Abstract
The mechanisms of hepatitis C virus (HCV) replication remain poorly understood, and the cellular factors required for HCV replication are yet to be completely defined. CD81 is known to mediate HCV entry. Our study uncovered an unexpected novel function of CD81 in the HCV life cycle that is important for HCV RNA replication. HCV replication occurred efficiently in infected cells with high levels of CD81 expression. In HCV-infected or RNA-transfected cells with low levels of CD81 expression, initial viral protein synthesis occurred normally, but efficient replication failed to proceed. The aborted replication could be restored by the transient transfection of a CD81 expression plasmid. CD81-dependent replication was demonstrated with both an HCV infectious cell culture and HCV replicon cells of genotypes 1b and 2a. We also showed that CD81 expression is positively correlated with the kinetics of HCV RNA synthesis but inversely related to the kinetics of viral protein production, suggesting that CD81 may control viral replication by directing viral RNA template function to RNA replication. Thus, CD81 may be necessary for the efficient replication of the HCV genome in addition to its role in viral entry.
Hepatitis C virus (HCV) infection affects about 170 million people worldwide. Chronic HCV infection is an important cause of liver diseases, leading to cirrhosis and hepatocellular carcinoma (2, 18). The therapy for chronic HCV infection to date is suboptimal and associated with many side effects (12, 13). The mechanisms of HCV replication and persistent infection remain poorly understood (3, 31).
HCV carries a positive- and single-stranded RNA genome consisting of approximately 9,600 nucleotides (nt) (36). HCV encodes 10 proteins and exploits cellular factors for replication (24, 32, 35, 41). However, many crucial host factors required for HCV RNA replication remain undefined. The HCV RNA genome, like other positive-stranded RNA viruses, serves as templates for both viral protein translation and RNA replication (4, 15, 28), which are expected to be asynchronous in vivo, as the template pool is constantly replenished from ongoing HCV infection and replication (4). However, a coordinated translation/transcription process would be predicted if the use of HCV RNA as a template is subjected to cellular factor control that directs HCV RNA for specific template functions and synchronizes the translation/transcription process. CD81 has diverse functions in various biological processes (23, 39, 48) and is known to mediate HCV entry (10, 30, 34, 49). CD81 was recently suggested to play a role in postentry events (8). In this study we identified CD81 as a key cellular factor required for efficient HCV RNA replication, and inefficient RNA replication occurs in HCV-infected or RNA-transfected cells with low levels of CD81. Our data also showed that the utilization of HCV RNA as templates for viral protein synthesis and RNA synthesis is mutually exclusive and suggested that HCV RNA template function for RNA replication may be subjected to CD81 control.
MATERIALS AND METHODS
Plasmids.
The HCV JFH1 full cDNA sequence cloned into plasmid pUC19 (pJFH-1) and the JFH1 subgenomic replicon plasmid (pSGR-JFH1/Luc) were described previously (46). Plasmid pJ6/JFH1-p7Rluc2A (20) was a gift of Charlie Rice (Rockefeller University). pHCV-FLuc-3′-UTR, containing the firefly luciferase gene driven by the HCV internal ribosome entry site (IRES), was provided by Michael Niepmann (40). The human CD81 gene-carrying plasmid (pCDM8hCD81) (33) was a gift of Shoshana Levy (Stanford University, Stanford, CA). The CD81 mutant construct was generated by PCR-based mutagenesis that converted the methionine coding codon (AGT) to a stop codon (TAG) in the reading frame. The primers used were sense primer ATC AAG TAC CTG CTC TTC GTC TAG AAT TTC GTC TTC TGG CTG and antisense primer TGT TCT TGA GCA CTG AGG TGG TCA AAG CAG. The lack of CD81 expression was verified by CD81 staining after transfection with this construct.
Cell lines and culture.
Huh7.5 cells (7) maintained in our laboratory contain less than a 50% CD81-positive population. To obtain cell populations with different percentages of CD81-positive cells, three populations were selected from Huh7.5 cell by fluorescence-activated cell sorter (FACS) sorting: a CD81 high-level population (CD81-H), with 90% CD81-positive cells; CD81 low-level population 1 (CD81-L1), with 10% CD81-positive cells; and CD81 low-level population 2 (CD81-L2), with no detectable CD81 expression. All three populations were subjected to the same culture conditions as those used for the Huh7.5 cells. The cells were expanded, cryopreserved, and thawed for subsequent experiments. Continuous passages of these cells were avoided because of concern for altered phenotypes during long-term culture. Typically, these cells were not passaged more than 10 times after thawing. CD81 expression profiles for the three populations were monitored by FACS for CD81 expression after the reestablishment of cryopreserved cells and during experiments. The phenotype of CD81 expression remained stable during this short-term culturing. For FACS analysis, cells were trypsinized and then washed with magnetic cell sorting (MACS) buffer containing phosphate-buffered saline (PBS), bovine serum albumin (BSA), EDTA, and 0.09% azide (pH 7.2) (Miltenyi Biotec Inc., Auburn, CA). Approximate 106 cells were suspended in 100 μl MACS buffer, mixed with 10 μl fluorescein isothiocyanate (FITC)-conjugated anti-human CD81 (JS-81; BD Pharmingen) or 10 μl FITC-conjugated mouse IgG1 as an isotype control (BD Pharmingen), incubated on ice for 20 min, washed twice with MACS buffer, and suspended in 400 μl MACS buffer for FACS analysis. FACS for Sr-B1 expression was performed with mouse monoclonal anti-human CLA-1 (BD Transduction Laboratories).
The Huh7-25 clone and subgenomic HCV replicon cell lines Huh7/Rep-Feo (genotype 1b) and pSGR-JFH1-C4/1 (genotype 2a) were described previously (1, 25, 43). Another subgenomic HCV JFH1 replicon cell line, SGR-JFH1-FLuc/Neo, expressing FLuc-neo as the reporter, was generated similarly. All replicon cells were maintained in the presence of 500 μg/ml of G418 (Invitrogen).
siRNA and transfection.
The CD81 small interfering RNA (siRNA) pool (siGenome SMARTpool Duplex catalog numbers D-017257-01, -03, -04, and -05) and nontarget control 2 siRNA (catalog number D-001810-02-05) were purchased from Dharmacon Inc. siRNA transfection was carried out in 12-well plates with Oligofectamine reagents (Invitrogen), and the final siRNA concentration was 50 nM.
Production of HCV pseudovirus and HCV JFH1 virus and viral inoculation.
The HCV pseudovirus (H77 strain) was a gift of C. M. Rice (Rockefeller University). HCV pseudoparticle (HCVpp) infection and measurement of infectivity were described previously (14). For the transfection of JFH1 HCV RNA, cells were plated onto a 6-well plate at 1.5 × 105 cells per well overnight. Cells were washed twice with PBS after the medium was removed, and 1 ml Opti-MEM (Invitrogen) was added. Transfection mix containing 1,000 μl Opti-MEM, 12 μl DMRIE-C (1,2-dimyristyloxypropyl-3-dimethylhydroxy ethyl ammonium bromide-cholesterol) (Invitrogen), and 10 μg in vitro-transcribed JFH1 RNA was then added to cells and incubated for 4 h. Opti-MEM was then replaced with regular Dulbecco's modified Eagle's medium (DMEM). Media containing high titers of HCV (from day 3 to day 7) were collected as an initial inoculum. For HCV infection, Huh7.5 cells were plated onto 6-well plates at 1.5 × 105 cells per well overnight. The cells were inoculated with cell culture-derived HCV (HCVcc) as described above. The inoculum was removed at day 1 postinoculation (p.i.), and the infected cells were transferred onto a 10-cm dish at day 3 p.i. The media were collected and pooled together from days 4 to 7. The medium containing high-titer HCV was aliquoted and stored at −80°C as an inoculum. The inoculum was used at a multiplicity of infection (MOI) of 200:1 (based on the number HCV RNA genome copies per cell). In our experience, one infectious unit is equivalent to about 200 to 500 HCVcc RNA genome copies.
HCV RNA and core antigen assays.
Trypsinized cells were washed with PBS once, and RNA was extracted by using Trizol (Invitrogen). RNA was resuspended in 50 μl diethyl pyrocarbonate (DEPC)-H2O, and the final concentration was adjusted at 0.1 μg RNA/μl. Three microliters of RNA was used for quantitative reverse transcription (RT)-PCR (q-RT-PCR), and HCV RNA copies were expressed as copies per μg RNA. For the extraction of RNA in the medium, 250 μl medium was mixed with 750 μl Trizol LS (Invitrogen) according to instructions provided by the manufacturer. RNA was resuspended in 50 μl DEPC-H2O. Three microliters of RNA was used for q-RT-PCR, and HCV RNA copies were normalized as copies per ml medium. HCV RNA q-RT-PCR was described previously (42). The primers and TaqMan probe used for CD81 RNA quantification were sense primer AGG GCT GCA CCA AGT GC, antisense primer TGT CTC CCA GCT CCA GAT A, and TaqMan probe sequence 5′-6-carboxyfluorescein (6-FAM)-CAA GTA CCT GCT CTT CGT CTT CAA-6-carboxytetramethylrhodamine (TAMRA)-3′. For minus-strand RNA detection we applied a method described previously by Lanford et al. (27). A sense primer (GCG TTA GTA TGA GTG TCG TAC AGC, at nt 87 to 110 of the 5′ untranslated region [UTR] of the JFH1 genome) was used to synthesize cDNA from the minus-strand RNA templates at 70°C by rTth DNA polymerase (Applied Biosystems). The reverse transcription activity of rTth was blocked by adding buffer chelating MnCl2, and the DNA polymerase activity was facilitated by including MgCl2-containing buffer after the RT reaction. q-RT-PCR was performed for the detection of total HCV RNA.
For intracellular core determinations, approximately 300,000 cells were suspended with 100 μl lysis buffer (pH 7.5) (20 mM Tris, 1% NP-40, 1% Na deoxycholate, 0.1% SDS, and 1× protease inhibitor cocktail) and incubated on ice for 20 min. The supernatant was transferred into a new tube after a brief spin to remove cell debris. Ten microliters of supernatant was diluted 10-fold for core protein enzyme-linked immunosorbent assay (ELISA) (Ortho). The total amount of intracellular core protein was expressed as attomoles per well. A similar amount of uninfected cells at each time point was harvested and prepared in the same way as that for the infected cells for a negative control in ELISA tests. For extracellular core determinations, 5 μl of culture supernatant was diluted 20-fold and used for core ELISA. The core protein in the medium was expressed as attomoles/ml.
Single-cell-based q-RT-PCR assay.
The assay for single-cell HBVcc cDNA quantification was described previously (50, 51). Briefly, trypsinized cells were suspended in DMEM and counted. The initial cell concentration in the suspension was approximately 105 cells/ml. The cell suspension was then subjected to two steps and 100-fold dilutions with buffer containing 150 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 10 mM NaCl. The cell suspension concentration was then further adjusted to 100 cells per ml. Ten microliters of cell suspension containing approximately a single cell was manually distributed into each of the 96 wells of the plate. Ten microliters of proteinase K solution was added to each well (final concentration, 2 mg/ml) and incubated at 50°C for 60 min, and proteinase K was inactivated at 75°C for 15 min. Ten microliters of solution was transferred from each well onto a new plate. One plate was used for HCV RNA, and the other was used for CD81 RNA q-RT-PCR.
RESULTS
Divergent levels of HCV RNA in various CD81-expressing cells after HCV infection.
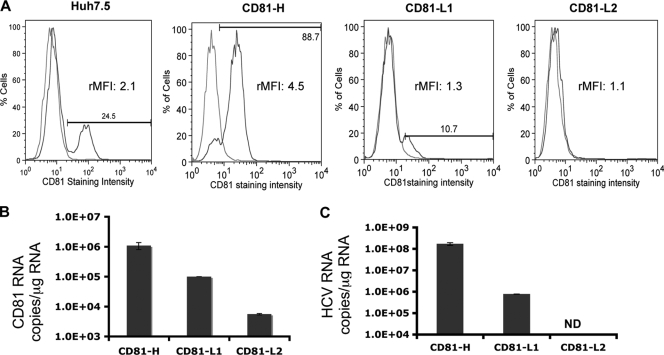
To investigate HCV infection and replication efficiency in cells with different CD81 expression levels, two cell populations, CD81-high (CD81-H) and CD81-low 1 (CD81-L1), containing 90% and 10% CD81-positive cells, respectively, were first isolated from Huh7.5 cells through cell sorting. A third cell population containing barely detectable CD81 expression was isolated from CD81-L1 cells and designated CD81-L2. The difference in CD81 expression levels detected by FACS was also confirmed by CD81 RNA quantification among three cell lines (Fig. 1A and B). CD81-H and CD81-L1 cells not only differ by the percentages of CD81-positive cells but also differ in CD81 expression intensities by relative mean fluorescence intensities (rMFIs) (up to 4-fold difference) (Fig. 2B). The CD81 expression profiles of the various cell lines determined by FACS were similar to those found in a study reported previously, in which Huh7-derived clones with variable CD81 expression levels were selected (1). CD81 expression levels did not increase appreciably after the permeabilization of cells before staining, indicating that most of the expression was on the cell surface (see Fig. S1 in the supplemental material). No major difference in SR-BI expression levels was detected among the parental Huh7.5 cell line and its derived cell lines (Fig. S2). Each cell population was inoculated with the same-size inoculum (MOI of 200:1 based on the number of HCV genome copies per cell). The intracellular HCV RNA level was determined at day 6 postinoculation (p.i.) by quantitative RT-PCR (q-RT-PCR). As shown in Fig. 1C, a >200-fold difference in HCV RNA levels was found between CD81-H and CD81-L1 cells, whereas no HCV RNA was detectable in CD81-L2 cells (detection limit of 103 copies/ml). The difference was initially thought to result from the difference in the percentages of CD81-positive cells, assuming that only cells with detectable cell surface CD81 expression levels could be infected by HCV (1). It would take multiple rounds of infection to have all 90% of the CD81-positive cells infected in CD81-H cells. We reasoned that a 9-fold difference in CD81-positive cells might not explain the >200-fold difference in HCV RNA levels between CD81-H and CD81-L1 cells and the even bigger difference between CD81-H and CD81-L2 cells, suggesting that the infected CD81-H cells might support higher HCV replication in addition to the difference in the number of infected cells.
FIG. 1.
CD81 expression and HCV RNA levels among the three Huh7.5 cell populations. CD81 cell surface expression and CD81 RNA levels of the three cell lines were monitored regularly, and representative profiles are shown. (A) Percentages of CD81-positive cells detected by FACS (blue, CD81; red, isotype IgG1). The relative mean fluorescence intensity (rMFI) (ratio of the CD81 mean fluorescence intensity over the IgG1 control mean fluorescence intensity in the same cell population) of each cell population is shown. (B) CD81 RNA expression level determined by q-RT-PCR. (C) Intracellular HCV RNA level in CD81-H, CD81-L1, and CD81-L2 cells detected by q-RT-PCR at day 6 p.i. with JFH1 HCVcc. ND, not detectable. Mean values (triplicates) ± standard deviations (SD) are shown. Data from one of three independent experiments are shown.
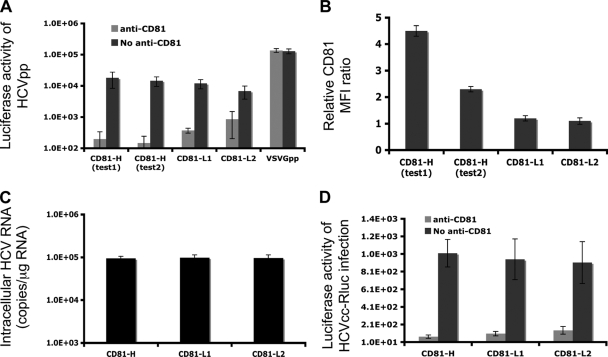
FIG. 2.
Similar efficiencies of viral entry among the three Huh7.5 cell populations. (A) Luciferase activities measured at 48 h after HCVpp inoculation. Shown are mean values (triplicates) ± SD. (B) The CD81 relative mean fluorescence intensities (MFI) among the three cell populations were monitored. Significant differences in rMFI among the cell populations exist (P = 0.0084 for CD81-H in test 1 versus test 2 and P = 0.0026 and P = 0.00056 for CD81-H in test 1 versus CD81-L1 and versus CD81-L2, respectively). (C) Intracellular HCV RNA level determined at 4 h p.i. by q-RT-PCR. Cells were harvested by 8 min of trypsin treatment that removes almost all bound viruses but not internalized viruses (data not shown). (D) Luciferase activities measured at 24 h postinfection with HCVcc containing the luciferase gene generated from pJ6/JFH1-p7Rluc2A. Shown are mean values (triplicate) ± SD. Data from one of three independent experiments are shown.
Additional role of CD81 in HCV RNA replication other than viral entry.
There are two possible explanations (or a combination of both) for the higher HCV RNA level in infected CD81-H cells: CD81-H cells support more efficient HCV entry, or CD81-H cells support more efficient HCV RNA replication. To investigate these two possibilities, CD81-H, CD81-L1, and CD81-L2 cells were tested for HCV entry efficiency by the HCVpp assay (6, 14). The three cell lines did not show any significant differences in HCVpp infectivity (Fig. 2A) despite a 4-fold difference in the CD81 rMFI (Fig. 2B) among the populations tested. It is noteworthy that the CD81 expression level in CD81-positive cells fluctuates periodically. CD81 expression levels showed a 2-fold reduction in rMFI (Fig. 2B) in the retesting of CD81-H cells with HCVpp infection. No corresponding reduction of HCVpp entry efficiency was observed. HCVpp infectivity was significantly suppressed by anti-CD81 antibodies in all cells tested, indicating that CD81 is indeed important for viral entry (Fig. 2A). Although the three cell lines did not show major differences in HCVpp infectivity, the CD81-L2 cells did show somewhat lower luciferase activities in the HCVpp assay than the CD81-H cells, suggesting that the very low level of CD81 may be affecting viral entry.
To further investigate viral entry efficiency, each cell population was inoculated with HCVcc. HCV-infected cells were harvested by trypsin treatment and extensive washing to eliminate surface-bound virus (8) at 4 h p.i., and the intracellular HCV RNA level was determined. There was no difference in HCV RNA levels among the three populations (Fig. 2C).
The HCVcc entry data were confirmed by infection with HCVcc containing the luciferase reporter gene (20). The luciferase activity was measured at 24 h p.i., and no difference was detected among the three cell populations tested (Fig. 2D). Both HCVpp and HCVcc data suggest that cells with high CD81 levels do not facilitate more HCV entry. This finding prompted us to consider that high levels of CD81 expression might not be sufficient to increase viral entry efficiency if other cellular factors (37) involved in HCV entry, like SR-BI (5), were limiting (see Fig. S2 in the supplemental material). It was shown previously that HCVcc or HCVpp can enter into cells with low levels of CD81 (1, 26, 52), and cells with different CD81 levels have no noticeable differences in initial HCV infection (26). It is conceivable that a threshold of CD81 expression is necessary for viral entry and that beyond this threshold, more efficient viral entry does not occur. This observation of a CD81 threshold level was already suggested previously (26). Thus, our data suggest that the high level of HCV RNA in CD81-H cells may result from a more efficient HCV RNA replication.
To study whether cells with higher CD81 levels support more efficient RNA replication, we investigated whether HCV RNA replication efficiency is correlated with the CD81 level at the single-cell level. Both HCV RNA and CD81 RNA levels were determined for the same cells harvested on days 3 and 6 p.i. by a single-cell-based RNA quantification technique (50, 51). Figure 3A shows the distribution patterns of CD81 RNA and HCV RNA copies per cell at day 6 p.i. The highest level of CD81 RNA per cell was 3,600 copies, while the lowest was under 100 copies. HCV replication efficiencies differed significantly among individual cells. The highest level of HCV RNA per cell was 89,600 copies. Average HCV RNA copies per cell increased by 13-fold, from 320 to 4,300 copies in cells with an average of 400 CD81 RNA copies between days 3 and 6 p.i. (Fig. 3B). For cells with an average of 1,800 CD81 RNA copies, the average number of HCV RNA copies increased by more than 200-fold, from 130 to 35,000 copies, over the same time period (Fig. 3C). The relationship between CD81 RNA and HCV RNA levels in the same individual cells was further analyzed. As shown in Fig. 3C, there was a positive correlation between CD81 RNA and HCV RNA levels in the same cells on day 6 p.i. (P < 0.0001), suggesting that HCV RNA replication efficiency is dependent on the CD81 level in individual cells. In contrast, the percentage of HCV-infected cells, as demonstrated by a single-cell assay, did not show a corresponding difference between high-level and low-level CD81-expressing cell populations (Fig. 3D). In fact, a somewhat higher percentage of lower-level CD81-expressing cells was positive for HCV RNA.
FIG. 3.
Single-cell correlation of HCV RNA and CD81 levels. Huh7.5 cells were infected with HCVcc and subjected to a single-cell-based q-RT-PCR assay. (A) Distribution of CD81 and HCV RNA copies/cell measured at day 6 p.i. for individual cells. (B) HCV and CD81 RNA levels were determined on days 3 and 6. Cells were divided into those with 100 to 1,000 (top) and those with 1,000 to 10,000 (bottom) CD81 copies/cell and plotted for their corresponding numbers of HCV RNA copies/cell. (C) Correlation of HCV RNA with CD81 RNA copy numbers of individual cells on day 6 p.i. (r = 0.58133 and P < 0.0001, determined by use of Microcal Software Inc.). (D) Percentage of HCV infection in cells with different CD81 levels. Infected cells were harvested at day 3 p.i., and HCV RNA levels and CD81 RNA copy numbers in the same cells were determined. The percentage of infection in each group was calculated by dividing the number of cells with detectable HCV RNA by the number of the total cells in each of three populations that contained <100, 100 to <1,000, and >1,000 copies of CD81 RNA per cell.
CD81-L cells do not support efficient viral replication after viral protein synthesis.
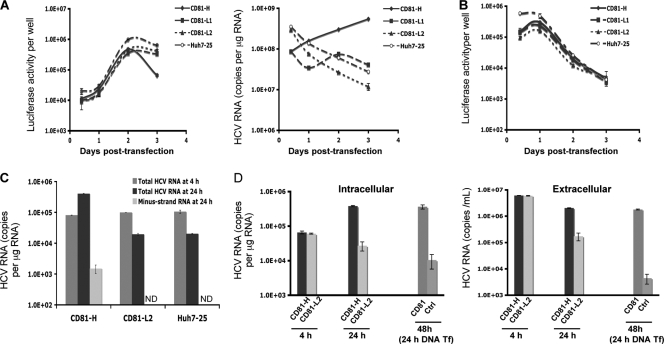
To determine whether CD81-L cells support efficient HCV RNA replication, CD81-H, CD81-L1, and CD81-L2 cells were transfected with JFH1 subgenomic replicon RNA containing the luciferase gene. The cells were harvested for determinations of luciferase activities and HCV RNA levels at various time points. As shown in Fig. 4A (left), there were no differences in luciferase activities among all cells tested up to day 2 posttransfection. However, efficient HCV RNA replication was detected for CD81-H but not CD81-L cells, as reflected by the decreasing HCV RNA levels in CD81-L and Huh7-25 cells (Fig. 4A, right). To exclude the possibility that the requirement of high levels of CD81 for HCV RNA replication is a unique feature of the Huh7.5 cell lines that we selected, a different Huh7 cell clone (Huh7-25) with little CD81 expression (1) (see Fig. S4A in the supplemental material) was tested, and a result was obtained that was similar to that for the CD81-L2 cells. Equally efficient protein translation in CD81-L1 and CD81-L2 cells was further demonstrated by the transfection of a construct containing the luciferase reporter gene under the control of the HCV IRES (40) (Fig. 4B).
FIG. 4.
Protein synthesis and RNA replication among cell populations with different CD81 levels. (A) Luciferase activities (left) and HCV RNA levels (right) measured at different time points after JFH1 subgenomic replicon RNA (pSGR-JFH1/Luc) transfection (Tf) of Huh7.5 cells. Shown are mean values (triplicates) ± SD. (B) Luciferase activities of Huh7.5 cells after transfection with a construct containing the luciferase gene driven by the HCV IRES (pHCV-FLuc-3′-UTR). Mean values (triplicates) ± SD are shown. (C) Intracellular total and minus-strand RNA levels at 24 h after pSGR-JFH1/Luc RNA transfection. Cells were transfected and harvested at 4 and 24 h. The total and minus-strand HCV RNA levels were determined by q-RT-PCR. The minus-strand HCV RNA assay has a detection limit of about 500 to 1,000 copies/μg of total RNA. ND, not detected. (D) Intracellular and extracellular RNA levels at 24 h after transfection with JFH1 genomic RNA. CD81-H and CD81-L2 cells were first transfected with JFH1 RNA, and cells and medium were harvested 4 and 24 h later for determinations of HCV RNA levels. Another set of CD81-L2 cells was transfected with a CD81 expression plasmid at 24 h after JFH1 RNA transfection and harvested 24 h later (48 h after JFH1 RNA transfection) for HCV RNA determinations as described above. Mean values ± SD are shown. Data from one of two independent experiments are shown.
We investigated the difference in the RNA replication efficiencies during the first 24-h period after cells were transfected with JFH1 subgenomic RNA or JFH1 genomic RNA. As shown in Fig. 4C and D, a 10-fold difference or more in intracellular or extracellular HCV RNA levels was detected between transfected CD81-H and CD81-L2 cells. In addition, HCV minus-strand RNA was detected only in the CD81-H cells. These data indicate that these cells support protein synthesis equally after RNA transfection, but cells with low levels of CD81 do not support efficient HCV RNA replication. It also raises the interesting possibility that HCV protein reporter constructs, like luciferase, may not necessarily reflect the HCV RNA replication status in certain situations.
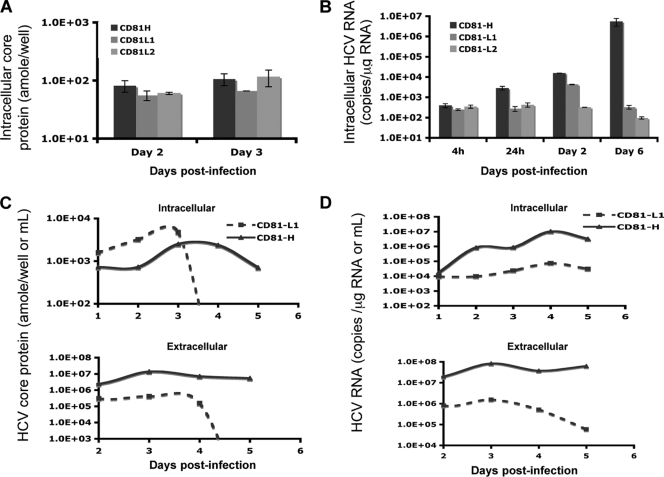
Viral protein synthesis and RNA replication were also tested with infected cells. Intracellular core protein and HCV RNA levels were analyzed at different time points after infection of CD81-H, CD81-L1, and CD81-L2 cells. Consistent with data shown in Fig. 4A, the core protein levels were similar among infected CD81-H, CD81-L1, and CD81-L2 cells on days 2 and 3 (Fig. 5A), suggesting that CD81-L cells are capable of supporting viral protein synthesis after viral entry. The newly synthesized core protein detected by ELISA was further verified by core-staining-positive cells in these cells. As many as 27% of CD81-L2 cells became core positive on day 3 (see Fig. S3 in the supplemental material) but not on day 1 (data not shown), suggesting that the detected core protein was newly synthesized in infected CD81-L2 cells. However, little HCV RNA replication occurred in infected CD81-L2 cells, while only modest HCV RNA replication was detected for CD81-L1 cells (Fig. 5B). These data suggest that efficient HCV RNA replication did not occur in CD81-L cells despite active viral protein synthesis. In contrast, efficient HCV RNA replication occurred in CD81-H cells with HCV RNA levels 10-fold and 1,000-fold greater than those of CD81-L1 cells at 24 h and on day 6 p.i., respectively (Fig. 5B), suggesting that viral replication proceeds efficiently in CD81-H cells.
FIG. 5.
HCV core protein and HCV RNA levels in the three CD81 cell populations. (A) Intracellular core protein levels of infected CD81-H, CD81-L1, and CD81-L2 cells were determined by ELISA at days 2 and 3 p.i. (B) Intracellular HCV RNA levels of the three cell lines were measured by q-RT-PCR. (C and D) Intracellular (top) and extracellular (bottom) core (C) and HCV RNA (D) levels at various time points p.i. were measured. Mean values ± SD are shown. Data from one of three independent experiments are shown.
The difference in HCV RNA replication levels between CD81-L1 and CD81-H cells was further demonstrated by analyzing the distribution of newly synthesized core protein. A larger amount of core protein was detected intracellularly in CD81-L1 cells from days 1 to 3 p.i., while more core protein was detected extracellularly in CD81-H cells during the same time period (Fig. 5C), suggesting that CD81-L1 cells were infected by HCV and supported viral protein synthesis. However, the synthesized core protein was mainly intracellular and accumulated to a higher level in CD81-L1 cells. In contrast, the synthesized core protein in CD81-H cells was efficiently exported to the medium, probably in the form of virions, as a result of active HCV RNA replication. This observation was further supported by the kinetics of intracellular and extracellular HCV RNA levels after infection. Both intracellular and extracellular HCV RNA levels in CD81-H cells were 100-fold higher than those in CD81-L1 cells later in the infection (Fig. 5D).
Ectopic expression of CD81 leads to efficient HCV RNA replication in CD81-L cells.
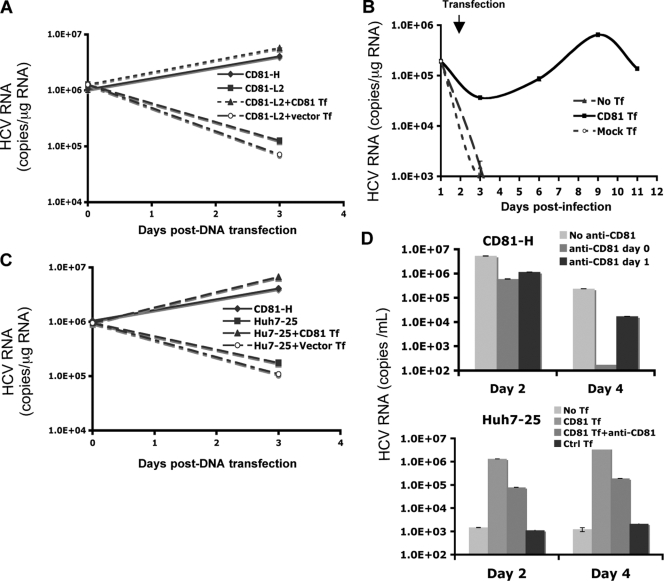
To test directly whether high levels of CD81 are required for efficient HCV RNA replication, CD81-L2 cells were first transfected with the JFH1 subgenomic replicon or JFH1 genomic RNA and then transfected with a CD81 expression plasmid or vector 1 day later. As shown in Fig. 4D and 6A, CD81-L2 cells showed decreasing HCV RNA levels after HCV RNA transfection, suggesting a lack of efficient HCV RNA replication. In contrast, following CD81 DNA transfection, CD81-L2 cells showed significantly increasing HCV RNA levels, similar to CD81-H cells, suggesting that HCV RNA replication was restored in CD81-L2 cells after CD81 transfection. To further explore the role of CD81 in viral replication, CD81-L2 cells were first infected with HCV, and after infection, cells were washed extensively and replated onto 6-well plates on day 1 p.i. The replated cells were then transfected with a CD81 expression plasmid on day 2 p.i. Cells were harvested at various time points to determine HCV RNA levels. As shown in Fig. 6B, infected CD81-L2 cells did not support HCV RNA replication at all. In contrast, CD81-L2 cells transfected with CD81 after inoculation demonstrated a rapid increase in HCV RNA levels. The kinetics of the increase suggest that this early increase likely occurs at the replication level. It is unlikely that this rapid increase is the effect of CD81 transfection on HCV entry because CD81-mediated HCV entry should occur within 1 h of infection (11, 17). However, the later increase probably represents the additional effect of enhanced CD81-dependent viral spread. These data provide direct evidence that a high level of CD81 is required for efficient HCV RNA replication and that the inability of HCV to replicate efficiently in CD81-L2 is a result of low levels of CD81 expression. This experiment also supports the above-described finding that HCV can enter into CD81-low cells but fails to initiate replication efficiently.
FIG. 6.
Transient expression of CD81 restores HCV RNA replication in CD81-L2 and Huh7-25 cells. (A) JFH1 subgenomic HCV RNA (pSGR-JFH1/Luc) was transfected into CD81-H or -L2 cells in triplicate followed by the transfection of a CD81 expression plasmid (CD81-L2+CD81 Tf) or a control plasmid (CD81-L2+Vector Tf) 24 h later. HCV RNA was harvested, and levels were determined immediately and 2 days after CD81 transfection. (B) CD81-L2 cells were infected with HCVcc and 24 h later were transfected with a CD81 expression plasmid. The amount of intracellular HCV RNA was then quantified at various time points. No Tf, no transfection; Control Tf, transfection with a control plasmid. (C) JFH1 subgenomic HCV RNA was transfected into Huh7-25 cells in triplicate, followed by the transfection of a CD81 expression plasmid (Huh7-25+CD81 Tf) or a control plasmid (Huh7-25+Vector Tf) 24 h later. HCV RNA was harvested, and levels were determined immediately and 2 days after CD81 transfection. (D) CD81-H (top) and Huh7-25 (bottom) cells were infected with HCVcc, and Huh7-25 cells were transfected with a CD81 expression plasmid 24 h later. Both cells were treated with anti-CD81 antibody at various time points (anti-CD81 day 0, anti-CD81 antibody was added together with HCVcc; anti-CD81 day 1, CD81 antibody was added on day 1 p.i; CD81 Tf, HCVcc infection followed by transfection with CD81 1 day later; CD81 Tf+anti-CD81, HCVcc infection followed by transfection of CD81 1 day later with the addition of anti-CD81). HCV RNA levels in the culture supernatant were determined at various time points. Data from one of three independent experiments are shown.
Huh7-25 cells were also tested in this replicon transfection experiment. As shown in Fig. 6C, JFH1 subgenomic replicon RNA replicated efficiently only in Huh7-25 cells transfected with CD81 DNA but not in cells with vector DNA or no DNA transfection, confirming that the Huh7-25 clone behaves like CD81-L2 cells in these experiments. Like CD81-L2 cells, Huh7-25 cells showed little virus production after infection with JFH-1 HCVcc. However, robust viral production was detected in infected Huh7-25 cells that were later transfected with the CD81 expression plasmid (Fig. 6D). To determine that CD81-mediated entry did not account for this increase, CD81-blocking antibody was added to the medium after transfection. CD81-H cells were used as a control and showed that the addition of CD81 antibodies after infection substantially inhibited viral spread (Fig. 6D, top), suggesting that the recently reported CD81-independent cell-to-cell transmission (45, 47) does not play a major role here. In contrast, Huh7-25 cells transfected with CD81 showed a substantial increase (2 logs) in HCV RNA levels despite the addition of CD81 antibodies on the day of CD81 transfection (Fig. 6D, bottom). A further increase (1 log) without the addition of CD81 antibodies after CD81 transfection was noted, probably due to the effect on viral spread.
Modulation of HCV replication by CD81 in HCV replicon cells.
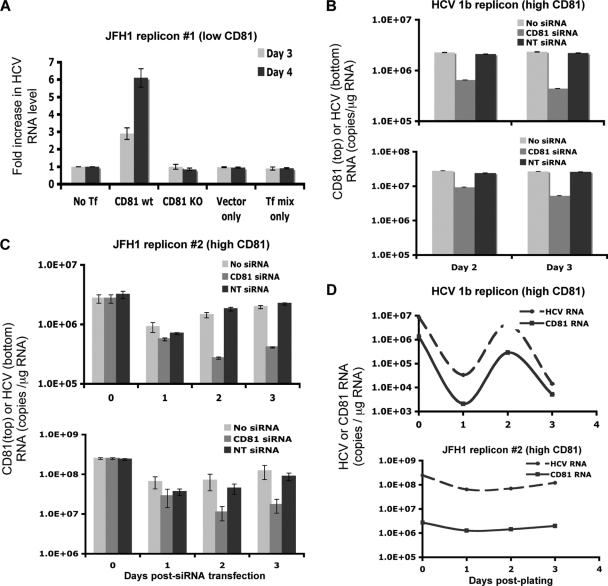
To further study the effect of CD81 expression on HCV replication, several subgenomic replicon cells (25, 43) were studied. Since subgenomic replicon cells harbor HCV RNA sequences covering only the NS3-NS5 region, infectious virus is not produced and viral spread does not occur. The HCV RNA level in a JFH1 subgenomic replicon cell line with low CD81 levels (pSGR-JFH1-C4/1) (25) (see Fig. S4B in the supplemental material) was studied by transiently transfecting a CD81 expression plasmid. After CD81 transfection, a 3- to 6-fold increase in HCV RNA levels was observed for this cell line (Fig. 7A). Control transfection with an empty vector or a CD81 mutant had no effect on HCV replication.
FIG. 7.
Modulation of the CD81 level changes HCV RNA replication efficiency. (A) JFH1 subgenomic replicon cells with low CD81 levels (pSGR-JFH1-C4/1) were transfected with a CD81 expression plasmid, and HCV RNA levels were determined on days 3 and 4 after transfection. The fold increase in the HCV RNA level after transfection is shown. CD81 KO, CD81 plasmid with a mutated start codon. (B and C) Two HCV subgenomic replicon cell lines, Huh7/Rep-Feo (B) and SGR-JFH1-FLuc/Neo (C), both of which have high levels of CD81 expression, were treated with CD81 siRNA to silence CD81 expression. CD81 RNA (top) and HCV RNA (bottom) levels were determined 2 and 3 days later. NC, negative control (nontargeted 2) siRNA. (D) CD81 and HCV RNA levels were determined in 1b (top) and JFH 2 (bottom) subgenomic replicon cells at various time points after plating. Mean values ± SD are shown. Data from one of three independent experiments are shown.
Two other subgenomic replicon cell lines, Huh7/Rep-Feo (genotype 1b) (43) and SGR-JFH1-FLuc/Neo (genotype 2a), had high levels of CD81 expression (see Fig. S4C and S4D in the supplemental material) and were treated with CD81 siRNA (Dharmacon) to reduce CD81 expression levels by 80% or more. The HCV RNA level was reduced by 4- to 6-fold in CD81 siRNA-transfected cells, whereas control siRNA transfection had no effect (Fig. 7B and C). The data showed that the replications of both JFH1 genotype 2a and 1b RNAs are CD81 dependent. The lowering of the HCV RNA level as a result of the specific reduction in the level of CD81 by siRNA confirmed the causative relationship between the CD81 level and the HCV replication efficiency.
CD81-dependent HCV RNA replication was further illustrated by the kinetics of HCV RNA and CD81 RNA levels in genotype 1b and 2a subgenomic replicon cells with high levels of CD81 expression. CD81 levels in those cells fluctuated, and so did the HCV RNA levels. A positive correlation between the two kinetics was observed (Fig. 7D), suggesting the importance of high levels of CD81 for efficient HCV replication.
CD81 in HCV RNA template function.
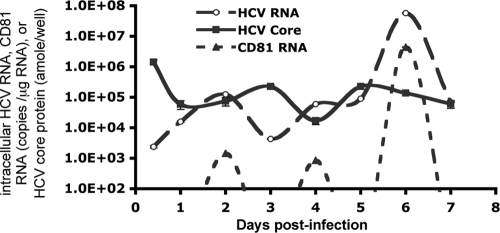
Like other positive-strand RNA viruses, the use of the same viral RNA as templates by viral protein translation and RNA transcription in the HCV life cycle should be mutually exclusive. The experimental data supporting this notion for positive-strand RNA viruses were generated from an in vitro cell-free system (4, 15). The template pool is constantly replenished as in vivo infection proceeds, and the two processes are expected to be asynchronous. However, a clear pattern for a single dominant template function at a time would also be predicted if the template function is subjected to control by a cellular factor(s) allowing coordinated protein translation and RNA transcription. Indeed, we observed a pattern for the mutually exclusive use of HCV RNA as templates for viral protein translation versus RNA replication in infected cell cultures when the kinetics of HCV RNA and core protein levels were analyzed. As shown in Fig. 8, the kinetics of the intracellular HCV RNA and core protein levels were opposite at each time point: an increasing intracellular HCV RNA level was accompanied by a decreasing core protein level and vice versa. Our data not only support the mutually exclusive use of HCV RNA for each template function in vivo but also suggest that cellular factors may direct HCV RNA to one template function at a time. When the kinetics of the intracellular HCV RNA, core protein, and CD81 levels were analyzed, changing CD81 RNA levels were positively correlated with the changing HCV RNA level but were inversely related to the core protein level (Fig. 8). The positive and inverse correlations among CD81, HCV RNA, and viral protein kinetics suggest that CD81 may be one of the cellular factors directing HCV RNA to the replication process. The viral protein level appears normal while the HCV RNA level is decreasing when the CD81 level is low, suggesting that templates can still be used for viral protein translation but not for RNA synthesis. This is probably why no difference in the luciferase activities or core protein levels in CD81-L cells from those in CD81-H was detected at the early phase of transfection or infection. Thus, CD81 may control HCV RNA replication, possibly through directing HCV RNA template function toward RNA replication.
FIG. 8.
Relationship of intracellular HCV RNA and core protein levels to CD81 levels during viral replication. CD81-H cells were infected with HCV and harvested at various time points. Intracellular HCV RNA, CD81 RNA, and core protein levels were determined. amole/well, attomoles/well. Mean values ± SD are shown. Data from one of two independent experiments are shown.
DISCUSSION
CD81 is known to mediate viral entry in HCV infection (10, 30, 34, 49) and was also implicated in the cell-to-cell transmission of HCV infection (45, 47). Our study showed that significant differences existed in HCV RNA levels after HCV infection among CD81-H, CD81-L1, and CD81-L2 populations and could not be explained simply by the CD81 entry function. This observation prompted us to investigate whether CD81 is required for additional steps in the HCV life cycle, such as RNA replication. Using a variety of techniques and cell lines, we uncovered a novel function of CD81 in the HCV life cycle that is important for HCV RNA replication. CD81 is a tetraspanin family member and is enriched in the lipid rafts of membranous compartments of the cell, where HCV RNA replication is believed to take place (9, 16). The requirement for CD81 participation by the HCV replication process can be facilitated by the physical proximity of CD81 to the HCV replicating site.
To explain our data and the proposed dual functions of CD81 in the context of HCV infection and replication, we reason that a low threshold amount of CD81 is required for the HCV entry function but that a much higher level of CD81 is necessary for efficient HCV replication subsequent to viral entry. Koutsoudakis et al. (26) previously reported that about 70,000 CD81 molecules in a cell appear to be the threshold for viral entry. Our data suggest that the three cell lines with very different levels of CD81 allow similar viral entry but appear to support divergent efficiencies of HCV replication that correlate well with CD81 levels. It is interesting that CD81-L2 cells, despite having very low levels of CD81, can still support viral entry although at a somewhat lower level than the higher-level-CD81-expressing cells. The CD81 expression level of the L2 cells is probably just around the “threshold level” for viral entry.
Our data provide evidence for the mutually exclusive use of HCV RNA as templates for either RNA replication or protein synthesis in infected cell cultures. Two lines of evidence support that the use of HCV RNA for RNA replication is subjected to cellular factor control, such as CD81. One line of evidence is the absence of efficient RNA replication after viral protein translation in HCV-infected and RNA-transfected CD81-L1 and -L2 cells, suggesting that RNA replication could not occur efficiently when the CD81 level was low. The other evidence is that a clear pattern for one dominant template function at a time was shown for infected CD81-H cells, suggesting that there is a coordinated process that directs HCV RNA molecules toward RNA replication function. It is likely that cellular factors are involved in directing viral RNA molecules toward two distinct template functions. On the other hand, viral protein synthesis is negatively correlated with CD81 and HCV RNA levels. These data suggest that the template function of HCV RNA is controlled by cellular factors like CD81, which directs HCV RNA toward its replication function instead of protein translation. However, it is not clear how CD81 exerts this function.
CD81 may assist directly in the assembly of the HCV replicase complex, including NS5B, contributing to viral RNA replication. Alternatively, CD81 may be linked indirectly to a cellular pathway that is critical for efficient viral replication. In a recent study, Brazzoli et al. showed that the engagement of CD81 during HCV infection activates the Raf/MEK/extracellular signal-regulated kinase (ERK) signaling cascade and that this pathway affects postentry events of the HCV life cycle, presumably at the replication step (8). Further experiments are needed to elucidate the molecular mechanism of this novel CD81 function.
CD81 was previously reported to have diverse functions in biological process. For instance, CD81 is implicated in the metastasis of cancer cells (21). CD81 can influence the adhesion, morphology, activation, proliferation, and differentiation of B, T, and other cells (22). In parasite infections, hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity (39). CD81 was also implicated in the modulation of infectivity, enhancement of viral gene expression, and promotion of virus assembly, budding, and cell-to-cell spread in the HIV life cycle (19, 38, 44). The identification of this novel CD81 function in HCV replication indicates that CD81 plays a more pleiotropic role in the HCV life cycle besides its well-defined role in viral entry. Our data suggest that CD81 has dual functions in HCV infection: a low threshold level of CD81 required for viral entry and a higher level of CD81 necessary for efficient HCV RNA replication. The dependence of HCV replication on CD81 creates an inherent vulnerability for HCV replication. Thus, CD81 functions could be explored for potential therapeutic development because of the multiple roles of CD81 in HCV infection, as was explored in a recent study of the Alb-uPA/SCID mouse model engrafted with human hepatocytes (29).
Supplementary Material
Acknowledgments
We thank Matthew Evans and Charlie Rice for providing the J6/JFH1p7Rluc2A plasmid and H77 HCVpp and Huh 7.5 cells, Shoshana Levy for the human CD81 plasmid, Michael Niepmann for the pHCV-FLuc-3′-UTR plasmid, Takanobu Kato and Takaji Wakita for the Huh7-25 and SGR-JFH1-C4/1 cell lines, and Naoya Sakamoto for the Huh7/Rep-Feo cell line.
This study was supported by the Intramural Research Program of NIDDK, National Institutes of Health.
Footnotes
Published ahead of print on 20 January 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akazawa, D., T. Date, K. Morikawa, A. Murayama, M. Miyamoto, M. Kaga, H. Barth, T. F. Baumert, J. Dubuisson, and T. Wakita. 2007. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J. Virol. 81:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, M. J. 2007. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 13:2436-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 4.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, B., C. Logvinoff, C. M. Rice, and J. A. McKeating. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 6.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazzoli, M. A. Bianchi, S. Filippini, A. Weiner, Q. Zhu, M. Pizza, and S. Crotta. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82:8316-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claas, C., C. S. Stipp, and M. E. Hemler. 2001. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 276:7974-7984. [DOI] [PubMed] [Google Scholar]
- 10.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wölk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 12.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967-972. [DOI] [PubMed] [Google Scholar]
- 13.Feld, J. J., and T. J. Liang. 2005. HCV persistence: cure is still a four letter word. Hepatology 41:23-25. [DOI] [PubMed] [Google Scholar]
- 14.Flint, M., C. Logvinoff, C. M. Rice, and J. A. McKeating. 2004. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J. Virol. 78:6875-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haberstroh, A., E. K. Schnober, M. B. Zeisel, P. Carolla, H. Barth, H. E. Blum, F. L. Cosset, G. Koutsoudakis, R. Bartenschlager, A. Union, E. Depla, A. Owsianka, A. H. Patel, C. Schuster, F. Stoll-Keller, M. Doffoël, M. Dreux, and T. F. Baumert. 2008. Neutralizing host responses in hepatitis C virus infection target viral entry at postbinding steps and membrane fusion. Gastroenterology 135:1719-1728. [DOI] [PubMed] [Google Scholar]
- 18.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 19.Jolly, C., and Q. J. Sattentau. 2007. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J. Virol. 81:7873-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Naour, F., M. André, C. Greco, M. Billard, B. Sordat, J. F. Emile, F. Lanza, C. Boucheix, and E. Rubinstein. 2006. Profiling of the tetraspanin web of human colon cancer cells Mol. Cell. Proteomics 5:845-857. [DOI] [PubMed] [Google Scholar]
- 22.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 23.Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5:136-148. [DOI] [PubMed] [Google Scholar]
- 24.Li, Q., A. L. Brass, A. Ng, Z. Hu, R. J. Xavier, T. J. Liang, and S. J. Elledge. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U. S. A. 106:16410-16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 26.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanford, R. E., C. Sureau, J. R. Jacob, R. White, and T. R. Fuerst. 1994. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology 202:606-614. [DOI] [PubMed] [Google Scholar]
- 28.Lott, W. B., S. S. Takyar, J. Tuppen, D. H. Crawford, M. Harrison, T. P. Sloots, and E. J. Gowans. 2001. Vitamin B12 and hepatitis C: molecular biology and human pathology. Proc. Natl. Acad. Sci. U. S. A. 98:4916-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meuleman, P., J. Hesselgesser, M. Paulson, T. Vanwolleghem, I. Desombere, H. Reiser, and G. Leroux-Roels. 2008. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 48:1761-1768. [DOI] [PubMed] [Google Scholar]
- 30.Molina, S., V. Castet, L. Pichard-Garcia, C. Wychowski, E. Meurs, J. M. Pascussi, C. Sureau, J. M. Fabre, A. Sacunha, D. Larrey, J. Dubuisson, J. Coste, J. McKeating, P. Maurel, and C. Fournier-Wirth. 2008. Serum-derived hepatitis C virus infection of primary human hepatocytes is tetraspanin CD81 dependent. J. Virol. 82:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453-463. [DOI] [PubMed] [Google Scholar]
- 32.Ng, T. I., H. Mo, T. Pilot-Matias, Y. He, G. Koev, P. Krishnan, R. Mondal, R. Pithawalla, W. He, T. Dekhtyar, J. Packer, M. Schurdak, and A. Molla. 2007. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 45:1413-1421. [DOI] [PubMed] [Google Scholar]
- 33.Oren, R., S. Takahashi, C. Doss, R. Levy, and S. Levy. 1990. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 10:4007-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 35.Randall, G., M. Panis, J. D. Cooper, T. L. Tellinghuisen, K. E. Sukhodolets, S. Pfeffer, M. Landthaler, P. Landgraf, S. Kan, B. D. Lindenbach, M. Chien, D. B. Weir, J. J. Russo, J. Ju, M. J. Brownstein, R. Sheridan, C. Sander, M. Zavolan, T. Tuschl, and C. M. Rice. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 37.Sabahi, A. 2009. Hepatitis C virus entry: the early steps in the viral replication cycle. Virol. J. 6:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato, K., J. Aoki, N. Misawa, E. Daikoku, K. Sano, Y. Tanaka, and Y. Koyanagi. 2008. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J. Virol. 82:1021-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silvie, O., E. Rubinstein, J. F. Franetich, M. Prenant, E. Belnoue, L. Rénia, L. Hannoun, W. Eling, S. Levy, C. Boucheix, and D. Mazier. 2003. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 9:93-96. [DOI] [PubMed] [Google Scholar]
- 40.Song, Y., P. Friebe, E. Tzima, C. Jünemann, R. Bartenschlager, and M. Niepmann. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 80:11579-11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tai, A. W., Y. Benita, L. F. Peng, S. S. Kim, N. Sakamoto, R. J. Xavier, and R. T. Chung. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 43.Tanabe, Y., N. Sakamoto, N. Enomoto, M. Kurosaki, E. Ueda, S. Maekawa, T. Yamashiro, M. Nakagawa, C. H. Chen, N. Kanazawa, S. Kakinuma, and M. Watanabe. 2004. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon-alpha. J. Infect. Dis. 189:1129-1139. [DOI] [PubMed] [Google Scholar]
- 44.Tardif, M. R., and M. J. Tremblay. 2005. Tetraspanin CD81 provides a costimulatory signal resulting in increased human immunodeficiency virus type 1 gene expression in primary CD4+ T lymphocytes through NF-kappaB, NFAT, and AP-1 transduction pathways. J. Virol. 79:4316-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timpe, J. M., Z. Stamataki, A. Jennings, K. Hu, M. J. Farquhar, H. J. Harris, A. Schwarz, I. Desombere, G. L. Roels, P. Balfe, and J. A. McKeating. 2008. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47:17-24. [DOI] [PubMed] [Google Scholar]
- 46.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Kräusslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witteveldt, J., M. J. Evans, J. Bitzegeio, G. Koutsoudakis, A. M. Owsianka, A. G. Angus, Z. Y. Keck, S. K. Foung, T. Pietschmann, C. M. Rice, and A. H. Patel. 2009. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90:48-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yunta, M., and P. A. Lazo. 2003. Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell. Signal. 15:559-564. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, Y. Y., B. H. Zhang, D. Theele, S. Litwin, E. Toll, and J. Summers. 2003. Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proc. Natl. Acad. Sci. U. S. A. 100:12372-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Y. Y., D. P. Theele, and J. Summers. 2005. Age-related differences in amplification of covalently closed circular DNA at early times after duck hepatitis B virus infection of ducks. J. Virol. 79:9896-9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.