Abstract
MicroRNAs (miRNAs) are small regulatory RNAs with many biological functions and disease associations. We showed that in situ hybridization (ISH) using conventional formaldehyde fixation results in significant miRNA loss from mouse tissue sections, which can be prevented by fixation with 1–ethyl–3–(3–dimethylaminopropyl) carbodiimide (EDC) that irreversibly immobilizes the miRNA at its 5' phosphate. We determined optimal hybridization parameters for 130 locked nucleic acid (LNA) probes by recording nucleic acid melting temperature during ISH.
Keywords: immunohistochemistry, heart, liver, brain, dendrites, methanal, paraformaldehyde, water–soluble carbodiimide, phosphoramidate, crosslink, non–coding RNA, small RNA, locked nucleic acid probe
Association studies are rapidly linking miRNAs1 with cancer and neurological disorders2. miRNAs show specific expression and function in specialized cell types3, emphasizing the need to define cell–type–specific miRNA expression patterns. The most common method for visualizing gene expression in specific cell types is in situ hybridization (ISH). In our laboratory, conventional ISH worked for highly abundant miRNAs, however examining less abundantly expressed miRNAs often yielded inconsistent or negative results. In zebrafish whole–mount ISH for miRNAs were robust4, whereas for miRNAs in D. melanogaster embryos ISH was e mostly unsuccessful5. miRNA ISH in tissue sections6–10 showed some results that supported association of miRNA to disease11,12, or define expression patterns in multiple cell types present in the brain13 and eye14 however lower expressed miRNAs were not detected14. [AU: correct as edited? Please keep the parallelism started in the previous sentence: zebrafish – successful, fly – not successful; tissue-sections – semi-successful? beginning to be successful? In reference 14, the authors describe difficulty with several miRNA ISH.] The technical difficulties in miRNA ISH also led to development of transgenic15 or cell sorting methods3 that monitor cell–type–specific miRNA expression.
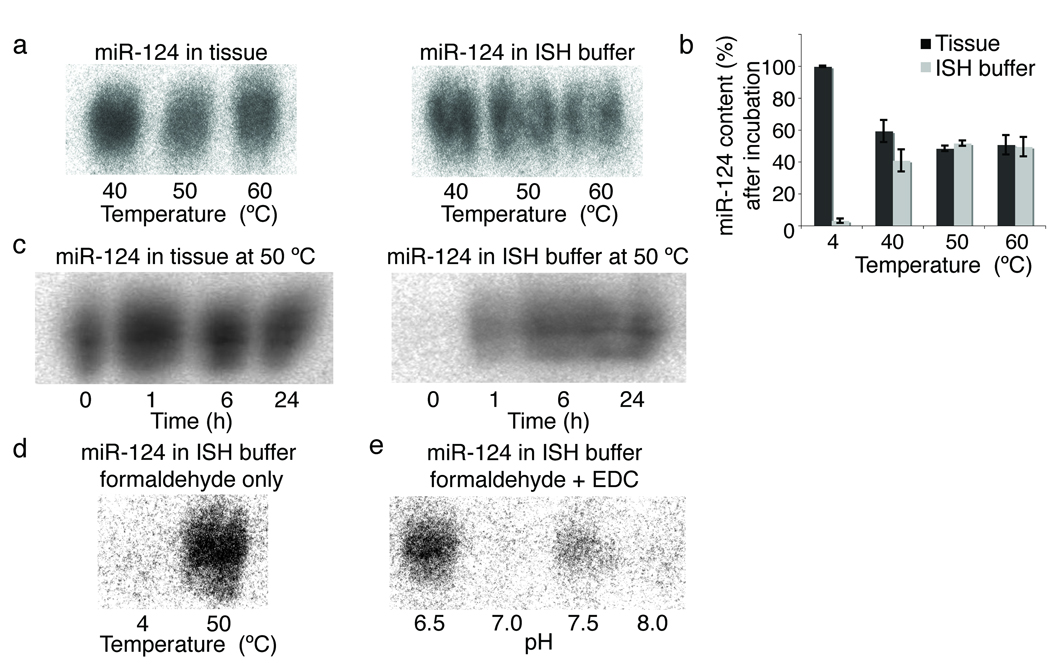
To better understand the technical challenges associated with miRNA ISH, we investigated the importance of miRNA fixation and probe hybridization. For fixation of proteins and nucleic acids in tissues, a solution containing 3.7% formaldehyde (10% formalin) is commonly used16. Formaldehyde protein crosslinks and nucleic acid base modifications are reverted by incubation at elevated temperature and/or in the presence of proteinase K [Au note: either high temperature or proteinase K can weaken formalin crosslinks.]17. While this reversal is necessary for probe hybridization, it creates the problem of miRNA release and diffusion out of the tissue sections. To examine the extent of miRNA escape from tissue during ISH, we exposed conventionally fixed brain sections to treatments in the course of ISH, such as proteinase K and overnight hybridization in buffer, then isolated RNA from both fractions to examine the highly expressed neuronal miR–124 by Northern blotting. [AU correct as edited? It was not clear what a mock ISH is and it is easier to follow if you state your intent and describe what you did. How long was the buffer on the sections?] At hybridization temperatures above 40 °C, at least 50% of miR–124 initially present in the tissue section accumulated in the buffer as early as 1 h (Supplementary Fig. 1a–c). At 4 °C, the hybridization buffer showed no signal for miR–124 (Supplementary Fig. 1d), which would suggest that RNA–protein crosslinks remained intact at lower temperatures.
To prevent the loss of miRNAs, we added an additional miRNA fixation step that uses the miRNA 5’ phosphate end1, which does not react with fomaldehyde16. The water–soluble 1–ethyl–3–(3–dimethylaminopropyl) carbodiimide, EDC, reacts with phosphate and condenses it with amino groups in the protein matrix to form stable linkages18,19. When formaldehyde–fixed specimens were additionally treated with EDC, miR–124 no longer escaped from sections, and only trace amounts of miR–124 were detected in the ISH buffer (Supplementary Fig. 1e). EDC treatment alone, without prior formaldehyde fixation failed to retain miRNAs in tissues (data not shown).
The second critical factor affecting ISH is probe hybridization. Probes that bind to miRNAs with high thermodynamic stability such as DNA probes containing locked nucleic acid (LNA) residues are well suited for miRNA ISH4,20. The melting of LNA–miRNA duplexes can be observed experimentally by UV spectrophotometry20. Thermodynamic analysis of LNA–modified deoxynucleotide duplexes led to models that predict their melting temperatures (TM) for any LNA–RNA pair21. [AU please add a reference here, reference added.] Unfortunately the TMs for LNA–RNA pairs formed during ISH cannot be predicted accurately, as the programs do not consider the presence of formamide denaturant which influences the melting of the LNA–RNA pair. which modifies the RNA? [AU please state how the formamide influences the Tm]. To derive optimal hybridization conditions, the melting profiles of 130 miRNA–LNA probe pairs were measured in the presence of formamide containing ISH solution, in the place of sodium cacodylate buffer[please state how your measurements differ from those shown in ref 20, yes in formamide] (Supplementary Table 1). Since measuring melting profiles individual probe sets are costly, requiring 37-fold more probe than the ISH experiment, we provide a correction factor for the prediction program using our experimentally determined data set, and by manipulating the salt parameter of LNA–RNA TM prediction programs to 50 mM NaCl instead of the actual 750 mM salt contained in the ISH buffer[what was the rational for changing the ‘salt parameter’, also please define what that is, concentration? nature of the salt?], we obtained estimates for the hybridization temperature in formamide–containing ISH buffer (Supplementary Table 1). We generated our LNA probe set based on miRNAs expressed with a clone frequency of at least 0.04% based on small RNA library sequencing from 5 regions of the mouse brain (Supplementary Table 2).
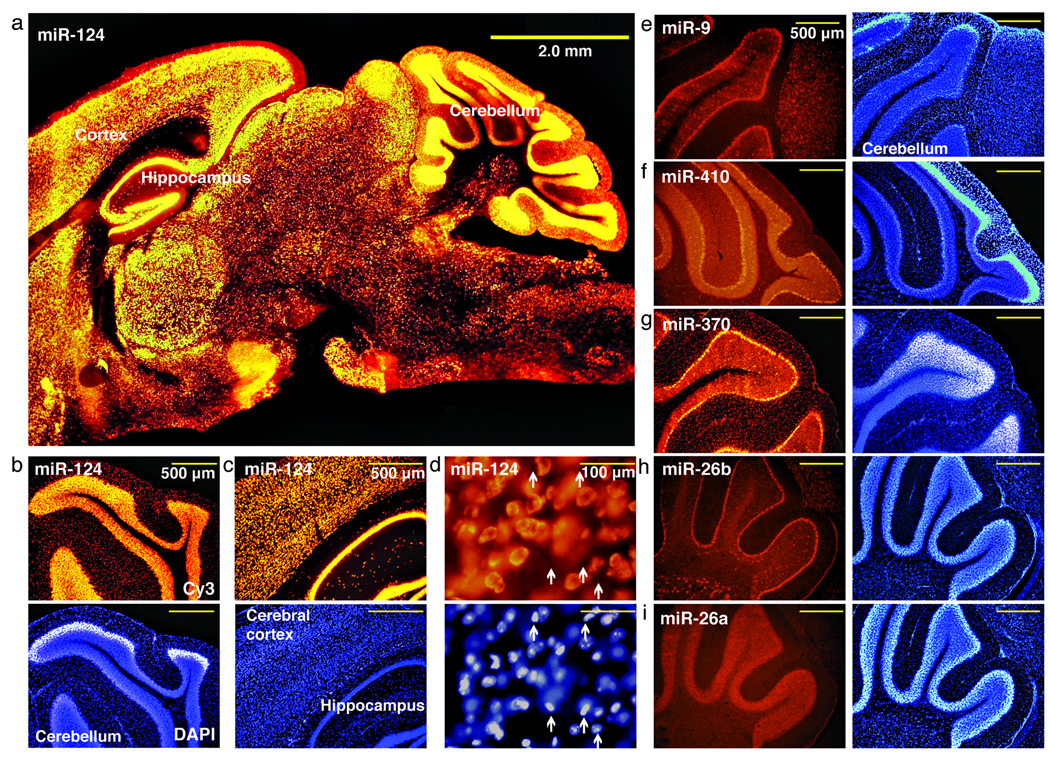
To determine the range of the ISH sensitivity, we conducted ISH for miRNAs identified at varying frequencies from small RNA cloning libraries in mouse brain; miR–9, 9.3%; miR–124, 8.8%; miR–26a, 3.3%; miR–26b, 0.25%; miR–370, 0.14%; miR–130a, 0.12%; and miR–410, 0.07% (Supplementary Table 2). Tissue sections were fixed with formaldehyde + EDC and then ISH performed (Supplementary Methods). We determined the effect of formaldehyde + EDC versus formaldehyde fixation alone by conducting ISH for the most abundant miR–124 and the 73–fold less abundant miR–130a. ISH images show thatthe signals for miR–124 were improved moderately in the formaldehyde + EDC-fixed sample, whereas miR–130a signals were only detectable in formaldehyde + EDC–fixed samples (Supplementary Figure 2). Since the abundant miR–124 presumably retained enough miRNA to nearly saturate the signal, the improvements remained modest, but for lowly expressed miRNAs, loss by diffusion hindered its detection in formaldehyde alone samples. miR–124 is mostly detected in neuronal cells present in different regions of the brain (Fig. 1a–d and Supplementary Fig. 3) and predominately localized in the cytoplasm, not the nucleus. Another highly expressed miRNA in brain, miR–9, also preferentially localizes in neurons, and is particularly enriched in the Purkinje cell layer (Fig. 1e). We also observed robust ISH signal for the less abundant miRNAs tested (Fig. 1f–i), with unique miRNA distributions. Interestingly, miR–26a and miR–26b, which differ by 2 nucleotides (C11U, C21U) and originate from different clusters, showed differential expression patterns in the mouse cerebellum (Fig. 1h,i). The absolute signal intensities for the miRNAs tested were not directly correlated with the abundance of miRNAs and were likely attributed to differences in kinetics of probe hybridization. Nevertheless, for a given probe, the signal intensities accurately reflected miRNA differential expression.
Fig. 1.
Visualization of miRNAs expressed at different levels in the mouse brain. (a) Low magnification images of nervous system specific miR–124 (orange) shows broad expression [is it possible to say that expression is restricted to neurons from these low mag pictures, Previous studies show miR-124 is restricted to the nervous system (ref #3) and based on dual labeling of miR-124 by ISH and labeling neurons with a specific antibody via immunostain, we show miR-124 is mostly present in neurons (Supplementary Fig. 3)]. (b–c) High magnification images of miR–124 demonstrate ubiquitous expression in the neurons of the cerebellum (top, orange), (b) cerebral cortex and hippocampus (c). (d) Higher magnification images show that miR–124 signals are not present in all cells, marked with arrows. Bottom panels show with 4’, 6–diamino–2–phenylindole dihydrochloride (DAPI) stain. (e) Fluorescence images of mouse brain sections probed for highly expressed miR–9 (, red) localized in Purkinje cells of the cerebellum. miR-410 (f) and miR–370 (g), have intermediate expression. miRNAs differing by 3 nucleotides, miR–26b (h) and miR–26a (i) are differentially expressed. Panels on the right show DAPI stain (e-i). Scale bars, 500 µm (f–i).
To examine probe hybridization specificity, we selected miR–124 and introduced mismatches in the probe central regions. The ISH was performed at constant temperature 20 °C below the TM of the fully complementary miR–124 probe. The probes with the greatest difference in TM showed the least signal and central mismatches abolished detection (Supplementary Table 3 and Supplementary Fig. 4). Interestingly, signal strength observed for mismatched probes with minor reduction in TM dropped disproportionately, again indicative of altered kinetics of probe hybridization.
Instead of a mismatch approach to control for probe specificity, conducting ISH using two probes directed against the mature miRNA and the opposing fragment in the miRNA duplex, known as the miRNA* sequence3 and/or polycistronic transcripts comprising several miRNAs that expressed in a cluster[AU correct or please define what clustered members are] may be useful to help rule out signals derived from cross–hybridization. We probed for cistronically expressed miRNAs that are distinct in sequence for colocalization, including members in the mir–99a/mir–125b–1/let–7c–1 cluster. The ISH for let–7c and miR–99a reveal mostly identical patterns that showed superimposable band–like patterns (Supplementary Fig. 4g). Other let–7 family members also show similar expression, however, the signal observed in the band–like region of the cortex likely originated from let–7c due to colocalization of miR–99a. We also tested the miRNA* sequences as a specificity control for miRNA ISH signals. We examined miR–140 and its complementary miR–140* sequence, expressed at relative clone frequency of 5 to 1, respectively. The ISH with both probes revealed superimposible expression in the cortex (Supplementary Fig. 4h).
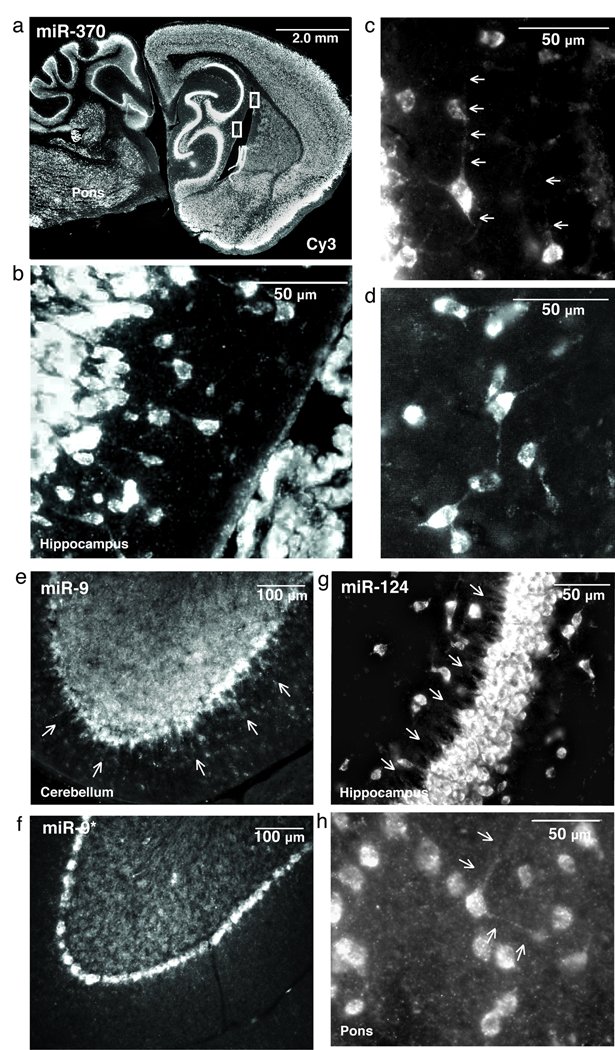
miRNAs are known to localize to subcellular compartments for local regulation of mRNA, possibly to the dendrites of neurons2,22. We tested several miRNAs for expression in dendrites including miR–370 (Fig. 2a–d), miR–9 (Fig. 2e), and miR–124 (Fig. 2g,h), which extended up to 50 µm from the cell body. Interestingly, compared to miR-9, the 20–fold less expressed miR–9*, did not reveal localization in dendrites (Fig. 2f). Together, these data demonstrate this method detects miRNAs in subcellular compartments.
Fig. 2.
Formaldehyde + EDC–fixed sections show miRNA localized in the dendrites of neurons. (a) Low magnification fluorescent images demonstrate a robust and broad distribution of miR–370 expression (white) with intense staining in the hippocampus (inset). (b,c) High magnification images show a single neuron in the CA1 region of the hippocampus for which miR–370 is localized both in the cell body and the dendrites (arrows) (c,d), represent inset (a). miR–370 localization extends 50 µm from the cell body (b), and 30 µM in other neurons (c,d). (e) The mature miRNA miR–9 expressed in dendrites of Purkinje cells in the cerebellum, yet the miR–9* (f) was absent in dendrites. (g) Pyramidal cells in the hippocampus (arrows) show staining of miR–124 in hippocampus dendrites and in neurons located in the pons.
Finally, to broaden this technique for sections not amenable to fluorescent imaging, we compared the pigment detection system, Nitro blue tetrazolium chloride with 5–Bromo–4–chloro–3–indolyl phosphate, toluidine salt (NBT/BCIP), [please define] for miR–370 in the brain to fluorescent detection and observed similar staining (Supplementary Fig. 5). We also tested the pigment detection for miR–122 and miR–126–3p in the mouse liver, miR–1 in the mouse heart, and observed cell–type–specific staining (Supplementary Fig. 6). Furthermore, we combined the detection of proteins by immunohistochemistry with miRNA ISH (Supplementary Fig. 3), although immunostaining that preceded EDC fixation reduced the signal strength for the miRNA ISH.
In summary, eliminating miRNA diffusion by introducing an irreversible crosslink between the 5’ phosphate of the miRNA and protein side chains significantly improves miRNA retention in tissues. The advantage of EDC–based phosphoramidate–linked miRNA is that the sample can be exposed to higher temperatures for a longer time, resulting in a more complete reversion of the formaldehyde induced nucleobase modification of the miRNAs and less interference with probe hybridization. Furthermore, the melting temperature determined for many probe-miRNA duplexes under ISH conditions more accurately defined ISH hybridization temperatures. At present, we showed that different tissues and detection systems are compatible with our approach. This method paves the way for reliable disease association studies and the potential use of miRNA ISH expression analysis as a diagnostic tool for biopsy material.
T.T. is a co-founder and scientific advisor of Alnylam Pharmaceuticals and a scientific advisor of Regulus Therapeutics.
Supplementary Material
Acknowledgements
We are grateful to the Memorial Sloan Kettering Sequencing Core for 454 sequencing. We thank S. Juranek, N. Renwick, J. McManus, H. Yamahachi, and T. Farazi for comments on the manuscript. We would like to thank L. Fleming, for the technical assistance and C. D. Gilbert for his guidance. T.T. was supported by an Irma T. Hirschl Career Scientist Award, J.T.G.P. supported by Kirschstein-NRSA fellowship and the project was partly funded by NIH grants GM073047, EY18082-01A2 andMH080442.
In the interest of space and format that is required for figure 1 I suggest moving fig. 1 to Supplementary Materials as new SI figure 1 (please change references in the text) Changes were made, figure 1 has been move to Supplementary material.
As noted in the Checklist I forwarded to you, we require 6 bandwidth around a cropped gel which would make fig. 1 very large – it would take space that is – in my view – better devoted to fig. 2 and fig. 3. The whole text and figures have to fit on 3 print pages We made the appropriate changes.
Fig. 3.
Footnotes
Author Contributions
J.T.G.P., S.H.R., and C.S.L. designed LNA–modified probes, conducted the mouse ISH experiments, and recorded ISH images with microscopy. C.S.L. recorded the melting profiles for the miRNA/LNA–probe duplexes. C.S.L., C.L., D.H., and A.M. conducted LNA probe synthesis. S.H.R. sectioned all tissues. J.T.G.P dissected the mouse brain sections for ISH and M.H. and A.M. prepared RNA for large–scale small RNA sequencing. J.L. advised on chemical conditions used for the miRNA cross–linking. M.Z. and P.B. designed and carried out the small RNA annotation. J.T.G.P. and T.T. wrote the manuscript.
References
- 1.Farazi TA, et al. Development. 2008;135(7):1201. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 2.Kosik KS. Nat. Rev. Neurosci. 2006;7(12):911. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 3.Landgraf P, et al. Cell. 2007;129(7):1401. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kloosterman WP, et al. Nat. Methods. 2006;3(1):27. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 5.Sokol NS, et al. Genes Dev. 2005;19(19):2343. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson PT, et al. RNA. 2006;12(2):187. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silahtaroglu AN, et al. Nat. Protoc. 2007;2(10):2520. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 8.Thompson RC, et al. Methods. 2007;43(2):153. doi: 10.1016/j.ymeth.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuovo GJ. Methods. 2008;44(1):39. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Bak M, et al. RNA. 2008;14(3):432. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sempere LF, et al. Cancer Res. 2007;67(24):11612. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 12.Wang WX, et al. J. Neurosci. 2008;28(5):1213. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer A, et al. J. Exp. Med. 2007;204(7):1553. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan DG, et al. Mol. Vis. 2006;12:1175. [PubMed] [Google Scholar]
- 15.Mansfield JH, et al. Nat. Genet. 2004;36(10):1079. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 16.Feldman MY. Prog. Nucleic Acids Res. Mol. Biol. 1973;13(1) doi: 10.1016/s0079-6603(08)60099-9. [DOI] [PubMed] [Google Scholar]
- 17.Masuda N, et al. Nucleic Acids Res. 1999;27(22):4436. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tymianski M, et al. Cell calcium. 1997;21(3):175. doi: 10.1016/s0143-4160(97)90042-7. [DOI] [PubMed] [Google Scholar]
- 19.Pall GS, et al. Nucleic Acids Res. 2007;35(8):e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur H, et al. Biochemistry. 2008;47(4):1218. doi: 10.1021/bi700996z. [DOI] [PubMed] [Google Scholar]
- 21.Allawi HT, et al. Biochemistry. 1997;36(34):10581. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 22.Kye MJ, et al. RNA. 2007;13(8):1224. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.