Abstract
Very limited evidence has been reported to show human adaptive immune responses to the 2009 pandemic H1N1 swine-origin influenza A virus (S-OIV). We studied 17 S-OIV peptides homologous to immunodominant CD4 T epitopes from hemagglutinin (HA), neuraminidase (NA), nuclear protein (NP), M1 matrix protein (MP), and PB1 of a seasonal H1N1 strain. We concluded that 15 of these 17 S-OIV peptides would induce responses of seasonal influenza virus-specific T cells. Of these, seven S-OIV sequences were identical to seasonal influenza virus sequences, while eight had at least one amino acid that was not conserved. T cells recognizing epitopes derived from these S-OIV antigens could be detected ex vivo. Most of these T cells expressed memory markers, although none of the donors had been exposed to S-OIV. Functional analysis revealed that specific amino acid differences in the sequences of these S-OIV peptides would not affect or partially affect memory T-cell responses. These findings suggest that without protective antibody responses, individuals vaccinated against seasonal influenza A may still benefit from preexisting cross-reactive memory CD4 T cells reducing their susceptibility to S-OIV infection.
The outbreak of H1N1 swine-origin influenza A virus (S-OIV) in April 2009 has raised a new threat to public health (5, 6). This novel virus (with A/California/04/09 H1N1 as a prototypic strain) not only replicated more efficiently but also caused more severe pathological lesions in the lungs of infected mice, ferrets, and nonhuman primates than a currently circulating human H1N1 virus (9). Similarly, human patients with influenza-like illness who tested negative for S-OIV had a milder clinical course than those who tested positive (13). Another major concern is the lack of immune protection against S-OIV in the human population. Initial serum analysis indicated that cross-reactive antibodies to this novel viral strain were detected in only one-third of people over 60 years of age, while humoral immune responses in the population under 60 years of age were rarely detected (3, 8). In addition, vaccination with recent seasonal influenza vaccines induced little or no cross-reactive antibody responses to S-OIV in any age group (3, 8).
Only a few studies address whether preexisting seasonal influenza A virus-specific memory T cells cross-react with antigenic peptides derived from S-OIV (7). In the absence of preexisting cross-reactive neutralizing antibodies, it is likely that T-cell-mediated cellular immunity contributes to viral clearance and reduces the severity of symptoms, although virus-specific T cells cannot directly prevent the establishment of infection (10). Greenbaum and colleagues recently compared published T-cell epitopes for seasonal influenza viruses with S-OIV antigens (Ags) using a computational approach (7). Several seasonal H1N1 epitopes were found to be identical to S-OIV sequences. This implies that seasonal flu-specific memory T cells circulating in the peripheral blood of vaccinated and/or previously infected individuals are able to recognize their S-OIV homologues.
The first objective of this study was to determine the extent of cross-reactivity of seasonal H1N1 influenza A virus-specific CD4 T cells with S-OIV epitopes, especially those less conserved peptide sequences. We chose 17 immunodominant DR4-restricted T-cell epitopes derived from a seasonal H1N1 strain, compared the binding of these epitopes and their S-OIV homologous peptides to DR4, tested the ability of S-OIV peptides to drive seasonal influenza virus-specific T-cell proliferation in vitro, and estimated the frequency of S-OIV cross-reactive T cells in the periphery of noninfected donors. We found that most homologous S-OIV peptides were able to activate seasonal H1N1 virus-specific CD4 T cells. The second objective was to compare the antigen dosage requirement to activate those T cells. By assessing the alternations in the functional avidities (of T cells to the cognate peptide and S-OIV homologue) due to amino acid differences in S-OIV peptides, we showed how those cross-reactive CD4 T cells differentially responded to the antigenic peptides derived from seasonal H1N1 virus or S-OIV. This study leads to the conclusion that previous exposure to seasonal H1N1 viral antigens will generate considerable levels of memory CD4 T cells cross-reactive with S-OIV.
MATERIALS AND METHODS
Human subjects.
Peripheral blood used to identify seasonal H1N1 influenza A viral epitopes was obtained from six HLA-DR0401+ donors recruited in 2007 and 2008 in Seattle, WA. S-OIV-specific T-cell responses were studied in 11 HLA-DR0401+ volunteers recruited between June and August 2009. Samples from two of these donors were used to generate Ag-specific T-cell lines, six samples were used for ex vivo tetramer staining to estimate the Ag-specific T-cell frequency, and the samples from the remaining three donors were used for in vitro functional studies. A brief questionnaire was conducted when the blood sample was collected to confirm that every donor had received trivalent seasonal influenza vaccines with the past 5 years. There was no clinical history of S-OIV infection in any donor. The study was approved by the Institutional Review Board (IRB) of Benaroya Research Institute (BRI, Seattle, WA). HLA typing was conducted by the BRI sequencing and genotyping core facilities. All HLA-DR0401+ subjects were healthy volunteers of Caucasian descent and were recruited with informed consent for these studies.
Peptides, production of recombinant class II major histocompatibility complex (MHC), and epitope mapping.
For epitope mapping, partially overlapping peptide panels covering the seasonal influenza A/New Caledonia/20/99 (H1N1) virus hemagglutinin (HA) and neuraminidase (NA) and A/New York/348/03 (H1N1) virus polymerase PB1 were provided by BEI Resources (Manassas, VA). Peptide panels covering seasonal influenza A/Puerto Rico/8/34 (H1N1) virus nuclear protein (NP), M1 matrix protein (MP), and biotinylated reference peptide for indirect peptide binding assays were purchased from Mimotopes (Clayton Victoria, Australia). For assessing seasonal H1N1 and S-OIV specific T-cell responses, peptides derived from A/New Caledonia/20/99 (H1N1) and A/California/04/2009 (H1N1) influenza viruses were purchased from Sigma (St. Louis, MO). The procedure for recombinant HLA-DR0401 protein production used Drosophila S2 cell expression system (Invitrogen, Carlsbad, CA) and affinity chromatography. The procedure for immunodominant CD4 T-cell epitope identification using tetramer-guided epitope mapping has been described extensively in previous studies (12, 14, 17). Briefly, freshly isolated CD4 T cells (2 × 106/well in a 48-well plate) were stimulated with pooled influenza A virus peptides (five peptide for each pool and 10 μg/ml for each peptide) in the presence of adherent cells from autologous peripheral blood mononuclear cells (PBMCs) in T-cell culturing medium (14, 17) supplemented with 10% pooled human serum for 7 days and expanded with 5% human interleukin-2 (IL-2) (Hemagen, Columbia, MA) for another 7 days. On day 14, a fraction of T cells were stained with pooled peptide tetramers and analyzed by flow cytometry. Cells from pools with positive staining were analyzed again with individual peptide tetramers to identify the peptide epitope.
Indirect peptide binding assay.
Nonbiotinylated DR0401 proteins were diluted into 150 mM citrate-phosphate buffer (pH 5.4) containing 7.5 mg/ml of n-octyl-β-d-glucopyranoside and 1 mM Pefabloc (Sigma, MO). The final concentration of DR0401 protein was 4 μg/ml. Nonbiotinylated target peptides were incubated with DR0401 protein at final concentrations ranging from 0.01 μM to 10 μM for 1 h at 37°C, followed by an additional 16 h of incubation in the presence of 0.01 μM biotinylated reference peptide (HA306, PKYVKQNTLKLAT). The binding reaction was stopped by adding an equal volume of 50 mM Tris-Cl buffer (pH 8.0). The DR molecules were then immobilized on 96-well plates coated with anti-HLA-DR monoclonal antibody L243. The amount of biotinylated reference peptide-bound DR0401 was quantified using a europium-streptavidin detection system on a Victor2 microtiter plate reader (Perkin-Elmer, Waltham, MA). The concentrations of target peptides required to inhibit 50% of maximal biotinylated reference peptide binding were retrieved from regression curves fitted by a sigmoidal dose-response equation provided by Prism software (GraphPad, San Diego, CA).
Ag-specific T-cell lines and proliferation assay.
Total CD4 T cells were isolated from PBMCs using the Dynal CD4 isolation kit (Invitrogen) and stimulated in vitro with 10 μg/ml of target peptide in the presence of adherent cells from autologous PBMCs in T-cell culturing medium (14, 17) supplemented with 10% pooled human serum for 7 days and expanded with 5% human IL-2 (Hemagen) for another 7 days. On day 14, the T-cell culture was stained with tetramers. The Ag-specific T cells were separated from rest of the T cells by cell sorting and further expanded/maintained by coculturing with 1 μg/ml phytohemagglutinin (PHA) in the presence of irradiated (50 Gy) PBMCs. T-cell proliferation was performed by incubating the Ag-specific T-cell line (10,000 cells/well) in the presence of irradiated autologous PBMCs or dendritic cells (20,000 cells/well) and 10 μg/ml target peptide or vaccine (Fluzone by Aventis Pasteur, 2004 to 2005 formula) in triplicate in a 96-well round-bottom plate for 72 h. During the last 12 h of incubation, cells were pulsed with [3H]thymidine (1 μCi/well). Cells were then harvested, and thymidine incorporation was determined using a Microbeta TriLux 1450 scintillation counter (Perkin-Elmer). Dendritic cells were generated by culturing adherent cells from PBMCs in the presence of 50 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) and 1,000 U/ml IL-4 (eBioscience, San Diego, CA) as previously described (2). The vaccine was extensively dialyzed into 1× phosphate-buffered saline (PBS) before it was used to stimulate T cells.
Ex vivo tetramer staining to determine the frequency of Ag-specific T cells.
For each donor, we collected 200 to 250 ml peripheral blood with informed consent under an approved IRB protocol. Freshly isolated PBMCs (20 × 106 cells) were stained with 8 μl (500 μg/ml) of phycoerythrin (PE)-conjugated tetramers for 120 min at room temperature in the dark. Fluorescein isothiocyanate (FITC)-conjugated anti-CD45RA, peridinin chlorophyll protein (PerCP)-conjugated anti-CD14 and anti-CD19, and allophycocyanin (APC)-conjugated anti-CD4 (OKT4) (BD Biosciences) antibodies were added to PBMC aliquots for 20 min at 4°C. After two cycles of washing, the cells were incubated with 40 μl anti-PE microbeads (Miltenyi Biotec) for 20 min at 4°C. After another cycle of washing, cells were resuspended in 1 ml of running buffer, and 1/40 of this cell suspension was reserved as “a preenrichment” aliquot for direct flow cytometric analysis to estimate the total number of CD4 T cells. The remaining labeled cells were loaded onto a Miltenyi MS column already placed in the magnet, washed, and eluted according to the manufacturer's protocol for enrichment of PE-conjugated tetramer-positive cells. The “postenrichment” and “preenrichment” cells were incubated with 20 ml of ViaProbe (BD Biosciences) for 15 min at 4°C. All events in the “postenrichment” and “preenrichment” tubes were analyzed on a FACSCalibur (BD Biosciences). Cells were gated on the CD4+ CD14− CD19− ViaProbe− subset in the live gate by using FlowJo (TreeStar Inc., Ashland, OR). Tetramer-positive T cells from the “postenrichment,” including both CD45RA+ and CD45RA− cells, were enumerated. In conjunction with total estimated CD4 T-cell inputs, the frequency was calculated and normalized to number of Ag-specific CD4 T cells per 1 × 106 CD4 T cells (see Fig. S1 in the supplemental material).
ELISPOT.
Purified CD4+ T cells were stimulated in vitro with 10 μg/ml of target peptide in the presence of adherent cells obtained from autologous PBMCs for 7 days and expanded with human IL-2 for another 7 days. On day 14 the frequency of cytokine-secreting CD4 T cells was quantified using the human gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) kit according to the manufacturer's protocols (eBioscience). In brief, T cells were washed with 1× PBS and seeded onto a Millipore polyvinylidene difluoride (PVDF) multiscreen plate (Fisher Scientific) precoated with anti-human IFN-γ ELISPOT capture antibody in triplicate. Each well contained 10 × 103 T cells, 50 × 103 T cell-depleted autologous PBMCs as antigen-presenting cells, and serial diluted target peptide. The plate was incubated at a 37°C in a 5% CO2 incubator for 16 to 20 h prior to washing and developing with 3-amino-9-ethylcarbazole (AEC) substrate (Vector Laboratories, Burlingame, CA) according to the manufacturer's protocol. Development reactions were monitored under a dissection microscope and stopped by addition of distilled water after approximately 10 min. The plate was scanned using a CTL-ImmunoSpot 55 MicroAnalyzer (Cellular Technology Ltd., Shaker Heights, OH), and the number of spot-forming cells (SFC) per well was calculated using ImmunoSpot 5.0 Professional analysis software. The detection sensitivity level for quantifying SFC was calculated using the SmartWell algorithm (15). For each plate, multiple wells containing various concentrations of peptide were chosen to determine the range of spot size. As recommended by the manufacturer's instructions and a previous study (15), at least 300 SFC were required to avoid mistakenly excluding oversized (strong stimulus due to high peptide concentration) and undersized (weak stimulus due to low peptide concentration) spots. Using this setting, spot sizes typically ranged from 0.001 to 0.1 mm2. To avoid enumerating artifacts at well edges, only 95% of each well was counted for spots, and the results were subsequently normalized to 100% automatically by the software. The frequencies of SFC against peptide concentrations were further analyzed with Prism 4.0 software to calculate a nonlinear regression dose-dependent curve fitted by a sigmoidal dose-response equation. The best-fit 50% effective concentration (EC50) value and corresponding 95% confidence intervals were calculated for each curve.
Dual-tetramer staining.
CD4 T cells (5 × 106) from 2-week in vitro cultures (the same T cells used for ELISPOT assay) were stained with 2 ml PE-conjugated tetramer (loaded with seasonal H1N1 peptide) and 2 ml PE-Cy5-conjugated tetramer (loaded with S-OIV peptide) at 37°C for 1 h. FITC-conjugated anti-CD4 (OKT4) (eBioscience) was then added to the cell suspension for a 30-min incubation at 4°C. Cells were washed twice, and the staining results were collected on a FACSCalibur. To reveal the dual-tetramer staining results, cells were gated on CD4+ and PE-tetramer+ subset using FlowJo.
Statistical analysis.
To calculate P values for ELISPOT results, an F test was used to evaluate the difference between EC50 values of seasonal flu virus and S-OIV peptides. An unpaired two-tailed t test was used to evaluate the difference of SFC correlated with maximal antigen concentration.
RESULTS
Identification of immunodominant DR0401-restricted seasonal H1N1 influenza A virus-specific epitopes.
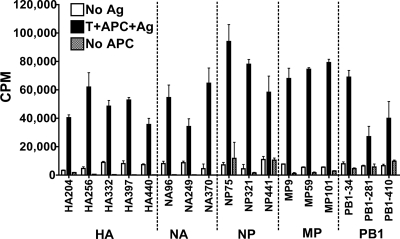
We used tetramer-guided epitope mapping (14, 17) to identify immunodominant (in >50% individuals by our arbitrary definition) DR4-restricted CD4 T-cell epitopes derived from seasonal H1N1 influenza A viruses (see Fig. S2 in the supplemental material). In total, 94 peptides derived from hemagglutinin (HA), 78 peptides from neuraminidase (NA), 61 peptides from nuclear protein (NP), 30 peptides from M1 matrix protein (MP), and 126 peptides from polymerase PB1 were tested. Seventeen epitopes were identified: five from HA, three from NA, three from NP, three from MP, and three from PB1 (see Table S1 in the supplemental material). One NP-derived epitope (NP321) and one MP-derived epitope (MP59) were also previously identified by other groups using different methods (1, 11). CD4 T-cell lines specific to these epitopes responded to trivalent vaccines containing seasonal H1N1 viral antigens (Fig. 1). These responses to whole vaccine confirmed that all 17 epitopes are naturally processed and presented.
FIG. 1.
Seasonal influenza virus-specific CD4 T cells respond to naturally processed epitopes. [3H]thymidine incorporation results for 17 individual seasonal influenza virus-specific T-cell lines cocultured with dendritic cells derived from autologous monocytes in the presence of medium (white bars) or 10 μg/ml trivalent seasonal influenza virus vaccine (black bars) are shown. Results for T-cell lines incubated with vaccine in the absence of antigen-presenting cells are also shown (gray bars). Error bars represent 1 standard deviation.
Comparing seasonal H1N1 peptides with S-OIV homologues for peptide binding affinity and antigenicity.
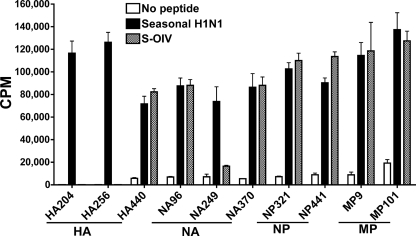
It was unknown whether CD4 T cells specific for seasonal influenza virus epitopes would cross-react with S-OIV. To address this question, we evaluated the peptide binding affinities and antigenicities of the peptides derived from an S-OIV strain (A/California/04/09 H1N1) and a common seasonal H1N1 strain (A/New Caledonia/20/99 H1N1). All NP, MP, and PB1 epitopes identified from the various H1N1 strains mentioned above are identical to the corresponding A/New Caledonia/20/99 sequences, except for MP9, which has one amino acid difference. However, the MP9 epitope from A/New Caledonia/20/99 H1N1 was found in virtually every DR4 donor and elicited robust T-cell responses (data not shown). As shown in Table 1, seven S-OIV peptides (sHA332, sHA397, sNP75, sMP59, sPB1/34, sPB1/281, and sPB1/410) are identical to their seasonal homologues. These S-OIV peptides can be expected to bind class II MHC and stimulate T cells in a fashion identical to that for the seasonal flu virus homologues. For the 10 S-OIV peptides that differed from their seasonal strain homologues by at least one amino acid, four S-OIV sequences (sNA96, sNA370, sNP441, and sMP101) bound HLA-DR0401 with a binding affinity similar to that of the corresponding seasonal flu virus epitope. The remaining six S-OIV sequences (sHA204, sHA256, sHA440, sNA249, sNP321, and sMP9) bound with binding affinities at least 2-fold less than those of their seasonal homologues. Despite the fact that binding affinities were altered by the amino acid difference between S-OIV and seasonal H1N1 peptides, when we restimulated those seasonal influenza virus-specific CD4 T-cell lines (the same T-cell lines shown in Fig. 1; seven T-cell lines specific to conserved peptides were not tested) in vitro, we found that all but two S-OIV peptides, i.e., sHA204 and sHA256, retained their ability to drive T-cell proliferation (Fig. 2). Together, these data suggest that 15 of the 17 S-OIV peptides are able to cross-react with seasonal flu virus-specific T cells.
TABLE 1.
Sequence alignment and peptide binding affinity of seasonal influenza virus (A/New Caledonia/20/99 H1N1) and S-OIV
| Epitope no. | Epitopea | Sequenceb | IC50 (μM)c | RBAd |
|---|---|---|---|---|
| 1 | HA204 | QRALYHTENAYVSVVS | 0.54 | 1 |
| sHA204 | -QS--QNADT--F-G- | 13.02 | 24 | |
| 2 | HA256 | IIFEANGNLIAPWYAFA | 3.61 | 1 |
| sHA256 | -T---T---VV-R---- | 121 | 33.5 | |
| 3 | HA332 | TGLRNIPSIQSRGLFGAIA | 1.81 | |
| sHA332 | ------------------- | - | ||
| 4 | HA397 | SVIEKMNTQFTAVGKE | 4.61 | |
| sHA397 | ---------------- | - | ||
| 5 | HA440 | ELLVLLENERTLDFHDS | 4.34 | 1 |
| sHA440 | -------------Y--- | 10.3 | 2.37 | |
| 6 | NA96 | GWAIYTKDNSIRIGSKG | 0.72 | 1 |
| sNA96 | -----S----V------ | 0.42 | 0.58 | |
| 7 | NA249 | GAASYKIFKIEKGKVTK | 1.46 | 1 |
| sNA249 | -Q------R-----IV- | 9.72 | 6.66 | |
| 8 | NA370 | GFEMIWDPNGWTDTDS | 1.01 | 1 |
| sNA370 | ------------G--N | 0.83 | 0.82 | |
| 9 | NP75 | RNKYLEEHPSAGKD | 11.84 | |
| sNP75 | -------------- | - | ||
| 10 | NP321 | NPAHKSQLVWMACNSAAFED | 0.22 | 1 |
| sNP321 | -------------H-----I | 5.57 | 25.3 | |
| 11 | NP441 | RAEIIKMMESARPEEVSFQ | 1.73 | 1 |
| sNP441 | -T-V-R-----K--DL--- | 1.09 | 0.63 | |
| 12 | MP9 | TYVLSIVPSGPLKAEIAQRL | 6.23 | 1 |
| sMP9 | ------I------------- | 46.64 | 7.49 | |
| 13 | MP59 | ILGFVFTLTVPSERG | 0.04 | |
| sMP59 | --------------- | - | ||
| 14 | MP101 | RKLKREITFHGAK | 0.73 | 1 |
| sMP101 | K------------ | 0.81 | 1.11 | |
| 15 | PB1/34 | TGTGYTMDTVNRTHQ | 1.24 | |
| sPB1/34 | --------------- | - | ||
| 16 | PB1/281 | KLANVVRKMMTNSQDTE | 1.33 | |
| sPB1/281 | ----------------- | - | ||
| 17 | PB1/410 | GMFNMLSTVLGVSILNLGQ | 0.14 | |
| sPB1/410 | ------------------- | - |
Peptides from seasonal influenza virus (A/New Caledonia/20/99 H1N1) have no prefix; peptides from S-OIV (A/California/04/09 H1N1) have an “s” prefix.
-, identical amino acid residue.
IC50, concentration required by a target peptide to inhibit 50 % binding of the reference peptide. —, not measured.
RBA, relative binding affinity (relative IC50 in reference to that of the seasonal flu virus peptide).
FIG. 2.
Seasonal influenza virus-specific CD4 T cells respond to S-OIV peptides. [3H]thymidine incorporation for seasonal flu virus-specific T-cell lines restimulated with 10 μg/ml of peptides derived from the seasonal influenza virus strain (black bars), S-OIV (hatched bars), or medium control (white bars), in the presence of irradiated (50 Gy) autologous PBMC, are shown. Error bars represent 1 standard deviation.
Ex vivo detection of both seasonal flu virus- and S-OIV-specific CD4 T cells in the peripheral blood of healthy individuals.
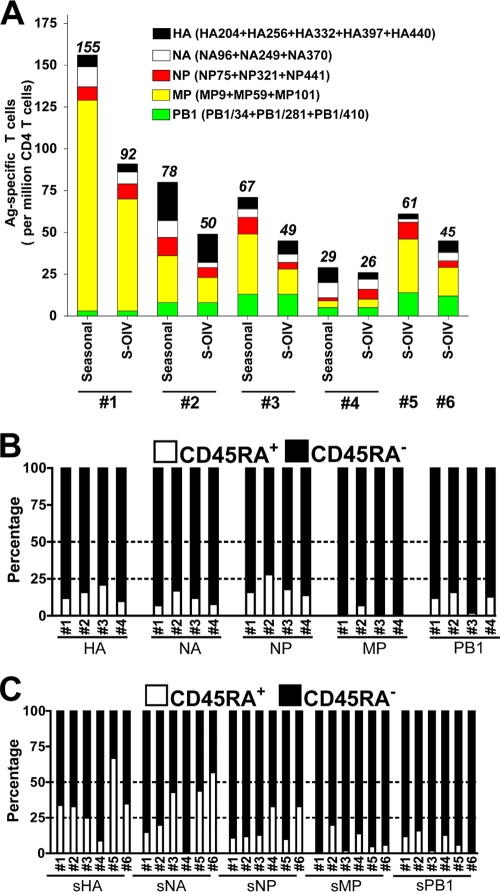
Given the high degree of cross-reactivity observed in S-OIV peptides (15 out of 17), we next tested whether S-OIV-reactive T cells could be detected in the peripheral blood of healthy DR4 individuals using direct ex vivo tetramer staining (illustrated in Fig. S1 in the supplemental material). Due to the relatively low ex vivo frequencies of Ag-specific T cells and the limited availability of PBMCs from each donor, the evaluation was designed to target pools of epitopes rather than each individual epitope. Seasonal flu virus-specific CD4 T-cell frequency analysis was also performed when sufficient PBMCs were available (for four donors). T cells responsive to S-OIV epitopes could be detected ex vivo (Fig. 3A), but the overall frequencies were lower than that of seasonal (s) flu virus-responsive T cells (∼75%). T cells specific for (s)MP and (s)PB1 were predominant. The high percentages of CD45RA− Ag-specific T cells (Fig. 3B) indicate that most subjects were previously primed with antigens in vivo. Because the donors had not been exposed to S-OIV, the presence of these S-OIV tetramer positively staining memory T cells is necessarily the result of previous seasonal flu virus infection or vaccination. The numbers of (s)HA-, (s)NA-, and (s)NP-responding cells were relatively low compared to those of (s)MP- or (s)PB1-responding T cells (Fig. 3A). Although the overall frequencies for sHA and sNA were lower than those for seasonal flu virus-specific T cells from the same donors, the percentages of CD45RA+ cells increased slightly in S-OIV tetramer-positive populations compared to seasonal flu virus tetramer-positive populations (Fig. 3B and C), suggesting the presence of a naïve T-cell repertoire specific for novel S-OIV antigenic peptides.
FIG. 3.
Ex vivo detection of seasonal flu virus-specific and S-OIV-specific CD4 T cells. (A) Number of influenza virus-specific CD4 T cells per 1 × 106 CD4 T cells estimated by ex vivo tetramer staining. Each colored bar segment represents the sum of the indicated Ag-specific T cells. Cells were gated on CD19− CD14− CD4+ tetramer+ populations. The number above each bar represents the total number of Ag-specific T cells investigated. (B and C) Relative frequencies of CD45RA+ and CD45RA− subsets within seasonal influenza virus (B)- and S-OIV (C)-specific T cells. The cells were gated on CD19− CD14− CD4+ tetramer+ populations.
Functional avidity of seasonal H1N1 virus-specific T cells that cross-react with S-OIV epitopes.
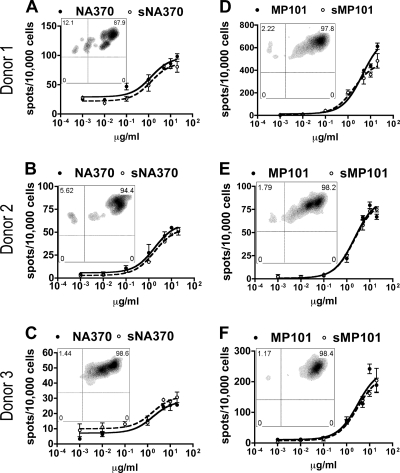
The observation of S-OIV tetramer-positive T cells ex vivo suggested that these cells are potentially responsive to S-OIV challenge. We next examined whether the amino acid differences in S-OIV peptides would affect the quality of the T-cell response. Two S-OIV peptides, sNA370 and sMP101, elicited a full-scale cross-reaction. The dose-response curves for these epitopes and their seasonal flu virus epitopes nearly overlapped (Fig. 4). No significant differences in maximal SFC values were observed for these epitopes (Table 2), indicating that the majority of seasonal flu virus-specific T cells responded to S-OIV homologues as well. The EC50 values for these S-OIV peptides were no different from those for seasonal flu virus peptides (Table 2), suggesting that the functional avidities of T cells responding to these two S-OIV peptides were similar to those for seasonal H1N1 virus-derived cognate peptides. Dual-tetramer staining assays also indicated that the Ag-specific T cells could be stained simultaneously with tetramers loaded with seasonal influenza virus or S-OIV peptides.
FIG. 4.
S-OIV peptides elicit full-scale cross-reaction. IFN-γ ELISPOT results from three HLA-DR0401+ donors in response to (s)NA370 (A to C) and (s)MP101 (D to F) are shown. Each plot represents the results from one donor for a single seasonal flu virus epitope (closed circles, solid lines) and the S-OIV homologue (open circles, dashed lines). Two-week in vitro (seasonal H1N1 virus peptide)-primed T cells were used as responders. The number of spots per 10,000 cells is plotted against the concentration of peptide. Error bars represent 1 standard deviation. The embedded fluorescence-activated cell sorter (FACS) density plot shows the dual-tetramer staining (of the same T cells used for ELISPOT), where the y axis represents seasonal H1N1 virus peptide-loaded tetramer and the x axis represents S-OIV homologue peptide-loaded tetramer. Gated populations represent the CD4+ and seasonal flu tetramer+ cells.
TABLE 2.
In vitro functional avidity analysis of T-cell responses against cognate seasonal flu virus epitopes and corresponding S-OIV homologues
| Peptide | Donor | EC50 (95% CI), μg/mla |
Maximum SFC/10,000 cells, mean ± SDb |
||||
|---|---|---|---|---|---|---|---|
| Seasonal H1N1 virus | S-OIV | P (F test) | Seasonal H1N1 virus | S-OIV | P (t test) | ||
| (s)NA370 | 1 | 1.255 (0.588-2.679) | 1.308 (0.634-2.702) | 0.9385 | 98 ± 5 | 81 ± 9 | 0.0566 |
| 2 | 1.167 (0.659-2.067) | 2.887 (0.537-15.54) | 0.2841 | 51 ± 4 | 53 ± 16 | 0.8769 | |
| 3 | 0.823 (0.320-2.118) | 3.175 (0.939-10.74) | 0.0681 | 26 ± 1 | 31 ± 4 | 0.0717 | |
| (s)MP101 | 1 | 4.341 (2.93-6.43) | 1.854 (0.972-3.535) | 0.0461 | 612 ± 28 | 483 ± 64 | 0.1211 |
| 2 | 1.911 (1.055-3.463) | 2.524 (1.957-3.255) | 0.3019 | 67 ± 3 | 75 ± 3 | 0.1056 | |
| 3 | 3.216 (1.291-8.011) | 2.813 (1.606-4.929) | 0.8125 | 189 ± 3 | 204 ± 40 | 0.6876 | |
| (s)MP9 | 1 | 0.035 (0.018-0.067) | 0.162 (0.115-0.227) | 0.0001 | 669 ± 11 | 672 ± 64 | 0.9536 |
| 2 | 0.414 (0.298-0.576) | 1.795 (1.009-3.192) | 0.0002 | 86 ± 5 | 81 ± 4 | 0.4319 | |
| 3 | 0.101 (0.065-0.156) | 1.689 (1.443-1.975) | <0.0001 | 357 ± 13 | 316 ± 11 | 0.0826 | |
| (s)NP441 | 1 | —c | — | — | — | — | — |
| 2 | 5.335 (3.499-8.135) | Incalculabled | Incalculable | NAe | NA | NA | |
| 3 | 2.034 (1.124-3.680) | 11.26 (5.808-21.82) | 0.0002 | 92 ± 1 | NA | NA | |
| (s)HA440 | 1 | 1.830 (1.144-2.928) | 6.080 (2.561-14.43) | 0.0463 | 250 ± 23 | 139 ± 8 | 0.0014 |
| 2 | 0.670 (0.435-1.030) | 0.838 (0.550-1.278) | 0.5083 | 301 ± 25 | 163 ± 12 | 0.0203 | |
| 3 | 3.055 (2.445-3.819) | 5.081 (2.367-10.91) | 0.1072 | 112 ± 5 | 61 ± 16 | 0.0058 | |
| (s)NA96 | 1 | 0.578 (0.417-0.800) | 1.133 (0.575-2.233) | 0.1188 | 399 ± 15 | 122 ± 5 | <0.0001 |
| 2 | 0.135 (0.108-0.170) | 3.296 (1.909-5.691) | <0.0001 | 584 ± 9 | 214 ± 14 | <0.0001 | |
| 3 | 0.295 (0.165-0.526) | 0.914 (0.330-2.531) | 0.1666 | 221 ± 23 | 89 ± 16 | 0.0046 | |
| (s)NA249 | 1 | 0.078 (0.037-0.168) | 0.409 (0.201-0.832) | 0.0083 | 138 ± 12 | 113 ± 13 | 0.0654 |
| 2 | 0.076 (0.042-0.136) | Incalculable | Incalculable | 85 ± 3 | 36 ± 11 | 0.0272 | |
| 3 | 0.134 (0.062-0.291) | 1.682 (0.817-3.465) | <0.0001 | 61 ± 2 | 42 ± 1 | 0.0069 | |
| (s)NP321 | 1 | 0.190 (0.106-0.341) | 0.656 (0.406-1.060) | 0.0129 | 322 ± 11 | 194 ± 19 | 0.0142 |
| 2 | 0.865 (0.494-1.514) | 2.721 (1.305-5.675) | 0.0187 | 169 ± 10 | 92 ± 17 | 0.0025 | |
| 3 | 0.283 (0.190-0.423) | 2.170 (0.768-6.137) | 0.0057 | 251 ± 8 | 47 ± 5 | <0.0001 | |
EC50, peptide dosage required for 50 % of maximal T-cell response, represented as the best-fit value followed by the 95% confidence interval (CI).
SFC read from the wells with the highest peptide concentrations.
Unable to be calculated because the possible value is more than the highest peptide concentration tested.
NA, not available due to an unsaturated response.
—, no measurement due to the lack of a detectable response.
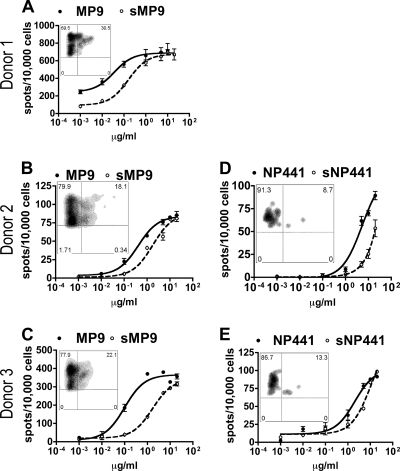
Seasonal flu virus-specific T cells also cross-reacted with sMP9 and sNP441 peptides. However, the dose-response curves suggested that excessive S-OIV peptides were required to reach a full-scale response (Fig. 5). At low Ag concentrations, the curves for S-OIV peptides were segregated from seasonal flu virus peptides, indicating that the response to S-OIV peptide was weaker than that to the seasonal H1N1 virus peptide. No significant differences in maximal SFC values were observed between these two S-OIV peptides and their corresponding seasonal flu virus counterparts (Table 2). Significantly increased EC50 values confirmed that T cells were less sensitive to S-OIV peptides than cognate seasonal flu virus peptides (Table 2). Dual-tetramer staining showed that the staining for the S-OIV tetramers was compromised, suggesting that the interactions between the T-cell receptors (TCRs) and S-OIV agonists were weak.
FIG. 5.
S-OIV peptides require an excessive dosage to induce full-scale cross-reaction. IFN-γ ELISPOT results from three HLA-DR0401+ donors for (s)MP9 (A to C) and two donors for (s)NP441 (D and E) are shown. Each plot represents the results from one donor for the seasonal flu virus epitope (closed circles, solid lines) and the S-OIV homologue (open circles, dashed lines). Two-week in vitro (seasonal H1N1 peptide)-primed T cells were used as responders. The number of spots per 10,000 cells is plotted against the concentration of peptide. Error bars represent 1 standard deviation. The embedded FACS density plot shows the dual-tetramer staining (of the same T cells used for ELISPOT), where the y axis represents seasonal H1N1 virus peptide-loaded tetramer and the x axis represents S-OIV homologue peptide-loaded tetramer. Gated populations represent the CD4+ and seasonal flu tetramer+ cells.
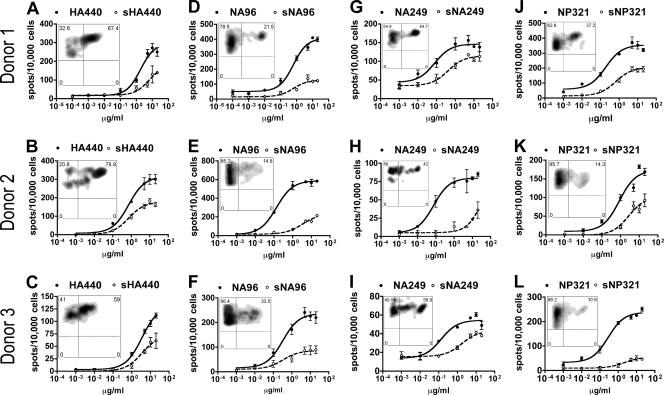
Dosage curves for sHA440, sNA96, sNA249, and sNP321 suggested partial cross-reactivity for these S-OIV peptides (Fig. 6). The maximal SFC values triggered by S-OIV were lower than those for seasonal flu epitopes (Table 2). In addition, only a subset of T cells was simultaneously stained by the two tetramers. The EC50 values for sHA440 and sNA96 were not significantly higher than those for their seasonal flu virus homologues, except for an (s)NA96 response in one donor (donor 2 in Table 2). These data suggest that the dosage requirement for triggering the cross-reactive T-cell subpopulation is similar for seasonal flu virus and S-OIV homologous peptides. For both sNA249 and sNP321, the EC50 values were significantly increased in comparison to those for NA249 and NP321, showing that seasonal flu virus-specific T cells were less sensitive to these two S-OIV agonistic peptides than to cognate seasonal flu virus peptides. Finally, neither dual-tetramer staining nor functional assays showed that HA204- and HA256-specific T cells responded to their S-OIV peptides.
FIG. 6.
S-OIV peptides induce partial cross-reaction. IFN-γ ELISPOT results from three HLA-DR0401+ donors for (s)HA440 (A to C), (s)NA96 (D to F), (s)NA249 (G to I), and (s)NP321 (J to L) are shown. Each plot represents the results from one donor for a single seasonal flu virus epitope (closed circles, solid lines) and the S-OIV homologue (open circles, dashed lines). Two-week in vitro (seasonal H1N1 virus peptide)-primed T cells were used as responders. The number of spots per 10,000 cells is plotted against the concentration of peptide. Error bars represent 1 standard deviation. The embedded FACS density plot shows the dual-tetramer staining (of the same T cells used for ELISPOT), where the y axis represents seasonal H1N1 virus peptide-loaded tetramer and the x axis represents S-OIV homologue peptide-loaded tetramer. Gated populations represent the CD4+ and seasonal flu tetramer+ cells.
DISCUSSION
Several previous reports showed that the presence of memory T-cell responses against H5N1 avian flu virus in healthy individuals resulted from seasonal flu vaccination (10, 14). By the same token, we postulated that previous seasonal influenza virus infection or vaccination might also generate memory T cells cross-reactive with S-OIV antigens. This study aimed to determine how common seasonal H1N1 virus-specific CD4 T cells cross-react with S-OIV antigens and to define the quality of these cross-reactive responses. Our investigation targeted 17 DR4-restricted epitopes from surface (HA and NA) and internal (NP, MP, and PB1) antigens of a seasonal H1N1 virus strain (A/New Caledonia/20/99) recommended by WHO for flu vaccine from 2000 through 2007 (4). T cells specific for these epitopes are present in virtually every vaccinated DR4 individual according to our epitope mapping results, suggesting a dominant role for these epitope-specific T cells in building up adaptive immune responses against influenza virus infection. It was thus tempting to examine whether these dominant antiviral T cells were able to respond to homologous S-OIV peptides. A wide range of cross-reactive memory T cells in individuals who had not been exposed to S-OIV would suggest that people might still benefit from seasonal flu vaccination, even without substantial protective antibody responses.
Several assays were used to determine the extent of cross-reactivity of seasonal H1N1 virus-specific CD4 T cells with S-OIV homologues. Peptide binding affinity measurements allowed us to draw conclusions about the expected presentation of seasonal influenza virus and S-OIV peptides by DR0401. An in vitro proliferation assay using highly enriched Ag-specific T-cell lines revealed that cross-reactive S-OIV peptides were not limited to those conserved antigenic sequences. Ex vivo tetramer staining provided us with the ability to estimate the relative abundance of T cells specific to or cross-reactive with individual viral antigens. We found that (s)MP and (s)PB1 were the major targets of the antiviral CD4 T-cell repertoire (Fig. 3). In fact, both antigens were the most conserved among different H1N1 strains and even among different influenza A virus subtypes. Repeated vaccination or infection by these strains could be expected to prime immune responses and boost memory T cells. Therefore, it was not surprising that nearly all MP- and PB1-specific T cells were CD45RA− memory cells (Fig. 3). T cells recognizing (s)HA, (s)NA, and (s)NP were also detected. Although the T cells specific for these antigenic peptides were presented at reduced frequencies, they would likely still elicit immune responses to S-OIV infection.
We also focused on addressing the quality of memory CD4 T cells responding to S-OIV, particularly whether the magnitude of the response would be negatively impacted by the amino acid differences in S-OIV peptides. Different experimental approaches were used to solve this puzzle. Dual-tetramer staining (utilizing two tetramers conjugated with different fluorescent labels loaded with corresponding seasonal influenza virus or S-OIV peptides) allowed us to “visualize” the cross-recognition of S-OIV epitopes by the TCRs of polyclonal T cells specific for seasonal H1N1 virus peptides. For the same T-cell population, a strong staining result for the first tetramer (of seasonal influenza A virus peptide) but a weak staining result for the second tetramer (of homologous S-OIV peptide) could reveal a reduced affinity of peptide-MHC-TCR interactions. The ELISPOT assay, performed over a range of antigen dosages, provided a tool to compare the functional avidities (15) of T-cell responses to seasonal virus and S-OIV sequences. Together, these assays allowed us to assess the impact of the amino acid differences in S-OIV sequences on antigen presentation, TCR engagement, and the overall functional response. The results of our assays (with cells from three different donors) were consistent and indicated different levels of cross-reactivity. Two S-OIV epitopes (sNA370 and sMP101) were fully cross-reactive. In general, these epitopes not only had binding affinities that were similar to those of the corresponding seasonal influenza virus sequences but also had similar affinity for TCR interaction. The impact of amino acid differences in these S-OIV sequences on the functional response was little. Similar to those peptides conserved between S-OIV and seasonal H1N1 virus (HA332, HA397, NP75, MP59, PB1/34, PB1/281, and PB1/410), sNA370 and sMP101 would induce similar or even increased functional responses compared to their seasonal influenza virus homologues. For sMP9 and sNP441, high antigen doses were required to elicit a functional response comparable to that of their seasonal flu virus homologues. The (amino acid) substitution negatively affected sMP9 in binding DR0401. For sNP441, it appeared that the substitution did not interfere with peptide binding to MHC but affected the affinity of the peptide-MHC-TCR interaction. Several S-OIV epitopes (sHA440, sNA96, sNA249, and sNP321) were able to activate only a subset of seasonal flu virus-specific T cells, consistent with dual-tetramer staining results. The maximal functional response was reduced and could not be recovered by increased antigen concentration (as indicated by ELISPOT results). The substitutions might abolish the productive interactions with a portion of the seasonal flu virus-specific TCR repertoire. These results of in vitro T-cell assays suggested that six S-OIV agonistic peptides (sMP9, sNP441, sHA440, sNA96, sNA249, and sNP321) could still drive preexisting memory T-cell expansion but that the sensitivity of the responses was decreased.
Although we addressed only the cross-reactivity of CD4 T cells restricted to DR0401, the prevalence of cross-reactive CD8 and CD4 T cells with other DR restriction could also be evaluated using similar ex vivo and in vitro analysis methodologies. These memory CD8 and CD4 cells resulting from previous seasonal influenza virus infection or vaccination can potentially play a role in attenuating the course of S-OIV-induced disease.
In summary, we have demonstrated the presence of memory T cells responding to S-OIV epitopes in healthy individuals who had not encountered S-OIV. Previous exposure to seasonal H1N1 viruses, either through natural infection or through vaccination, presumably led to the generation of these memory T cells. The functional avidity analysis also revealed the antigen dosage requirement to activate these memory T cells in vitro. It might provide a premise to hypothesize about the in vivo activities of these T cells. A recent publication from Steel and colleagues showed that a prior exposure to H1N1 and H3N2 seasonal influenza A virus strains provided partial immunity against S-OIV infection in guinea pigs (16). In consensus with the results from those animal models, our findings provide an explanation for this type of partial immunity in humans.
Supplementary Material
Acknowledgments
This work was supported by NIH contracts HHSN272200900043C and HHSN266200400028C (to W.W.K).
We thank K. Arumuganathan for cell sorting, Theresa Gates and Diana Sorus for reviewing the manuscript, and Christine Chan, Kevin Criste, and Thien-Son Nguyen for arranging and collecting blood samples. We are also grateful to all donors.
Footnotes
Published ahead of print on 13 January 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Assarsson, E., H. H. Bui, J. Sidney, Q. Zhang, J. Glenn, C. Oseroff, I. N. Mbawuike, J. Alexander, M. J. Newman, H. Grey, and A. Sette. 2008. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 82:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barchet, W., J. D. Price, M. Cella, M. Colonna, S. K. MacMillan, J. P. Cobb, P. A. Thompson, K. M. Murphy, J. P. Atkinson, and C. Kemper. 2006. Complement-induced regulatory T cells suppress T-cell responses but allow for dendritic-cell maturation. Blood 107:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. 2009. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 58:521-524. [PubMed] [Google Scholar]
- 4.Daum, L. T., L. C. Canas, C. B. Smith, A. Klimov, W. Huff, W. Barnes, and K. L. Lohman. 2002. Genetic and antigenic analysis of the first A/New Caledonia/20/99-like H1N1 influenza isolates reported in the Americas. Emerg. Infect. Dis. 8:408-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 6.Fraser, C., C. A. Donnelly, S. Cauchemez, W. P. Hanage, M. D. Van Kerkhove, T. D. Hollingsworth, J. Griffin, R. F. Baggaley, H. E. Jenkins, E. J. Lyons, T. Jombart, W. R. Hinsley, N. C. Grassly, F. Balloux, A. C. Ghani, N. M. Ferguson, A. Rambaut, O. G. Pybus, H. Lopez-Gatell, C. M. Alpuche-Aranda, I. B. Chapela, E. P. Zavala, D. M. Guevara, F. Checchi, E. Garcia, S. Hugonnet, and C. Roth. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenbaum, J. A., M. F. Kotturi, Y. Kim, C. Oseroff, K. Vaughan, N. Salimi, R. Vita, J. Ponomarenko, R. H. Scheuermann, A. Sette, and B. Peters. 2009. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc. Natl. Acad. Sci. U. S. A. 106:20365-20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, K., V. Veguilla, X. Lu, W. Zhong, E. N. Butler, H. Sun, F. Liu, L. Dong, J. R. Devos, P. M. Gargiullo, T. L. Brammer, N. J. Cox, T. M. Tumpey, and J. M. Katz. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361:1945-1952. [DOI] [PubMed] [Google Scholar]
- 9.Itoh, Y., K. Shinya, M. Kiso, T. Watanabe, Y. Sakoda, M. Hatta, Y. Muramoto, D. Tamura, Y. Sakai-Tagawa, T. Noda, S. Sakabe, M. Imai, Y. Hatta, S. Watanabe, C. Li, S. Yamada, K. Fujii, S. Murakami, H. Imai, S. Kakugawa, M. Ito, R. Takano, K. Iwatsuki-Horimoto, M. Shimojima, T. Horimoto, H. Goto, K. Takahashi, A. Makino, H. Ishigaki, M. Nakayama, M. Okamatsu, K. Takahashi, D. Warshauer, P. A. Shult, R. Saito, H. Suzuki, Y. Furuta, M. Yamashita, K. Mitamura, K. Nakano, M. Nakamura, R. Brockman-Schneider, H. Mitamura, M. Yamazaki, N. Sugaya, M. Suresh, M. Ozawa, G. Neumann, J. Gern, H. Kida, K. Ogasawara, and Y. Kawaoka. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, L. Y., L. A. Ha do, C. Simmons, M. D. de Jong, N. V. Chau, R. Schumacher, Y. C. Peng, A. J. McMichael, J. J. Farrar, G. L. Smith, A. R. Townsend, B. A. Askonas, S. Rowland-Jones, and T. Dong. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linnemann, T., G. Jung, and P. Walden. 2000. Detection and quantification of CD4(+) T cells with specificity for a new major histocompatibility complex class II-restricted influenza A virus matrix protein epitope in peripheral blood of influenza patients. J. Virol. 74:8740-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak, E. J., A. W. Liu, G. T. Nepom, and W. W. Kwok. 1999. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J. Clin. Invest. 104:R63-R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Padilla, R., D. de la Rosa-Zamboni, S. Ponce de Leon, M. Hernandez, F. Quinones-Falconi, E. Bautista, A. Ramirez-Venegas, J. Rojas-Serrano, C. E. Ormsby, A. Corrales, A. Higuera, E. Mondragon, and J. A. Cordova-Villalobos. 2009. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N. Engl. J. Med. 361:680-689. [DOI] [PubMed] [Google Scholar]
- 14.Roti, M., J. Yang, D. Berger, L. Huston, E. A. James, and W. W. Kwok. 2008. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 180:1758-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Standifer, N. E., E. A. Burwell, V. H. Gersuk, C. J. Greenbaum, and G. T. Nepom. 2009. Changes in autoreactive T cell avidity during type 1 diabetes development. Clin. Immunol. 132:312-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steel, J., P. Staeheli, S. Mubareka, A. Garcia-Sastre, P. Palese, and A. C. Lowen. 2010. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J. Virol. 84:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, J., N. A. Danke, D. Berger, S. Reichstetter, H. Reijonen, C. Greenbaum, C. Pihoker, E. A. James, and W. W. Kwok. 2006. Islet-specific glucose-6-phosphatase catalytic subunit-related protein-reactive CD4+ T cells in human subjects. J. Immunol. 176:2781-2789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.