Abstract
p53, p63 and p73 are members of the p53 protein family involved in regulation of cell cycle, apoptosis, differentiation and other critical cellular processes. Here we investigated the contribution of the entire p53 family in chemotherapeutic drug response in gastrointestinal tumors. Real-time PCR and immunohistochemistry revealed complexity and variability of expression profiles of the p53 protein family. Using colon and esophageal cancer cells, we found that the integral transcription activity of the entire p53 family, as measured by the reporter analysis, associated with response to drug treatment in studied cells. We also found that p53 and p73, as well as p63 and p73, bind simultaneously to the promoters of p53 target genes. Taken together, our results support the view that the p53 protein family functions as an interacting network of proteins and show that cellular responses to chemotherapeutic drug treatment are determined by the total activity of the entire p53 family, rather than p53 alone.
Keywords: p73, p63, p53, colorectal tumor, esophageal tumor
Introduction
Colorectal carcinomas (CRC), carcinomas of the colon and rectum, are the most common gastrointestinal cancer in the world. Chemotherapy, together with surgery and radiation therapy, is a major modality of CRC treatment (1). In the late 1980s the introduction of 5-fluorouracil (5-FU)-based adjuvant chemotherapy for resectable colon cancer reduced mortality by as much as 30% (2). 5-FU, as well as other chemotherapeutic drugs commonly used in CRC treatment at the present time, induce DNA damage and strongly activate p53.
Extensive studies of p53 have demonstrated its fundamental role in multiple cellular stress responses including DNA damage in the colon. It is generally accepted that functional loss of p53 is a late event in colon tumor progression that reveals itself as a late adenoma-carcinoma transition. p53 mutations have been described in 40-50% of colorectal carcinomas (3). However, despite numerous studies, the prognostic role of p53 mutations in chemotherapeutic drug response remains to be controversial (4).
p53 is the founding member of a protein family, which also includes p63 and p73. These proteins have significant structural similarities to p53 and share similar activities, such as regulation of apoptosis, cell cycle arrest, and cellular senescence (5). Extensive homology exists in the transactivation, DNA-binding, and oligomerization domains of p73/p63 and p53. The highest similarity is found in the DNA-binding domain, in which p63 and p73 share approximately 60% amino acid identity with p53 (6). Evolutionally, this domain is the most conserved, suggesting that the regulation of transcription plays a pivotal role in an array of functions attributed to the p53 family. p63 and p73 are expressed as a set of isoforms. Full-length products of the TP63 and TP73 genes contain the transactivation domain (TA domain) and were termed TA isoforms. Due to the alternative intragenic promoter, N-terminal splicing and translation from the internal ribosome entry site, different N-terminally truncated isoforms are produced by the same genes (7). A general name for these proteins is ΔN (or ΔTA) because they lack the TA domain at the N-terminus. TA and ΔN isoforms also vary by the structure of their C-termini, as a result of extensive splicing (7). It is generally considered that TA isoforms exhibit p53-like properties activating transcription of most of the p53-target genes involved in apoptosis and cell cycle regulation, while ΔN isoforms act as potent dominant-negative inhibitors of TA isoforms and p53.
In the present study, we investigated, for the first time, how the entire interactive network of the p53 family contributes to cellular response to chemotherapeutic drug treatment in gastrointestinal tumors.
Materials and Methods
Cancer tissues, tissue array and immunohistochemistry
After the Institutional Review Board approval, 104 colorectal adenocarcinoma cases and 14 non-neoplastic colon epithelia samples resected at Vanderbilt University Medical Center were histologically verified and representative regions were selected for inclusion in tissue microarray. Characterization of tumor specimen is summarized in Supplemental Tables 1 and 2.
Immunohistochemical staining was performed using p73 IHC Antibody from Bethyl Laboratories, ΔNp73 from Imgenex, p63(4A4) from Santa Cruz Biotechnology, and p53(DO-1) from Calbiochem (8). Specificity of staining was verified by omitting a primary antibody step in the protocol. Immunohistochemical results were evaluated for intensity and staining frequency in nuclear and cytoplasmic compartments. The intensity of staining was graded as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The frequency was graded according to the percentage of positive cells. Total nuclear and cytoplasmic scores were calculated by multiplying the intensity score by the percentage of positive cells.
Cell culture, transfections, retroviral infections and siRNA
LIM1215, SW480, HCT8, isogenic HCT116 p53+/+, and HCT116 p53−/− cells were obtained from Dr. R Coffey (Vanderbilt University, Nashville, TN). TE1, TE-7 and TE11 human esophageal carcinoma cell lines were kindly provided by Dr. J Katz and Dr. A Rustgi (University of Pennsylvania, Philadelphia, PA). Isogenic RKO cell lines were obtained from Dr. Vogelstein of Johns Hopkins University. Cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen), supplemented with 10% fetal bovine serum.
For inhibition of p73, either lentiviral transduction with shRNA or transfection with siRNA were used. Both approaches targeted the same p73 sequence (5′-GCAATAATCTCTCGCAGTA-3′) found in all p73 isoforms. shRNA was delivered using the pSicoR lentivirus system (9), which was kindly provided by Dr. Pietenpol of Vanderbilt University (10). p73 and p63 specific siRNA were purchased from IDT (Coralville, IA) and Dharmacon, respectively. As controls for RNAi experiments, negative control siRNA (Ambion) and GFP shRNA in pSicoR were used.
Cells were transfected with Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Roche, Indianapolis, IN) reagents following the manufacturers’ protocols.
Vectors, antibodies, and real-time PCR
Plasmids expressing human p53, TAp73α, TAp73β, ΔNp73α, ΔNp73β, ΔNp63α, and luciferase reporter constructs PG13 and MG15 have been described previously (11-13). Human TAp63γ expression vector was kindly provided by Dr. Pietenpol (Vanderbilt University). Antibodies to the following proteins were used: Fas (C-20), p63 (4A4) and non-specific mouse IgG from Santa Cruz Biotechnology; TAp73 from Bethyl Laboratories; p53 (DO-1), p73 (Ab-2) and p21 (Ab-1) from Calbiochem; ΔNp73 and NOXA from Imgenex; PUMA (ab9643) from Abcam Inc; non-specific rabbit IgG from Jackson ImmunoResearch; actin and BAX from Cell Signaling.
RNA extraction, reverse transcription and quantitative PCR have been described previously (14). Data are presented as average ±s.d. Supplementary Table 3 summarizes the sequences of primers used for detection of the p53 family.
Luciferase reporter assay and integral transcription activity
Luciferase activity was measured using a Dual-Luciferase Reporter Assay kit (Promega), as described earlier (14). All experiments were performed in triplicate. Data are presented as average ±s.d.
To determine the integral transcription activity of the p53 family, cells were co-transfected with a reporter (pRL-TK) expressing the Renilla luciferase and either PG13 or MG15 reporter plasmids at a molar ratio PG13(MG15):(pRL-TK)=9:1 for 24h. Cells were next treated with 1 uM 5-FU for additional 24h.
Detection of COOH-terminal splice variants of p73
Nested RT-PCR reaction spanning the mid-region of exon 10 to the end of exon 14 of p73 was used for detection of the p73 COOH-terminal splice variants as described previously (15). Following primer sets were used: external: GCCGGGAGAACTTTGAGATC and TCCTTGATGGGCTGCTTGC at p73 cDNA positions 1192-1211 and 1993-1975, respectively; internal: CAGCCACTGGTGGACTCCTATC and TAGTCGGGCCCTGCTTCAG at p73 cDNA positions 1257-1278 and 1771-1752, respectively.
MTT, apoptosis and cell cycle analysis
Cells were treated with different concentrations (0.1uM - 1mM) of 5-FU for 72hs and then cellular response to the drug treatment was assessed using the MTT cell viability assay (ATCC) according to the manufacturer’s protocol. Dose-response curves were generated and IC50 values were calculated using PRISM software (GraphPad). Apoptosis and cell cycle analyses have been described earlier (16, 17).
DNA affinity immunoblotting (DAI)
DAI has been described previously (18). In brief, 500ug of total protein collected from cells that were either treated with 2.5uM 5-FU or left untreated, were then incubated with 20ng of 5′-biotinylated DNA probe for 30 min at room temperature. DNA-protein complexes were precipitated with NeutrAvidin agarose beads (Fisher, Waltham, MA), washed, eluted, and analyzed by Western blotting with p53 (DO-1), p63 (4A4), TAp73 (Bethyl Laboratories) and ΔNp73 (Imgenex) antibody.
5′-biotinylated p21 and wtPUMA probes were generated by PCR using human genomic DNA as a template with the following primer sets: p21 (5′-biotin-AGCCTCCCTCCATCCCTAT-3 ′ a n d 5 ′-CCCTTCCTCACCTGAAAACA-3′), n / s ( 5′-biotin-TAGCTGGGAAGCTGGGACTA-3 ′ a n d 5 ′-GGTTTCCTTGCCCTAAAAGG-3′), and wtPUMA (5′-biotin-CCCAGTCAGTGTGTGTGTCC -3′ and 5′-CCCCCGCGTGACGCTAC-3′.
To generate mtPUMA probe, four point mutations were introduced into the p53 binding site of wtPUMA (CTG(C→G)AA(G→T)TCCTGA(C→A)TT(G→A)TCC) using the Quick Change mutagenesis kit (Stratagene).
Chromatin immunoprecipitation (ChIP) and re-ChIP
ChIP analysis was performed using the ChIP assay kit (Upstate Biotechnology), as described earlier (19). To detect bindings of p53, p63 and p73, specific antibodies against p53 (DO-1), p63 (4A4), and TAp73 (Bethyl Laboratories) were used. As a negative control, immunoprecipitation was performed with rabbit or mouse non-specific IgG. DNA released from the precipitated protein-DNA complexes was amplified by PCR using the following primers: p21 (5′-ACCTTTCACCATTCCCCTAC-3′, 5′-GCCCAAGGACAAAATAGCCA-3′), and PUMA (5′-TGTCCATGGTGTGGATTTGCG-3′, 5′-AGACACCGGGACAGTCGGACA-3′). Locations of primers used for ChIP analyses are shown in Supplementary Figure 2.
Re-Chip analysis was performed to assess the simultaneous binding of p53, p63 and p73 to their target promoters. Protein-DNA complexes generated in the first round of chromatin precipitation were washed, incubated with 10mM DTT for 30 min at 37°C, diluted (x40), and then subjected to the second round of immunoprecipitation and PCR.
Statistical analysis
Statistical analysis was performed using the Student t-test. Results were expressed as average values ± s.d. The difference was considered significant if p<.05. For statistical analyses of immunohistochemical staining Wilcoxon rank sum test, Spearman rank correlation and Kruskal-Wallis one-way analysis of variance by ranks were used.
The Wilcoxon rank sum test was used for statistical analysis of the integral transcription activity (PG-13/MG-15) and 5-FU sensitivity (Supplementary table 4). To assess correlations between chemotherapeutic drug response (IC50) and either p53 status or integral transcription activity, the analysis of variance (ANOVA) was used. In order to meet the assumption of normality and reduce the influence of extremely high values, the analyses were performed with log-transformed values of IC50 and PG13/MG15.
Results
p53 family is expressed in tumors as a complex of isoforms
In order to assess the expression profiles of different p73 and p63 isoforms in primary colon adenocarcinomas we performed immunohistochemical analysis, as described in the Material and Methods section using p73- and p63-specific antibodies in 104 colorectal adenocarcinomas and 14 normal colon specimen collected at Vanderbilt University Medical Center. Among informative cases, elevated TAp73 immunoreactivity was detected in 52 of 87 tumors (60%). It was primarily confined to the nuclei of cancer cells; staining of surrounding tissues were negative (Fig. 1A, panel 2). Normal mucosa was only positive in 1 of 12 cases. We found a statistically significant increase (p=.00186) in p73 nuclear staining in tumor samples compared to non-neoplastic mucosa. Interestingly, high nuclear staining correlated with a decreased vascular invasion (p=.026). Cytoplasmic TAp73 staining was observed in 23 of 87 tumors (26%) and in 1 of 12 (8%) normal cases. This pattern of expression was significantly lower in mucinous tumors (p=.015).
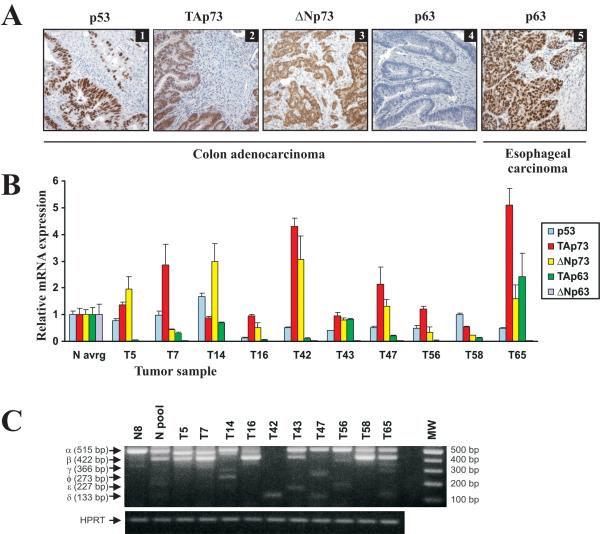
Figure 1.
Protein and mRNA expression of the p53 family in gastrointestinal tumors. A. Representative immunostainings for p53, TAp73, ΔNp73, and p63 in colon adenocarcinomas (panels 1, 2, 3, and 4, respectively) and p63 in esophageal squamous carcinoma (panel 5). p63 antibody (4A4, Santa Cruz Biotechnology) used in this staining recognizes both TA and DN isoforms of p63. B. The bar graph represents quantitative real-time RT-PCR analyses of p53, TA and ΔN transcripts of p63 and p73 mRNAs in primary colon adenocarcinomas, compared to the average levels of 17 normal colon tissues. Data were normalized to HPRT1 mRNA expression (mean ± s.d.; n=2). Expression of p53, p63 and p73 isoforms in normal tissues was arbitrarily set at 1. C. Expression of the COOH-terminal splice variants of p73 in 10 colon adenocarcinomas and normal colon mucosa. N pool - pooled sample from 14 normal colon tissues; N8 - a representative normal specimen.
ΔNp73 staining was found in both the nucleus and cytoplasm of colon epithelia cells in 28% (24/87) of tumors (Fig.1A, panel 3). All normal cases were negative for ΔNp73. Immunohistochemistry revealed low levels of p63 isoforms in colon tumors (Fig. 1A, panel 4); only 3 of 90 tumor cases were found to be positive for nuclear p63. As a positive control for p63 staining, squamous cell carcinomas of the esophagus, which are known for high levels of p63 (20), were used (Fig. 1A, panel 5). An increased p53 staining in the nuclei of tumor cells was detected in 53% (46/87) of colon tumors. A representative staining for p53 is shown in Figure 1A (panel 1). This elevated p53 immunoreactivity indicates the presence of p53 mutations.
Using real-time PCR, we also analyzed the expression profiles of p53, p63 and p73 mRNA in a subset of these tumors (Fig. 1B). Similar to TMA analysis, we found elevated levels of TAp73 and ΔNp73 mRNA compared to the average expression in non-neoplastic colon tissues. Using nested PCR, as described previously (15), we determined the spectrum of C-terminal splice variants of p73 (Fig. 1C). Although multiple isoforms of p73 (γ, δ, φ and ε) were detected, the most prevalent were alpha and beta. The levels of p63 isoforms were low (except for one case) or undetectable in both normal and tumor samples (Fig. 1B). We also did not find significant changes in p53 mRNA (Fig. 1B).
Taken together, our data show complex expression profiles of the p53 family in gastrointestinal tumors. p73 and p53 proteins, and in some cases p63, are co-expressed in tumor tissues from the same patient (Fig. 1B). Notably, each cancer patient is characterized by a unique expression profile of the p53 family isoforms.
p73 and p63 are involved in chemotherapeutic drug response
As a prelude to defining the role of the p53 family in gastrointestinal tumors, we first sought to characterize the p73 and p63 isoforms in our model systems of interest. We ectopically expressed p73 and p63 isoforms in H1299 cells, which lack p53 and express low levels of endogenous p63 and p73 proteins. Using the p53-specific firefly luciferase reporter PG13-LUC and Western blotting, we found that all transfected TA isoforms activate the reporter (Fig. 2A, upper panel) and induce expression of p53 target genes NOXA, p21 and FasR (Fig. 2A, lower panel). The induction levels were dependent on an isoform; TAp73α was found to be less active than TAp73β and TAp63γ. Interestingly, some ΔN isoforms were also able to activate reporter and target genes expression. ΔNp63α showed a significant induction of NOXA, p21 and FasR. ΔNp73β transfection was sufficient to induce NOXA expression and activate reporter. However, when ΔN isoforms were co-expressed with p53, they inhibited activation of p53 reporter and target genes transcription, which were induced by p53 (Fig. 2B). Similar results were obtained in RKO colon cancer cell line (data not shown). These findings suggest that p73 and p63 isoforms have similar transcription properties to p53 and induce target genes that have been shown to be up-regulated by p53; however, these effects are dependent on isoform properties as well as an expression context of other p53 family members.
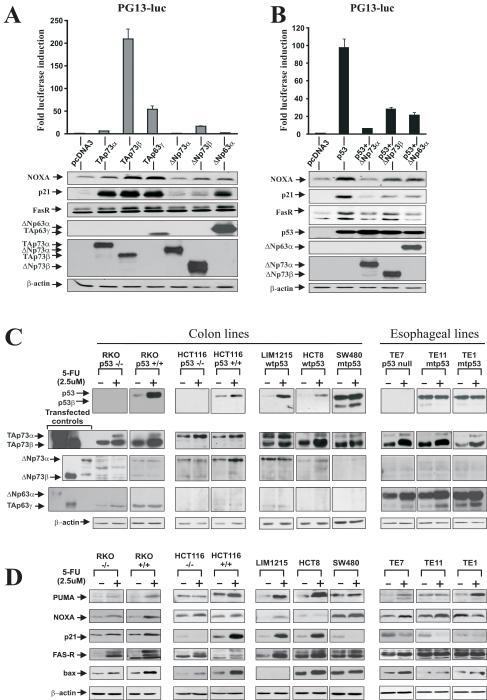
Figure 2.
Isoforms of p73 and p63 regulate p53-target genes. A. H1299 cells were co-transfected with PG13LUC luciferase reporter and indicated isoforms of p73 and p63. B. Same as (A), except H1299 cells were co-transfected with p53. Luciferase activity (upper panels) and protein expression of p53 transcription targets (Western blotting, lower panels) were analyzed 24h after transfection. Expression of transfected p53, p63 and p73 constructs was confirmed by Western blotting using p53 (DO-1), p63 (4A4) and p73 (Ab-2) antibodies. C. Protein expression of endogenous p53, p63 and p73 isoforms in the indicated cell lines. Protein lysates were prepared from control untreated cells (−) and those treated with 2.5 μM 5-FU (+) for 24h and analyzed by Western blotting. D. Protein expression of p53 targets, PUMA, NOXA, p21/WAF1, FasR, and BAX after treatment with 5-FU. Protein lysates were prepared from control cells (−) and those treated with 2.5 uM 5-FU (+) for 48h and analyzed by Western blotting.
Next, we analyzed the expression of endogenous proteins and their role in chemotherapeutic drug response. To evaluate the expression of the p73 and p63 isoforms, we employed colon cancer cell lines that express wild-type or mutant p53. We also took advantage of recently generated isogenic RKO and HCT116 cell lines that differ only in their p53 expression (21). Similar to primary colon tumors, colon cancer cell lines express multiple isoforms of p73 and low or undetectable levels of p63 (Fig. 2C). Therefore, three additional esophageal cell lines, TE1, TE7 and TE11, derived from esophageal carcinomas were added to our analysis. Esophageal tumors are known for high expression of p63 (20). Consistent with this, we found high levels of both ΔN and TA isoforms of p63 in all esophageal cell lines (Fig. 2C).
To assess the response to chemotherapeutic drug treatment we analyzed changes in p53, p73, p63 and their target genes after treatment with chemotherapeutic drugs. 5-FU was selected as a prototypical agent, as it is commonly used in clinic. As expected, in cell lines expressing wild-type p53 (HCT8, LIM1215, RKO, and HCT116), levels of endogenous p53 protein were significantly increased following the 5-FU treatment (Fig. 2C). An increase of mutant p53α and p53β isoforms was detected in the SW480 cell line. Notably, both TAp73α and TAp73β proteins were found to be strongly up-regulated by 5-FU in all tested cells (Fig. 2C). In esophageal cell lines, endogenous TAp63γ and ΔNp63α isoforms were also increased compared to untreated control.
To evaluate changes in p53-dependent transcription, we next compared the protein levels of p53 transcription targets PUMA, NOXA, p21/Waf1, FasR, and BAX in 5-FU treated and untreated control cells (Fig. 2D). These proteins are involved in cell cycle regulation, apoptosis and are directly activated by p53. We found marked induction of the p53 targets in treated cells expressing wild-type p53. However, we also observed up-regulation of these proteins in p53-deficient cells, although at a lesser extent than that of cells expressing wild-type p53. For instance, TE7 cells, which lack the expression of p53 and strongly activate TAp73α and β, TAp63γ and ΔNp63α, displayed a significant increase in the endogenous levels of PUMA, NOXA, FasR, and BAX in response to 5-FU (Fig. 2D).
In addition, 5-FU treatment induced all analyzed target genes in p53 knockout cell line RKO p53−/−; however, at a lesser extend than in parental isogenic RKO p53+/+ (Fig. 2D). Similarly, up-regulation of p53 targets was also found in the isogenic p53-null cell line HCT116, implicating p73 in the regulation of the p53 targets. Some p53 targets were also up-regulated in p53-deficient TE1, TE11 and SW480 cells (Fig. 2D).
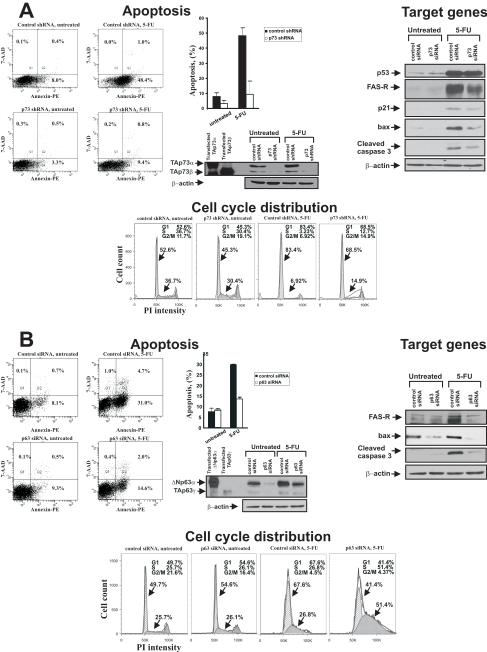
To directly assess the role of p73, we employed RNAi-mediated silencing that specifically targets p73 isoforms (Fig. 3A, left panel) and does not affect p53 (Fig. 3A, right panel). As shown in Figure 3A, stable transfection of the p73 shRNA significantly inhibits the 5-FU-induced apoptosis in RKO (p53 +/+) colon cancer cells, which express high levels of endogenous TAp73 (Fig. 2C). Using Western blotting, we analyzed the expression of p53 target genes in these cells. FasR, p21, and BAX proteins were inhibited by p73 siRNA (Fig. 3A, right panel). While in these cells inhibition of endogenous p73 has significant effect on 5-FU sensitivity and p53-dependent transcription, we still observed up-regulation of p53 target genes and 3-fold induction of apoptosis after drug treatment in the p73-deficient cells. This is likely explained by the activity of endogenous wtp53. Our data also show that not only apoptosis but also cell cycle arrest is affected by inhibition of p73 activity. We found that suppression of p73 in RKO cells with shRNA protects cells from 5-FU-induced cell cycle arrest in the G1 phase, as measured by Flow Cytometry (FACS) analysis (Fig. 3A, bottom panel). Similarly, inhibition of p73 with a dominant-negative DDp73 mutant (22) or specific siRNA against TAp73 isoforms inhibited the cytotoxic effect of 5-FU in RKO cells (data not shown).
Figure 3.
p73 and p63 are involved in cellular response to 5-FU treatment. A. Left panel: RKO cells were transduced with lentivirus expressing either negative control or shRNA against p73 isoforms and then treated with 0.25mM 5-FU for 4h. Apoptosis was measured by FACS using staining with Annexin V-PE and 7-AAD (BD Biosciences, CA). Bar graph represents data obtained in two independent experiments; percentage of cells in early apoptosis (AnnexinV-positive, 7-AAD-negative) is shown. Western blot with a TAp73 specific antibody (Bethyl Laboratories) was performed to demonstrate the effect of shRNA on endogenous p73. Right panel: Cells were transfected with either negative control or p73 siRNA for 48h and then treated with 25 uM 5-FU for additional 48h. Western blot analysis showed that p73 siRNA suppresses 5-FU-induced activation of p53 target genes, FasR, p21, and BAX. p73 siRNA also inhibited accumulation of cleaved caspase 3. The immunoblot with p53 (DO-1) antibody revealed that endogenous p53 level was not affected by p73 siRNA. Bottom panel: Cell cycle distribution was analyzed by FACS in RKO cells transfected with either negative control or p73 shRNA and then treated either with 25 uM 5-FU or vehicle control for 48h. Data analysis was performed using FloJo (Tree Star) software. A significant increase of G1 arrested cells and a decrease in ones in the S phase were found after 5-FU treatment of control siRNA transfected cells. In contrast, cells transfected with p73 siRNA were less sensitive to G1 cell cycle arrest. B. Left panel: TE7 cells, which express high levels of p63, were transfected with either p63 siRNA or non-specific control RNA and then treated with 0.25 mM 5-FU for 18h. Apoptosis was measured as described in (A). Inhibition of p63 by specific siRNA led to a significant decrease of 5-FU-induced apoptosis. The bar graph shows the percentage of cells in early apoptosis and represents the results of two independent experiments. The Western blot analysis with p63 (4A4) antibody confirmed the down-regulation of endogenous TA and ΔN p63 proteins in TE7 cells after p63-specific siRNA transfection. The p63 siRNA did not have a significant effect on p73 isoforms (Supplementary Figure 1). Right panel: TE7 cells were transfected with either control or p63 siRNA for 48h and treated with 25 uM 5-FU for additional 48h. Western blot analysis showed that p63 siRNA inhibits accumulation of p53 family target genes FAS-R and BAX, as well as cleaved caspase 3. Bottom panel: TE7 cells were transfected with either negative control or p63-specific siRNA and then treated with 15 uM 5-FU for 48h. Cell cycle was analyzed by FACS.
To inhibit p63 activity we used p63-specific siRNA in TE7 cells, which express high levels of p63 isoforms. Suppression of p63 with siRNA led to a decrease in sensitivity to 5-FU induced apoptosis (Fig. 3B, right panel) and expression of p53 targets, BAX and FasR (Fig. 3B, left panel), compared to control scrambled RNA transfected cells. In contrast to RKO cells, we did not observe any significant effect of p63 siRNA on cell cycle arrest and p21 levels, suggesting that the effect of p73 and p63 inhibition depends on cellular context (Fig. 3B and data not shown).
Thus, not only p53, but also p73 and p63 are accumulated in response to chemotherapeutic treatment, induce p53 transcription targets, and significantly contribute to chemotherapeutic drug response in cancer cells.
p53, p63 and p73 simultaneously bind to the promoters of the p53 target genes following drug treatment
To investigate the mechanisms of functional interactions between p53, p73 and p63, we assessed their binding to the PUMA(BBC3) and p21(CDKN1A) gene promoters that play critical roles in DNA damage-induced apoptosis and cell cycle regulation. TE7, TE11 and isogenic HCT116 cell lines were selected, as they express p73/p63 and have null (TE7), mutant (TE11) and wild-type (HCT116) p53. As a first step, we performed DNA affinity immunoblotting (DAI), a sensitive assay for quantitative measurement of protein-DNA binding (18). Native DNA sequences (~200 bp) containing p53-response elements (p53-RE) derived from the human p21 and PUMA promoters were used as specific probes. As controls, we generated non-specific probes: mutated PUMA (mtPUMA) with point mutations in the p53-RE and intronic sequence of p21 gene (n/s).
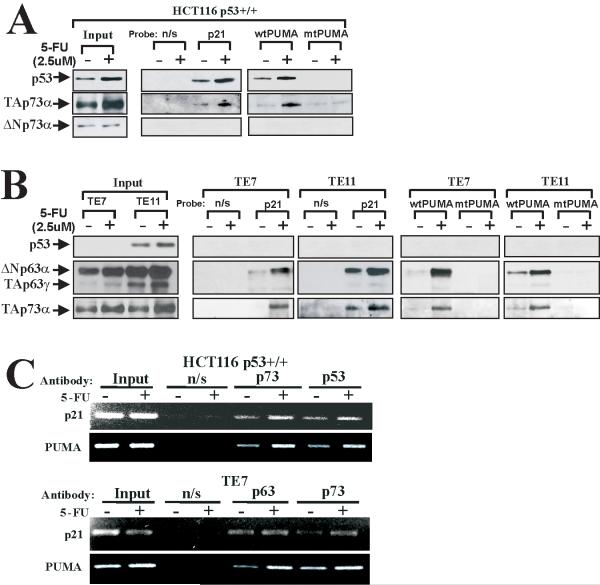
After analyzing the DNA-bound proteins from HCT116, TE7 and TE11 cells we found strong specific binding of TAp73 isoform to PUMA and p21 probes, which was increased after 5-FU treatment (Figs. 4A, B). We did not observe any significant binding to the control probes suggesting that p73 specifically interacts with the promoters. These interactions are likely mediated by p53-RE, as p73 failed to bind to the PUMA probe containing mutant p53-RE (Fig. 4A, B). As expected, endogenous p53 was also bound to the promoter probes in the HCT116 cell line (Fig. 4A). Similar to TAp73, this binding was increased in response to drug treatment. In another wild-type p53 cell line, LIM1255, binding of p53 and TAp73 was also detected (data not shown). In both p53-null TE7 and mutant p53 TE11 cell lines, we found strong specific binding of TAp63γ and ΔNp63α as well as TAp73 to the p21 and PUMA probes (Fig. 4B) that were significantly increased after 5-FU treatment. We did not detect any binding of mutant p53 or ΔNp73.
Figure 4.
Multiple isoforms of p63 and p73 bind to the promoters of p53 target genes. A. DNA affinity immunoblotting (DAI) analysis of HCT116 p53+/+ cells that were either mock treated (−) or incubated with 2.5uM 5-FU for 24h (+). Input lanes show expression of indicated proteins in cell lysates analyzed by Western blotting. B. DAI analyses of TE7 and TE11 cells treated with 2.5 uM 5-FU (+) or left untreated (−). C. ChIP analyses of p53, p63 and p73 binding to the p21 and PUMA promoters in HCT116 p53+/+ (upper panel) and TE7 (lower panel) cells treated as described in (A). As a negative control, non-specific IgG (n/s) were used.
To confirm these data and investigate the binding of endogenous p53, p63 and p73 proteins to the native p21 and PUMA promoters, we performed chromatin immunoprecipitation assay (ChIP). Consistent with DAI, we found increased interactions of p53, p73 and p63 with promoters of p21 and PUMA genes after 5-FU treatment (Fig. 4C). Taken together, these findings show that not only p53, but also various isoforms of p63 and p73 bind to the promoters of target genes in response to chemotherapeutic treatment with 5-FU.
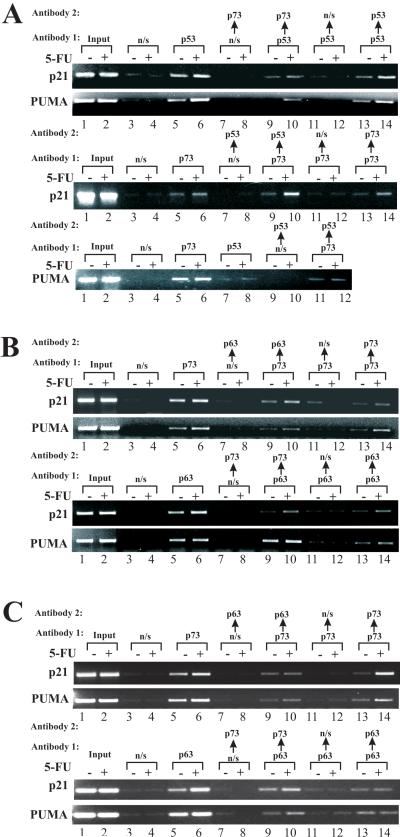
Analyzing the promoter binding, we noticed that both TAp73 and wild-type p53, as well as TAp73 and p63, effectively bind to the same promoters. To assess whether these proteins bind simultaneously we performed re-ChIP analysis, a sequential chromatin immunoprecipitation, using two different antibodies. Using p53 and p73 antibodies, we found that p53 and p73 bind alongside the PUMA and p21 promoters in HCT116 cells (Fig. 5A, lanes 9, 10). This co-binding was significantly increased after 5-FU treatment. However, when unspecific antibodies were employed for either the first or second round of precipitations, no binding was detected (Fig. 5A, lanes 7, 8, 11, 12). Specificity of this analysis was further confirmed by reciprocal experiments (Fig. 5A-C, bottom panels) and with the same antibodies for both immunoprecipitation rounds (Figs. 5A-C, lanes 13, 14). Using the same techniques, we also found that p63 and p73 bind together to the PUMA and p21 promoters in TE7 cells (Fig. 5B). In TE11 cells, which express mutant p53, p73 and p63, also simultaneously bind to the p21 and PUMA promoters suggesting that mutant p53 does not affect binding of the other members of the p53 family in these cells (Fig. 5C). Thus, our data show that p53, p73 and p63 interact at the levels of target gene promoters.
Figure 5.
Re-ChIP assay revealed simultaneous binding of p53 and p73 (HCT116 p53+/+), and p63 and p73 (TE7 and TE11 cells) to the promoters of p21 and PUMA genes. Chromatin was precipitated with Antibody 1 and re-immunoprecipitated with Antibody 2. As negative controls, unspecific antibodies (n/s) were used in the first or second rounds of ChIP. A. Upper panel: Re-ChIP was performed in HCT116 p53+/+ cells, as described in the Materials and Methods section. p53 and p73 antibodies were used for the first and second rounds of immunoprecipitations, respectively. Lower panel: Reciprocal re-ChIP experiment in HCT116 p53+/+ cells. B. Upper panel: Re-ChIP in TE7 cells using p73 and p63 antibodies for the first and second rounds of immunoprecipitations, respectively. Lower panel: Reciprocal re-ChIP experiment in TE7 cells. C. Same as (B), except TE11 cells were used.
Cell response to chemotherapy depends on the integral transcription activity of the p53 family
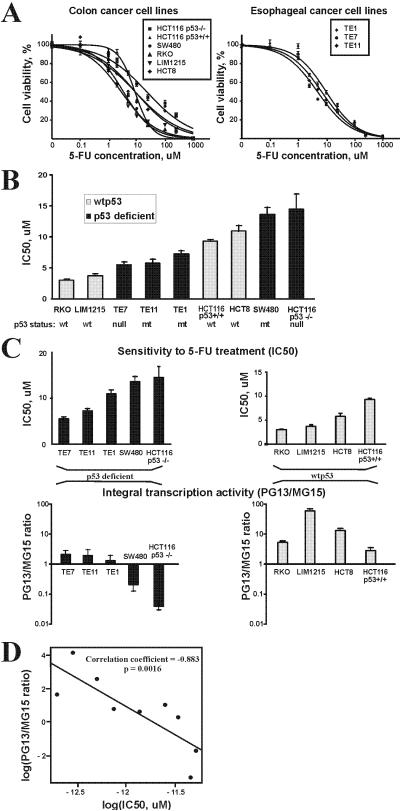
We next analyzed associations between the activity of the p53 family and sensitivity to 5-FU treatment. We first assessed the role of p53, as it plays a critical role in cellular response to chemotherapeutic drugs. Using dose-response curves we determined the sensitivity (IC50) of all studied colon and esophageal cell lines to 5-FU treatment (Fig. 6A) and evaluated its association with the functional status of p53 (Fig. 6B). Although cell lines that express wild-type p53 tend to be more sensitive to drug treatment, our analysis failed to find statistically significant correlations between p53 status and cellular response to drug treatment (p=.3).
Figure 6.
Integral transcription activity of the entire p53 family correlates with response to 5-FU treatment. A. Dose-response curves for 5-FU are shown. The indicated cell lines were treated with different concentrations of 5-FU for 72h and cytotoxicity was measured by MTT assay. B. Comparison of sensitivity to 5-FU treatment with the p53 status. Wild-type cell lines are shown in black and p53-deficient in grey. No correlation between p53 status and cellular response (IC50) was found (p=.3). C. Upper panel: Sensitivity of indicated cell lines to 5-FU treatment. IC50 values are shown. Lower panel: Integral transcription activity of the p53 family was calculated as a ratio of relative activities of PG13 and MG15 luciferase reporters after treatment with 1uM 5-FU. Total activity of the p53 family was significantly correlated with sensitivity to 5-FU treatment (p<.05). Cell lines were divided into two groups based on the p53 status. D. The scatter plot illustrates correlation between log transformed values of IC50 and integral transcription activity. Linear regression line is shown.
As the cellular response to 5-FU does not correlate with the p53 status in our experiments, we evaluated an alternative approach. We hypothesize that integral (total) transcription activity of the entire p53 family is a better predictor of cell response to drug treatment than commonly used static expression or mutation analyses. To determine the integral transcription activity we used luciferase reporter PG13-LUC because it can measure activity, not only of p53 but also p73 and p63 isoforms (Fig. 2A and B). The integral transcription activity was calculated as a ratio between the activation of PG13 and MG15 luciferase reporters (relative reporter activation) following the 5-FU treatment. The latter reporter contains the p53 consensus binding sites inactivated by mutations. Normalization of PG13 reporter activation to MG15 allowed us to eliminate transcription effects unrelated to the p53 family and compare p53-dependent transcription in different cell lines. We found significant association between the sensitivity of cell lines to 5-FU treatment and the integral transcription activity measured by reporter analysis (Fig. 6C and D, p=.0016). Thus, the predicting power of the integral transcription activity for evaluation of drug response is significantly better than p53 alone. This shows that interaction between members of the p53 family at the transcription levels has a profound effect on chemotherapeutic drug response in tumor cells.
Discussion
This is the first comprehensive study of the p53 family in gastrointestinal tumors. In this study, we investigated the interactions between members of the p53 family and their roles in cellular response to treatment with commonly used chemotherapeutic agent 5-FU. Our data demonstrated that activities of the p53 family are more tightly intertwined than previously thought. We showed, for the first time, that p53, p73 and p63 bind simultaneously to target gene promoters in tumor cells treated with DNA-damaging drug 5-FU. These associations to promoters are determined by expression profiles of the p53 family in tumor cells. Using re-ChIP assay and DAI, we found that p53 and TAp73 isoforms bind simultaneously to PUMA and p21 promoters in HCT116 colon cancer cells. Whereas in p63 expressing esophageal cancer cells, TA isoforms of p63 and p73, as well as ΔNp63α, are associated with PUMA and p21 promoters. p63 and p73 isoforms were present at the promoters in p53 deficient cells suggesting that they can partially compensate for the lack of p53.
Our data also show that p73 and p63 are expressed as a complex set of isoforms in cancer cell lines and primary tumors. In colon cancers, we found increased co-expression of TAp73 and ΔNp73 compared to normal tissue and low levels of all analyzed p63 isoforms (Fig. 1A and B). Interestingly, cancer tissues tend to exhibit a more complex expression pattern of the COOH-terminal isoforms. p73 γ, δ, φ and ε isoforms were primarily detected in tumors, but not in normal tissues (Fig. 1C). Notably, mRNA expression profiles for p63 and p73 isoforms were unique for each analyzed patient (Fig. 1B, C). Significant variability was also observed in cancer-derived colon and esophageal cell lines.
Consistent with previous reports on other tumor types (23, 24), we confirmed that p73 and p63 contribute to the up-regulation of p53 transcription targets and p53-dependent apoptosis in colon and esophageal tumors (Fig. 3). We found the up-regulation of multiple p53 transcription targets in p53-deficient cells after treatment with 5-FU cells, although it occurs at a lesser extent than in wild-type p53 cells (Fig. 2D). Specific inhibition of p73 and p63 with siRNA led to inhibition of p53 target genes, apoptosis and cell cycle arrest induced by 5-FU. Thus, p73 and p63 can, at least in part, compensate for p53 deficiency. However, this does not exclude the inhibitory role of mutant p53 on TAp73 and TAp63 as previously reported (25-27). In our experiments, p53-deficient cells were generally less sensitive to 5-FU treatment.
Using reporter assay and Western blotting, we analyzed transcription activities of TA and ΔN isoforms of p73 and p63 that commonly express in colon and esophageal tumors. Previously, it has been reported by us and others that TA isoforms are transcriptionally active and pro-apoptotic, whereas ΔN isoforms are inhibitory and anti-apoptotic (7, 28, 29). Moreover, different COOH-splice variants have varied transcription activities (30-34). Our analysis support these data; however, it also shows that the effects of isoforms is contextual (i.e. dependent on the expression of other isoforms). For instance, when ΔNp73β co-expresses with p53, it inhibits the p53 transcription activity, but alone it activates p53 reporter and induces p53 target genes p21, NOXA and FasR (Fig. 2B). Combined, these data show that activation of p53-dependent transcription is defined by isoform expression profile of p53, p73 and p63, their relative activity and molecular ratios between different isoforms. It also suggests that response to chemotherapeutic drug treatment with 5-FU is defined by total activity of the p53 family.
The next question we asked was whether total activity of the p53 family correlates with cellular response to 5-FU treatment. One potential way to determine the total activity of the p53 family is to assess the expression levels of the p53 family isoforms. However, as shown above, p53, p73 and p63 are expressed as a complex set of isoforms (at least 30 isoforms are currently known (7)). These isoforms interact with each other in a complex fashion as shown in our study as well as by others (35). Therefore, it is difficult to make a conclusion based solely on expression profiles of the p53 family. Another alternative approach is to assess the expression level or activity of p53 transcriptional targets. Though potentially valuable, this approach has its own set of hurdles. All p53 target genes have complex transcriptional regulation and are affected by multiple regulatory pathways. One example is the p21/Waf1(CDKN1A) gene that regulated in a p53-dependent and -independent manner (36). Therefore, it is difficult to dissect the contribution of the p53 pathway in the regulation of their expression. In addition, the vast multiplicity of p53 targets makes it difficult to select a relevant set of targets.
Based on these data, we hypothesize that the total transcription activity will be a better predictor of cellular response to 5-FU. We introduced the term “integral transcription activity” defined as total transcription activity measured by reporter analysis. Although our analysis revealed variability between cell lines, we found that the integral transcription activity of the entire p53 family significantly correlates with cellular response to 5-FU (p=.0016). Interestingly, this correlation was more obvious in p53-deficient cells than in its wild-type counterparts. The reason for this difference is unclear, but non-transcriptional mechanisms (i.e. a direct effect of p53 at mitochondria) may be a factor (37).
In our studies we did not find correlation between p53 status and cellular response to drug treatment (p=.3). A recent large-scale meta-analyses in colorectal tumors also concluded that the predicting value of p53 mutations does not reach the level of clinical usefulness and fails to correlate with a treatment response (4, 38). It can be explained, at least in part, by current inadequate methodologies for detecting p53 abnormalities, immunohistochemistry and sequencing analysis that provide limited information on the activity of p53. Another complicating factor that was partly addressed in our studies is that p53 can be affected by multiple factors and inhibited by mechanisms that are not identified in mutational analyses (17, 39, 40). Our data suggest that the integral functional activity of the entire p53 family, as a measurement of cellular response to treatment, may help to overcome these problems. Additional studies are needed to confirm this in vivo.
Taken together, our work demonstrates that interactions between the members of the p53 family have a profound effect on chemotherapeutic drug response in tumor cells.
Supplementary Material
Acknowledgments
We thank Drs. El-Rifai, Pietenpol, and Beauchamp for their interest and valuable discussions.
Grant support: National Cancer Institute grants NIH CA108956 and NIH CA129655
Abbreviations
- 5-FU
fluorouracil (5-Fluoro-2,4(1H,3H)-pyrimidinedione)
- CRC
colorectal carcinoma
- ChIP
chromatin immunoprecipitation
References
- 1.Waters JS, Ross PJ, Popescu RA, Cunningham D. New approaches to the treatment of gastrointestinal cancer. Digestion. 1997;6:508–19. doi: 10.1159/000201494. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society . Colorectal Cancer Facts & Figures Special Edition 2005. American Cancer Society; Atlanta: 2005. [Google Scholar]
- 3.Hamroun D, Kato S, Ishioka C, Claustres M, Beroud C, Soussi T. The UMD TP53 database and website: update and revisions. Hum Mutat. 2006;1:14–20. doi: 10.1002/humu.20269. [DOI] [PubMed] [Google Scholar]
- 4.Munro AJ, Lain S, Lane DP. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer. 2005;3:434–44. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaika AI, El-Rifai W. The role of p53 protein family in gastrointestinal malignancies. Cell Death Differ. 2006;6:935–40. doi: 10.1038/sj.cdd.4401897. [DOI] [PubMed] [Google Scholar]
- 6.Melino G, Lu X, Gasco M, Crook T, Knight RA. Functional regulation of p73 and p63: development and cancer. Trends Biochem Sci. 2003;12:663–70. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Vilgelm A, El-Rifai W, Zaika A. Therapeutic prospects for p73 and p63: rising from the shadow of p53. Drug Resist Updat. 2008;4-5:152–63. doi: 10.1016/j.drup.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong SM, Cho H, Moskaluk CA, Yu E, Zaika AI. p63 and p73 expression in extrahepatic bile duct carcinoma and their clinical significance. J Mol Histol. 2007;3:167–75. doi: 10.1007/s10735-007-9084-7. [DOI] [PubMed] [Google Scholar]
- 9.Ventura A, Meissner A, Dillon CP, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;28:10380–5. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol. 2008;19:5951–64. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaika A, Irwin M, Sansome C, Moll UM. Oncogenes induce and activate endogenous p73 protein. J Biol Chem. 2001;14:11310–6. doi: 10.1074/jbc.M005737200. [DOI] [PubMed] [Google Scholar]
- 12.Vilgelm A, Wei JX, Piazuelo MB, et al. DeltaNp73alpha regulates MDR1 expression by inhibiting p53 function. Oncogene. 2008;15:2170–6. doi: 10.1038/sj.onc.1210862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–92. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomkova K, Belkhiri A, El-Rifai W, Zaika AI. p73 isoforms can induce T-cell factor-dependent transcription in gastrointestinal cells. Cancer Res. 2004;18:6390–3. doi: 10.1158/0008-5472.CAN-04-2176. [DOI] [PubMed] [Google Scholar]
- 15.Zaika AI, Kovalev S, Marchenko ND, Moll UM. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999;13:3257–63. [PubMed] [Google Scholar]
- 16.Wei J, O’Brien D, Vilgelm A, et al. Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology. 2008;5:1412–23. doi: 10.1053/j.gastro.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaika AI, Slade N, Erster SH, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;6:765–80. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Asch H, Kulesz-Martin MF. Functional quantification of DNA-binding proteins p53 and estrogen receptor in cells and tumor tissues by DNA affinity immunoblotting. Cancer Res. 2001;14:5402–6. [PubMed] [Google Scholar]
- 19.Tomkova K, El-Rifai W, Vilgelm A, Kelly MC, Wang TC, Zaika AI. The gastrin gene promoter is regulated by p73 isoforms in tumor cells. Oncogene. 2006;44:6032–6. doi: 10.1038/sj.onc.1209610. [DOI] [PubMed] [Google Scholar]
- 20.Hara T, Kijima H, Yamamoto S, et al. Ubiquitous p63 expression in human esophageal squamous cell carcinoma. Int J Mol Med. 2004;2:169–73. [PubMed] [Google Scholar]
- 21.Sur S, Pagliarini R, Bunz F, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;10:3964–9. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irwin M, Marin MC, Phillips AC, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;6804:645–8. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 23.Gressner O, Schilling T, Lorenz K, et al. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;13:2458–71. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr. Chemosensitivity linked to p73 function. Cancer Cell. 2003;4:403–10. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 25.Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;2:1438–49. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;5:1874–87. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strano S, Fontemaggi G, Costanzo A, et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;21:18817–26. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- 28.Lin KW, Nam SY, Toh WH, Dulloo I, Sabapathy K. Multiple stress signals induce p73beta accumulation. Neoplasia. 2004;5:546–57. doi: 10.1593/neo.04205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiewe T, Theseling CC, Putzer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem. 2002;16:14177–85. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, Nozell S, Xiao H, Chen X. DeltaNp73beta is active in transactivation and growth suppression. Mol Cell Biol. 2004;2:487–501. doi: 10.1128/MCB.24.2.487-501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller M, Schilling T, Sayan AE, et al. TAp73/Delta Np73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ. 2005;12:1564–77. doi: 10.1038/sj.cdd.4401774. [DOI] [PubMed] [Google Scholar]
- 32.De Laurenzi V, Costanzo A, Barcaroli D, et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;9:1763–8. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scaruffi P, Casciano I, Masiero L, Basso G, Romani M, Tonini GP. Lack of p73 expression in mature B-ALL and identification of three new splicing variants restricted to pre B and C-ALL indicate a role of p73 in B cell ALL differentiation. Leukemia. 2000;3:518–9. doi: 10.1038/sj.leu.2401698. [DOI] [PubMed] [Google Scholar]
- 34.Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene. 1999;35:4993–8. doi: 10.1038/sj.onc.1202817. [DOI] [PubMed] [Google Scholar]
- 35.Flores ER, Tsai KY, Crowley D, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;6880:560–4. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 36.Zeng YX, Somasundaram K, el-Deiry WS. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet. 1997;1:78–82. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- 37.Zaika A, Marchenko N, Moll UM. Cytoplasmically “sequestered” wild type p53 protein is resistant to Mdm2-mediated degradation. J Biol Chem. 1999;39:27474–80. doi: 10.1074/jbc.274.39.27474. [DOI] [PubMed] [Google Scholar]
- 38.Vincenzi B, Cesa AL, Santini D, et al. Predictive factors for response to chemotherapy in colorectal cancer patients. Crit Rev Oncol Hematol. 2004;1:45–60. doi: 10.1016/j.critrevonc.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 39.O’Brate A, Giannakakou P. The importance of p53 location: nuclear or cytoplasmic zip code? Drug Resist Updat. 2003;6:313–22. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Ramos YF, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 2001;5:1839–42. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.