Abstract
Epitopes shared by the vaccinia and variola viruses underlie the protective effect of vaccinia immunization against variola infection. We set out to identify a subset of cross-reactive epitopes using bioinformatics and immunological methods. Putative T-cell epitopes were computationally predicted from highly conserved open reading frames from seven complete vaccinia and variola genomes using EpiMatrix. Over 100 epitopes bearing low human sequence homology were selected and assessed in HLA binding assays and in T-cell antigenicity measurements using PBMCs isolated from Dryvax-immunized subjects. Experimental validation of computational predictions illustrates the potential for immunoinformatics methods to identify candidate immunogens for a new, safer smallpox vaccine.
Keywords: smallpox, T-lymphocyte epitopes, epitope mapping
1. Introduction
Over 200 years ago, Edward Jenner developed vaccination against variola virus using the related poxvirus vaccinia, enabling a worldwide effort that culminated in the eradication of smallpox 1979. In wake of the September 11, 2001 terror attacks, fears of deliberate dissemination of variola in an unprotected world population prompted the United States government to stockpile vaccine for the civilian population. Licensed smallpox vaccines, such as Dryvax (Wyeth) and WetVax (Aventis Pasteur) effectively protect against infection but are contraindicated for about 20% of the US population because they are associated with a broad range of adverse events. For example, dermal complications including vaccinia necrosum, a progressive skin condition with case-fatality rates of 75% to 100% among persons with cellular immunodeficiency, were observed during the global campaign to eradicate smallpox [1]. Eczema vaccinatum, a complication among eczema patients, was associated with case-fatality rates of up to 10% overall and 30% to 40% in children younger than two years of age. Moreover, inadvertent inoculation may result in wider spread when vaccinia is transferred from the vaccination site to another location on the vaccinee or to another person [2]. As a result, smallpox vaccination is contraindicated in persons who have eczema, active acute, chronic, or exfoliative skin conditions that disrupt the epidermis, HIV/AIDS, autoimmune conditions, cancer, radiation treatment, or immunodeficiencies.
We set out to design a safer smallpox vaccine that will provide protection to a greater proportion of the US population. Because epitopes provide the minimal essential information needed to trigger a protective immune response, epitope-driven vaccines represent a logical approach to vaccine development that obviates the risks inherent in live vaccines. Our approach to discovering T-cell epitopes is based on the advent of fully sequenced poxvirus genomes coupled with the availability of immunoinformatics tools that can rapidly identify potentially immunogenic and protective poxvirus sequences. In light of the well known fact that vaccinia immunization protects against variola infection, our studies focused on the subset of epitopes that are common to both poxvirus strains (Figure 1). Here, we report the results of the first steps in the vaccine design process whereby computationally identified epitopes were validated in vitro and ex vivo in order to select epitopes to be later tested as a prototype vaccine in a human leukocyte antigen (HLA) transgenic mouse model.
Figure 1.
In silico approach to identification of smallpox vaccine candidates. Immunoinformatics tools identify potential T-cell epitopes from large viral genome datasets, such as variola and the related vaccine strain vaccinia. Here, informatics methods were used to delineate the intersection of vaccinia and variola immunogenic epitope sets, which contain sequences that provide T cell-mediated protection against variola. Conserved sequences among 3 variola and 4 vaccinia genomes were assessed for immunogenic potential using the T-cell epitope mapping algorithm EpiMatrix. We selected 110 epitopes for vaccine design, of which 50 were promiscuous Class II HLA epitopes, 40 were Class I HLA A2 and 20 were Class I B7. 60% of the epitopes were derived from regulatory factors, 24% from hypothetical and unknown proteins and16% from structural proteins.
2. Materials and Methods
2.1. Immunoinformatics
Seven complete poxvirus genomes were downloaded from GenBank: four vaccinia strains (Tian Tian, Accession AF095689; Western Reserve, Accession AY243312; Copenhagen, Accession M35027; Ankara, Accession U94848) and three variola (Variola major, strain India 1967, Accession X69198; Variola major, strain Bangladesh 1975, Accession L22597; Variola minor, strain Garcia 1966, Accession Y16780).
EpiMatrix, a matrix-based epitope mapping algorithm, was used to identify Class I and II HLA epitopes in vaccinia and variola open reading frame (ORF) sequences [3,4,5]. For Class I epitope identification, 9-mer and 10-mer sequences were scored for potential binding to six supertype Class I alleles (A*0101, A*0201, A*0301, A*2402, B*0702 and B*4403 alleles) that cover >90% of humans in five major human population groups [6]. For Class II epitope identification, potential binding of 9-mer sequences was scored for eight archetypical Class II alleles (DRB1*0101, *0301, *0401, *0701, *1101, *1301 and *1501) that are expected to cover over 95% of any given human population [7]. EpiMatrix raw scores were normalized with respect to a score distribution derived from a very large set of randomly generated peptide sequences. The resulting “Z” scores are normally distributed and directly comparable across alleles. Any peptide scoring above 1.64 on the “Z” scale (approximately the top 5% of any given peptide set) has a significant chance of binding to the MHC molecule for which it was predicted. Peptides scoring above 2.32 on the scale (the top 1%) are extremely likely to bind; most well known T-cell epitopes fall within this range of scores.
Vaccinia/variola ORF and epitope homology were evaluated using Conservatrix, a sequence alignment algorithm that searches a dataset for matching segments. Criteria for conservation significance were application dependent and are described in the Results section. Epitopes were evaluated for homology with human sequences using the BLAST algorithm [8]. Clusters with no more than 7 matches per 9-mer frame were selected for further study.
For ORFs conserved across all vaccinia/variola strains, HLA Class II epitopes were further analyzed for clustering and among all other ORFs, extended immunogenic consensus sequences (ICS) were developed. Regions of HLA Class II epitope density were discovered using ClustiMer, an algorithm that uses a statistical function to identify sequences which contain more predicted epitopes than would be found by chance alone [9,10]. Clusters whose sum of significant EpiMatrix Z-scores exceeded a value of 10 after subtracting the expected sum of scores for a random sequence of equal length, were considered for further study. ICS were built by EpiAssembler, an algorithm that maximizes epitope density in a 20-25 amino acid stretch by assembling potentially immunogenic 9-mers to be identically positioned as they are in their native protein sequences [11].
2.2. Peptide Synthesis
Peptides were manufactured using 9-fluoronylmethoxycarbonyl (Fmoc) chemistry by SynPep (Dublin, CA) and by New England Peptide (Gardner, MA). Peptides were purified to >80% as ascertained by analytical reversed phase HPLC. Peptide mass was confirmed by MALDI-TOF mass spectrometry.
2.3. HLA Binding Assays
2.3.1. Class I Assay
Class I A2 and B7 peptides were assayed for HLA binding using a quantitative “sandwich” ELISA, as described previously [12]. HLA class I A2 or B7 molecules were incubated at a concentration of ∼2 nM together with 25 nM human β2 microglobulin (β2m) and an increasing concentration of test peptides at 18°C for 48 h. HLA molecules were then captured on a 96-well plate coated with the pan-specific anti-human MHC class I mouse monoclonal antibody W6/32, and HLA–peptide complexes incubated with horseradish peroxidase-conjugated anti-human β2m conformational-specific polyclonal detection antibody (Dako P0174) and signal enhancer (Dako Envision). Plates were developed by colorimetric reaction and absorbances measured at 450 nm using a Victor2 Multilabel ELISA reader. Based on a standard curve, absorbance measurements were converted to the concentration of HLA–peptide complexes using a standard curve, and plotted against the concentration of test peptide used in the assay. The concentration of peptide required to half-saturate the HLA was determined. At the limiting HLA concentration used, the half-saturation value approximates the equilibrium dissociation constant value (KD).
2.3.2. Class II Assay
Class II HLA binding assays were performed as previously described [13]. Briefly, in 96-well plates, non-biotinylated test peptide at 100 μM competed for binding to purified DR1 (50 nM) against biotinylated influenza hemagglutinin 306-318 standard peptide (0.1 μM) for 24 hours at 37°C. DR1 molecules were then captured on ELISA plates using pan anti-Class II antibodies (L243, anti-HLA-DR), developed by addition of streptavidin-europium and read on a time-resolved fluorescence (TRF) plate reader. Percent inhibition of biotinylated peptide binding was calculated. Peptides that inhibited competitor by >50% were considered DRB1*0101 binders.
2.4. Human PBMC T Cell Assay
2.4.1. Study Subjects
Twenty-two healthy adults, ages 18 to 29 years and vaccinated with Dryvax, were recruited for blood draws at the Saint Louis University Center for Vaccine Development. Donors were vaccinated between two and three years before blood draws. Donor HLA types (Class I and II) were determined using One Lambda Micro SSPTM High Resolution HLA class I and II kits at the Hartford Hospital Transplant Immunology Laboratory. Human subject studies were performed in accordance with NIH regulations and with the approval of the Independent Review Consulting (EpiVax) and Saint Louis University institutional review boards.
2.4.2. PBMC Isolation and Culture
PBMCs were isolated from whole blood by centrifugation over a Ficoll cushion. PBMCs were seeded in 12-well tissue culture plates at 10×106 cells/well and stimulated with pools of Class I or Class II peptides in RPMI supplemented with 10% human AB serum, L-glutamine, gentamicin (Invitrogen), at 37°C under a 5% CO2 atmosphere. 10 U/mL IL-2 and 20 ng/mL IL-7 (R&D Systems) were added to each of the wells. Cells were fed every 2 days by half media replacement containing the same concentration of cytokines. Seven to twenty days post-stimulation, PBMCs were collected and washed in preparation for antigen re-stimulation to measure cytokine secretion measurements by enzyme-linked immunospot (ELISpot) assay.
2.4.3. ELISpot Assay
Interferon-gamma ELISpot assays were performed using kits purchased from Mabtech and performed according to the manufacturer's specifications. Individual target peptides were added at 10 μg/mL to triplicate wells containing 250,000 PBMCs (in RPMI1640 with 10% human AB serum) and incubated for twenty to forty-eight hours at 37°C under a 5% CO2 atmosphere. Triplicate wells were plated with phytohemagglutinin (PHA; 10 μg/mL) and CEF peptide pool (2 μg/mL) as positive controls and six wells with no peptide were used for background determination. Results were recorded by ZellNet Consulting, Inc. using a Zeiss high resolution automated ELISpot reader system and companion KS ELISpot software. In general, responses are considered positive if the number of spots is at least two times background and greater than 20 spots per one million cells over background (1 response over background per 50,000 PBMCs). Results are recorded as the average number of spots over background and adjusted to spots per one million cells seeded.
3. Results
3.1. In silico epitope mapping
3.1.1. Class I HLA
1,472 open reading frames from 4 vaccinia and 3 variola virus genomes were computationally screened for conserved Class I MHC epitopes using EpiMatrix (see Methods for details). First, each protein sequence was parsed into 9-mer and 10-mer sequences, each overlapping the next by 8 or 9 amino acids, respectively, for a total of 369,394 9-mers and 367,922 10-mers. Using Conservatrix to discover unique, identical peptides conserved across all vaccinia and variola strains, we narrowed down the Class I smallpox immunome to 27,158 9-mers and 26,287 10-mers. Each of these peptides was then scored for Class I HLA motif matches to the A*0101, A*0201, A*0301, A*2402, B*0702 and B*4403 alleles. More than 1000 EpiMatrix hits (Z-score > 1.64; top 5% of scores) per allele were discovered (data not shown). The top 100 hits for each allele were subject to a BLAST search against the human genome to exclude epitopes that may be recognized as self (with a cutoff of no more than 7 identities in a 9-mer sequence), and the top 40 A2 and 20 B7 peptides in a list of ascending human homology were selected for experimental validation (Figures 2 and 3).
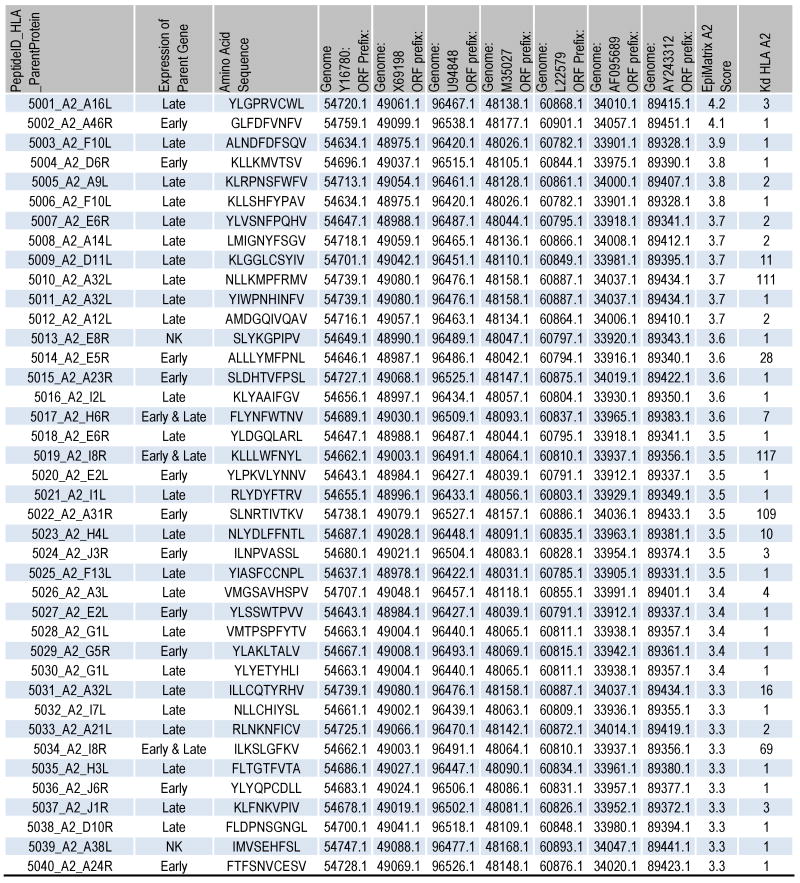
Figure 2.
Characteristics of selected A2 peptides. The peptide IDs and amino acid sequences are shown followed by their gene expression temporality. Here, “early and late” refers to early and late post-replication phase expression. The accession numbers for the corresponding ORFs within 3 variola and 4 vaccinia genomes are listed. In addition, the EpiMatrix A2 Z-score and KD (nM) are presented.
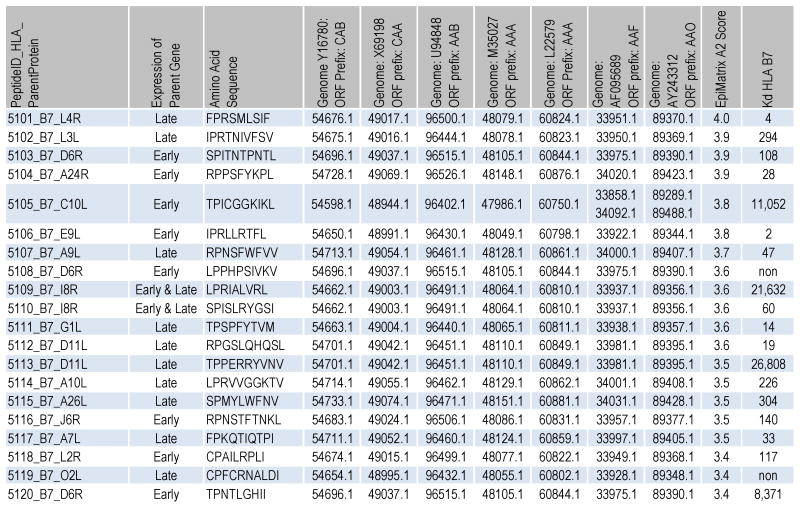
Figure 3.
Characteristics of selected B7 peptides. The peptide IDs and amino acid sequences are shown followed by their gene expression temporality. Here, “early and late” refers to early and late post-replication phase expression. The accession numbers for the corresponding ORFs within 3 variola and 4 vaccinia genomes are listed. In addition, the EpiMatrix HLA B7 Z-score and KD (nM) are shown.
3.1.2. Class II HLA
Two strategies to identify conserved and immunogenic Class II MHC epitopes were pursued to maximize the likelihood of discovering protective vaccine immunogens. First, an ORF-by-ORF sequence comparison was performed with the Copenhagen vaccinia strain selected as the standard for alignment because it contains the most ORFs of all the strains under consideration. 107 of the 262 ORFs in Vaccinia Copenhagen had matching ORFs in all six alternate strains with at least 80% identity within the first 200 amino acids. These ORFs in Vaccinia Copenhagen were computationally screened using EpiMatrix and ClustiMer to identify epitope dense regions containing sequences predicted to bind multiple Class II HLA alleles (DRB1*0101, *0301, *0401, *0701, *1101, *1301 and *1501). 272 epitope clusters were identified, each bearing at least 90% sequence identity across all seven strains and a cluster score of 15 or above. The sequences were then analyzed by the BLAST algorithm for human homology. Epitope clusters were ranked first by lowest human homology with no more than 7 matches in a 9-mer frame accepted, and then by cluster score. The top 24 epitope clusters were selected for in vitro confirmation (Figure 4). In addition, a 25th cluster was selected for maximal potential immunogenicity, regardless of human homology.
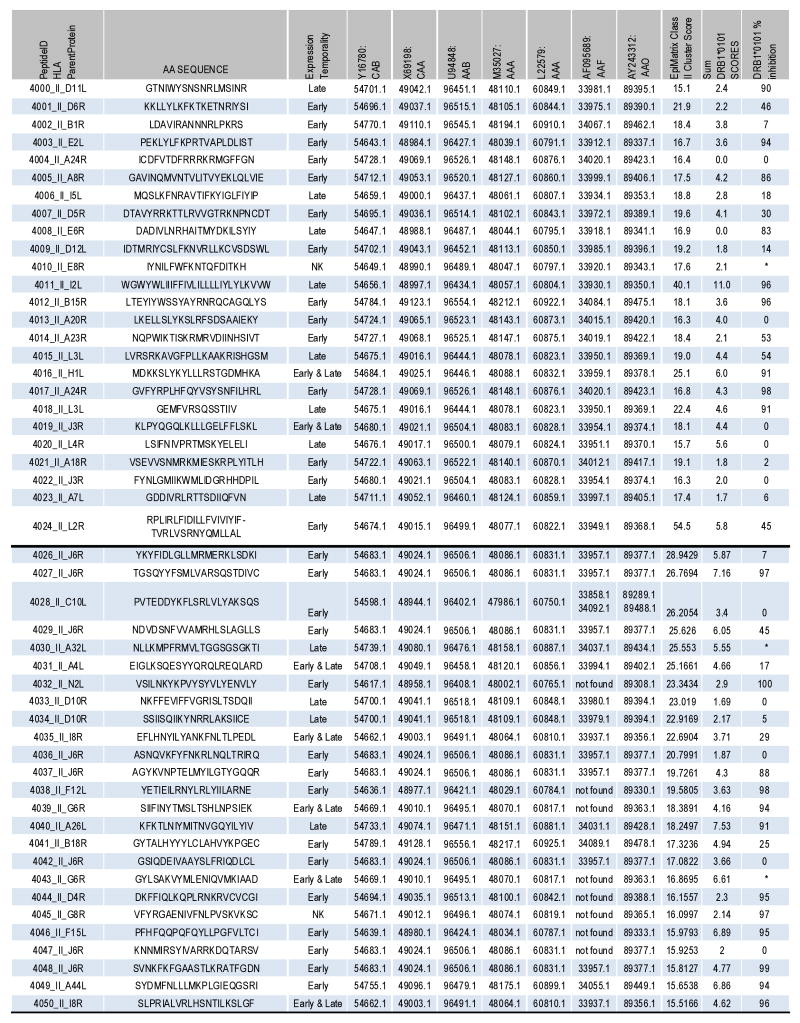
Figure 4.
Characteristics of selected Class II peptides. The peptide IDs and amino acid sequences are shown followed by their gene expression temporality. Here “early and late” refers to early and late post-replication phase expression. The accession numbers for the corresponding ORFs within 3 variola and 4 vaccinia genomes are listed. The top 25 epitopes were discovered in the first Class II epitope screen and the bottom 25 in the second (see Results). In addition, the EpiMatrix Class II cluster score, the cluster sum of DRB1*0101 scores and % inhibition of competitor peptide binding to DRB1*0101 at 100 μM epitope peptide are shown. Asterisks refer to peptides that were not assayed for binding.
In a second, separate computational screen, ORFs excluded from the investigation above were analyzed using Conservatrix to find identical 9-mers in at least six strains, where minimally three were vaccinia-derived and two variola. 5,781 peptides were discovered, each then scored for binding affinity to a panel of 8 HLA Class II alleles (see above) using EpiMatrix. 786 unique 9-mers were EpiMatrix hits and subsequently input into the EpiAssembler algorithm to identify sets of overlapping, conserved and promiscuous epitopes, termed immunogenic consensus sequence” (ICS) T helper epitopes. 74 ICS with cluster scores greater than 15 were identified and analyzed for human homology using BLAST. Epitope clusters were ranked first by lowest human homology, as above, and then by cluster score. The top 25 ICS were selected for in vitro validation (Figure 4).
3.2. In vitro validation of computational predictions
3.2.1. Class I HLA binding assay
EpiMatrix-predicted epitopes were assessed for their HLA binding potential in binding assays using soluble HLA. Affinities of HLA*A2 and *B7 epitope peptides for their respective HLA were assessed. We found that 100% of the 40 selected A2 epitopes identified by EpiMatrix bound A2 (Figures 2 and 5). 31 bound with very high affinity (1-5 nM KD), 4 with high affinity (6-25 nM KD) and 5 at moderate affinity (26-500 nM KD). Of 20 B7 peptides assayed, 14 (70%) bound B7, 2 with very high affinity, 2 with high affinity, and 10 with moderate affinity (Figures 3 and 5).
Figure 5.
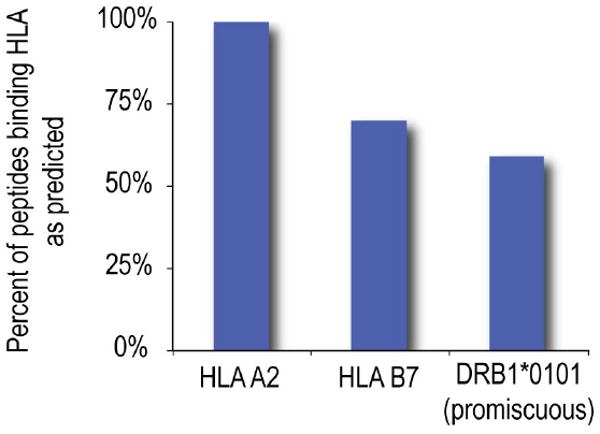
In vitro validation of computationally identified epitopes. Predicted epitopes were assayed for binding to individual HLA alleles, A2, B7 and DRB1*0101. The percent of predicted epitopes that bound these alleles is shown. Class II epitopes were selected for predicted binding to multiple HLA alleles (see Methods), not solely to DRB1*0101.
3.2.2. Class II HLA binding assay
Class II epitopes, at a peptide concentration of 100 μM, were screened for binding HLA DRB1*0101 in a competition binding assay using soluble HLA. Percent inhibition of competitor peptide was used to estimate test peptide affinity. 21 of 50 peptides bound with high affinity (75%-100% inhibition), 2 peptides with moderate (50%-75% inhibition) and 5 peptides with weak affinity (30%-50%). In total, 28/50 (56%) of peptides tested bound DR1, as expected of a set of sequences that were predicted to cover a HLA diverse population, not only DRB1*0101 carriers.
3.3. Ex vivo validation of computational predictions
We validated EpiMatrix-predicted epitopes in measurements of antigen-specific T-cell responses in 22 human subjects, ages 18-29 years old, who received Dryvax 2 to 3 years before blood draw. PBMCs were stimulated with pools of smallpox epitopes for 7-20 days and re-stimulated with individual epitopes and epitope pools in an IFNγ ELISpot assay for 20-48 hours. Response frequency among subjects ranged from 10 to 100% for individual Class II epitopes. Responses were observed to 41 of 50 (82%) Class II epitopes, with an average of 36% positive responses per subject (Figure 6 and Figure 7, top). All subjects exhibited a robust response to pooled Class II peptides. Responses plotted according to gene expression temporality [14,15] reveal no preponderance of T-cell reactivity in a single group of antigens.
Figure 6.
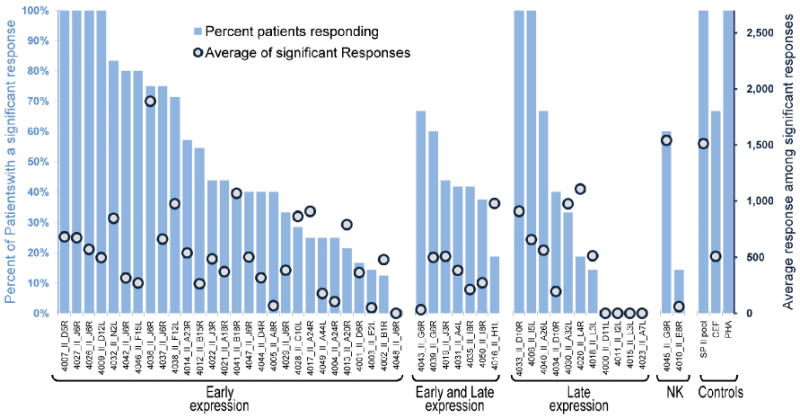
Ex vivo validation of computationally identified Class II HLA epitopes. Predicted epitopes were assayed for T cell reactivity by IFN-γ ELISpot assay using PBMCs isolated from Dryvax-vaccinated donors. Epitopes are grouped according to their timing of expression as parts of whole proteins in the poxvirus life cycle. The “early and late” designation refers to early and post-replication late expression. The percentage of subjects who responded to individual epitopes in descending order (bars) and the average spot forming cells per million PBMCs for each epitope (circles) are illustrated.
Figure 7.
IFN-γ ELISpot responses to predicted Class I and Class II epitopes in Dryvax vaccinated human subjects. The numbers of spot forming cells (over background) per million PBMCs that secrete IFN-γ in response to individual and pooled Class I and Class II epitopes, as well as PHA and CEF are presented. For simplicity, non-significant results are denoted by n/s and missing data are omitted (not tested, NT). Significant results are highlighted in black. An ELISpot response was considered positive if two criteria were met: (1) spot-forming cells per million PBMC were at least 20 over background; (2) spot-forming cells (SFC) per million PBMC were at least two-fold over background. Column headers: human subject ID code, average responses and percent of subjects responding. Row labels: peptide ID. Epitopes are grouped according to their timing of expression as parts of whole proteins in the poxvirus life cycle.
For Class I epitopes, antigenicity was detected for 17 of 40 (43%) HLA-A*0201 (Figure 7, bottom left) and 5 of 20 (25%) B*0702 epitopes (Figure 7, bottom right). Per subject, responses to A2 epitopes averaged 7% and to B7 epitopes 10%.
4. Discussion
Using immunoinformatics methods, we scanned vaccinia- and variola-conserved sequences for HLA Class I and Class II epitopes and then validated the predictions in HLA binding assays and in humans vaccinated with Dryvax. Our approach to epitope prediction was to discover the intersection of vaccinia and variola genome sequences that give rise to CD8 and CD4 T cell-mediated protection against variola as conferred by vaccination with vaccinia. CD8 responses play an important role in containing orthopoxvirus infections and may be a critical correlate of protection after re-exposure [16,17]. CD4 responses are critical for robust CD8 T cell proliferation and function and for their differentiation into memory cells in vaccinia infection [18,19]. Moreover, CD4 responses provide required help to B cells to produce antibodies that are necessary and sufficient to protect against orthopoxvirus challenge [20]. Hence, we set out to discover potential HLA Class I and Class II variola/vaccinia protective determinants using computational methods as a high throughput method for scanning large orthopoxvirus genomes without bias to time of expression or protein function.
4.1. CD4+ T-cell epitopes
We identified 50 HLA Class II epitopes conserved in vaccinia and variola genomes. Half were predicted with a requirement that all epitopes be conserved in all genomes analyzed and the other half with the more relaxed requirement that sequences be conserved in at least 3 vaccinia and 2 variola genomes only out of a total of seven genomes analyzed. We discovered that >80% of the epitopes were antigenic, with multiple responses observed in all Dryvax vaccinees tested, illustrating the effectiveness of a predictive approach. Unexpectedly, we found that, with the exception of the Tian Tian strain, the ICS sequences were more highly conserved, contained more EpiMatrix hits and produced greater numbers of spot forming cells in interferon-gamma ELISpot assays than the sequences derived from the more traditional alignment based-approach. Higher ELISpot numbers may be attributed to the relatively higher concentration of high scoring 9-mers in the ICS epitopes, to greater sequence conservation or to both factors.
In two related studies, Koelle and co-workers used a non-predictive, experimental screen to identify CD4+ T cell-antigenic open reading frames and/or protein fragments in the vaccinia proteome [21,22]. As in those studies, we observed a broad CD4+ T cell response to vaccinia antigens expressed both early and late in infection, with a majority of the responses to proteins expressed at the early stage. Because the predictive approach triages sequences for ex vivo validation, only 29 open reading frames are represented by the selected Class II promiscuous epitopes in this study. Among these 29, 23 (79%) were observed by Koelle and co-workers to be antigenic. Considering that the predictive approach significantly limits the part of an open reading frame that is used to measure antigenicity, this finding illustrates the advantage immunoinformatics provides for rapidly identifying antigenic open reading frames and focusing in on a segment that is immunoreactive. Notably, only one of these open reading frames (L4R) is recognized by 100% of subjects in the non-predictive study, whereas here, 3 of 16 subjects (19%) responded to 4020_II_L4R. While the predictive method is useful for selecting immunogenic proteins, it is important to note that a single epitope from a highly immunogenic protein will not necessarily be recognized by all individuals tested. Differences in the frequency of recognition of a single epitope versus the large protein sequences evaluated by Koelle and co-workers are possibly due to the limited number of L4R epitope sequences assayed in the present study, the limited number of subjects in the studies (5 in [21], 12 in [22], 16 here), the HLA types of the subjects, or differences in the methods used for measuring antigenicity (proliferation vs. ELISpot).
4.2. CD8+ T-cell epitopes
We selected 40 A2 and 20 B7 supertype epitopes conserved in the vaccinia and variola genomes using immune-informatics methods and evaluated them for T cell reactivity ex vivo. Of the 60 epitopes tested, 17 A2 epitopes and 5 B7 epitopes elicited IFN-gamma responses in ex vivo ELISpot; thus, one in three predicted sequences was confirmed. Only one of these 22 Class I restricted antigenic epitopes, 5019_A2_I8R, has been reported beforehand [23]. This is especially noteworthy as several published reports identify A2 vaccinia epitopes [23,24,25,26]. Particularly, Sette and co-workers performed an extensive study of HLA Class I restricted vaccinia responses using immuno-informatics methods [24]. Starting with a list of approximately 2000 A2 and B7 supertype epitopes, 14 A2 and 5 B7 epitopes were found to be antigenic in Dryvax vaccinees, a ratio of roughly one in 100. None of these epitopes are identical to the sequences reported here, although two are found in common open reading frames (E2L, A2; J6R, B7). In a study similar in design, Kazura and co-workers reported 6 new A2 epitopes reactive in persons vaccinated against smallpox. Two are common to protein antigens reported here (G1L, I8R) and one is identical in sequence, as mentioned above. Factors that may have contributed to a lack of concordance between these studies include the different epitope prediction tools used by each group and the limited numbers of subjects sampled. Nevertheless, like the previously published studies, the breadth of vaccinia-induced immune response is shown here by ex vivo responses for epitopes derived from 18 different open reading frames. In addition, we note that the number of epitopes recognized in this study is far fewer than we observed in previous epitope mapping studies of HIV using EpiMatrix [27]. This may be a natural outcome of the larger size of the genome, compared to the genome of HIV, which gives rise to more complex CD8+ T-cell epitope hierarchies [28]. Alternatively, class I epitopes may not be as critical for protection from poxviruses as previously believed. We plan to evaluate the relative contributions of class I and class II epitopes to protection from vaccinia in a challenge study.
4.3. Future studies
The broader goal of this study is to identify epitopes for incorporation into a new smallpox vaccine that is safer than previously licensed smallpox vaccines. The use of epitopes overcomes potential safety concerns associated with vaccinating with live vaccinia virus. In addition, multiple epitopes derived from more than one antigen can be packaged into a relatively small delivery vehicle. Furthermore, epitope-based vaccines appear to be capable of inducing more potent responses than whole protein vaccines [29], and they sidestep the propensity for the immune system to focus on a single immunodominant epitope by simultaneously targeting multiple dominant and subdominant epitopes [30,31]. This latter feature is particularly significant because of the great breadth of the antiviral response [21,22,32]. It should be noted that the field of immuno-informatics is new, and few epitope-driven vaccines for infectious pathogens have reached the stage of efficacy trials in humans, although several have been shown to be effective in animal models.
In summary, we are in the process of developing an epitope-based vaccine based on intersecting sets of epitopes derived from the variola and the vaccinia genomes; this article describes our progress along the pathway to that goal. We believe that our immunoinformatics-driven smallpox vaccine development approach may have several advantages over other approaches: (1) rapidity (the mapping of epitopes and confirmation using human PBMC was accomplished in less than 18 months); (2) safety (the entire protein is not used, thus the recipient is not exposed to live vaccinia virus, which may be associated with side effects); and (3) broad immunogenicity (delivery of multiple epitopes, derived from multiple proteins, recognized in the context of many different MHC). We also believe that the methods described here, which will lead to the development of a multi-epitope smallpox vaccine, may be a step in the right direction for the development of a range of safer, more effective biodefense vaccines.
Acknowledgments
Supported by NIH R43AI058376 (ADG).
Footnotes
Disclosures: Two of the contributing authors, Anne S. De Groot and William D. Martin are senior officers and majority shareholders at EpiVax, Inc., a privately-owned vaccine design company located in Providence, RI. Dr. De Groot is also a faculty member at the University of Rhode Island and Brown University. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosenthal SR, Merchlinsky M, Kleppinger C, Goldenthal KL. Developing new smallpox vaccines. Emerg Infect Dis. 2001 Nov-Dec;7(6):920–6. doi: 10.3201/eid0706.010602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemper AR, Davis MM, Freed GL. Expected adverse events in a mass smallpox vaccination campaign. Eff Clin Pract. 2002 Mar-Apr;5(2):84–90. [PubMed] [Google Scholar]
- 3.Schafer JR, Jesdale BM, George JA, Kouttab NM, De Groot AS. Prediction of well-conserved HIV-1 ligands using a matrix-based algorithm, EpiMatrix. Vaccine. 1998;16:1880–4. doi: 10.1016/s0264-410x(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 4.De Groot AS, Jesdale BM, Szu E, Schafer JR. An interactive web site providing MHC ligand predictions: Application to HIV research. AIDS Res Hum Retroviruses. 1997;13:539–541. doi: 10.1089/aid.1997.13.529. [DOI] [PubMed] [Google Scholar]
- 5.De Groot AS, Rayner J, Martin W. Modeling the immunogenicity of the therapeutic proteins using T cell epitope mapping. In: Brown F, Suis AM, editors. Immunogenicity of therapeutic biological products Developments in biologicals. Vol. 112. Basel: Karger; 2003. pp. 71–80. [PubMed] [Google Scholar]
- 6.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and –B polymorphism. Immunogenetics. 1999;50(November 3–4):201–12. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 7.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–73. [PubMed] [Google Scholar]
- 8.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Meister GE, Roberts CG, Berzofsky JA, De Groot AS. Two novel T cell epitope prediction algorithms based on MHC-binding motifs; comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine. 1995;13:581–91. doi: 10.1016/0264-410x(94)00014-e. [DOI] [PubMed] [Google Scholar]
- 10.De Groot AS, Berzofsky JA. From genome to vaccine—new immunoinformatics tools for vaccine design. Methods. 2004;34:425–8. doi: 10.1016/j.ymeth.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 11.De Groot AS, Marcon L, Bishop EA, Rivera D, Kutzler M, Weiner DB, Martin W. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine. 2005 Mar 18;23(1718):2136–48. doi: 10.1016/j.vaccine.2005.01.097. [DOI] [PubMed] [Google Scholar]
- 12.Sylvester-Hvid C, Kristensen N, Blicher T, Ferre H, Lauemoller SL, Wolf XA, Lamberth K, Nissen MH, Pedersen LO, Buus S. Establishment of a quantitative ELISA capable of determining peptide - MHC class I interaction. Tissue Antigens. 2002 Apr;59(4):251–258. doi: 10.1034/j.1399-0039.2002.590402.x. [DOI] [PubMed] [Google Scholar]
- 13.Reijonen H, Kwok WW. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods. 2003 Mar;29(3):282–8. doi: 10.1016/s1046-2023(02)00350-x. [DOI] [PubMed] [Google Scholar]
- 14.Assarsson E, Greenbaum JA, Sundström M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):2140–5. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt JF, Stunnenberg HG. Sequence and transcriptional analysis of the vaccinia virus HindIII I fragment. J Virol. 1988 Jun;62(6):1889–97. doi: 10.1128/jvi.62.6.1889-1897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buller RM, Palumbo GJ. Poxvirus pathogenesis. Microbiol Rev. 1991 Mar;55(1):80–122. doi: 10.1128/mr.55.1.80-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulginiti VA, Papier A, Lane JM, Neff JM, Henderson DA. Smallpox vaccination: a review, part II. Adverse events. Clin Infect Dis. 2003;37:251–271. doi: 10.1086/375825. [DOI] [PubMed] [Google Scholar]
- 18.Bevan MJ. Helping the CD8+ T-cell response. Nat Rev Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 19.Bachmann MF, Wolint P, Schwarz K, Oxenius A. Recall proliferation potential of memory CD8+ T cells and antiviral protection. J Immunol. 2005;175:4677–4685. doi: 10.4049/jimmunol.175.7.4677. [DOI] [PubMed] [Google Scholar]
- 20.Edghill-Smith Y, Golding H, Manischewitz J, King LR, Scott D, Bray M, Nalca A, Hooper JW, Whitehouse CA, Schmitz JE, et al. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005;11:740–747. doi: 10.1038/nm1261. [DOI] [PubMed] [Google Scholar]
- 21.Jing L, Chong TM, Byrd B, McClurkan CL, Huang J, Story BT, Dunkley KM, Aldaz-Carroll L, Eisenberg RJ, Cohen GH, Kwok WW, Sette A, Koelle DM. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. J Immunol. 2007 May 15;178(10):6374–86. doi: 10.4049/jimmunol.178.10.6374. [DOI] [PubMed] [Google Scholar]
- 22.Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol. 2008 Jul;82(14):7120–34. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrout ND, McHugh MM, Tisch DJ, Moormann AM, Brusic V, Kazura JW. Long-term T cell memory to human leucocyte antigen-A2 supertype epitopes in humans vaccinated against smallpox. Clin Exp Immunol. 2007 Aug;149(2):265–73. doi: 10.1111/j.1365-2249.2007.03401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci U S A. 2005 Sep 27;102(39):13980–5. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drexler I, Staib C, Kastenmuller W, Stevanović S, Schmidt B, Lemonnier FA, Rammensee HG, Busch DH, Bernhard H, Erfle V, Sutter G. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc Natl Acad Sci U S A. 2003 Jan 7;100(1):217–22. doi: 10.1073/pnas.262668999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidney J, Grey HM, Kubo RT, Sette A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol Today. 1996 Jun;17(6):261–6. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 27.Identification of immunogenic HLA-B7 “Achilles' heel” epitopes within highly conserved regions of HIV. De Groot AS, Rivera DS, McMurry JA, Buus S, Martin W. Vaccine. 2008 Jun 6;26(24):3059–71. doi: 10.1016/j.vaccine.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kastenmuller W, Gasteiger G, Gronau JH, Baier R, Ljapoci R, Busch DH, Drexler I. Cross-competition of CD8+ T cells shapes the immunodominance hierarchy during boost vaccination. J Exp Med. 2007 Sep 3;204(9):2187–98. doi: 10.1084/jem.20070489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishioka GY, Fikes J, Hermanson G, Livingston B, Crimi C, Qin MS, del Guercio MF, Oseroff C, Dahlberg C, Alexander J, et al. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J Immunol. 1999;162:3915. [PubMed] [Google Scholar]
- 30.Oukka M, Manuguerra JC, Livaditis N, Tourdot S, Riche N, Vergnon I, Cordopatis P, Kosmatopoulos K. Protection against lethal viral infection by vaccination with nonimmunodominant peptides. J Immunol. 1996;157:3039. [PubMed] [Google Scholar]
- 31.Tourdot S, Oukka M, Manuguerra JC, Magafa V, Vergnon I, Riche N, Bruley-Rosset M, Cordopatis P, Kosmatopoulos K. Chimeric peptides:a new approach to enhancing the immunogenicity of peptides with low MHC class I affinity: application in antiviral vaccination. J Immunol. 1997;159:2391. [PubMed] [Google Scholar]
- 32.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006 Jul;24(7):817–9. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]