range of ionic strengths. Environ. Microbiol. Rep. 1:71-77. [DOI] [PubMed] [Google Scholar]
- 44.Lubbers, M. W., N. R. Waterfield, T. P. Beresford, R. W. Le Page, and A. W. Jarvis. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 46.Nunes-Düby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int. family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oren, A., G. Bratbak, and M. Heldal. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143-149. [DOI] [PubMed] [Google Scholar]
- 48.Pfeiffer, F., A. Broicher, T. Gillich, K. Klee, J. Mejia, M. Rampp, and D. Oesterhelt. 2008. Genome information management and integrated data analysis with HaloLex. Arch. Microbiol. 190:281-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pietilä, M. K., S. Laurinavičius, J. Sund, E. Roine, and D. H. Bamford. 2010. The ssDNA genome of novel archaeal virus HRPV-1 is enclosed in the envelope decorated with glycoprotein spikes. J. Virol. 84:788-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietilä, M. K., E. Roine, L. Paulin, N. Kalkkinen, and D. H. Bamford. 2009. An ssDNA virus infecting Archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 72:307-319. [DOI] [PubMed] [Google Scholar]
- 51.Porter, K., P. Kukkaro, J. K. Bamford, C. Bath, H. M. Kivelä, M. L. Dyall-Smith, and D. H. Bamford. 2005. SH1: a novel, spherical halovirus isolated from an Australian hypersaline lake. Virology 335:22-33. [DOI] [PubMed] [Google Scholar]
- 52.Porter, K., B. E. Russ, J. Yang, and M. L. Dyall-Smith. 2008. The transcription programme of the protein-primed halovirus SH1. Microbiology 154:3599-3608. [DOI] [PubMed] [Google Scholar]
- 53.Prangishvili, D., P. Forterre, and R. A. Garrett. 2006. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 4:837-848. [DOI] [PubMed] [Google Scholar]
- 54.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 55.Rohwer, F., and R. V. Thurber. 2009. Viruses manipulate the marine environment. Nature 459:207-212. [DOI] [PubMed] [Google Scholar]
- 56.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 57.Saren, A. M., J. J. Ravantti, S. D. Benson, R. M. Burnett, L. Paulin, D. H. Bamford, and J. K. Bamford. 2005. A snapshot of viral evolution from genome analysis of the Tectiviridae family. J. Mol. Biol. 350:427-440. [DOI] [PubMed] [Google Scholar]
- 58.Schleper, C., K. Kubo, and W. Zillig. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. U. S. A. 89:7645-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 60.Skipski, V. P., and M. Barclay. 1969. Thin-layer chromatography of lipids, p. 530-598. In J. M. Lowenstein (ed.), Lipids, vol. 14. Academic Press, Inc., New York, NY. [Google Scholar]
- 61.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 62.Stewart, C. R., S. R. Casjens, S. G. Cresawn, J. M. Houtz, A. L. Smith, M. E. Ford, C. L. Peebles, G. F. Hatfull, R. W. Hendrix, W. M. Huang, and M. L. Pedulla. 2009. The genome of Bacillus subtilis bacteriophage SPO1. J. Mol. Biol. 388:48-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weigel, C., and H. Seitz. 2006. Bacteriophage replication modules. FEMS Microbiol. Rev. 30:321-381. [DOI] [PubMed] [Google Scholar]
- 64.Vestergaard, G., M. Häring, X. Peng, R. Rachel, R. A. Garrett, and D. Prangishvili. 2005. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336:83-92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.