Abstract
The nervous and immune systems consist of complex networks that have been known to be closely interrelated. However, given the complexity of the nervous and immune systems of mammals, including humans, the precise mechanisms by which the two systems influence each other remain understudied. To cut through this complexity, we used the nematode Caenorhabditis elegans as a simple system to study the relationship between the immune and nervous systems using sophisticated genetic manipulations. We found that C. elegans mutants in G-protein coupled receptors (GPCRs) expressed in the nervous system exhibit aberrant responses to pathogen infection. The use of different pathogens, different modes of infection, and genome-wide microarrays highlighted the importance of the GPCR NPR-1 in avoidance to certain pathogens and in the regulation of innate immunity. The regulation of innate immunity was found to take place at least in part through a mitogen-activated protein kinase signaling pathway similar to the mammalian p38 MAPK pathway. Here, the results that support the different roles of the NPR-1 neural circuit in the regulation of C. elegans responses to pathogen infection are discussed.
Keywords: neurons, innate immunity, infection, inflammation, G-protein coupled receptors, GPCR, npr-1
Introduction
A wealth of data indicates that the nervous system receives inputs from infected local sites and integrates them to coordinate appropriate immune responses.1–3 It has also been postulated that the immune system may function as a “sixth sense”, recognizing microorganisms and microbial toxins that cannot be seen, heard, tasted, touched, or smelled.2 However, given the complexity of the nervous and immune systems of mammals, including humans, the precise mechanisms by which the two systems influence each other remain understudied. While the mammalian nervous system contains hundreds of billions of neurons, the simple nervous system of C. elegans only contains 302 neurons and 56 glial cells, which represent 37% of all somatic cells in an adult hermaphrodite. The simplicity of the C. elegans nervous system, together with the great deal of information about the morphology and synaptic connectivity of every neuron in the adult organism, provide an excellent system to study neural circuits that regulate behaviors and different organismal processes, including innate immunity.
C. elegans is a 1 millimeter long nematode that in the laboratory is cultivated on agar plates containing Escherichia coli. In nature, C. elegans lives in soils and composts rich in microorganisms, and, while it lacks adaptive immunity, it has evolved mechanisms to respond to different microorganisms in different ways. For example, C. elegans is capable of sensing some bacterial compounds and avoid certain pathogens.4, 5 The nematode can also respond to pathogen exposure with an inducible innate immune system that comprises conserved effectors such as antimicrobial peptides, lectins, lysozymes.6–10 A recent study indicates that the C. elegans nervous system may not only regulate avoidance to certain pathogens but also innate immune responses that are independent of the behavioral avoidance to pathogens.11
Mutants in GPCR-encoding genes exhibit aberrant responses to pathogen infection
GPCRs constitute the largest family of transmembrane signaling proteins that are present in the cell surface of all multicellular organisms where they regulate host physiological processes including homeostasis, metabolism, neurotransmission, cardiovascular function, and immune function. Thus, GPCRs represent the most widely targeted pharmacological protein class, accounting for the targets of about one-third of all approved drugs.12, 13 To study the role of GPCRs in the regulation of innate immune response, the susceptibility of forty C. elegans strains carrying mutations in GPCRs to the human opportunistic pathogen Pseudomonas aeruginosa strain PA14 was studied. Depending on the statistical analysis used, three or five mutants exhibited enhanced resistance to P. aeruginosa while only one or two mutants exhibited enhanced susceptibility to P. aeruginosa.11
The strain that consistently exhibited enhanced susceptibility to P. aeruginosa-mediated killing was npr-1(ad609), which carries a loss-of-function allele of a gene that encodes NPR-1, a G protein-coupled receptor related to mammalian neuropeptide Y receptors.14 In nature, NPR-1 is found in two allelic forms that differ in a single amino acid at position 215, NPR-1(215V) and NPR-1(215F).14 The NPR1(215V) allele, which is found in the standard laboratory strain N2, has high activity whereas the NPR-1(215F) allele has low activity.15, 16 The lifespan of npr-1(ad609) animals grown on agar plates covered with killed E. coli or P. aeruginosa was not different than that of N2 animals,11 indicating that npr-1(ad609) animals are deficient in host-defense responses to live P. aeruginosa.
Hyperoxia avoidance overrides avoidance to P. aeruginosa in animals with low NPR-1 activity
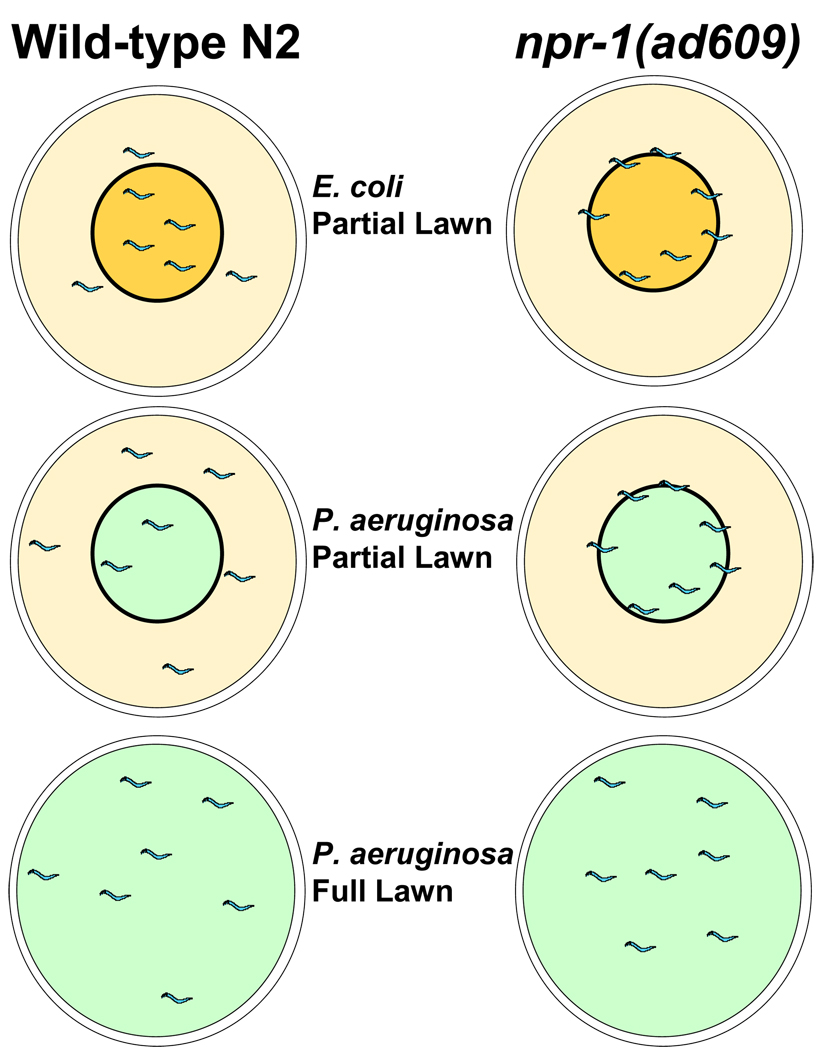
Genetic studies indicate that npr-1 is part of a neural circuit that integrates behavioral responses to environmental oxygen, bacteria, and other animals.15, 17–21 While N2 animals avoid oxygen levels above 10% when E. coli is absent and fail to avoid high oxygen in the presence of E. coli, npr-1(ad609) animals have strong hyperoxia avoidance in the absence or presence of E. coli.18 As a result, npr-1(ad609) animals show a preference for regions in the bacterial lawn rich in metabolically active bacteria that lower oxygen levels15 (Figure 1). When npr-1(ad609) animals are grown at high densities, they also tend to aggregate into feeding groups to decrease local oxygen concentrations.18
Figure 1. Hyperoxia avoidance overrides avoidance to P. aeruginosa.
P. aeruginosa strain PA14 is known to negatively affect the attraction response of C. elegans to E. coli OP50, which is the food source of nematodes in the laboratory. As a consequence, animals grown on agar plates containing lawns of P. aeruginosa PA14 spend more time outside the bacteria lawn than animals grown on plates containing lawns of E. coli. Moreover, npr-1(ad609) animals have a strong preference for low oxygen concentrations such as those that exist at the edge of the bacterial lawn, and spend more time on the bacteria lawn than wild-type N2 animals. To expose npr-1(ad609) and N2 animals to comparable amounts of P. aeruginosa, full lawns of P. aeruginosa are used.
Since C. elegans naturally exhibits an avoidance behavior towards P. aeruginosa, 5, 22, 23 it is possible that the reduced lifespan of npr-1(ad609) animals infected by P. aeruginosa is due to a higher exposure to bacteria. As illustrated in Figure 1, while N2 animals can freely enter and exit the P. aeruginosa lawn, npr-1(ad609) animals spend most of the time on the border of the lawn and in full contact with bacteria.11 C. elegans spends less time on the P. aeruginosa lawn than on the E. coli lawn. To allow npr-1(ad609) animals to move more freely on the plates, the infections were performed at 8% oxygen, a favorable oxygen environment that suppresses most behavioral phenotypes of npr-1 mutants. Under 8% oxygen, npr-1(ad609) animals do not exhibit a preference for regions in the agar plates with lower oxygen levels such as the bacterial border, and are capable of leaving the P. aeruginosa lawn. At this low oxygen level, npr-1(ad609) animals are more resistant to P. aeruginosa-mediated killing than at 21% oxygen, indicating that the preference for low oxygen has a deleterious effect on C. elegans survival. Additional experiments using agar plates that were either partially covered in P. aeruginosa (Figure 1, partial lawn) or completely covered in P. aeruginosa (Figure 1, full lawn) highlighted the importance of avoidance to P. aeruginosa.11 While N2 animals grown on full lawns of P. aeruginosa died at a higher rate than N2 animals grown on partial lawns of P. aeruginosa, npr-1(ad609) animals were equally susceptible to P. aeruginosa when grown on full lawns or partial lawns.11 Since C. elegans does not avoid P. aeruginosa strains PAK1 or PA01,23 it would be interesting to study the susceptibility of npr-1(ad609) animals to those strains.
Avoidance to P. aeruginosa is not the only determinant of pathogen susceptibility in npr-1 animals
Even under 8% oxygen or on full lawns of P. aeruginosa, two conditions that control for pathogen avoidance, npr-1(ad609) animals were found to be significantly more susceptible to P. aeruginosa-mediated killing than N2 animals.11 After our study was published, another group of investigators published data that confirm the importance of avoidance to P. aeruginosa and show small residual differences between N2 and npr-1(ad609) animals under conditions that control for avoidance to P. aeruginosa.24 A caveat of these experiments is the use of the drug 5-fluorodeoxyuridine (FUdR) in the assays. FUdR is an inhibitor of DNA synthesis that is used to prevent progeny formation to avoid the transfer of the nematodes to fresh plates every day.25 FUdR could be affecting the virulence of P. aeruginosa as well as different aspects of C. elegans physiology that may contribute to susceptibility to P. aeruginosa. For example, P. aeruginosa-infected animals become immobile and die, and in a high number of cases, animals become laden with eggs and embryos hatched internally.26 Since FUdR prevents progeny formation, and embryo hatching from gravid adults increases their susceptibility to P. aeruginosa,26 the use of the drug eliminates a component of P. aeruginosa-mediated killing.
The enhanced susceptibility to P. aeruginosa of npr-1(ad609) animals compared to that of N2 animals, when infections were performed at 8% oxygen or on full lawns of P. aeruginosa,11 indicate that pathogen avoidance cannot account for all of the difference between the two C. elegans strains. To provide further evidence that npr-1(ad609) animals succumb to pathogen infection faster than N2 animals by mechanisms that are independent of pathogen avoidance, we used the pathogen Salmonella enterica. As in mammalian hosts, a small inoculum of S. enterica is capable of establishing a persistent infection in C. elegans.27 In contrast to P. aeruginosa-mediated killing, S. enterica-mediated killing does not require constant exposure to bacteria and cannot be prevented by transferring the infected animals to plates containing E. coli. Indeed, it is known that 5 hours of exposure to S. enterica is sufficient to cause a similar rate of killing to the one obtained when C. elegans is in contact with S. enterica during the entire assay.27 In addition, it has been demonstrated that S. enterica does not elicit an avoidance behavior.28 Thus, the observed increased susceptibility of npr-1(ad609) to S. enterica,11 is consistent with a role of NPR-1 in the regulation of immune responses that are independent of pathogen avoidance.
The NPR-1 neural circuit regulates innate immunity
Several transcriptional profiling analyses have demonstrated that C. elegans responds to pathogen attack by differentially regulating the expression of conserved immune effectors including antimicrobial peptides, lysozymes, and lectins. Some of the genes that are markers of innate immunity are under the regulation of the TGF-β-related gene dbl-1,6, 29 the FOXO transcription factor DAF-1630, the intestinal GATA transcription factor ELT-27, 8, and MAPK pathways9, 31. To identify possible genes and pathways involved in innate immunity that may be regulated by NPR-1, we performed a full-genome microarray analysis.11 The data show that most of the genes that are misregulated in npr-1(ad609) animals correspond to markers of innate immune responses that are regulated by DBL-1, DAF-16, and the conserved PMK-1/P38 MAPK signaling pathway11. A significant enrichment in intestinally-expressed genes was also found11. Differential expression of genes that are markers of innate immunity may be a consequence of different exposure to pathogens. However, npr-1(ad609) and N2 animals were infected on plates completely covered by P. aeruginosa, ruling out this possibility. Another independent comparison of gene expression between N2 animals and animals carrying a polymorphism in npr-1 support the idea that NPR-1 regulates the expression of immune genes in a manner that is independent of pathogen avoidance32.
The difference in pathogen susceptibility between the commonly used N2 strain and the wild isolate CB4856 has been attributed to a polymorphism in npr-1.24 A genome-wide microarray study involving CB4856 supports the idea that NPR-1 regulates the expression of immune genes in a manner that is independent of pathogen avoidance.32 The microarray analysis shows that the pattern of expression of immune genes over time differs between N2 and CB4856. In addition, some of the immune genes are solely expressed in either N2 or in CB4856.32 Since N2 only avoids oxygen levels above 10% when E. coli is absent and fails to avoid high oxygen in the presence of E. coli,18 it is unlikely that a different exposure to E. coli could account for differences in the expression of innate immunity genes observed between N2 and CB4856. In addition, since genes that are markers of C. elegans innate immunity were identified essentially by comparing the expression profile of animals infected with pathogens to that of animals exposed to E. coli, different exposure to E. coli should not result in misregulation of immune genes.
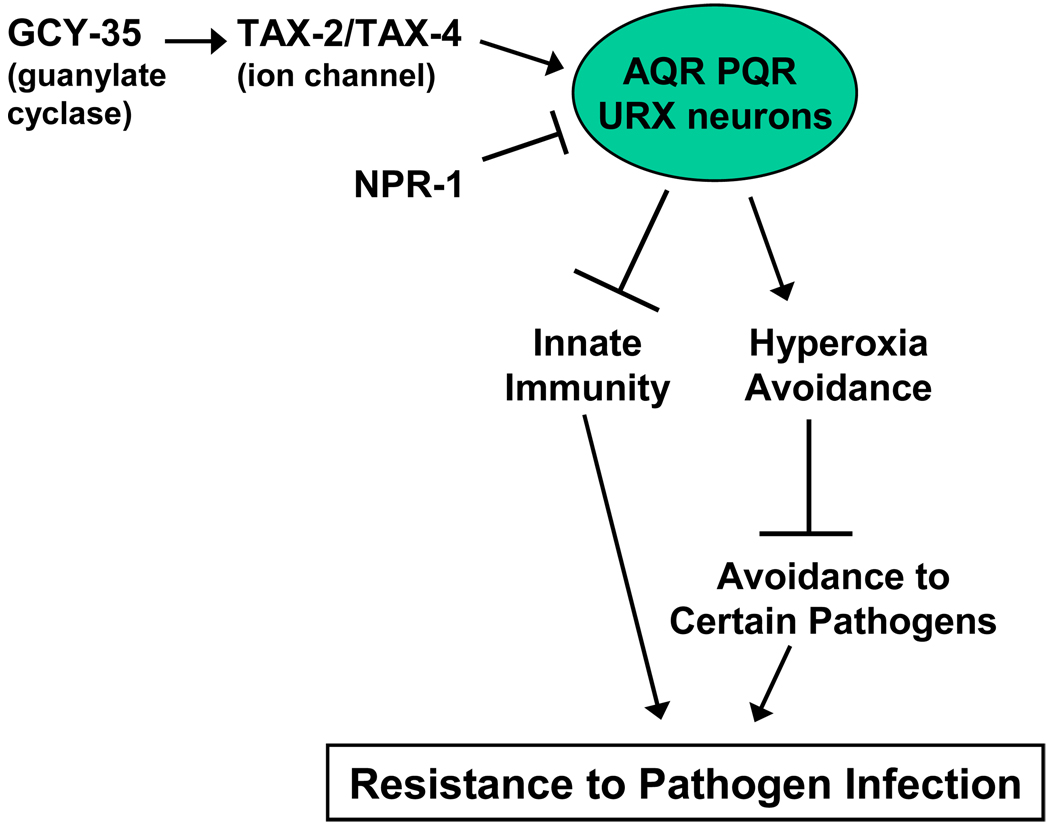
Genetic studies have shown that the NPR-1 inhibition of neural activation requires the soluble guanylyl cyclase GCY-35 and TAX-2 and TAX-4, which are two subunits of a cGMP-gated-ion-channel.14, 19, 33 Consistent with the idea that NPR-1 regulates innate immunity, it was found that a gcy-35 mutation in npr-1(ad609) animals rescues the altered expression of 10 genes that are markers of C. elegans immune response. In addition, the observed low levels of active PMK-1 in npr-1(ad609) animals compared to N2 animals grown on E. coli further indicate that NPR-1 regulates innate immune responses that are independent of pathogen avoidance (Figure 2).
Figure 2. Model of the role of the NPR-1 neural circuit in resistance to pathogen infection in C. elegans.
NPR-1 inhibits the activity of at least AQR, PQR, URX neurons that suppress pathogen resistance by at least two mechanisms. While GCY-35, TAX-2, and TAX-4 are required for the activation of AQR, PQR and URX neurons, NPR-1 inhibits their activity. When the inhibitory function of NPR-1 is affected, AQR, PQR and URX neurons promote hyperoxia avoidance which suppresses pathogen avoidance and results in a reduced resistance to infection. Additionally, when the NPR-1-mediated inhibition of AQR, PQR and URX neurons is affected, innate immunity is suppressed.
Conclusions
Several lines of evidence indicate that animals deficient in the activity of NPR-1 are more susceptible to the human pathogen P. aeruginosa strain PA14 due to two factors: decreased pathogen avoidance and decreased innate immune responses. The use of different oxygen concentrations, different bacteria, and different modes of infection provide additional evidence that the NPR-1 neural circuit regulates, at least in part, innate immune responses in C. elegans. C. elegans neurons are known to express numerous secreted peptides of the TGF beta family, the insulin family, and neuropeptide families.34–39 This myriad of secreted factors has the potential to act at a distance to modulate various physiological processes by regulating the function of neural and non-neural cells throughout the animal. Taken together, the studies discussed here show that specific genes and neurons in the nervous system of C. elegans control immune responses, indicating that cell non-autonomous signals from different neurons may act on non-neural tissues to regulate innate immunity. It is plausible that the role of NPR-1 in innate immunity, together with the postulated trade-off between dispersal and competitive ability,40 conspire to maintain npr-1 polymorphisms.
Acknowledgements
A.A. is funded by The Whitehead Scholars Program and NIH grant GM070977.
Footnotes
NO CONFLICT-OF-INTEREST AND FINANCIAL DISCLOSURE
References
- 1.Sternberg EM. Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nat Rev Immunol. 2006;6:318–328. doi: 10.1038/nri1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson J. The inflammatory reflex--introduction. J Intern Med. 2005;257:122–125. doi: 10.1111/j.1365-2796.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 3.Tracey KJ. The inflammatory reflex. Nature. 2002;420:853–859. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- 4.Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 6.Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 7.Kerry S, Tekippe M, Gaddis NC, Aballay A. GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS ONE. 2006;1:e77. doi: 10.1371/journal.pone.0000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322:460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 13.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 14.de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 15.Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature. 2002;419:925–929. doi: 10.1038/nature01170. [DOI] [PubMed] [Google Scholar]
- 16.Rogers C, Reale V, Kim K, Chatwin H, Li C, Evans P, et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci. 2003;6:1178–1185. doi: 10.1038/nn1140. [DOI] [PubMed] [Google Scholar]
- 17.de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature. 2002;419:899–903. doi: 10.1038/nature01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 19.Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol. 2005;15:905–917. doi: 10.1016/j.cub.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol. 2006;16:649–659. doi: 10.1016/j.cub.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 21.Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 2006;4:e274. doi: 10.1371/journal.pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beale E, Li G, Tan MW, Rumbaugh KP. Caenorhabditis elegans senses bacterial autoinducers. Appl Environ Microbiol. 2006;72:5135–5137. doi: 10.1128/AEM.00611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laws TR, Atkins HS, Atkins TP, Titball RW. The pathogen Pseudomonas aeruginosa negatively affects the attraction response of the nematode Caenorhabditis elegans to bacteria. Microb Pathog. 2006;40:293–297. doi: 10.1016/j.micpath.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandhi S, Santelli J, Mitchell DH, Stiles JW, Sanadi DR. A simple method for maintaining large, aging populations of Caenorhabditis elegans. Mech Ageing Dev. 1980;12:137–150. doi: 10.1016/0047-6374(80)90090-1. [DOI] [PubMed] [Google Scholar]
- 26.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 28.Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–109. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mochii M, Yoshida S, Morita K, Kohara Y, Ueno N. Identification of transforming growth factor-beta- regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc Natl Acad Sci U S A. 1999;96:15020–15025. doi: 10.1073/pnas.96.26.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 31.Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, Aroian RV. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A. 2004;101:10995–11000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capra EJ, Skrovanek SM, Kruglyak L. Comparative developmental expression profiling of two C. elegans isolates. PLoS ONE. 2008;3:e4055. doi: 10.1371/journal.pone.0004055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung BH, Arellano-Carbajal F, Rybicki I, de Bono M. Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr Biol. 2004;14:1105–1111. doi: 10.1016/j.cub.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann N Y Acad Sci. 1999;897:239–252. doi: 10.1111/j.1749-6632.1999.tb07895.x. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci U S A. 2001;98:14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 39.Schackwitz WS, Inoue T, Thomas JH. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 40.Gloria-Soria A, Azevedo RB. npr-1 Regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 2008;18:1694–1699. doi: 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]