Abstract
The crystal structure of the dimerization domain of rabies virus phosphoprotein was determined. The monomer consists of two α-helices that make a helical hairpin held together mainly by hydrophobic interactions. The monomer has a hydrophilic and a hydrophobic face, and in the dimer two monomers pack together through their hydrophobic surfaces. This structure is very different from the dimerization domain of the vesicular stomatitis virus phosphoprotein and also from the tetramerization domain of the Sendai virus phosphoprotein, suggesting that oligomerization is conserved but not structure.
Rabies virus is a negative-strand RNA virus of the Rhabdovirus family. Its genomic RNA is encapsidated by numerous copies of the viral nucleoprotein (N) that binds to the sugar phosphate backbone of the RNA with a stoichiometry of nine nucleotides per N protomer (1). RNA replication and transcription take place on this N-RNA template (2) and are catalyzed by the viral RNA-dependent RNA polymerase (L). L binds to the N-RNA template with the help of the polymerase cofactor, the phosphoprotein (P) (7). The phosphoproteins of the Rhabdoviridae and of the Paramyxoviridae are oligomers (5, 8); P of the Rhabdoviridae (rabies virus and vesicular stomatitis virus [VSV]) form dimers (6, 9), whereas P of the Paramyxoviridae (Sendai virus) form tetramers (21). These phosphoproteins are modular proteins that have an N-terminal domain that keeps newly produced N (called N0) in a soluble, RNA-free form, a central oligomerization domain and a C-terminal domain that binds to N-RNA. The three domains are connected by two intrinsically disordered regions (10, 13). The atomic structures of the oligomerization domains of the phosphoproteins of VSV and Sendai virus have been determined, and it was found that these structures were quite different (6, 21). Because the secondary structure prediction of the oligomerization domain of rabies virus P was different again from that of Sendai virus and VSV (10), we decided to determine its structure.
The sequence analysis of rabies virus P suggests that the oligomerization domain stretches from amino acid residues 92 to 131 (10). The DNA corresponding to residues 91 to 133 was cloned into a pET22b vector and expressed in the Escherichia coli BL21 (DE3-RIL) strain as a His tag fusion protein. The protein was purified by nickel resin and size-exclusion chromatography (S75). In solution the protein behaves as a dimer with a molecular mass of 13.9 kDa (monomer, 6,501 Da) (10). Monomers or higher-order oligomers have never been observed.
The protein was crystallized at 20°C in 50 mM sodium cacodylate (pH 6.5)-10 mM MgSO4 plus 2 M (NH4)2SO4. Diffraction data on frozen crystals were obtained on beam lines ID14-4 and BM14 (ESRF, Grenoble, France). The crystals diffracted to a 1.5-Å resolution and belong to space group I4122 with one dimer in the asymmetric unit. The phases were solved by the Sulfur-MAD method (Table 1) . Integration and scaling of the data were performed with XDS (12). Three methionine sites were found with the automated software Auto-RickShaw (17). An initial 2.5-Å resolution model was built, which was then used for molecular replacement with MOLREP (22) using the native data set. ARP-wARP (18) was used to add water molecules. The model was improved by hand and finally refined using REFMAC5 (16), and the quality of the model was checked with PROCHECK (14) (Table 1).
TABLE 1.
Data collection and refinement statistics
| Parameter | Anomalous data |
Native crystal | |
|---|---|---|---|
| Set 1 | Set 2 | ||
| General data | |||
| X-ray source | BM14 | BM14 | ID14-4 |
| Wavelength (Å) | 1.771 | 0.885 | 0.976 |
| Space group | I4122 | I4122 | I4122 |
| Cell dimensions | |||
| a (Å) | 43.15 | 43.23 | 43.29 |
| b (Å) | 43.15 | 43.23 | 43.29 |
| c (Å) | 190.90 | 191.44 | 192.02 |
| α = β = γ (°) | 90.00 | 90.00 | 90.00 |
| Resolution range (Å)a | 25-2.11 (2.19-2.11) | 25-2.0 (2.07-2.0) | 25.0-1.5 (1.56-1.49) |
| Completeness (%)a | 97.6 (86.7) | 97.9 (96.3) | 93.6 (90.6) |
| Rsym (I) (%)a,b | 5.7 (31.7) | 5.5 (30.8) | 5.1 (42.5) |
| I/σIa | 66.75 (11.53) | 85.80 (21.81) | 20.36 (4.75) |
| Total reflectionsa | 428,072 (24,834) | 478,480 (45,631) | 133,743 (13,727) |
| Unique reflectionsa | 9,704 (904) | 6,451 (595) | 17,851 (1,755) |
| Multiplicitya | 44.11 (27.47) | 74.17 (76.69) | 7.49 (7.82) |
| Refinement statistics | |||
| R-factor (%) | 19.9 | ||
| Rfree (%) | 23.5 | ||
| rmsd from ideal values | |||
| Bond length (Å) | 0.02 | ||
| Bond angle (°) | 1.46 | ||
| Mean B-factor (Å2) | 22.4 | ||
| Ramachandran plot | |||
| Most favored regions (%) | 100 | ||
Values in parentheses are for the highest-resolution shell. rmsd, root mean square deviations.
Rsym (I) = [Σhkl Σi|<Ihkl> − Ihkl,i|]/[Σhkl Σi|Ihkl|], where i is the number of reflection hkl.
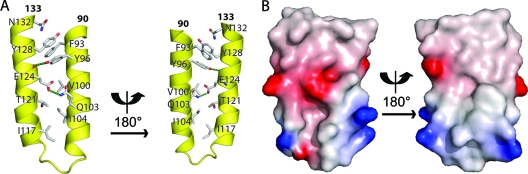
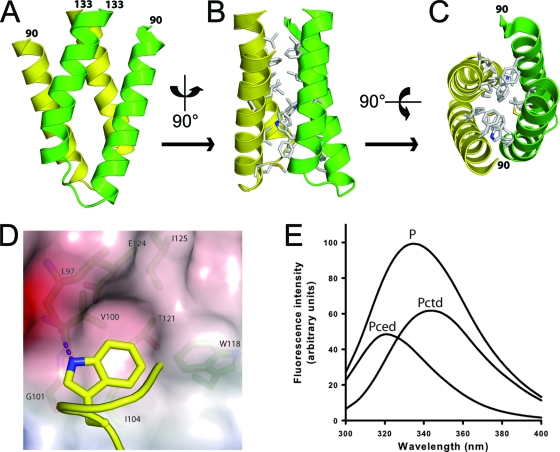
Figure 1A shows two views of the monomer. Residues 90 to 109 form an N-terminal α-helix linked to a C-terminal helix (residues 114 to 133) by a loop (residues 110 to 113). The two helices are kept together by an extensive hydrophobic intramonomer interface, as indicated in Fig. 1A, completed by hydrogen bonds between Y96 and the main chain carbonyl of E124 and between Q103 and E124 (Fig. 1A). The two helices form a rather flat surface with a charged and a hydrophobic face (Fig. 1B). The dimer is formed by the interaction of the two hydrophobic faces, the two monomers crossing with an angle of about 46° (Fig. 2A and B), close to the value of 50° typically encountered in helix-to-helix packing in globular proteins, but different from the angle of 20° observed in four-helix bundles (4). The surface of the interface shows a conspicuous bulge next to a hydrophobic cavity (Fig. 1B). The bulge is formed by W118 and the cavity by L97, V100, G101, and T121 (Fig. 2D). When the two monomers lock together in the dimer, both tryptophans insert into the opposite cavities. The relative positions of W118 and the cavity contribute to or define the crossing angle. Residue W118 is deeply buried within the hydrophobic dimer interface, as shown by the large blue shift of the fluorescence emission spectrum of the dimerization domain (λmax = 318 nm), compared to the emission spectrum of the C-terminal domain (containing W186 and W265; maximum at 344 nm) and to that of the full-length protein (maximum at 333 nm) (Fig. 2E). Other residues involved in the dimer interface are (chain A-chain B): M108-M108, D98-V122, I125/V122-L97, I104-F114, V129-F93/V129, and F93-V126, most of which are not involved in the intramonomer interface. The hydrogen bonds contributing to the dimer interface are (main chain interactions in italics): L97-W118, W118-L97, E123-Q94, and S119-Q94, the last two mediated by water molecules. In general, the N-terminal helix of one monomer interacts with the C-terminal helix of the other monomer (Fig. 2C).
FIG. 1.
Structure of the monomer in the dimerization domain of the rabies virus phosphoprotein. (A) Front and back views of the helical hairpin monomer showing residues in the intramonomer interface. The amino acids forming the hydrophobic core holding the two helices together are indicated, as well as two hydrogen bonds (green stippled lines). (B) Surface potential (from −5 kT/e [red] to +5 kT/e [blue]) of the hydrophilic (left) and hydrophobic (right) surfaces of the monomer; the molecules have the same orientation as in panel A.
FIG. 2.
Structure of the dimerization domain of the rabies virus phosphoprotein. (A and B) Front and side views of the dimer. Residues in the interface are depicted in panel B. (C) Top-down view of the dimer showing that the N-terminal helix of one monomer interacts mainly with the C-terminal helix of the other. (D) Tryptophan 118 lodging in the hydrophobic cavity of the opposite monomer. Also indicated is the hydrogen bond between W118 and the main chain carbonyl oxygen of L97. (E) Fluorescence emission spectra of the intact phosphoprotein (P), the isolated C-terminal domain (PCTD), and the dimerization domain (PCED), showing the large blue shift of W118.
Some of the amino acids in the hydrophobic interface were mutated to alanine, and the effects on dimerization were measured with the yeast two-hybrid system (10). Mutation to Ala of Y128 in the intramonomer interface or of different residues in the subunit interface (F114A, W118A, and I125A) led to loss of dimerization. An estimation of the contribution of each of these residues to the overall stability of the dimerization domain was obtained by using the FoldX web server (20). The predicted free-energy difference between the mutant and the wild-type proteins (ΔΔG) is large for each of these mutants (Y128A, 4.9 kcal mol−1; F114A, 10.0 kcal mol−1; W118A, 9.6 kcal mol−1; and I125A, 6.3 kcal mol−1). The Y129A mutation also in the dimerization interface reduced but did not eliminate dimerization (calculated ΔΔG of 4.1 kcal mol−1).
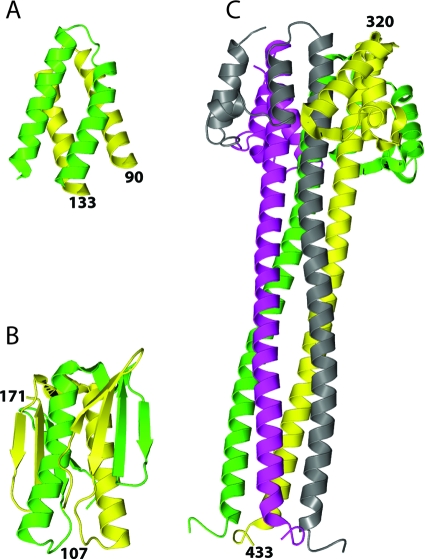
The structure of the oligomerization domain of the rabies virus phosphoprotein is very different from those of VSV and Sendai virus (Fig. 3). The VSV monomer consists of an α-helix preceded and followed by a two-stranded β-sheet. In the dimer (Fig. 3B), the two helices interact mainly through a hydrophobic cluster at their N-terminal ends. At both sides of the helices, the N-terminal sheet of one subunit forms a four-stranded sheet with the C-terminal sheet of the other subunit, stabilizing the dimer further by adding main chain hydrogen bonds. There are additional hydrophobic interactions between the two helices and the sheets (6). The crystal packing of the VSV dimerization domain suggests how higher-order oligomers might be formed, but this is not obvious from the packing of the rabies virus domain. The Sendai domain (Fig. 3C) forms a tetrameric coil that is stabilized by four times three N-terminal helices forming a small hydrophobic core (21). The largest functional difference between the three oligomerization domains is that in the rabies virus P dimer the N-terminal end that is linked to the N0 binding domain is positioned next to the C-terminal end that is linked to the N-RNA binding domain, whereas in both the VSV and the Sendai virus structures the N- and C-terminal ends are at opposite ends of the oligomerization domain. However, this difference may not have biological consequences because for all three phosphoproteins the N0 and the N-RNA binding domains are connected to the oligomerization domain by extensive disordered regions (10).
FIG. 3.
Comparison of the oligomerization domains of the phosphoproteins of rabies virus, VSV, and Sendai virus. (A and B) Dimerization domains of the rabies virus and VSV phosphoproteins, respectively. (C) Tetramerization domain of Sendai virus P.
There is no sequence similarity between the three oligomerization domains, but sometimes structural conservation can be recognized between proteins with different sequences, as was the case for the N-RNA binding domains of the rabies virus and VSV phosphoproteins (15, 19). In contrast, there is no structural conservation of the oligomerization domains. Similarly, the structures of the N-RNA binding domains of rabies virus and VSV P are very different from those of measles and Sendai virus (3, 11). For these phosphoproteins it seems that their modular organization consisting of three functional domains separated by two disordered regions is conserved rather than the structure of the functional domains.
The coordinates have been deposited in the RCSB PDB as PDB ID code 3L32.
Acknowledgments
This study was supported by a grant from the French ANR (ANR-07-001-01; ANRAGE).
We thank Giuseppe Zaccai (Institut Laue Langevin, Grenoble, France), Laurence Serre (PSB), and Nicolas Tarbouriech, Hassan Belrhali, and Nicolas Martinelli (UVHCI) for extensive discussions and help; Francine Gérard for the fluorescence spectroscopy measurements; and the Partnership for Structural Biology in Grenoble for the excellent structural biology environment.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Albertini, A. A. V., A. K. Wernimont, T. Muziol, R. B. G. Ravelli, C. R. Clapier, G. Schoehn, W. Weissenhorn, and R. W. H. Ruigrok. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:357-360. [DOI] [PubMed] [Google Scholar]
- 2.Arnheiter, H., N. L. Davis, G. Wertz, M. Schubert, and R. A. Lazzarini. 1985. Role of the nucleocapsid protein in regulating vesicular stomatitis virus RNA synthesis. Cell 41:259-267. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard, L., N. Tarbouriech, M. Blackledge, P. Timmins, W. P. Burmeister, R. W. H. Ruigrok, and D. Marion. 2004. Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution. Virology 319:201-211. [DOI] [PubMed] [Google Scholar]
- 4.Chothia, C., M. Levitt, and D. Richardson. 1981. Helix-to-helix packing in proteins. J. Mol. Biol. 145:215-250. [DOI] [PubMed] [Google Scholar]
- 5.Curran, J., R. Boeck, N. Lin-Marq, A. Lupas, and D. Kolakofsky. 1995. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214:139-149. [DOI] [PubMed] [Google Scholar]
- 6.Ding, H., T. J. Green, S. Lu, and M. Luo. 2006. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 80:2808-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson, S. U., and M. Schubert. 1987. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 84:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, Y., and J. Lenard. 1995. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 14:1240-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gérard, F. C. A., E. de Almeido Ribeiro, Jr., A. A. V. Albertini, I. Gutsche, G. Zaccai, R. W. H. Ruigrok, and M. Jamin. 2007. Unphosphorylated Rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry 46:10328-10338. [DOI] [PubMed] [Google Scholar]
- 10.Gérard, F. C. A., E. de Almeido Ribeiro, Jr., C. Leyrat, I. Ivanov, D. Blondel, S. Longhi, R. W. H. Ruigrok, and M. Jamin. 2009. Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388:978-996. [DOI] [PubMed] [Google Scholar]
- 11.Johansson, K., J. M. Bourhis, V. Campanacci, C. Cambillau, B. Canard, and S. Longhi. 2003. Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278:44567-44573. [DOI] [PubMed] [Google Scholar]
- 12.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 13.Karlin, D., F. Ferron, B. Canard, and S. Longhi. 2003. Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 84:3239-3252. [DOI] [PubMed] [Google Scholar]
- 14.Laskowski, R. A., M. W. Mac Arthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 15.Mavrakis, M., A. A. McCarthy, S. Roche, D. Blondel, and R. W. H. Ruigrok. 2004. Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J. Mol. Biol. 343:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murshudov, G. N. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 17.Panjikar, S., V. Parthasarathy, V. S. Lamzin, M. S. Weiss, and P. A. Tucker. 2005. Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D 61:449-457. [DOI] [PubMed] [Google Scholar]
- 18.Perrakis, A. R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 19.Ribeiro, E. A., Jr., A. Favier, F. C. A. Gérard, C. Leyrat, B. Brutscher, D. Blondel, R. W. H. Ruigrok, M. Blackledge, and M. Jamin. 2008. Solution structure of the C-terminal N-RNA binding domain of the vesicular stomatitis virus phosphoprotein. J. Mol. Biol. 382:525-538. [DOI] [PubMed] [Google Scholar]
- 20.Schymkowitz, J., J. Borg, F. Stricher, R. Nys, F. Rousseau, and L. Serrano. 2005. The FoldX web server: an online force field. Nucleic Acids Res. 33:W382-W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarbouriech, N., J. Curran, R. W. H. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nature Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]
- 22.Vagin, A., and A. Teplyakov. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. 229:105-124. [DOI] [PubMed] [Google Scholar]