Abstract
Cancer cells preferentially metabolize glucose by aerobic glycolysis, characterized by increased lactate production. This distinctive metabolism involves expression of the embryonic M2 isozyme of pyruvate kinase, in contrast to the M1 isozyme normally expressed in differentiated cells, and it confers a proliferative advantage to tumor cells. The M1 and M2 pyruvate-kinase isozymes are expressed from a single gene through alternative splicing of a pair of mutually exclusive exons. We measured the expression of M1 and M2 mRNA and protein isoforms in mouse tissues, tumor cell lines, and during terminal differentiation of muscle cells, and show that alternative splicing regulation is sufficient to account for the levels of expressed protein isoforms. We further show that the M1-specific exon is actively repressed in cancer-cell lines—although some M1 mRNA is expressed in cell lines derived from brain tumors—and demonstrate that the related splicing repressors hnRNP A1 and A2, as well as the polypyrimidine-tract-binding protein PTB, contribute to this control. Downregulation of these splicing repressors in cancer-cell lines using shRNAs rescues M1 isoform expression and decreases the extent of lactate production. These findings extend the links between alternative splicing and cancer, and begin to define some of the factors responsible for the switch to aerobic glycolysis.
Keywords: aerobic glycolysis, cancer, RNA splicing
Cancer cells exhibit a metabolic phenotype characterized by increased glycolysis with lactate generation, regardless of oxygen availability—a phenomenon termed the Warburg effect. Recent work demonstrated that expression of the type II isoform of the pyruvate-kinase-M gene (PKM2, referred to here as PK-M) is a critical determinant of this metabolic phenotype, and confers a selective proliferative advantage to tumor cells in vivo (1). This finding adds to the growing body of evidence that alterations in alternative prem-RNA splicing play important roles in different aspects of cancer progression (2, 3).
Pyruvate-kinase (PK) is the enzyme that catalyzes the final step in glycolysis, generating pyruvate and ATP from phosphoenolpyruvate and ADP (4). The resulting pyruvate can be converted to lactate or it can be incorporated into the tricarboxylic acid (TCA) cycle to drive oxidative phosphorylation. PK is encoded by two paralogous genes, each of which is alternatively spliced, such that four PK isoforms are expressed in mammals. The L and R isozymes, derived from the PKLR gene, show tissue-specific expression in the liver and red-blood cells, respectively. They have different first exons, defined by tissue-specific promoters (5). The PKM2 (PK-M) gene consists of 12 exons, of which exons 9 and 10 are alternatively spliced in a mutually exclusive fashion to give rise to the PK-M1 and PK-M2 isoforms, respectively (6). Exons 9 and 10 each encode a 56 amino acid variable segment that confers distinctive properties to the regulation and activity of PK-M1 and PK-M2 enzymes; as a result, PK-M1 is constitutively active, whereas the activity of PK-M2 is allosterically regulated by fructose-1,6-bisphosphate levels and interaction with tyrosine-phosphorylated proteins (1).
The ability of PK-M2 activity to be regulated is thought to provide a mechanism for cells to control the availability of metabolites for anabolic processes, and to confer a proliferative advantage during tumorigenesis (7). Despite increasing evidence demonstrating the significance of PK-M2 isoform expression in cancer-cell metabolism and tumorigenesis, the mechanisms governing alternative splicing of the PK-M gene are not understood. PK-M2 is expressed in a range of cancer cells, as well as in fetal and undifferentiated adult tissues, whereas PK-M1 is expressed predominantly in terminally differentiated tissues (1, 8).
Alternative splicing involving pairs of mutually exclusive exons represents only ∼2% of all alternative splicing events in human genes (9). In terms of gene structure, the regulated exons in such genes are often closely related in sequence—as in the case of PK-M exons 9 and 10—indicating that they originally arose by exon duplication (10). Well characterized examples of this alternative splicing pattern in mammals include the tropomyosins, fibroblast growth factor receptor 2, and α-actinin [reviewed in (11)]. Although some of the trans-acting factors involved in the recognition of individual mutually exclusive exons in these genes have been identified, it remains unclear how these pairs of exons are coordinately regulated in a way that maintains their mutually exclusive properties (11).
We have begun to dissect the molecular mechanisms underlying PK-M2 alternative splicing regulation. Previous work implicated the splicing-repressor paralogs PTB and nPTB in the repression of exon 9 (12). In addition, other splicing repressors, such as hnRNP A1 and hnRNP A2, and activators, such as SF2/ASF, have been implicated in oncogenic transformation (2, 3), and hence could potentially play a role in PK-M alternative splicing. Here we characterize the expression of PK-M isoforms at the mRNA and protein level in primary tissues and cancer-cell lines, as well as during terminal differentiation of muscle cells in culture. We show that hnRNP A1/A2, in addition to PTB, repress the use of exon 9, such that knocking down expression of these splicing repressors allows expression of PK-M1, accompanied by a decrease in lactate production.
Results
Relative PK-M1 and PK-M2 Expression in Tissues and Cell Lines Correlates with hnRNP A1/A2 and PTB Expression.
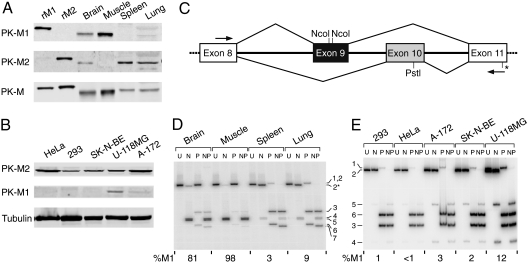
Proliferating cells and cancer cells preferentially express PK-M2 over PK-M1 at the protein level (1, 13). To determine if the expressed protein isozymes are a direct reflection of differences in alternative splicing—as opposed to, e.g., being affected by mRNA stability or translational control—we measured the levels of mRNA and protein isoforms expressed in various tissues and cell lines. Representative organs/tissues isolated from adult mice were perfused with saline, and total protein and RNA were isolated and analyzed. The relative expression of PK-M1 and PK-M2 protein isoforms was organ/tissue-specific: Brain and skeletal muscle preferentially expressed PK-M1, whereas spleen and lung expressed mainly PK-M2, as detected by Western blotting with isoform-specific antibodies (Fig. 1A). As previously reported (1), several human cell lines expressed PK-M2 protein with no detectable PK-M1 (Fig. 1B). Among the cell lines tested, however, two brain-tumor-derived cell lines, namely U-118MG and A-172 glioblastoma cells, expressed detectable levels of PK-M1 protein, in addition to PK-M2, reminiscent of the protein pattern in mouse brain, where both isoforms are also readily detectable (cf. Fig. 1B, Fig. 1A).
Fig. 1.
Protein and transcript expression patterns of pyruvate kinase M1/M2 isoforms in cells and tissues. (A) Total adult-mouse organ/tissue homogenates were used for Western blotting with the indicated antibodies. rM1 and rM2: Flag-tagged purified recombinant human PK isoforms. (B) Total cell lysates of five human cancer-cell lines were used for Western blotting with the indicated antibodies. (C) Primers annealing to exon 8 and exon 11, respectively, were used to amplify mouse or human PK-M transcripts. The alternative exons that encode the distinctive segments of PK-M1 and PK-M2 are indicated in (black) and (gray), respectively. To distinguish between PK-M1 (exon 9 included) and PK-M2 (exon 10 included) isoforms, the PCR products were cleaved with NcoI, PstI, or both. There is an additional NcoI site (*) 11 bp away from the 3′ end of mouse exon 11. (D) Mouse organs were freshly dissected and perfused with saline. Total RNA was analyzed by radioactive RT-PCR followed by digestion with NcoI (N), PstI (P), or both enzymes (NP), plus an uncut control (U). Numbered bands are as follows: 1: Uncut M1 (502 bp); 2: uncut M2 (502 bp); 2*: M2 cleaved with NcoI in exon 11 (491 bp); 3: Pst1-cleaved M2 5’ fragment (286 bp); 4: NcoI-cleaved M1 5′ fragment (245 bp); 5: NcoI-cleaved M1 3’ fragment (240 bp); 6: PstI-cleaved M2 3’ fragment (216 bp); 7: PstI + NcoI-cleaved M2 3’ fragment (205 bp). The %M1 was quantified from band 1 (M1) and bands 3 and 6 (M2) in each P lane. (E) RT-PCR and restriction digest analysis of total RNA from the indicated human cell lines. The bands are numbered as for the mouse RT-PCR products, but the sizes are different because of the positions of the primers; the sizes are as follows: 1: 398 bp; 2: 398 bp; 3: 185 bp; 4: 144 bp; 5: 248 bp; 6: 213 bp. Note that the PK-M1 bands in the P and U lanes migrate slightly above the PK-M2 bands, which is also the case for the mouse PK-M1 transcripts.
To accurately measure the relative levels of PK alternatively spliced mRNA isoforms, we simultaneously detected both isoforms by radioactive RT-PCR with a single pair of primers corresponding to the flanking constitutive exons 8 and 11. Because exons 9 and 10 are identical in length (167 nt), the resulting cDNA amplicons were digested with restriction enzymes that cleave either exon 9 or 10 to distinguish the two isoforms (Fig. 1C; see also (14)). PK-M1 mRNA was the predominant isoform in striated muscle and brain—tissues that are enriched in terminally differentiated, nonproliferating cells. In contrast, PK-M2 mRNA was the major isoform in lung and spleen, presumably reflecting the abundance of proliferating cells in these organs. PK-M2 mRNA was invariably the major isoform in all cancer or transformed cell lines we assayed, consistent with the previous finding that PK-M2 expression at the protein level is strongly correlated with, and facilitates, proliferation and tumorigenesis (1)). However, PK-M1 mRNA was readily detectable in brain-tumor-derived cancer-cell lines, including glioblastoma (U-118MG and A-172) and neuroblastoma (SK-N-BE) cell lines. This finding is consistent with PK-M1 protein being detectable in U-118MG cells and to a lesser extent in A-172 cells (Fig. 1B). In general, we observed a strong correlation between PK-M1/M2 isoform ratios measured at the protein and mRNA levels, in both tissues and cell lines.
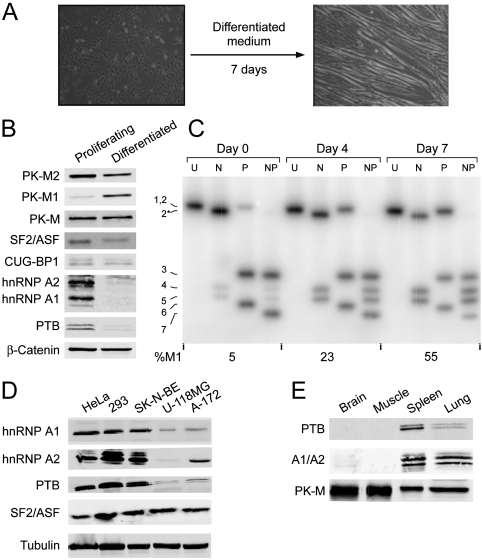
To facilitate studies of the mechanism underlying the PK-M2 to PK-M1 isoform switch during terminal differentiation, we examined several established systems for cell differentiation in culture. We found that proliferating mouse C2C12 myoblasts induced to terminally differentiate into myotubes (15) switched protein and mRNA isoform expression from almost exclusively PK-M2 to predominantly PK-M1 (Fig. 2). Consistent with PK-M2 being the predominant isoform in proliferating cells, and PK-M1 being the predominant isoform in muscle (Fig. 1A), this switch in PK-M isoform accompanied the morphological differentiation of the C2C12 myoblasts into myotubes (Fig. 2A). Normalizing to total PK-M protein expression, it is readily apparent that there was a pronounced increase in PK-M1 upon differentiation, at the expense of the PK-M2 isoform (Fig. 2B). Likewise, at the mRNA level, the proportion of the M1 isoform changed from 5% to 55% over a differentiation time course (Fig. 2C).
Fig. 2.
Expression of pyruvate-kinase isoforms and selected splicing factors in cell lines and tissues. (A) Differentiation of mouse C2C12 myoblasts into myotubes. The left field shows proliferating myoblasts, and the right field shows cells after seven days in differentiation medium, when most of the cells have fused into myotubes. (B) Western blot of proliferating myoblasts versus AraC-treated myotubes with the indicated antibodies against selected splicing factors, β-catenin, total PK-M, and PK-M1 or PK-M2. (C) Radioactive RT-PCR analysis of PK-M1 and PK-M2 expression in C2C12 cells over a differentiation time course. Bands are numbered as in Fig. 1D. (D) Total cell lysates of five human-tumor or transformed cell lines were used for Western blotting with the indicated specific antibodies. Tubulin was used as an internal control for loading. HeLa (cervical carcinoma); HEK293 (transformed embryonic kidney cells); SK-NB-E (neuroblastoma); U-118MG (glioma); A-172 (glioblastoma). (E) Mouse tissues were analyzed as in Fig. 1A, with the indicated antibodies.
We also measured the protein levels of representative alternative splicing factors, including some known to have oncogenic activities (3). In the C2C12 differentiation model, as well as in mouse or human tissues and cancer-cell lines, we observed a correlation between high levels of the alternative splicing factors hnRNP A1/A2 and PTB, and reduced expression of the PK-M1 isoform (Fig. 2B, D, and E). In contrast, there was little or no change in several other splicing factors, such as SF2/ASF and CUG-BP1 (Fig. 2B). U-118MG and A-172 cells, which expressed detectable levels of PK-M1, both at mRNA and protein levels (Fig. 1B, E), had less hnRNP A1/A2 and PTB compared to HeLa, HEK293, and SK-N-BE cells, whereas SF2/ASF was expressed at similar levels in these cell lines (Fig. 2D). Both hnRNP A/B and PTB (also known as hnRNP I) protein family members are well characterized splicing repressors, which led us to hypothesize that these factors might be partly responsible for repressing the use of exon 9 during prem-RNA splicing.
Blocking the 3′ Splice Site of Exon 10 Causes Abnormal Skipping of Both Exons 9 and 10.
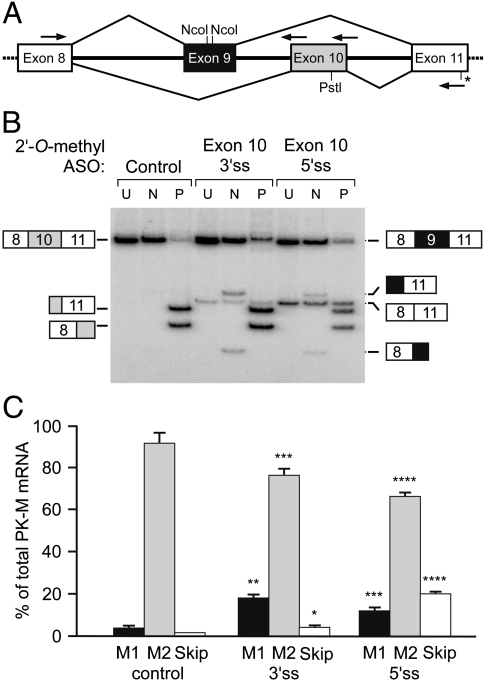
To determine if exon 9 is actively repressed in cancer-cell lines, or simply fails to compete effectively with exon 10, we blocked the 3′ or 5′ splice sites of exon 10 using 2′-O-methyl, phosphorothioate antisense oligonucleotides complementary to these regions (16) (Fig. 3A). The oligonucleotides were transfected into HEK293 cells and the endogenous PK mRNA isoforms were analyzed by radioactive RT-PCR (Fig. 3B). As expected, use of exon 10 was partially inhibited by the antisense oligonucleotides. However, in addition to increased use of exon 9 (PK-M1 isoform), we observed an abnormal mRNA arising from skipping of both mutually exclusive exons. These results indicate that even with reduced use of exon 10, there is residual repression of exon 9 use.
Fig. 3.
Exon 9 is partially rescued in HEK293 cells when exon 10 is blocked. (A) Schematic representation of the strategy used to block exon 10 with 2′-O-methyl antisense oligonucleotides. The two arrows above exon 10 denote the oligonucleotides complementary to the 3′ splice site and 5′ splice site regions. (B) Radioactive RT-PCR assay to measure PK-M1/PK-M2 levels after blocking each of the exon 10 splice sites with antisense oligonucleotides. An abnormal isoform arising from skipping of both exons 9 and 10 is indicated. (C) Quantitation of multiple experiments. (Error bars show s.d.; n = 3; p-values: = ∗0.007; = ∗∗0.005; = ∗∗∗0.004; = ∗∗∗∗0.001; Student’s paired t-test).
hnRNP Proteins Repress Exon 9 in a Gliobloastoma Cell Line.
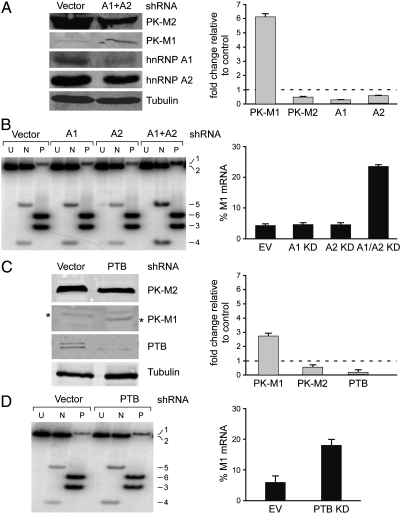
To address whether specific hnRNP proteins are responsible for repression of exon 9, we generated stable cell lines expressing shRNAs directed against hnRNP A1, A2, or PTB (Fig. 4). We chose A-172 glioblastoma cells for this analysis, both because they already express some PK-M1 (Fig. 1B, E) and because they tolerated simultaneous stable-knockdown of hnRNP A1 and A2, in contrast to other cells we examined. We achieved ∼65% knockdown of hnRNP A1 and ∼50% knockdown of hnRNP A2 (Fig. 4A). Although individual knockdown of these proteins—which are closely related in structure and function (17)—had little effect, the combined knockdown elicited a ∼6-fold increase in PK-M1 protein (Fig. 4A) and ∼5-fold increase in PK-M1 mRNA (Fig. 4B).
Fig. 4.
Repression of exon 9 by hnRNP proteins. (A) A-172 glioblastoma cells were transduced with retroviruses expressing hnRNPA1 and hnRNP A2 shRNAs. Total lysates were analyzed by Western blotting with the indicated antibodies. The histogram on the right shows the quantitation of multiple experiments by infrared-imaging (n = 4; error bars show s.d.; p = 0.02 (M1); p = 0.005 (M2); p = 0.05 (A1); p = 0.01 (A2); Student’s paired t-test) (B) Analysis of PK -MRNA transcripts from the cells in (A) using radioactive RT-PCR and NcoI or PstI digestion. Bands are numbered as in Fig. 1E. The histogram on the right shows the quantitation from several experiments (error bars show s.d.; n = 4; p = 10-4 for the A1/A2 double-knockdown; Student’s paired t-test) (C) and (D) As in (A) and (B) but using shRNAs against PTB (hnRNP I). (C) The * on each side indicates the band corresponding to PK-M1. The histogram on the right shows the quantitation (n = 4; error bars show s.d.; p = 0.03 (M1); p = 0.01(M2); p = 0.03 (PTB); Student’s paired t-test). (D) The histogram on the right shows the quantitation (error bars show s.d.; n = 3; p = 0.001; Student’s paired t-test).
PTB or PTB + nPTB siRNA-knockdown in HeLa cells was previously shown by quantitative 2D-gel proteomics to decrease total PK-M expression (12). Although the PK-M1 and PK-M2 spots could not be resolved, RT-PCR revealed a ∼4-fold increase in the PK-M1/M2 ratio (12). We stably expressed PTB shRNA in A-172 cells and performed Western blotting and radioactive RT-PCR analyses as above, and detected a ∼3-fold increase in PK-M1 protein and mRNA (Fig. 4C,D). Taken together, these results suggest that hnRNP A1/A2 and PTB directly or indirectly mediate active repression of exon 9 in cancer cells.
Knockdown of Splicing Repressors Inhibits Lactate Production in a Glioblastoma Cell Line.
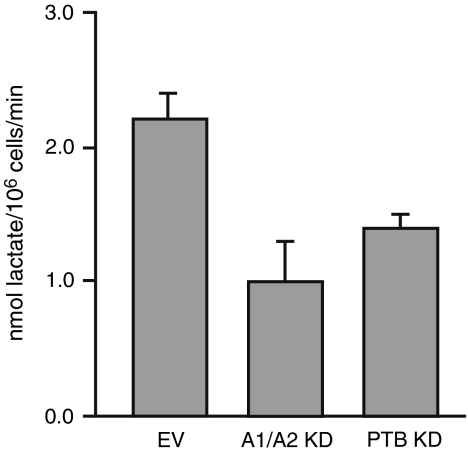
To test whether knockdown of hnRNP A1/A2 or PTB, which results in an increase in the PK-M1/PK-M2 protein ratio, is sufficient to affect cancer-cell metabolism, we measured the extent of lactate production in stable-knockdown versus control A-172 cells (Fig. 5). Remarkably, we observed a > 2-fold decrease in lactate production upon combined knockdown of hnRNP A1/A2, and a ∼1.5-fold reduction upon knockdown of PTB.
Fig. 5.
Effect of splicing-repressor knockdown on metabolism of glioblastoma cells. Lactate production was measured in A-172 glioblastoma cells transduced with empty vector or with combined shRNAs against hnRNP A1 and A2, or against PTB. Error bars show s.d.; n = 12 for EV control, and n = 6 for A1/A2 (p = 0.0034) and PTB (p = 0.014) knockdowns; Student’s paired t-test.
Discussion
We have demonstrated that downregulation of the splicing repressors hnRNP A1/A2 and PTB relieves repression of PK-M exon 9 inclusion, resulting in higher levels of PK-M1. Both types of hnRNP proteins have been implicated in cancer (2, 3, 18), consistent with their ability to promote expression of the protumorigenic PK-M2 isoform. hnRNP A1 and A2 are closely related proteins encoded by two separate genes (HNRNPA1 and HNRNPA2B1), which express additional minor isoforms (19); they are splicing repressors that typically, but not always, recognize exonic splicing silencer elements, and once bound to these elements they can spread along the RNA through cooperative interactions and interfere with the binding of spliceosomal components or activator proteins (20). PTB (hnRNP I) is also an abundant RNA-binding protein that binds to polypyrimidine tracts, such as those present at or upstream of 3′ splice sites, and it can regulate alternative splicing by creating a zone of silencing (21).
Previous analysis of PK-M splicing using a minigene suggested that inclusion of exon 10 is the default splicing pattern in proliferating cells (14). This is consistent with our observation that blocking either of the splice sites of exon 10 by means of antisense oligonucleotides leads to simultaneous skipping of both exons 9 and 10, rather than to efficient derepression of exon 9. We have shown that hnRNP A1/A2 and PTB are responsible for, or contribute to the repression of exon 9 in cells that express PK-M2. Although preferred motifs recognized by these repressors are known, we have not determined yet whether hnRNP A1/A2 and PTB regulate PK-M alternative splicing directly, and if so, where the relevant binding sites are located. However, two recent studies reported that PTB can be crosslinked in cells to several regions in intron 8 (22) and hnRNP A1 can bind in vitro to an RNA fragment encompassing the 5′ splice site of intron 9 (23). Further studies of cis-acting elements within and flanking both exon 9 and exon 10 will be necessary to uncover how individual cells achieve predominant or exclusive expression of only one of the two isoforms.
Considering that downregulation of hnRNP A1/A2 or PTB achieved only a three to fivefold increase over the low basal level of exon 9 inclusion, accompanied by a modest increase in PK-M1 protein with the persistence of PK-M2 protein, additional factors likely contribute to the usually tight control of PK-M alternative splicing. As in other instances of regulated alternative splicing [reviewed in (24)], combinatorial control by numerous RNA-binding proteins is likely operative for the PK-M gene.
The effects we observed on lactate production following combined knockdown of hnRNP A1/A2 are likely attributable to more than just the switch in PK-M isoform expression. hnRNP A1/A2 knockdown resulted in some PK-M1 protein expression, although cells continued to express appreciable amounts of PK-M2. A nearly complete switch of PK-M2 to PK-M1 using shRNA knockdown and isoform-specific rescue constructs resulted in at most a 30% decrease in lactate production (1). Thus, the > 2-fold reduction in lactate production we observed following combined hnRNP A1 + A2 knockdown probably reflects changes in alternative splicing that presumably occur in addition to those involving PK-M1/M2.
Aside from PK-M, additional genes important for cancer-cell metabolism are alternatively spliced. For instance, the bifunctional enzyme phosphofructokinase/bisphophatase B3 gene (PFKFB3) is a key regulator of glycolysis with preferential expression in cancer cells (25, 26). The kinase activity of this gene product generates fructose 2,6 bisphosphate (F2,6BP), which activates phosphofructokinase-1, a rate-limiting and regulatory control point of glycolysis, and indirectly leads to PK-M2 activation. In addition, HIF-1α and oncoproteins such as Ras activate PFKFB3, leading to increased F2,6BP in tumors (26). Multiple alternatively spliced isoforms of PFKFB3 exist, though their distinctive properties have not been characterized. There is cross-talk between cell-energy sensing and pyruvate-kinase regulation of this enzyme, suggesting that regulation of splicing to generate specific isoforms may be part of a larger metabolic program to promote proliferative metabolism (27). We speculate that hnRNP A1/A2 might modulate the expression of other alternatively spliced metabolic genes, in addition to PK-M, that collectively account for the large decrease in lactate production following double knockdown of hnRNP A1 and A2.
Materials and Methods
Cells and Transfections.
HeLa, HEK293, U-118MG, A-172, SK-N-BE, and C2C12 cells were grown in DMEM, supplemented with 10% (v/v) FBS, penicillin, and streptomycin. To induce differentiation, near-confluent C2C12 cultures were washed 3X with PBS and maintained for 7 d in medium containing 2% (v/v) horse serum, penicillin, and streptomycin. To select for a more homogenous differentiated culture, myotubes were treated for 4 d with 25 μM cytosine β-D-arabinofuranoside hydrochloride (AraC) (Sigma) beginning on day 7 after inducing differentiation. To generate stable transductant pools, A-172 cells were infected with LMP-puro or LMP-hygro retroviral vectors (3). The medium was replaced 24 h after infection, and starting 1 d later, infected cells were selected with puromycin (2 μg mL-1) for 3 d, or hygromycin (200 μg mL-1) for 7 d. In the case of double infections, cells were treated with hygromycin for 7 d after selection with puromycin for 3 d. Short hairpin RNA sequences were as follows: hnRNP A1—5′-TGCTGTTGACAGTGAGCGAAGGTTACAACAGATTTGTGAATAGTGAAGCCACAGATGTATTCACAAATCTGTTGTAACCTGTGCCTACTGCCTCGGA-3′; hnRNP A2—5′-TGCTGTTGACAGTGAGCGCGCCATGGGCTTCACTGTATAATAGTGAAGCCACAGATGTATTATACAGTGAAGCCCATGGCATGCCTACTGCCTCGGA-3′; PTB—5′-CGCGTCCCCGCAGTTGGAGTGACCTTACTTCAAGAGAGTAAGGTCACTTCAGCTGCTTTTTGGAAAT-3′.
Immunoblotting.
Cells were lysed in SDS and total protein concentration was measured. 30 μg of total protein from each lysate was separated by SDS-PAGE and transferred onto a nitrocellulose membrane. This was followed by blocking with 5% milk in Tris-buffered saline with Tween, probing with the indicated antibodies, and quantitation using an Odyssey infrared-imaging system (LI-COR Biosciences). Primary antibodies were: β-catenin (Abcam rAb 6302, 1∶4,000); tubulin (Genscript rAb, 1∶5,000); SF2/ASF (mAb AK96 culture supernatant, 1∶100 (28)); hnRNP A1 (Abcam mAb 4B10, 1∶1,000 or mAb UP1-55 culture supernatant (29)); hnRNP A2 (Abcam mAb DP3, 1∶1,000); hnRNP A1/A2 (A1/UP1-62 culture supernatant, 1∶20 (29)); PTB (mAb SH54 culture supernatant, 1∶100 (30) or Abcam rAb83897, 1∶1,000); CUG-BP1 (Abcam mAb 3B1, 1∶500); PK-M2 (rabbit, 1∶500) and PK-M1 (rabbit, 1∶2,000) (1); total PK-M (Abcam gAb6191, 1∶1,000). Secondary antibodies were IRdye 800 or 680 anti-rabbit, anti-mouse, or anti-goat (LI-COR Biosciences, 1∶10,000).
RT-PCR Assays.
2 μg of total RNA was extracted from freshly dissected, saline-perfused mouse tissues, and from cell lines, using Trizol reagent (Invitrogen). Contaminating DNA was removed by treatment with DNase I (Promega). Reverse transcription was carried out using ImPromp-II reverse transcriptase (Promega). Semiquantitative PCR using AmpliTaq polymerase (Applied Biosystems) was performed by including [α-32P]-dCTP in the reactions. The mouse- and human-specific primer sets anneal to exons 8 and 11, and their sequences are as follows: hPKMF: 5′-AGAAACAGCCAAAGGGGACT-3′; hPKMR: 5′-CATTCATGGCAAAGTTCACC-3′ and mPKMF: 5′-ATGCTGGAGAGCATGATCAAGAAGCCACGC-3′; mPKMR: 5′-CAACATCCATGGCCAAGTT-3′. After 22 amplification cycles, the reactions were separated into four aliquots for digestion with NcoI, PstI (New England Biolabs), both, or neither. The products were analyzed on a 5% native polyacrylamide gel, visualized by autoradiography, and quantitated on a FUJIFILM FLA-5100 phosphoirmager (Fuji Medical Systems) using Multi Gauge software Version 2.3 (Fujifilm). The %M1 mRNA was calculated using the GC-content-normalized intensities of the top undigested band (M1) and the bottom two digested bands (M2) in the PstI-digest lanes.
Antisense Oligonucleotides.
2′-O-methyl phosphorothioate oligonucleotides were purchased from TriLink Biotechnologies, and purity was confirmed by gel electrophoresis. 3′ splice site antisense oligonucleotide: CGGGCAATCTAGGGGAGCAAC; 5′ splice site antisense oligonucleotide: CCGCCTCCTACCTGCCAGAC. HEK293 cells were plated at 50,000 cells per well in 6-well plates in DMEM supplemented with 10% fetal calf serum, 500 units/mL penicillin, and 0.1 mg/mL streptomycin. After allowing cells to adhere, they were transfected with 500 nM oligonucleotide using Lipofectamine 2000 (Invitrogen). After 4 h at 37 °C, the transfecting mixture was replaced with fresh medium with serum. After 48 hr, the cells were washed three times with PBS, trypsinized, counted, and extracted RNA was analyzed by radioactive RT-PCR as described above.
Lactate Assays.
Lactate production was measured using a fluorescence-based assay kit (BioVision). Fresh medium was added to a 12-well plate of subconfluent cells and aliquots of media from each well were assessed 30 min later for the amount of lactate present. The cells were counted with a hemocytometer.
Acknowledgments.
We thank Chaolin Zhang, Martin Ackerman, Rotem Karni, Zuo Zhang, Yimin Hua, Mads Jensen, and Shuying Sun for helpful discussions and reagents. This work was supported by a grant from the Starr Cancer Consortium to A.R.K. and L.C.C. A Ruth L. Kirschstein National Research Service Award from the National Institutes of Health was received by C.V.C. A National Science Scholarship administered by the Agency for Science, Technology, and Research, Singapore was received by Z.W.
Footnotes
The authors declare a conflict of interest. L.C.C. and M.G.V.H. are associated with Agios Pharmaceuticals.
References
- 1.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 2.Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 5.Noguchi T, Yamada K, Inoue H, Matsuda T, Tanaka T. The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem. 1987;262:14366–14371. [PubMed] [Google Scholar]
- 6.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 7.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 8.Jurica MS, et al. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 9.Chacko E, Ranganathan S. Comprehensive splicing graph analysis of alternative splicing patterns in chicken, compared to human and mouse. BMC Genomics. 2009;10(Suppl 1):S5. doi: 10.1186/1471-2164-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 11.Smith CW. Alternative splicing—when two’s a crowd. Cell. 2005;123:1–3. doi: 10.1016/j.cell.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Seminars in Cancer Biology. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Takenaka M, et al. Alternative splicing of the pyruvate kinase M gene in a minigene system. Eur J Biochem. 1996;235:366–371. doi: 10.1111/j.1432-1033.1996.00366.x. [DOI] [PubMed] [Google Scholar]
- 15.Kislinger T, et al. Proteome dynamics during C2C12 myoblast differentiation. Mol Cell Proteomics. 2005;4:887–901. doi: 10.1074/mcp.M400182-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Sierakowska H, Agrawal S, Kole R. Antisense oligonucleotides as modulators of pre-mRNA splicing. Methods in Molecular Biology. 2000;133:223–233. doi: 10.1385/1-59259-215-5:223. [DOI] [PubMed] [Google Scholar]
- 17.Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 19.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okunola HL, Krainer AR. Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol Cell Biol. 2009;29:5620–5631. doi: 10.1128/MCB.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to positively or negatively regulate alternative splicing in mammalian cells. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2009. in press. [DOI] [PMC free article] [PubMed]
- 24.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 25.Van Schaftingen E, Hers HG. Phosphofructokinase 2: The enzyme that forms fructose 2,6-bisphosphate from fructose 6-phosphate and ATP. Biochem Biophys Res Commun. 1981;101:1078–1084. doi: 10.1016/0006-291x(81)91859-3. [DOI] [PubMed] [Google Scholar]
- 26.Atsumi T, et al. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–5887. [PubMed] [Google Scholar]
- 27.Bando H, et al. Phosphorylation of the 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase/PFKFB3 family of glycolytic regulators in human cancer. Clin Cancer Res. 2005;11:5784–5792. doi: 10.1158/1078-0432.CCR-05-0149. [DOI] [PubMed] [Google Scholar]
- 28.Hanamura A, Cáceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 29.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S, Deerinck TJ, Ellisman MH, Spector DL. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]