Abstract
The ORF50 gene of the varicella-zoster virus (VZV) encodes glycoprotein M (gM), which is conserved among all herpesviruses and is important for the cell-to-cell spread of VZV. However, few analyses of ORF50 gene expression or its posttranscriptional and translational modifications have been published. Here we found that in VZV-infected cells, ORF50 encoded four transcripts: a full-size transcript, which was translated into the gM, and three alternatively spliced transcripts, which were not translated. Using a splicing-negative mutant virus, we showed that the alternative transcripts were nonessential for viral growth in cell culture. In addition, we found that two amino acid mutations of gM, V42P and G301M, blocked gM's maturation and transport to the trans-Golgi network, which is generally recognized as the viral assembly complex. We also found that the mutations disrupted gM's interaction with glycoprotein N (gN), revealing their interaction through a bond that is otherwise unreported for herpesviruses. Using this gM maturation-negative virus, we found that immature gM and gN were incorporated into intracellularly isolated virus particles and that mature gM was required for efficient viral growth via cell-to-cell spread but not for virion morphogenesis. The virus particles were more abundant at the abnormally enlarged perinuclear cisternae than those of the parental virus, but they were also found at the cell surface and in the culture medium. Additionally, in the gM maturation-negative mutant virus-infected melanoma cells, typical syncytium formation was rarely seen, again indicating that mature gM functions in cell-to-cell spread via enhancement of syncytium formation.
Varicella-zoster virus (VZV) is a member of the human alphaherpesvirus subfamily that causes chickenpox and shingles (46, 47). In its genomic organization, VZV shows similarities to herpes simplex virus types 1 and 2 (HSV-1 and -2), with which it shares a number of homologous genes. In spite of the similarities, VZV has unique mechanisms for pathogenesis, including T-cell tropism and the formation of multinucleated polykaryocytes in skin infection (23), which are explained by differences in its gene expression.
The VZV genome is approximately 125 kb in size and contains at least 70 unique open reading frames (ORFs) (7). Among these ORFs, only the two transcripts from ORF42/45 and from ORFS/L are known to be generated from a spliced RNA (7, 19). VZV ORF42/45 is the homolog of the HSV-1 UL15 gene, which contains two large exons and is highly conserved among all herpesviruses, and ORFS/L is a unique gene carried by VZV.
The VZV envelope contains conserved glycoproteins common to all herpesviruses, including glycoproteins B, H, L, M, and N (gB, gH, gL, gM, and gN). The gMs are type III membrane proteins containing multiple (six to eight) transmembrane domains (TMDs). The gMs are highly conserved among the herpesviruses and are important for several steps in the viral life cycle, from penetration to egress, in all the subfamilies (1, 9, 18, 21, 26, 29, 35, 45, 48). Although the gMs in alphaherpesviruses are dispensable for viral replication in cell culture, their disruption reduces viral growth (1, 8, 9, 35). In contrast, the gMs encoded by human cytomegalovirus (HCMV) and Marek's disease virus serotype 1 are essential for viral replication (16, 45). In HCMV, the cytoplasmic tail of gM is required for trafficking during viral particle assembly at the assembly complex (22). Therefore, different herpesviruses seem to have various requirements for gM during viral replication. In addition, two recent reports showed the nuclear membrane targeting of HSV-1 gM; one was that the gM localized to the inner nuclear membrane (INM) and may be incorporated into perinuclear virions by budding through the INM (2), and the other showed that the gM was targeted to the nuclear membrane at an early phase of the infection independently of its known interaction partners (50).
In a number of herpesviruses, gM forms a complex with other viral proteins. One protein in the complex is gN, which is also conserved among all herpesviruses. gN is required for gM's transportation to the Golgi apparatus or to more distal compartments for viral assembly and egress (21, 24, 26-28). gM and gN form a complex that is covalently linked via a disulfide bond (18, 21, 25, 27, 48), and this bond is mediated between conserved cysteine residues located at the putative second extracellular loop of gM and at the position immediately adjacent to the amino-terminal boundary of the putative transmembrane domain of gN in HCMV (27). In HCMV, an additional, noncovalent linkage also contributes to the interaction, and this noncovalent linkage may be the core binding mechanism of the gM/gN complex (27). We recently reported that VZV gM is a glycosylated constituent of the virion, as reported for other herpesviruses, and functions in cell-to-cell spread (49). The VZV ORF9A gene, which encodes the gN homolog, has been reported to be nonessential for viral growth in cell cultures (38). However, the precise functions of VZV gM and gN have never been investigated.
For VZV, most studies of the mechanisms of virion assembly and egress, from primary envelopment to mature virion formation and involving the migration and processing of viral glycoproteins, have been performed using human embryo lung fibroblast (HELF) cells; a few reports provide detailed analyses using MeWo cells, as reviewed in reference 37. In MeWo cells, a viral deenvelopment and reenvelopment event, seen in HELF cells infected with VZV or other herpesviruses (see a review in reference 31), has not been observed. Therefore, the acquisition of viral glycoproteins is speculated to occur through fusion between Golgi apparatus-derived vesicles and vacuoles containing nascent enveloped particles from the endoplasmic reticulum (ER) (15, 17). A detailed electron microscopic examination of wild-type VZV infection in MeWo cells was performed, but the migration and processing of each glycoprotein which functions in primary or secondary envelopment were not examined with genetic support.
In this report, we found that the ORF50 gene encoded at least four different transcripts caused by alternative splicing, which were not required for viral growth in cell culture. We also found that VZV gM bound gN, forming a complex for the transportation of gM/gN, but the complex was not formed via the conserved disulfide bond. Two amino acid mutations of gM disrupted gM's maturation and the formation of the gM/gN complex. Interestingly, in cells infected with the gM maturation-negative mutant virus, immature gM was incorporated into intracellular particles, the enveloped particles were clearly detected at the enlarged perinuclear cisternae during late infection, and syncytium formation was rarely seen.
MATERIALS AND METHODS
Cells and viruses.
A human embryonic fibroblast cell line, MRC-5, was maintained as monolayer cultures in modified Eagle's minimal essential medium (MEM) (Nissui Pharmaceutical, Ueno, Tokyo) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL, Grand Island, NY) and used in the 26th to 32nd generations. A human melanoma cell line, MeWo, was grown in Dulbecco's modified Eagle medium (DMEM) (Gibco-BRL) supplemented with 10% FBS. MeWo cells stably expressing Cre recombinase, designated MeWo-Cre cells, were generated as follows. MeWo cells were transfected with pCX-Cre-neo using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, selected, and propagated in medium containing 500 μg/ml G418 (Nacalai Tesque, Kyoto, Japan). The parental VZV strain, Oka (pOka), which is the parental virus of recombinant pOka (rpOka) (32), rpOkaORF50AS(−), rpOkaORF50AS(−)Rev, rpOkagMim, and rpOkagMimRev, was propagated as described previously (13, 44). All the viruses were maintained in MeWo cells in DMEM with 3% FBS. The preparation of cell-free virus and purification of intracellular virus particles from the infected MeWo cells were described previously (41).
Virus particles secreted into the culture medium were also purified, as follows. The culture medium of VZV-infected MeWo cells 48 to 72 h after infection by cell-to-cell spread at a ratio of 1 infected cell to 10 uninfected cells was spun at 500 × g for 5 min at 4°C, and the supernatant was then subjected to ultracentrifugation at 52,000 × g for 2 h at 4°C. The pellet was dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 7.4) supplemented with protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), subjected to Histodenz (Sigma-Aldrich) gradient purification, and confirmed, as previously described (41).
Reverse transcriptase PCR (RT-PCR) and cloning for sequencing analysis.
Total RNA isolation, cDNA preparation from VZV-infected cells, and PCRs were carried out as described previously (41) using the Superscript III kit (Invitrogen) for cDNA synthesis. The PCR primer pairs used here were ORF50eccF-25 (5′-ACCGAATTCGTTGCTCTCCCCGGTGTTGG-3′) and ORF50xhoR (5′-ACCCTCGAGCTACTCCCACCCACTGTTTGATCG-3′). The PCR products amplified from cDNA were cloned into the TA cloning vector pCR-2.1-TOPO (Invitrogen), and each clone was sequenced.
Northern blot analysis.
Total RNAs from pOka-infected cells were separated by electrophoresis in a formaldehyde-1% agarose gel and transferred to a nylon transfer membrane (Hybond-N+) (GE Healthcare Bio-sciences, Piscataway, NJ). The membrane was prehybridized and hybridized in buffers containing 50% formaldehyde, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, 0.5% SDS, and 20 μg/ml denatured salmon sperm DNA. Hybridization was performed for 18 h at 42°C using 32P-labeled antisense oligonucleotides as probes. A Megalabel DNA 5′-end labeling kit (Takara, Otsu, Japan) was used for labeling. The oligonucleotide sequences used for the probes were ORF5051-75 (5′-CGCTTCCACCTCGGGTGTCGTGGTG-3′), ORF50341-370 (5′-ATACCGCCATGGCCATTGCCGTTAAAGCCG-3′), and ORF50904-928 (5′-AGGCCACGAGCGTTCCCAGGGACGG-3′). After hybridization, the membrane was washed and autoradiography was done using a FLA-7000 instrument (Fujifilm, Tokyo, Japan).
Plasmids.
pGEX-ORF50N was constructed to express an N-terminal ORF50 gene fragment corresponding to amino acids 2 to 35 in Escherichia coli, as described elsewhere (40). The primer pairs ORF50bamF (5′-ACCGGATCCGGAACTCAAAAGAAGGG-3′) and ORF50salR105 (5′-ACCGTCGACCCAATTAAGCGTATCC-3′) were used to amplify this region of the ORF50 gene from the pOka cDNA. The PCR products were inserted, in-frame, into the pGEX4T-1 bacterial expression vector (GE Healthcare Bio-Sciences) via the BamHI and SalI sites (underlined). The same procedure was used to construct pGEX-ORF9A. This DNA fragment (positions 61 to 147) of ORF9A was cloned into pGEX4T-1 via the EcoRI and SalI sites (underlined). The primer pairs were ORF9AecoF61 (5′-ACCGAATTCGACAGCAGTGGGGAGCCAAACTTTGC-3′) and ORF9AsalR147 (5′-ACCGTCGACCATTGATCCGTCGATATAAACTCC-3′). The full-length cDNA of ORF50 was amplified by PCR using the primer pairs ORF50eccF-25 and ORF50xhoR and cloned into the eukaryotic expression vector pCAGGS-MCS via the EcoRI and XhoI sites (underlined), resulting in pCAGGS/ORF50. The pCAGGS plasmid was kindly provided by Jun-ichi Miyazaki (Osaka University, Japan) (33). The ORF9A gene was amplified from the cDNA by PCR using the primer pairs ORF9AkpnF-20 (5′-ACCGGTACCTATTTACTGCCGTTTCC-3′) and ORF9AmycxhoR (5′-ACCCTCGAGCAGATCCTCTTCTGAGATGAGTTTTTGTTCCCACGTGCTGCGTAATACAG-3′) and cloned into pCAGGS-MCS via the KpnI and XhoI sites (underlined) to generate pCAGGS/ORF9A, containing a c-myc tag (5′-GAACAAAAACTCATCTCAGAAGAGGATCTG-3′) at the C terminus. A Cre recombinase-expressing plasmid, pCX-Cre, was a generous gift from Masaru Okabe (Osaka University, Japan). pCX-Cre-neo was generated as follows. A nucleotide fragment including the neomycin resistance gene was extracted from pMC1neopolyA (Stratagene, La Jolla, CA) via the SalI and XhoI restriction sites and ligated into SalI-digested pCX-Cre.
Antibodies.
To produce polyclonal antibodies (Abs) against the first ectodomain in the N terminus of gM, E. coli BL21 was transformed with pGEX-ORF50N, to express the recombinant fusion protein, glutathione S-transferase (GST)-ORF50N, which was purified and used to immunize a rabbit (Sigma Genosys, Hokkaido, Japan). The anti-ORF50N Abs were purified with GST-conjugated N-hydroxysuccinimide (NHS)-activated Sepharose (GE Healthcare Bio-Sciences) and GST-ORF50N-conjugated NHS-activated Sepharose. To produce anti-gN polyclonal Abs, E. coli BL21 was transformed with pGEX-ORF9A to express a GST-ORF9A fusion protein, which was purified with glutathione Sepharose 4B (GE Healthcare Bio-Sciences) and used to immunize rabbits (MBL, Nagoya, Japan). The anti-gN Abs were purified with NHS-activated Sepharose as described above, using a purified GST or GST-ORF9A fusion protein. An anti-gM Ab that recognizes the C-terminal region of gM was generated previously, and the anti-ORF16 Ab, anti-ORF49 Ab, and anti-gB-C Ab were previously described (41). The following Abs for cellular proteins were purchased: sheep anti-human TGN46 (AHP500G; AbD Serotec, Oxford, United Kingdom), anticalnexin mouse monoclonal Ab (MAb) (clone AF18; Abcam, Cambridge, United Kingdom), anti-lamin A/C mouse MAb (clone 14; BD Biosciences Pharmingen, San Diego, CA), and Myc tag mouse MAb (clone 9B11; Cell Signaling Technology, Inc., Danvers, MA). Alexa Fluor 488-labeled donkey anti-sheep immunoglobulin G (IgG), Alexa fluor 594-labeled donkey anti-rabbit IgG (Invitrogen) and Cy5-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were used as secondary Abs, and Hoechst33342 (Sigma-Aldrich) was used for nuclear staining.
Construction of mutant ORF50 expression plasmid.
The pCAGGS/ORF50 plasmid, including the full-length ORF50 gene, was used as a template to construct the ORF50 mutant. The maturation-negative gM mutant, pCAGGS/gMim, was generated by a QuikChange multisite-directed mutagenesis kit (Stratagene), in accordance with the manufacturer's recommendations, using the primers listed in Table 1. The resulting plasmid included the nucleotide mutations GT124CC and AGGA900CATG, in which the numbers indicate the first nucleotide position of the mutation.
TABLE 1.
Primers used for mutation of the ORF50 gene
| Primer | Sequencea | Amino acid mutation |
|---|---|---|
| ORF50 124gt-cc | 5′-ttg gat aat tca gCC gat gat gtt cac-3′ | 42 Valine to proline (V42P) |
| ORF50 125t-c | 5′-ttg gat aat tca ggC gat gat gtt cac-3′ | 42 Valine to alanine (V42A) |
| ORF50 448t-c | 5′-gct ccg gtg ttg Cat ggc tag ttg tag atc-3′ | |
| ORF50 900a-c | 5′-att ata tac acg tgt taa tCg gac cgt ccc-3′ | |
| ORF50 900agga-catg | 5′-att ata tac acg tgt taa tCA TGc cgt ccc-3′ | 301 Glycine to methionine (G301M) |
| ORF50 1101g-a | 5′-gca tat gtg tat cat cga caA aaa cgc ag-3′ |
Nucleotides that differ from those of wild-type ORF50 are shown in capital letters.
BAC mutagenesis by recA-mediated recombination.
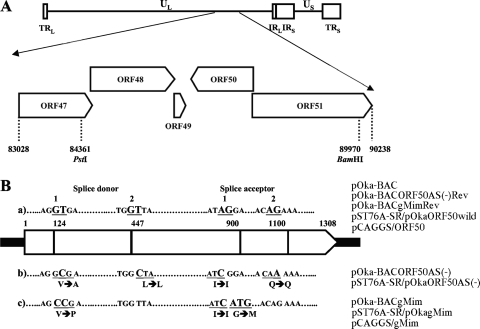
To generate mutant bacterial artificial chromosomes (BACs) for this study, we used the pOka-BACΔgM backbone, in which the ORF50 sequence was replaced with a kanamycin resistance gene (49), and recA-mediated recombination was performed to introduce multiple point mutations in ORF50. For the shuttle plasmid, 5.6 kbp of viral DNA, corresponding to nucleotide positions 84,361 to 89,970 of pOka, was excised from the pOka-BAC genome (32) using PstI and BamHI (see Fig. 4A) and cloned into the plasmid pST76A-SR, which carries the recA and sacB genes (kindly provided by Ulrich H. Koszinowski, Max von Pettenkofer Institut fur Virologie, Ludwig-Maximilians-Universitat Munchen, Germany) (16), designated pST76A-SR/pOkaORF50wild. This plasmid was used to make revertants of all the mutant BACs (see Fig. 4B-a).
FIG. 4.
Construction of ORF50 mutant plasmids and recombinant BAC genomes for ORF50 point mutants. (A) Location of ORF47 to ORF51 in the unique long region (UL) segment of the VZV strain pOka genome; terminal repeats (TR), unique short region (US), and internal repeats (IR) are indicated. PstI and BamHI recognition sites for cloning the gene region between both sites into the shuttle plasmid pST76A-SR for the revertant and mutant BACs are represented with their nucleotide position number in the pOka genome. (B) Schematic of the nucleotide mutations at or around the splice donor and acceptor sites with the amino acid substitutions in the ORF50 gene region. The BACs, shuttle plasmids, and expression plasmids are shown at right. The ORF50 gene is represented by the wide pointed rectangles, and splice donor and acceptor sequences are shown as underlined bold capital letters with the nucleotide position numbers, in panel a. In panels b and c, larger bold capital letters show the mutated nucleotides that differ from the wild-type ORF50 gene in panel a, and the underlined nucleotides indicate the in-frame codons, including silent mutations and amino acid changes, which are shown below the nucleotides.
The plasmids pST76A-SR/pOkaORF50AS(−) and pST76A-SR/pOkagMim (Fig. 4B-b and -c) were generated by using a QuikChange multisite-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's recommendations using mutation primers (Table 1) based on pST76A-SR/pOkaORF50wild (listed in Table 2). The recA-mediated allelic replacement in E. coli was done as described elsewhere (16).
TABLE 2.
Plasmids used for generation of revertant and mutant genomes
| Plasmid | Nucleotide mutationsa | Amino acid mutation(s) |
|---|---|---|
| pST76A-SR/pOkaORF50wild | ||
| pST76A-SR/pOkaORF50AS(-) | 125 T to C, 448 T to C, 900 A to C, 1101 G to A | V42A |
| pST76A-SR/pOkagMim | 124 GT to CC, 900 AGGA to CATG | V42P, G301M |
Number shows the first nucleotide position of the mutation.
Reconstitution of BAC cassette-free mutant and revertant viruses.
MeWo cells in one well of a six-well plate were transfected by Lipofectamine 2000 (Invitrogen) with 3 μg of BAC DNA, which had been purified using a Nucleobond BAC 100 kit (Machery-Nagel, Düren, Germany). After typical cytopathic effects (CPE) were seen in cells expressing green fluorescent protein (GFP), cell-free virus was prepared as described above and used to infect MeWo-Cre cells, to excise the BAC cassette containing the guanine phosphoribosyl transferase gene and gfp.
Western blot analysis.
Western blotting (WB) was performed as described previously (40).
Immunoprecipitation.
Abs were coupled with Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-Sciences) in phosphate-buffered saline (PBS) (pH 7.4) for 2 h at room temperature, washed three times with 0.2 M sodium borate buffer (pH 9.0), and cross-linked in 20 mM dimethyl pimelimidate-2HCl (Pierce Biotechnology, Rockford, IL) for 30 min at room temperature. The Ab-conjugated beads were washed once with 0.2 M ethanolamine (pH 8.0), rotated for 30 min to stop the cross-linking reaction, washed three times with PBS, and stored in PBS containing 0.05% NaN3 at 4°C. The cells were lysed in TNE buffer (0.01 M Tris-HCl [pH 7.4], 0.15 M NaCl, 1% NP-40, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) on ice for 30 min and centrifuged at 217,000 × g for 1 h; the supernatant was then collected. The cell lysates were precleared by Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-Sciences), incubated with the Ab-cross-linked beads for more than 4 h at 4°C, and washed five times in the lysis buffer to remove unbound material. The protein-Ab complex-bound beads were incubated in 0.1 M glycine (pH 2.5) to elute only the proteins from the protein-Ab-bead complexes, and the supernatant was collected by centrifugation as the immunoprecipitated proteins. The eluted proteins were neutralized by adding 1 M Tris-HCl (pH 9.2) and subjected to WB analysis.
Immunohistochemical analysis.
Indirect immunofluorescence (IF) assays were performed as described previously (41). All samples were examined on a Leica confocal microscope (model DMIRE2; Leica Microsystems, Wetzlar, Germany). All images were captured by serial z-stacks at intervals of 0.6 μm and analyzed by using the Leica confocal software (Leica Microsystems).
Analysis of replication kinetics and plaque sizes.
To analyze the growth kinetics of the recombinant viruses, an infectious center assay was performed on MeWo cells and MRC-5 cells as described previously (13). Images of the plaques were captured, and the area was calculated as described previously by using the ImageJ software program (41).
Syncytium formation analysis.
The recombinant virus-containing BAC sequence (rpOka-BAC and rpOka-BACgMim)-infected MeWo cells were spread at a ratio of 1 infected cell to 10 uninfected cells, and the images were captured every 24 h by a TE2000-U microscope (Nikon, Tokyo, Japan).
Electron microscopy.
Electron microscopy was performed as described previously (41). Briefly, the cells were fixed at 48 h postinfection (hpi) in 0.05 M cacodylate-buffered 2.5% glutaraldehyde solution, washed with 5% sucrose buffered with 0.1 M cacodylate, and postfixed in 0.1 M cacodylate buffered 1% osmium tetroxide at room temperature. The samples were block stained with a 0.5% aqueous solution of uranyl acetate for 1 h, dehydrated with a graded series of ethanol, and embedded in EPONmix. Ultrathin sections were cut with an ultramicrotome (Reichert-Nissei Ultracuts; Leica Microsystems, Wetzlar, Germany) and stained with uranyl acetate and lead citrate. The samples were observed with a Hitachi H7100 electron microscope (Hitachi High-Technologies).
RESULTS
Identification of novel splice variants of ORF50.
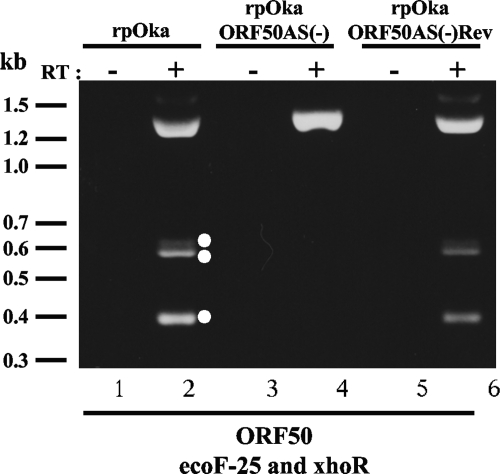
To analyze the expression of ORF50 mRNA in VZV-infected cells, MeWo cells were infected with the recombinant parental Oka strain (rpOka), total RNA was extracted, and RT-PCR was performed. Surprisingly, four bands were amplified by the primer set ORF50ecoF-25 and ORF50xhoR (see Fig. 2B-b) from the template with RT but not without RT (Fig. 1, lanes 2 and 1, respectively). A 1,332-bp band predicted from the ORF50 gene sequence was detected; the other three bands were around 600 bp and 400 bp (Fig. 1, lane 2). When we used stringent annealing temperatures (up to 64°C; data not shown), the other three PCR products were still amplified, but they were not amplified from genomic DNA (data not shown).
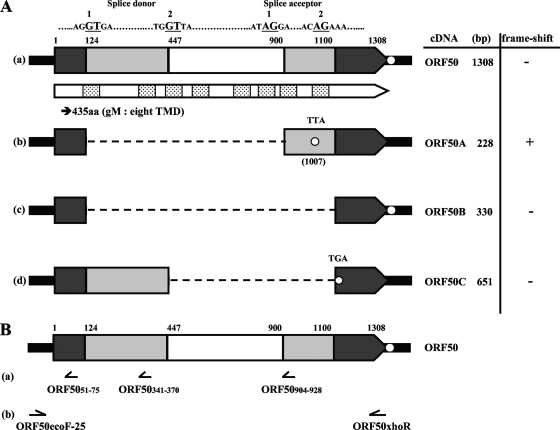
FIG. 2.
Schematic of ORF50 variants. (A) The full-length cDNA of ORF50 (a) and the cDNA variants (ORF50A, ORF50B, and ORF50C) (b, c, and d, respectively) are shown as wide, pointed rectangles. The sequences and nucleotide positions of the splice donor and acceptor sites are indicated as bold underlined capital letters just above the ORF50 cDNA. The spliced sequences in the cDNA variants are indicated as light and white boxes in ORF50 and as a dotted line in the cDNA variants. The dark boxes indicate the sequences common to all the transcripts from the ORF50 region. The white box indicates the intronic sequence common among the ORF50A, B-, and -C variants. The open circle shows each stop codon site. The predicted gene product of ORF50 is denoted by a narrow pointed rectangle just below the cDNA, and the boxes containing dots in panel a indicate the TMDs predicted by the TMHMM program. (B) The localization and direction of each probe used for Northern blot analysis (a) or the primer set used for PCR amplification (b) is shown below ORF50.
FIG. 1.
RT-PCR in the ORF50 gene region of rpOka, rpOkaORF50AS(−), and rpOkaORF50AS(−)Rev-infected MeWo cells and identification of novel transcripts from the ORF50 gene region. RNAs were extracted when full CPE was observed in each sample of virus-infected MeWo cells. RT-PCRs were carried out using a specific primer set for the ORF50 gene (lanes 2, 4, and 6), and PCRs without RT were carried out as a control for contamination with genomic DNA (lanes 1, 3, and 5). Molecular mass markers are shown at the left. White circles at the right of lane 2 indicate the spliced transcripts. The experiments were done twice independently, and one of them is shown here.
The PCR products were cloned into pCR2.1-TOPO (Invitrogen) and sequenced twice independently. As shown in Fig. 2A, three novel transcripts generated by alternative splicing were identified and named ORF50 variant A (ORF50A), ORF50 variant B (ORF50B), and ORF50 variant C (ORF50C). As we reported previously, the full-length ORF50 gene product predicted from the genomic DNA possesses eight TMDs (Fig. 2A-a) and corresponds to glycoprotein M (gM) (49). The newly identified variants were alternatively spliced at well-conserved splice donor and acceptor sequences (GT-AG) that we identified by analyzing the ORF50 sequence. The first splice donor started at nucleotide position 124 and the second splice donor at nucleotide (nt) 447; the first splice acceptor started at nt 900, and the second splice acceptor at nt 1,100 (Fig. 2A).
The ORF50A variant was produced by alternative splicing at splice donor 1 and splice acceptor 1. Posttranscriptional excision from nt 124 to 901 of ORF50 caused a frameshift in ORF50A (Fig. 2A-b). ORF50B, amplified as a 330-bp cDNA, resulted from the removal of the coding region from nt 124 to 1,101 within ORF50 (Fig. 2A-c). ORF50C, amplified as a 651-bp cDNA, resulted from alternative splicing between splice donor 2 and splice acceptor 2 and had a newly created TGA at nt 445 of ORF50 (Fig. 2A-d). All of the variants were also detected by RT-PCR following cloning and sequencing analyses of pOka-infected MRC-5 cells and of MeWo cells transfected with the ORF50 expression plasmid, pCAGGS/ORF50, indicating that the alternative splicing in the ORF50 gene region was highly stable independently of cell types and did not require any other viral factors. The prediction of the transmembrane topology was performed by the TMHMM software program, as described elsewhere (49).
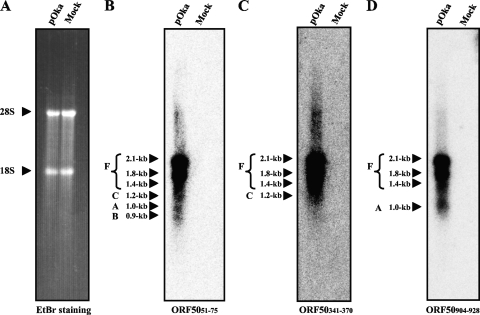
Detection of mRNA expression by Northern blot analysis.
To confirm the expression of the ORF50 mRNA isoforms in pOka-infected MeWo cells, we analyzed their expression on a Northern blot (Fig. 3). We used antisense oligonucleotides as the hybridization probes (Fig. 2B-a). One probe, ORF5051-75, located before splice donor 1, hybridized to six mRNA isoforms, 2.1 kb, 1.8 kb, 1.4 kb, 1.2 kb, 1.0 kb, and 0.9 kb (Fig. 3B). Another probe, ORF50341-370, located between splice donors 1 and 2, hybridized to four isoforms, 2.1 kb, 1.8 kb, 1.4 kb, and 1.2 kb (Fig. 3C), and probe ORF50904-928, located between splice acceptors 1 and 2, hybridized to four isoforms, 2.1 kb, 1.8 kb, 1.4 kb, and 1.0 kb (Fig. 3D). All of the probes, therefore, hybridized with the 2.1-kb, 1.8-kb and 1.4-kb mRNAs, and the mRNAs were abundant and detectable even with short exposure times (data not shown). Our analysis indicated that the full-length ORF50 mRNA described in Fig. 2A-a was derived from the 2.1-, 1.8-, and 1.4-kb mRNAs detected by the Northern blotting, and ORF50A, ORF50B, and ORF50C were derived from the 1.0-kb, 0.9-kb, and 1.2-kb mRNAs, respectively.
FIG. 3.
Northern blot analysis of ORF50 variants in pOka-infected MeWo cells. Total RNA was extracted from pOka-infected cells and mock-infected cells at 72 h postinfection (with full CPE in the infected cells) and separated on a formaldehyde-1% agarose gel. Ethidium bromide (EtBr) staining is shown for the size markers, and the 28S and 18S ribosomes are indicated by arrowheads (A). The blots were probed with three antisense oligonucleotides, whose positions are shown in Fig. 2B-a. The letters and size with an arrowhead or arrowheads grouped with a bracket at the left of each panel indicate the ORF50 variants predicted from each size. “F” indicates the ORF50 full-size cDNA without alternative splicing.
Novel splice variants of the ORF50 gene are not translated into proteins and are not essential for viral growth in cell culture.
To elucidate the functions of the alternatively spliced mRNAs and their products in infected cells, a mutant VZV BAC that was negative for alternative splicing (AS) in the ORF50 gene, named pOka-BACORF50AS(−), which contained one nt substitution in each splice donor and acceptor site, was generated by recA-mediated BAC mutagenesis using pST76A-SR/pOkaORF50AS(−) (Fig. 4B-b). Infectious virus, rpOkaORF50AS(−), was successfully reconstituted. The revertant of pOka-BACORF50AS(−), designated pOka-BACORF50AS(−)Rev, was also generated using pST76A-SR/pOkaORF50wild (Fig. 4B-a), based on pOka-BACORF50AS(−), and the revertant virus, rpOkaORF50AS(−)Rev, was obtained by the transfection of MeWo cells with the revertant BAC following the excision of the BAC cassette from rpOka-BACORF50AS(−)Rev.
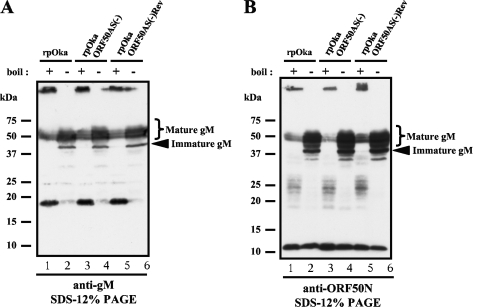
The alternative splicing of the ORF50 gene was successfully suppressed in rpOkaORF50AS(−)-infected cells as expected (Fig. 1, lane 4), and it was recovered in the revertant virus-infected cells (Fig. 1, lane 6; compared with the wild-type rpOka-infected cells, Fig. 1, lane 2). WB analysis of rpOka-infected MeWo cells revealed several bands that reacted with an anti-ORF50N Ab and an anti-gM Ab, whose epitopes are, respectively, in the first 34 amino acids of gM, excluding the first methionine, and its last 13 amino acids (Fig. 5A and B, lanes 1 and 2). The band found at the top of the gel in lane 1 of Fig. 5A or B was thought to be the result of the aggregation of gM proteins by boiling, while 18-kDa (Fig. 5A, lane 1) and 23- to 28-kDa (Fig. 5B, lane 1)bands were clearly detected even by the boiling. Therefore, the samples for gM detection by WB were prepared with and without boiling. By the boiling, a specific 18-kDa band reacted with anti-gM Ab and 23- to 28-kDa and 12-kDa bands reacted with anti-ORF50N Ab were apparently detected; therefore, these bands were thought to be derived from alternatively spliced mRNAs.
FIG. 5.
Comparison of protein expression from the ORF50 gene between rpOka, rpOkaORF50AS(−), and rpOkaORF50AS(−)Rev-infected MeWo cells. Each virus was propagated by cell-to-cell spread at a ratio of 1 infected cell to 10 uninfected cells, lysed when full CPE was observed (almost 72 hpi), and subjected to WB with anti-gM Ab (A) and anti-ORF50N Ab (B). All cells were lysed in SDS-PAGE sample buffer and either boiled (lanes 1, 3, and 5 of panels A and B) or prepared without boiling (lanes 2, 4, and 6 of panels A and B) to visualize all of the proteins translated from the ORF50 gene.
To examine protein expression, WBs of rpOkaORF50AS(−)- and rpOkaORF50AS(−)Rev-infected cells were performed (Fig. 5). Unexpectedly, there were no obvious differences in protein expression from the ORF50 gene among rpOka-, rpOkaORF50AS(−)-, and rpOkaORF50AS(−)Rev-infected cells, suggesting that the bands detected by boiling were not the products of spliced mRNAs. Furthermore, viral plaque formations were similar among these three viruses in both MeWo and MRC-5 cells. In MeWo cells, the mean areas for the plaques generated were not significantly different among rpOka (0.528 mm2; standard error of the mean [SEM], 0.017), rpOkaORF50AS(−) (0.535 mm2; SEM, 0.027), and rpOkaORF50AS(-)Rev (0.496 mm2; SEM, 0.017). As well, in MRC-5 cells, the mean areas for the plaques generated were not significantly different among rpOka (0.583 mm2; SEM, 0.033), rpOkaORF50AS(−) (0.581 mm2; SEM, 0.036), and rpOkaORF50AS(−)Rev (0.524 mm2; SEM, 0.032). In addition, the similar growth kinetics was seen among the viruses in the infectious center assay of both MeWo and MRC-5 cells (data not shown). These results indicated that the alternatively spliced OR50 transcripts were not translated and were dispensable for the VZV life cycle in cell culture. Since it is still unknown what the lower bands detected by anti-gM Ab (18 kDa) and anti-ORF50N Ab (12 kDa and 23 to 28 kDa) are, further analyses would be required.
Binding properties of gM and gN in VZV.
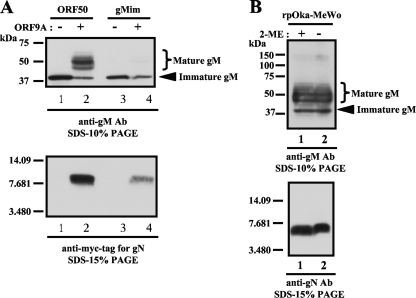
We next investigated the functions of gM in VZV-infected cells. We previously reported that at least one other viral factor was required for gM maturation (49). In herpesviruses, gN is a known binding partner of gM (21, 24, 26-28). Therefore, we examined the interaction of the gM and gN of VZV. The ORF9A gene was cloned into pCAGGS with a c-Myc tag at the C terminus (pCAGGS/ORF9A). The cotransfection of pCAGGS/ORF50 and pCAGGS/ORF9A into MeWo cells clearly resulted in the maturation of gM (Fig. 6A, upper panel, lane 2), as observed in rpOka-infected cells (Fig. 6B, upper panel, lane 1), and the expression of gN was confirmed by an anti-Myc-tag Ab (Fig. 6A, lower panel, lane 2) and an anti-gN Ab (data not shown). Thus, VZV gN was also the binding partner of VZV gM, and it enabled gM to mature without the presence of any other viral factors.
FIG. 6.
Binding properties of gM and gN in transiently transfected cells and rpOka-infected cells. (A) MeWo cells were transfected with pCAGGS/ORF50 (lanes 1 and 2) or pCAGGS/gMim (lanes 3 and 4) with (lanes 2 and 4) or without (lanes 1 and 3) pCAGGS/ORF9A. The cell lysates were subjected to WB with an anti-gM Ab (upper panel) and an anti-Myc-tag Ab to detect gN (lower panel). (B) The lysate of rpOka-infected MeWo cells was prepared with (lanes 1) or without (lanes 2) 2-mercaptoethanol (2-ME), and WB was performed with the anti-gM Ab (upper panel) and anti-gN Ab (lower panel).
We also confirmed the interaction between gM and gN in rpOka-infected cells. WB analysis under reducing or nonreducing conditions (Fig. 6B) and immunoprecipitation following WB (IP-WB) (see Fig. 9A and B lane 8) were performed with rpOka-infected cell lysates. Under reducing or nonreducing conditions, gM was mainly and equally detected as broad bands of 40 to 60 kDa (Fig. 6B upper panel). gN was detected as a single 7-kDa band and showed no difference in size between reducing and nonreducing conditions (Fig. 6B, lower panel). The interaction between gM and gN in rpOka-infected cells was verified by IP-WB, as shown in lane 8 of Fig. 9A and B. The anti-gM Ab precipitated both gM and gN from the rpOka-infected cell lysates (Fig. 9A and B, lane 8 in each). Since an anti-gN Ab was not available for IP, we could not confirm this finding by the reverse experiment.
FIG. 9.
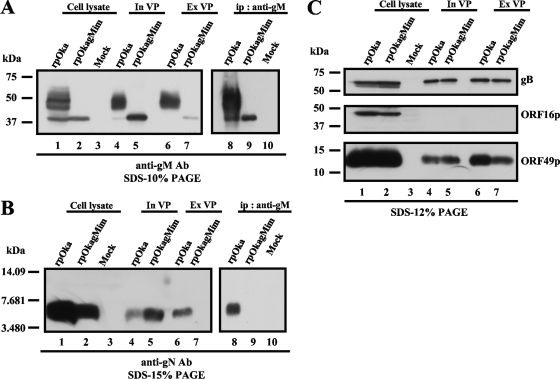
Incorporation of mutant gM into virus particles and the binding properties of mutant gM and gN. rpOka-, rpOkagMim-, and mock-infected cells (lanes 1, 2, and 3) were lysed in TNE buffer and subjected to WB with the anti-gM Ab (A), anti-gN Ab (B), anti-gB-C Ab (C), anti-ORF16 Ab (C), or anti-ORF49 Ab (C). Intracellular virus particles (In VP) of rpOka and rpOkagMim (lanes 4 and 5) were lysed in SDS-PAGE sample buffer and subjected to WB with the anti-gM Ab (A), anti-gN Ab (B), anti-gB-C Ab (C), anti-ORF16 Ab (C), or anti-ORF49 Ab (C). The extracellular virus particles (Ex VP) of rpOka and rpOkagMim (lanes 6 and 7) were lysed in SDS-PAGE sample buffer and subjected to WB with the anti-gM Ab (A), anti-gN Ab (B), anti-gB-C Ab (C), anti-ORF16 Ab (C), or anti-ORF49 Ab (C). The lysate of rpOka-, rpOkagMim-, and mock-infected cells was immunoprecipitated by the anti-gM Ab and subjected to WB with the anti-gM Ab (A) (lanes 8, 9, and 10) and anti-gN Ab (B) (lanes 8, 9, and 10). An equal amount of each sample was applied to each lane of the gels.
Our results suggested that gM and gN interacted with each other and that gM maturation could take place only through gM's interaction with gN. In addition, and in contrast to reports for other herpesviruses (18, 21, 25, 27, 48), the results obtained under reducing and nonreducing conditions showed that the VZV gM-gN interaction may not be supported by a disulfide bond between conserved cysteine residues.
gM and gN require each other for transportation in the secretory pathway.
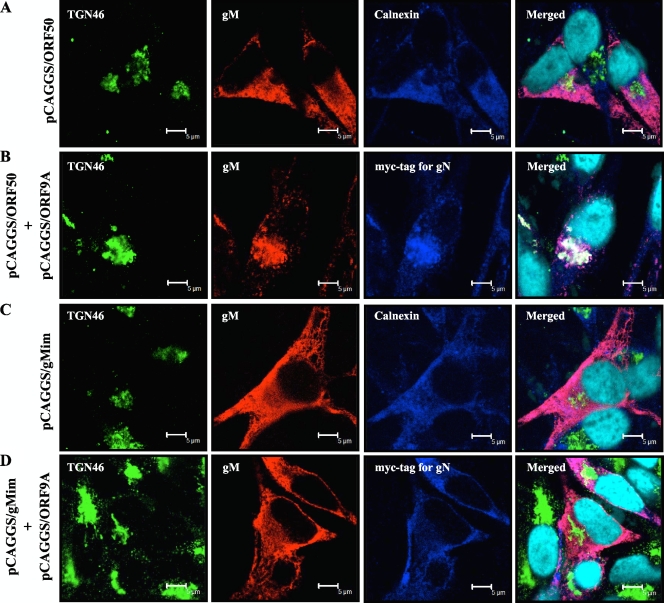
When gM was expressed alone in a transient expression system, gM localized predominantly at the ER and did not colocalize with either the Golgi apparatus marker (data not shown) or the trans-Golgi network (TGN) marker, TGN46 (Fig. 7A). The coexpression of gM and gN led to both of them accumulating at the juxtanuclear region with TGN46 (Fig. 7B), as seen in rpOka-infected cells (Fig. 8A).
FIG. 7.
Localization of wild-type or mutant gM in transiently transfected MeWo cells. MeWo cells were transfected with pCAGGS/ORF50 (A) or pCAGGS/gMim (C) or cotransfected with pCAGGS/ORF9A and pCAGGS/ORF50 or pCAGGS/gMim (B or D) and fixed in ice-cold acetone/methanol at 48 h posttransfection. The fixed cells were triply labeled for the TGN marker TGN46 (green), gM (red), and the ER marker calnexin (blue) (A and C) or gN with anti-Myc-tag Ab (blue) (B and D). Nuclei were stained with Hoechst 33342 (cyan). Scale bars = 5 μm.
FIG. 8.
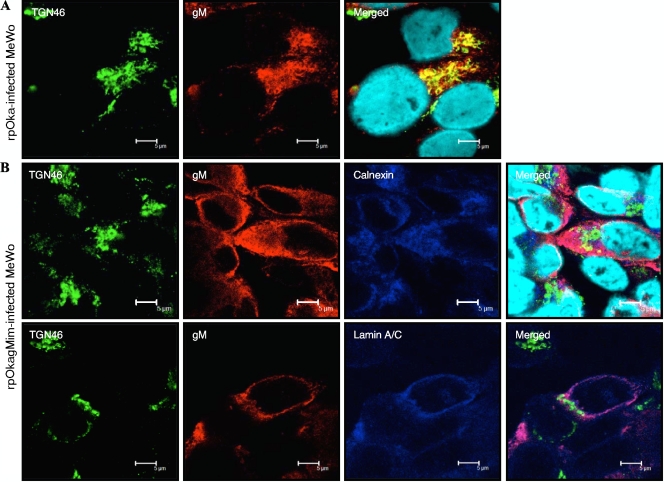
Localization of mutant gM in rpOkagMim-infected cells. MeWo cells were infected with rpOka (A) or rpOkagMim (B) cell-free virus at a multiplicity of infection (MOI) of 0.05 and fixed in ice-cold acetone-methanol at 48 hpi. The fixed cells were doubly (A) labeled for the TGN marker TGN46 (green) and gM (red) or triply (B) labeled for TGN46 (green), gM (red), and the ER marker calnexin (blue) (upper panel in B) or the nuclear membrane marker lamin A/C (blue) (lower panel in B). Nuclei were stained with Hoechst 33342 (cyan). Scale bars = 5 μm.
This result indicated that gM and gN localized to the ER after translation and bound each other there, and the steps were required for their transport along the secretory pathway, i.e., the Golgi apparatus and TGN.
Characterization of maturation-negative mutant gM in transiently expressing cells and infection with the mutant virus.
In the process of generating the alternative splicing-negative mutant plasmids, we also introduced several nucleotide mutations around the splice donors and acceptors that caused amino acid changes. First, all gM mutant plasmids were transfected with or without pCAGGS/ORF9A into MeWo cells to confirm the alternative splicing suppression, and only single-nucleotide substitution at all splice donors and acceptors was sufficient to suppress the alternative splicing, as shown in Fig. 1. Interestingly, in the series of experiments using the gM mutant and the gN expression plasmids, gM maturation was not seen in some gM mutants even when they were coexpressed with gN (data not shown). Among them, two amino acid changes, valine to proline at position 42 (V42P) and glycine to methionine at 301 (G301M), were found to be responsible for gM maturation. Therefore, pCAGGS/gMim with V42P and G301M mutations was generated (Fig. 4B-c) and used for further analyses.
Although the coexpression of wild-type gM and gN resulted in gM maturation and gM/gN transportation to the TGN (Fig. 6A, lanes 2, and Fig. 7B, respectively), the transfection of pCAGGS/gMim generated only the immature form of gM, regardless of the presence of gN (Fig. 6A, lanes 3 and 4). In these cells, the gM mutant and gN were targeted mainly to the ER (data not shown) but never transported to the TGN (Fig. 7D), as seen when cells were transfected with pCAGGS/gMim alone (Fig. 7C).
To characterize and elucidate the functions of immature and mature gM in infected cells, pOka-BACgMim was generated by recA-mediated mutagenesis (Fig. 4B-b), and rpOkagMim was reconstituted in MeWo cells. As shown previously (49) and in Fig. 8A, gM localized mainly to the TGN in wild-type virus-infected cells. In rpOkagMim-infected MeWo cells, mutant gM was located in the ER (Fig. 8B, upper panels) and partially at the nuclear membrane, which was identified by staining with anti-lamin A/C (Fig. 8B, lower panels), but it was never transported to the TGN (Fig. 8B).
WB analysis detected only immature gM in the rpOkagMim-infected cells (Fig. 9A, lane 2), and the total amount of the mutant gM in the cells was lower than that of gM in rpOka-infected cells (Fig. 9A, lane 1). Apparently reduced gN expression was detected in the mutant virus-infected cells (Fig. 9B lane 2) compared with that in rpOka-infected cells, although it was the same size as that in rpOka-infected cells (Fig. 9B, lane 1). The expression levels of gB, ORF16p, and ORF49p were the same in cells infected with either virus (Fig. 9C, lanes 1 and 2).
Mutant gM cannot form gM/gN complex, reducing the incorporation of gM and gN into extracellular particles.
To elucidate the role of the interaction between gM and gN, IP with the anti-gM Ab followed by WB with the anti-gM Ab (Fig. 9A, lanes 8 to 10) or anti-gN Ab (Fig. 9B, lanes 8 to 10) was performed using the lysates of rpOka-, rpOkagMim-, and mock-infected MeWo cells. The anti-gM Ab precipitated mature and immature gMs with gN from the rpOka-infected cells (Fig. 9A and B, lane 8). In the rpOkagMim-infected cells, the anti-gM Ab precipitated only immature gM (Fig. 9A, lane 9), and no gN was detected (Fig. 9B, lane 9).
Next, we purified intracellular virus particles which were obtained from infected MeWo cells and extracellular particles from the culture medium (see Materials and Methods). WB analysis showed that mature gM and gN were incorporated into both the intra- and extracellular virus particles, as expected (Fig. 9A and B, lanes 4 and 6). The purification of the virus particles was confirmed by the detection of ORF16p, which is predicted to encode the processivity subunit of DNA polymerase and to be a nonstructural protein (Fig. 9C, middle panel). To compare the incorporation of viral proteins into virus particles between rpOka and rpOkagMim, intracellular and extracellular particles were subjected to WB with an anti-gB-C Ab (Fig. 9C, upper panel) or the anti-ORF49 Ab (Fig. 9C, lower panel). The loading quantity was normalized to the expression levels of gB and the ORF49 protein (ORF49p), since both gB (Fig. 9C, upper panel, lanes 1 and 2) and ORF49p, one of the most abundant tegument proteins (Fig. 9C, lower panel, lanes 1 and 2), were expressed at the same level in rpOka- and rpOkagMim-infected cells. gB and ORF49p were also incorporated at the same level into the intracellular particles (see Fig. 9C, upper and lower panels, respectively, lanes 4 and 5 in each) and extracellular particles (see Fig. 9C, upper and lower panel, respectively, lanes 6 and 7 in each) in both viruses. However, under the same conditions, although the expression levels of immature gM and of gN in the rpOkagMim-infected cells were obviously lower than those of mature gM and gN in the rpOka-infected cells, as mentioned above, the quantity of gM incorporated into the intracellular particles of rpOkagMim was comparable to that of rpOka (Fig. 9A, lanes 5 and 4, respectively) and the quantity of gN incorporated into the intracellular particles of rpOkagMim was apparently more abundant than that of rpOka (Fig. 9B, lanes 5 and 4, respectively). In contrast, the extracellular particles of rpOkagMim contained a markedly lower quantity of immature gM (Fig. 9A, lane 7) than its intracellular particles (Fig. 9A, lane 5). Additionally, no gN was detected in the extracellular particles of rpOkagMim (Fig. 9B, lane 7).
These findings indicated that in rpOkagMim-infected cells, mutant gM could not form a complex with gN, owing to the amino acid mutations V42P and G301M, but immature gM and gN were independently incorporated into intracellular particles.
gM/gN complex functions in cell-to-cell spread by enhancing syncytium formation.
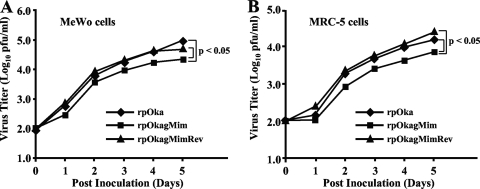
Viral growth was analyzed by a plaque size assay and an infectious center assay in both MeWo cells and MRC-5 cells. The rpOkagMim (0.159 mm2; SEM, 0.018 in MeWo cells, 0.177 mm2; SEM, 0.015 in MRC-5 cells) exhibited significantly reduced plaque formation compared with that of rpOka (0.633 mm2; SEM, 0.029 in MeWo cells, 0.680 mm2; SEM, 0.037 in MRC-5 cells) or rpOkagMimRev (0.620 mm2; SEM, 0.027 in MeWo cells, 0.622 mm2; SEM, 0.033 in MRC-5 cells) in both cell types (P < 0.01, Student's t test, for each), and it showed a reduced effect on viral growth compared with that of rpOka or rpOkagMimRev for both cell types (P < 0.05 in MeWo cells and P < 0.05 in MRC-5 cells) (Fig. 10A and B), as reported for rpOkaΔgM in our previous study (49).
FIG. 10.
Growth properties of recombinant viruses in MeWo cells and MRC-5 cells. Cells were infected with rpOka, rpOkagMim, or rpOkagMimRev (log10 2.0 PFU/well), harvested at the indicated time points, serially diluted, added to newly prepared cells on 6-well plates, and cultured for 5 days. The plaques were stained with crystal violet and counted. Each point represents the mean titer for two wells of one experiment. The experiments were done twice independently. Statistical significance was determined by Student's t test.
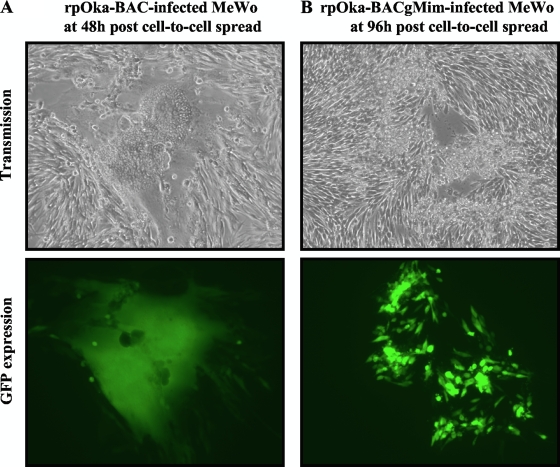
Since syncytium formation is the hallmark of VZV infection in MeWo cells, it was examined and compared among the three viruses. To visualize the area of infection clearly, the viruses containing a BAC cassette with the gfp gene were used in this experiment. In rpOka-BAC-infected MeWo cells, typical syncytium formation was observed within 24 h postinfection (hpi) by cell-to-cell spread and enhanced at 48 to 72 hpi (Fig. 11A), and the polykaryocytes detached from the dish by 96 hpi. In contrast, in rpOkagMim-infected MeWo cells, syncytium formation was rarely seen even at 96 hpi, and plaques caused by singly rounding cells with GFP expression were observed at 96 hpi (Fig. 11B, upper and lower panels).
FIG. 11.
Syncytium formation of gM mature-negative recombinant virus in MeWo cells. rpOka-BAC-infected (A) or rpOka-BACgMim-infected (B) MeWo cells with full CPE were propagated by cell-to-cell spread at a ratio of 1 infected cell to 10 uninfected cells, and syncytium formation (upper panels) and GFP expression (lower panels) were captured at 48 hpi (A) or 96 hpi (B). Magnification, ×100.
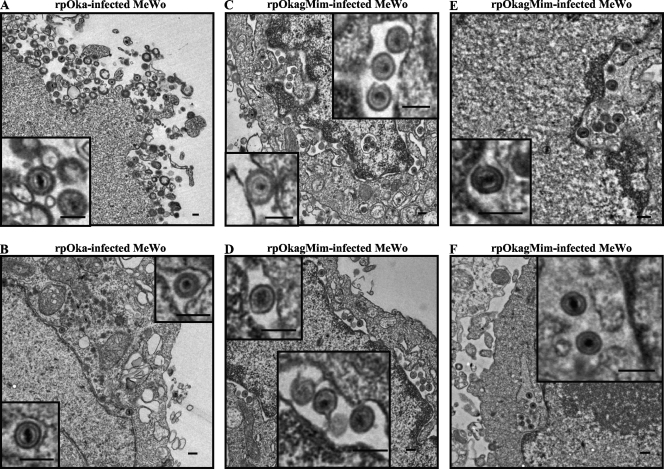
Virus particles were observed at the surfaces of both the rpOka- and rpOkagMim-infected cells at 48 hpi, although they were thought to be relatively more abundant in rpOka-infected cells than in rpOkagMim-infected cells (Fig. 12A and C). Despite the smaller amount of particles on the surfaces of rpOkagMim-infected cells, the mature particles were easily observed in the mutant-infected cells. Thus, virus particle formation or morphogenesis seemed not to be different between rpOka and rpOkagMim. On the other hand, in the cytoplasm of the rpOkagMim-infected cells, enveloped particles were easily and abundantly observed at the abnormally enlarged perinuclear cisternae and the ER (Fig. 12C to F), while only a few virus particles were observed at the perinuclear cisternae of the rpOka-infected cells (Fig. 12B).
FIG. 12.
Ultrastructural analysis of gM mature-negative recombinant viruses in MeWo cells. rpOka-infected (A and B) or rpOkagMim-infected (C, D, E, and F) MeWo cells with full CPE were propagated by cell-to-cell spread at a ratio of 1 infected cell to 10 uninfected cells, fixed at 48 to 72 h postinfection, and processed for electron microscopy. The small insets at the left side in panels A and C indicate the virus particles on the cell surface, and the other small insets in panels B to F indicate the virus particles in the perinuclear cisternae and ER. Bars = 200 nm.
DISCUSSION
Herpesviridae is one of the largest DNA virus families, and the genome consists of more than 70 open reading frames (up to 200 genes). Despite the high numbers of genes, most are reported not to be spliced; that is, their primary transcription product is the mature mRNA sequence, and the number of genes containing introns varies from a few to about 10% (36). The frequency of splicing events for alphaherpesviruses is thought to be less than that of other herpesviruses.
In this report, we identified at least three novel transcripts arising from the ORF50 gene. The transcripts are generated by alternative splicing but were not translated into proteins in vitro, and the function of this alternative splicing of the ORF50 gene was not elucidated.
Our current study also shows that VZV gM and gN form a complex and this step is required for gM maturation and transportation of the gM/gN complex through the secretory pathway, as also seen in Kaposi's sarcoma-associated herpesvirus (KSHV), Epstein-Barr virus (EBV), and human cytomegalovirus (HCMV) (21, 24, 26-28). Furthermore, we present the first evidence that valine at position 42 and glycine at position 301 of gM are the determinants for the formation of the gM/gN complex. In other herpesviruses, the binding of the gM/gN complex is mediated by a disulfide bond (18, 21, 25, 27, 48), and the cysteine residues that participate in this linkage are conserved in VZV gM and gN. Despite the conserved cysteines, however, VZV gM seems to bind with gN by a noncovalent linkage, as shown by our WB analysis of infected cells under nonreducing conditions and by IP-WB analysis. This was a great surprise for us, because the only herpesvirus that does not require this disulfide bond for the formation and transportation of the gM/gN complex or for efficient viral replication is HCMV, and although the existence of an additional, noncovalent linkage was proposed for HCMV, the residues involved in the proposed interaction were not elucidated (27).
Our data directly revealed that the core binding mechanism in the VZV gM/gN complex is through a noncovalent linkage mediated by valine at position 42 and by glycine at position 301. These two residues are located, respectively, at the predicted first and seventh TMDs of gM. It is possible that in the other herpesviruses, the disulfide bond simply supports this core interaction, although additively and strongly. In recent studies (3, 4, 34, 39, 42), a GXXXG motif for homodimeric or heterodimeric helix-helix formation at transmembrane domains was reported, and the glycine at position 301 of VZV gM is the first glycine in this motif, GPSLG. This motif is conserved at the predicted seventh TMD of gMs throughout human herpesviruses. The identification of the binding site in gN may elucidate in detail a ubiquitous interaction mechanism of the gMs and gNs of the herpesviridae.
VZV is unique among the herpesviruses in its ability to spread rapidly in vitro to adjacent cells by cell-to-cell expansion via syncytium formation, especially in MeWo cells. Transient expression studies suggest that VZV's fusogenic activity relies on gH and gL (10, 11) or gB and gE (30). In other herpesviruses, gM or the gM/gN complex inhibits this transfection-based membrane fusion, which can be mediated by viral fusogens besides herpesvirus glycoproteins (6, 20, 21). These findings have led to the suggestion that gM functions to direct the localization of viral envelope proteins to sites of secondary envelopment (6). There is no accurate method for evaluating the fusion activity of VZV-infected cells, but our data indicate that immature gM caused a small-plaque phenotype with reduced syncytium formation in MeWo cells. Therefore, VZV gM may function in cell-to-cell spread by enhancing the fusion activity of the other glycoproteins, especially in MeWo cells. In addition to our data, Ross et al. reported that disruption of the ORF9A gene in VZV results in reduced syncytium formation in MeWo cells (38), also indicating that gM/gN plays a role in syncytium formation in VZV-infected MeWo cells. Generally, in MRC-5 cells, syncytium formation is rarely observed even by pOka infection. However, the gM maturation-negative mutant virus also showed reduced plaque formation in MRC-5 cells, as seen in MeWo cells, suggesting that gM possibly functions in cell-to-cell spread even in MRC-5 cells. Further functional analyses would be required to elucidate the detailed mechanisms of cell-to-cell spread enhanced by gM.
Herpesvirus virion assembly and egress have been rather well documented in HSV, PRV, and HCMV (31), but only limited findings for VZV, especially in MeWo cells, have been reported (15, 17). In earlier studies, the assembly mechanisms of VZV were investigated in parallel with analyses of gB, gE, gH, gI, and gL because of their importance in and commonality among herpesviruses, and the VZV infectious cycle was carefully examined by electron microscopy, but these histological data were not backed up by genetic findings (12, 15). In our experiments using the mutant virus, mutant gM was never transported to the TGN, the recognized site of viral assembly, but it was still incorporated into particles, even extracellular particles. The detection of immature gM at the nuclear membrane and ER, the presence of enveloped particles at the enlarged perinuclear cisternae and the ER, and the incorporation of immature gM into intra- and extracellular particles suggest that there might be alternative pathways for viral assembly and egress, in addition to the well-documented route through the TGN.
In wild-type VZV infection of MeWo cells, similar amounts of mature gM were found in the nuclear and cytoplasmic fractions in a subcellular separation assay, while gB, gN, and ORF49p were detected mainly in the cytoplasmic fraction (data not shown). This finding indicates that mature gM may follow two possible routes. In one it is consistently transported to the nuclear membrane after it is glycosylated completely, and in another it is transported with gN to the TGN. In the gM maturation-negative virus, all of the immature gM may be transported to the nuclear membrane from the ER and incorporated into primary enveloped particles, which are secreted into the culture medium by an unidentified system. It was previously reported that viral glycoprotein-containing vesicles from the TGN fuse with nascent enveloped-particle-containing vacuoles from the ER and that final envelopment is achieved without the deenveloping and reenveloping process in VZV-infected MeWo cells (15), but the detailed mechanism of this phenomenon has not been identified.
In the present study, intracellular and extracellular particles containing immature gM, mature cleaved gB, and directly Golgi apparatus-derived membrane-targeted ORF49p were isolated from the same fraction of a 5 to 50% Histodenz gradient and were indistinguishable. By confocal microscopic analysis, the localization of mature gM with gB was less consistent than that of ORF49p with gB, even in pOka-infected MeWo cells, and gB and immature gM rarely colocalized in the rpOkagMim-infected MeWo cells (data not shown). These observations suggest that in wild-type virus infection, the route of gM's incorporation into virions may be qualitatively different from that of gB and other tegument proteins, and this difference may be more obvious in the gM maturation-negative mutant virus infection. Another possible explanation is that in wild-type virus infection, gB, gM, and ORF49p are incorporated into the same virions, but in the gM maturation-negative mutant virus infection, two kinds of virions are generated, one containing gB and ORF49p from the original route, in which the virions egress from the TGN-derived vesicles, and another containing immature gM from an alternative route, in which the virions directly egress from the ER. Further, detailed investigations of this phenomenon should elucidate novel mechanisms of VZV assembly and egress.
In VZV infection in vitro, it is widely accepted that infectious particles are mostly not secreted into the culture medium, although the release of VZV particles into the medium can still occur and has been detected in some studies (5, 14, 43). These extracellular particles have reduced infectivity (14) or are more often enveloped than the intracellular particles (43). In wild-type virus infection, the amounts of gB, ORF49p, and gM in the intra- and extracellular particles were indistinguishable, but the extracellular particles showed a reduced incorporation of ORF62p (data not shown). In contrast, here we showed that in gM maturation-negative mutant virus infection, less gM and no gN was incorporated into the extracellular particles compared with that for the intracellular particles. These findings also suggest that the gM maturation-negative mutant virus should be very useful for investigating mechanisms of virion assembly and egress and the roles of glycoprotein processing, especially gM processing, in VZV infection.
Our study reveals previously unidentified characteristics and functions of VZV gM. Further investigations will be required to analyze the function of each variant of the ORF50 mRNAs in VZV infection in vivo, as well as the detailed regulation mechanism of gM in cell-to-cell spread and the machinery for the incorporation of gM and gN into virus particles. These investigations will shed light on the VZV life cycle in its role as a human pathogen.
Acknowledgments
We thank Eiko Moriishi (National Institute of Biomedical Innovation, Osaka, Japan) for her technical assistance.
This study was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., E. Wills, R. J. Jacob, J. Pennington, and B. Roizman. 2007. Glycoprotein M of herpes simplex virus 1 is incorporated into virions during budding at the inner nuclear membrane. J. Virol. 81:800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosig, B., and D. Langosch. 1998. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 7:1052-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciczora, Y., N. Callens, F. Penin, E. I. Pecheur, and J. Dubuisson. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 81:2372-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook, M. L., and J. G. Stevens. 1968. Labile coat: reason for noninfectious cell-free varicella-zoster virus in culture. J. Virol. 2:1458-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517-3527. [DOI] [PubMed] [Google Scholar]
- 7.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67(Part 9):1759-1816. [DOI] [PubMed] [Google Scholar]
- 8.Dijkstra, J. M., A. Brack, A. Jons, B. G. Klupp, and T. C. Mettenleiter. 1998. Different point mutations within the conserved N-glycosylation motif of pseudorabies virus glycoprotein M result in expression of a nonglycosylated form of the protein. J. Gen. Virol. 79(Part 4):851-854. [DOI] [PubMed] [Google Scholar]
- 9.Dijkstra, J. M., N. Visser, T. C. Mettenleiter, and B. G. Klupp. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J. Virol. 70:5684-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duus, K. M., and C. Grose. 1996. Multiple regulatory effects of varicella-zoster virus (VZV) gL on trafficking patterns and fusogenic properties of VZV gH. J. Virol. 70:8961-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duus, K. M., C. Hatfield, and C. Grose. 1995. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology 210:429-440. [DOI] [PubMed] [Google Scholar]
- 12.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grose, C., D. M. Perrotta, P. A. Brunell, and G. C. Smith. 1979. Cell-free varicella-zoster virus in cultured human melanoma cells. J. Gen. Virol. 43:15-27. [DOI] [PubMed] [Google Scholar]
- 15.Harson, R., and C. Grose. 1995. Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J. Virol. 69:4994-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, F., and C. Grose. 1988. Role of cytoplasmic vacuoles in varicella-zoster virus glycoprotein trafficking and virion envelopment. J. Virol. 62:2701-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jons, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemble, G. W., P. Annunziato, O. Lungu, R. E. Winter, T. A. Cha, S. J. Silverstein, and R. R. Spaete. 2000. Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells. J. Virol. 74:11311-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 22.Krzyzaniak, M., M. Mach, and W. J. Britt. 2007. The cytoplasmic tail of glycoprotein M (gpUL100) expresses trafficking signals required for human cytomegalovirus assembly and replication. J. Virol. 81:10316-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ku, C. C., J. Besser, A. Abendroth, C. Grose, and A. M. Arvin. 2005. Varicella-zoster virus pathogenesis and immunobiology: new concepts emerging from investigations with the SCIDhu mouse model. J. Virol. 79:2651-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang, X., B. Chow, C. Raggo, and L. A. Babiuk. 1996. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J. Virol. 70:1448-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mach, M., B. Kropff, M. Kryzaniak, and W. Britt. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mach, M., K. Osinski, B. Kropff, U. Schloetzer-Schrehardt, M. Krzyzaniak, and W. Britt. 2007. The carboxy-terminal domain of glycoprotein N of human cytomegalovirus is required for virion morphogenesis. J. Virol. 81:5212-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean, C. A., S. Efstathiou, M. L. Elliott, F. E. Jamieson, and D. J. McGeoch. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72(Part 4):897-906. [DOI] [PubMed] [Google Scholar]
- 30.Maresova, L., T. J. Pasieka, and C. Grose. 2001. Varicella-zoster Virus gB and gE coexpression, but not gB or gE alone, leads to abundant fusion and syncytium formation equivalent to those from gH and gL coexpression. J. Virol. 75:9483-9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagaike, K., Y. Mori, Y. Gomi, H. Yoshii, M. Takahashi, M. Wagner, U. Koszinowski, and K. Yamanishi. 2004. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine 22:4069-4074. [DOI] [PubMed] [Google Scholar]
- 33.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 34.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 35.Osterrieder, N., A. Neubauer, C. Brandmuller, B. Braun, O. R. Kaaden, and J. D. Baines. 1996. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J. Virol. 70:4110-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellett, P. E., and B. Roizman (ed.). 2007. The family herpesviridae: a brief introduction. In Fields virology, 5th ed., vol. II. Lippincott Williams & Wilkins, Philadelphia, PA.
- 37.Quinlivan, M., and J. Breuer. 2006. Molecular studies of varicella zoster virus. Rev. Med. Virol. 16:225-250. [DOI] [PubMed] [Google Scholar]
- 38.Ross, J., M. Williams, and J. I. Cohen. 1997. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology 234:186-195. [DOI] [PubMed] [Google Scholar]
- 39.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 40.Sadaoka, T., K. Yamanishi, and Y. Mori. 2006. Human herpesvirus 7 U47 gene products are glycoproteins expressed in virions and associate with glycoprotein H. J. Gen. Virol. 87:501-508. [DOI] [PubMed] [Google Scholar]
- 41.Sadaoka, T., H. Yoshii, T. Imazawa, K. Yamanishi, and Y. Mori. 2007. Deletion in open reading frame 49 of varicella-zoster virus reduces virus growth in human malignant melanoma cells but not in human embryonic fibroblasts. J. Virol. 81:12654-12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senes, A., D. E. Engel, and W. F. DeGrado. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14:465-479. [DOI] [PubMed] [Google Scholar]
- 43.Shiraki, K., and M. Takahashi. 1982. Virus particles and glycoprotein excreted from cultured cells infected with varicella-zoster virus (VZV). J. Gen. Virol. 61(Part 2):271-275. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi, M., T. Otsuka, Y. Okuno, Y. Asano, and T. Yazaki. 1974. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet ii:1288-1290. [DOI] [PubMed] [Google Scholar]
- 45.Tischer, B. K., D. Schumacher, M. Messerle, M. Wagner, and N. Osterrieder. 2002. The products of the UL10 (gM) and the UL49.5 genes of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Gen. Virol. 83:997-1003. [DOI] [PubMed] [Google Scholar]
- 46.Weller, T. H. 1983. Varicella and herpes zoster. Changing concepts of the natural history, control, and importance of a not-so-benign virus. N. Engl. J. Med. 309:1362-1368. [DOI] [PubMed] [Google Scholar]
- 47.Weller, T. H. 1996. Varicella: historical perspective and clinical overview. J. Infect. Dis. 174(Suppl. 3):S306-S309. [DOI] [PubMed] [Google Scholar]
- 48.Wu, S. X., X. P. Zhu, and G. J. Letchworth. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the U(L)49.5 protein. J. Virol. 72:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamagishi, Y., T. Sadaoka, H. Yoshii, P. Somboonthum, T. Imazawa, K. Nagaike, K. Ozono, K. Yamanishi, and Y. Mori. 2008. Varicella-zoster virus glycoprotein M homolog is glycosylated, is expressed on the viral envelope, and functions in virus cell-to-cell spread. J. Virol. 82:795-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, J., C. H. Nagel, B. Sodeik, and R. Lippe. 2009. Early, active, and specific localization of herpes simplex virus type 1 gM to nuclear membranes. J. Virol. 83:12984-12997. [DOI] [PMC free article] [PubMed] [Google Scholar]