Abstract
This paper proposes a new nonlinear classifier based on a generalized Choquet integral with signed fuzzy measures to enhance the classification accuracy and power by capturing all possible interactions among two or more attributes. This generalized approach was developed to address unsolved Choquet-integral classification issues such as allowing for flexible location of projection lines in n-dimensional space, automatic search for the least misclassification rate based on Choquet distance, and penalty on misclassified points. A special genetic algorithm is designed to implement this classification optimization with fast convergence. Both the numerical experiment and empirical case studies show that this generalized approach improves and extends the functionality of this Choquet nonlinear classification in more real-world multi-class multi-dimensional situations.
Keywords: Choquet integral, signed fuzzy measure, classification, optimization, genetic algorithm
1. Introduction
Supervised classification is a procedure of constructing a mathematical model based on a training data set and using the model to assign a categorical class label to any new sample element. Essentially, this type of classification procedure is an optimization problem and has been widely applied in the pattern recognition and decision making literature. Classification methods, such as neural networks, decision trees, and nearest neighbor, have been studied extensively [1–7]. Nonlinear-integral based classification methods have recently gained more attention and encouraging results [8–11]. Our line of research concentrates on using the Choquet integral to conduct nonlinear classification [12, 13] and regression analyses [14–18]. Our core research in nonlinear Choquet classification is based on the theoretical development of Choquet integral [19] by Wang and Klir [20] and our subsequent research team [21–26]. Previously, we studied the applicability of Choquet integral in classification problems such as high-dimensional projection [12], and the algorithms for Choquet classification [27, 28]. To further advance our method, we realize that there are three issues yet to be solved. First, our previous research [12, 13] can solve the nonlinear classification problem only when the projection line is through the origin, which means that those projection lines not through the origin could not be identified, and therefore some classes with their actual boundaries on other projected locations in n-dimensional space cannot be properly classified. Second, our previous studies [12, 13] used discrete misclassification rates, where a predefined misclassification rate would be required each time in the classification process, which can be inaccurate or ineffective. In this paper, an automatic searching of the least misclassification rate using a continuous Choquet distance is addressed. Thirdly, our preliminary research [12, 13] has not yet found an effective way to penalize misclassified points which caused an unsolved optimization problem in practice, while in this study a penalty coefficient will be discussed to address this issue. Our contribution herein is to further generalize the functionality of nonlinear Choquet-integral based classification by solving the above three identified problems.
Literature indicates that the genetic algorithm is an effective approach to finding the optimal solution of a nonlinear classification problem [12, 29]. The genetic algorithm is a parallel random search technique widely applied in parameterized optimization problems, although it has been shown that its search speed is sometimes slow [27, 28, 30]. We studied different algorithms for Choquet classification. For example, compared to other algorithms such as neural networks, the advantage of the special genetic algorithm for Choquet integral avoids the risk of failing into a local minimum on the error surface, and its speed is also satisfactory [27, 28]. In this work, our specially designed genetic algorithm is further upgraded to accommodate the three newly identified issues for nonlinear Choquet classification.
Recently we proposed the Choquet classifier for linear models [15]. In [15] the classifier estimated a hyperplane to separate the given data in the feature space for a linear model. However, in the real world, the data are most likely to be linearly not separable. In this situation, nonlinear models are needed to enhance the classification power. A naive assumption is that the contribution from all the attributes is the sum of the contribution from each individual attribute. This consideration usually results in a power loss in classification models. If the interaction among attributes towards the classification is nonignorable, fuzzy measures (nonadditive measures) should be considered. When the nonadditive fuzzy measures are identified through the Choquet integral, the classifier becomes nonlinear [12, 13, 19, 20, 31].
In the following sections we first introduce the fuzzy measure used in our previous research and then the generalized Choquet integral used in this work. Sections 3 and 4 present our new Choquet-based nonlinear classification model and our upgraded special genetic algorithm to solve the above three identified issues. Then in Section 5 a numerical example is exhibited to illustrate the classification procedure in detail using artificial data. In Section 6 we demonstrate the performance and advantages of our proposed generalized approach in multi-class multi-dimensional situations using data from the UCI Machine Learning repository [32].
2. Fuzzy measures and Choquet integrals
The use of the Choquet integrals with respect to a signed fuzzy measure has been shown as an efficient approach to aggregate information from attributes via a nonadditive set function [22, 23, 25, 26]. Let X = {x1, …, xn} represent the attributes of the sample space and 𝒫(X) denote the power set of X. The signed fuzzy measure μ is defined as a set function
where μ(∅) = 0.
Let μi, i = 1, …, 2n − 1, denote the values of the set function μ on the nonempty sets in 𝒫(A), and f denote a given function, where f(x1), …, f(xn) represent the values of each attribute for one observation. The procedure of calculating the generalized Choquet integral is given in [14], summarized as follows. Let be a permutation of (x1, x2, …, xn) such that is in nondecreasing order. That is,
The Choquet integral with respect to fuzzy measure μ is defined as
where f(x0′) = 0 and (c) indicates Choquet integral. Let ω:X → [0, 1] be a nonnegative weight function on the attributes such that . In [12, 14] the weighted Choquet integral with respect to a nonadditive measure μ is defined by
where f is a nonnegative set function and μ(X) = 1.
In this paper, we generalize the weighted Choquet integral with respect to a nonadditive measure to a more comprehensive Choquet model, which is with respect to a nonadditive signed measure; that is, allowing the set function to take negative values and to be nonmonotone. Thus, a generalized weighted Choquet integral is expressed as
where signed measure μ is restricted to be regular (maxA⊂X|μ(A)| = 1). The parameters a = (a1, a2, …, an) and b = (b1, b2, …, bn), are n-dimensional vectors satisfying ai ∈ [0, ∞] with mini ai = 0 and |bi| ∈ [0, 1] with maxi|bi| = 1. We use this generalized Choquet model as a projection tool to reduce the complexity of the classification problem in an n-dimensional space [12, 25, 26]. We call a and b the matching vectors used to address the scaling and phase matching requirements of the feature attributes. In other words, matching vectors a and b are used to scale diverse units and ranges of the feature attributes with respective dimensions such that the signed measure μ can reflect the interaction appropriately. Also, with both scaling and phase matching parameters a and b, the projection line does not have to go through the origin. The simulation study in Section 5 will further demonstrate this function. Generally ϒ depends on f nonlinearly due to the nonadditivity of μ. For convenience,
are abbreviated by μ1, μ2, …, μn, μ12, μ13, …, respectively, hereafter.
3. A new nonlinear classification model
To simplify our theoretical illustration, 2-class classification based on Choquet integral is presented in detail, and the extension to multi-class classification is introduced at the end of this section.
We consider a 2-class nonlinear classification problem with classes A and A′. Suppose that the learning data consist of l sample points belonging to class A and l′ sample points belonging to class A′. Also, suppose that all of these sample points have the same feature attributes, x1, …, xn. Thus, the feature space is the n-dimensional Euclidean space ℝn. The j-th sample point in A, denoted by sj, is expressed as
while the j′–th sample point in A′ is similarly denoted by .
Now we want to find a Choquet hyperplane H determined by
| (1) |
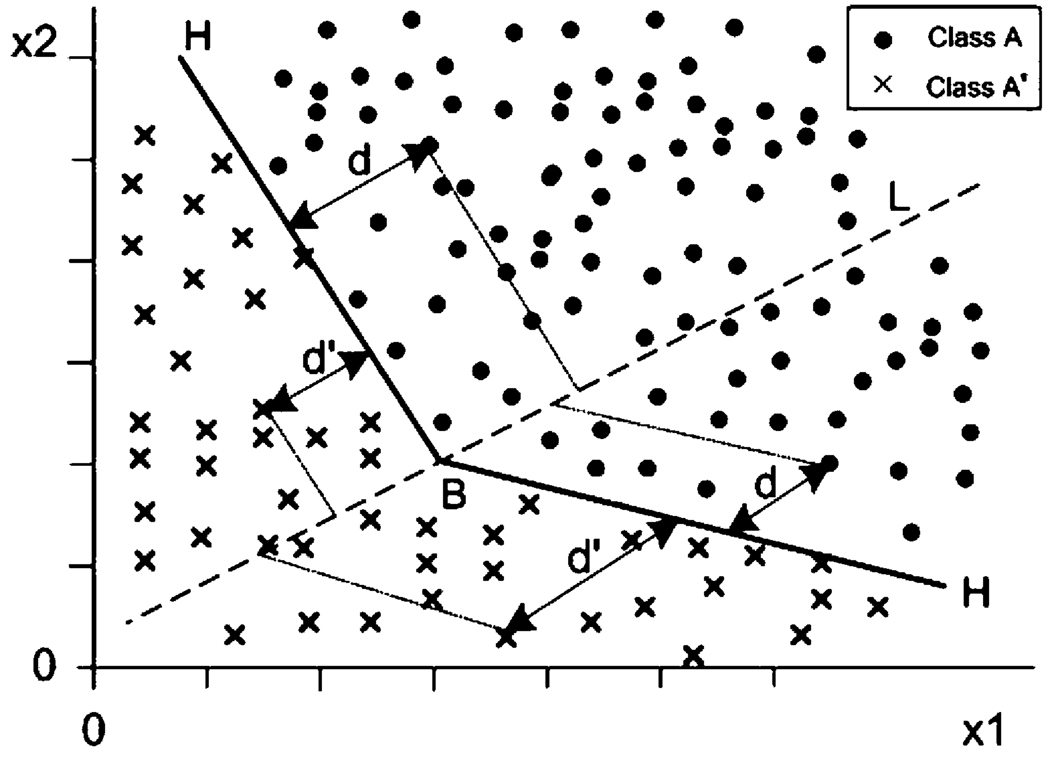
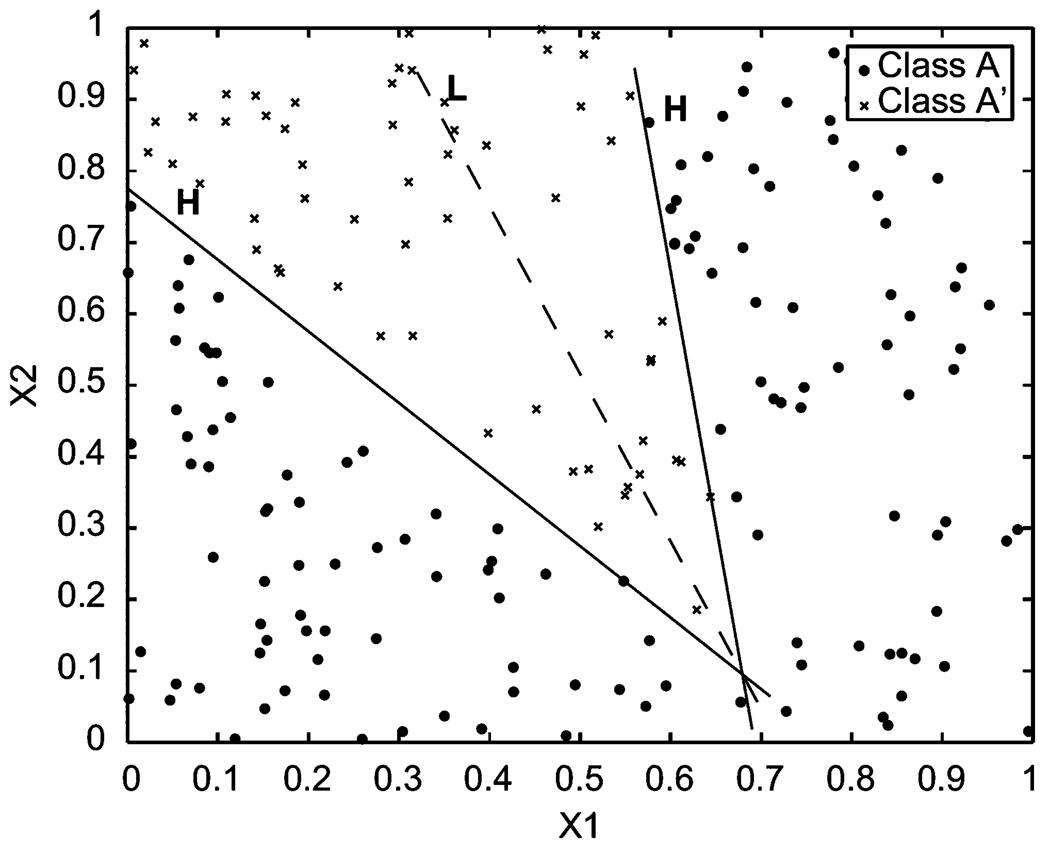
where B is an unknown real number. Without any loss of generality, we assume that all of these unknown parameters and B are in [−1, 1). A natural criterion to determine these parameters optimally is to maximize the total sum of signed distances of the learning sample points in the two classes from the respective side to the Choquet hyperplane H (see Figure 1).
Figure 1.
Two-dimensional data set projection based on Choquet integrals.
For example, on one side of H the signed distance dj from a sample point sj in A to H is the signed distance from the projection of sj paralleled with H on line L to the intersection (B) of H and L, which is equal to
From the other side of H, the signed distance of a sample point to H is just the signed distance from the projection of the point paralleled with H on line L to the intersection (B) of H and L, which is equal to
The projection paralleled with H onto L is a transformation identified by function
from the feature space to one-dimensional line L. That is, under this projection, any point
in the feature space has an image represented by the function (c)∫(a + bf) dμ, and the Choquet hyperplane H itself has an image represented by B. Thus, the total signed Choquet distance is
| (2) |
In this formula, the Choquet distance for those misclassified points will have a negative value. As to the optimization of Choquet hyperplane H (see Figure 1), we expect that the hyperplane H will be pushed to the opposite side as far as possible by the sample points from classes A and A′, respectively. In other words, H should be squeezed to an optimal position. In case there is a gap between classes A and A′, the Choquet hyperplane H as the classifying boundary should pass through the feature space along the gap. This means that the total signed Choquet distance D in (2) should be maximized. Such a criterion for determining the optimal hyperplane looks good. Unfortunately, it does not work well actually. In fact, if in the learning data set one class is larger than another, say l > l′, then class A has more power than class A′ to push hyperplane H to its opposite side infinitely such that the optimization problem has no solution. Thus, we must revise the above optimization model. Our previous research [12, 13] did not consider this issue and encountered this optimization problem in practice.
The revision can be realized by applying a large penalty coefficient to each misclassified sample point. Let
for j = 1, 2, …, l, and
for j′ = 1, 2, …, l′, where c > |l − l′| is a penalty coefficient and is usually taken as c = |l − l′| + 1. Then a penalized total signed distance is defined as
| (3) |
Thus, for a given learning sample data set with two classes, the unknown parameters a, b, μ, and B of hyperplane H as the classifying boundary can be determined by maximizing the penalized total distance Dc in expression (3). After determining the classifying boundary H expressed by Equation (1), for any new sample element sj = (fj (x1), …, fj (xn)), we classify s into class A if
and otherwise classify s into class A′.
The 2-class Choquet classification can easily be extended to multi-class classification where the boundary B will be expressed as a vector {b1, …, bk−1}. The element bk−1 in vector B denotes the projection point for the boundary of class k and class k − 1 on the projection real line L. Let s be the sample point and {A1, …, Ak} be the classes. Then the generalized Choquet multiclass classification can be deduced as follows:
4. A genetic algorithm
A specially designed genetic algorithm is applied to solve the optimization problem for this generalized Choquet-integral classification described in Section 3. First a population of classifiers (the chromosomes) is generated. These classifiers are each scored as to fitness using a fitness score based on Dc. The population is renewed by crossover and mutation operations, and the most fit are retained in the next generation. The components of the algorithm are outlined and explained as follows.
Coding and decoding. Unknown parameters μ1, μ2, …, matching vectors a and b, and B are coded as binary genes g1, g2, …, gN, and gN+1 (N = 2n − 1 + 2n). Thus, each gene is a bit string. The length of the bit string depends on the required precision for the solution. For example, if the required precision is 10−3, then each gene consists of ⌈log2(103)⌉ = 10 bits. Once the genes are generated, they are decoded by the formula ui = 2(gi − 0.5) for i = 1, 2, …, N; B̂ = 2(gN+1 − 0.5), etc.
Population and chromosomes. Each chromosome is a gene string, (g1, g2, …, gN+1). The population P consists of a large number of chromosomes. The number of chromosomes is called the size of the population and is denoted by p. The default value of p is 100.
- Chromosomes’ fitness. For each chromosome (g1, g2, …, gN+1), after decoding the genes, we may obtain the current parameter estimates u1, u2, …, un, â, b̂, and B̂, which represent a hyperplane H according to Equation (1). Then, based on the given learning data, the corresponding penalized total signed Choquet distance Dc from the sample points in the data set to the hyperplane H can be calculated by (3). The relative fitness of this chromosome in the current population is defined by
where(4)
and Dc(k) is the penalized total signed distances from the sample points in the data set to the Choquet hyperplane H(k) corresponding to the k-th chromosome in the current population. - Parents selection. Denoting the fitness of the k-th chromosome in the current population by F(k), we assign probability
to the k-th chromosome, k = 1, 2, …, p. Select two chromosomes at random from the population as the parents according to the probability distribution {pk|k = 1, 2, …, p}. Produce new chromosomes. According to a preset two-point probability distribution (α, 1 − α), choose a genetic operation via a random switch from mutation and crossover and then produce two new chromosomes. Repeat this procedure p/2 times to get p new chromosomes.
Renew population. Calculate the total signed distance of each new chromosome and add these p chromosomes to the current population. According to the total signed distance of these 2p chromosomes, delete the p worst from them and then form a new generation of the population.
Stopping controller. Repeat the above procedure to get the population generation by generation until the largest penalized total signed distance (which could achieve the least misclassification rate instead of the predefined misclassification rate used in the previous approaches [12, 13]). This largest distance is associated with the best chromosome in the population; it has not been significantly improved for w (with default value 10) consecutive generations. Here, “has not been significantly improved” means that the improvement Δ is less than 10−4 d(A, A′), where d(A, A′) is the distance between the centers of class A and class A′ in the learning data set.
After stopping, find the best chromosome in the last generation of population. Then, output the corresponding estimated values of parameters μ1, μ2, … μn, μ12, μ13, …, a, b, and B.
5. Simulations
We have implemented the algorithm shown in Section 4 using Microsoft Visual C)). All the functions are encapsulated into our CGenetic and CChoquet classes. Based on a training data set, the simulations were run on the Windows XP platform and regular PC desktop with AMD 1.6 GHZ CPU and 512M memory. It takes 1.5 min to stop and obtain the results.
To illustrate the classification procedure with numerical examples, we consider two data sets with known classification boundaries below: (a) where the projection line passes through the origin and (b) where the projection line does not pass through the origin.
The two-dimensional training data sets are generated by a random number generator and are separated into two classes by the straight line
where μ1, μ2, μ12, and B are pre-assigned separately. (In the examples, the data are uniformly distributed on the unit square.) Each sample point is labeled with class A if (c) ∫(a + bf ) dμ ≥ B; otherwise, (x1, x2) is labeled with class A′. In this way, 200 sample points are generated and labeled.
Running our classifier on the data for the two scenarios described below, we obtain the consecutive simulation results presented in Table 1 and Table 2, where G is the number of generations that have been created in the training procedure. The crossover probability in the simulation experiment was set to 0.9 and the mutation probability was 0.01.
Table 1.
Generations in training process for scenario (a) in Figure 2.
| G | A | A′ | μ12 | μ1 | μ2 | B | D |
|---|---|---|---|---|---|---|---|
| 1 | 150 | 40 | 0.1451 | 0.2369 | 0.5423 | 0.1021 | −2.7990 |
| 2 | 150 | 40 | 0.1451 | 0.2369 | 0.5423 | 0.1021 | −2.7990 |
| 3 | 153 | 42 | 0.1981 | 0.2244 | 0.5687 | 0.1189 | 0.6311 |
| 4 | 152 | 44 | 0.1336 | 0.1931 | 0.4901 | 0.0925 | 13.8546 |
| 5 | 152 | 44 | 0.1336 | 0.1931 | 0.4901 | 0.0925 | 13.8546 |
| 6 | 152 | 44 | 0.1336 | 0.1931 | 0.4901 | 0.0925 | 13.8546 |
| 7 | 152 | 44 | 0.1336 | 0.1931 | 0.4901 | 0.0925 | 13.8546 |
| … | … | … | … | … | … | … | … |
| 32 | 154 | 44 | 0.1410 | 0.1825 | 0.5501 | 0.0927 | 27.2005 |
| 33 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 34 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 35 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 36 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 37 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 38 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 39 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 40 | 155 | 45 | 0.1387 | 0.1800 | 0.5453 | 0.0916 | 27.4156 |
| 41 | 155 | 45 | 0.1388 | 0.1802 | 0.5453 | 0.0917 | 27.4177 |
| 42 | 155 | 45 | 0.1388 | 0.1802 | 0.5453 | 0.0917 | 27.4177 |
| 43 | 155 | 45 | 0.1388 | 0.1802 | 0.5453 | 0.0917 | 27.4177 |
| 44 | 155 | 45 | 0.1388 | 0.1802 | 0.5453 | 0.0917 | 27.4177 |
| 45 | 155 | 45 | 0.1388 | 0.1802 | 0.5453 | 0.0917 | 27.4177 |
| 46 | 155 | 45 | 0.1388 | 0.1802 | 0.5456 | 0.0917 | 27.4187 |
| 47 | 155 | 45 | 0.1389 | 0.1802 | 0.5456 | 0.0917 | 27.4189 |
| 48 | 155 | 45 | 0.1389 | 0.1802 | 0.5456 | 0.0917 | 27.4189 |
| 49 | 155 | 45 | 0.1389 | 0.1802 | 0.5460 | 0.0917 | 27.4211 |
| 50 | 155 | 45 | 0.1389 | 0.1802 | 0.5460 | 0.0917 | 27.4211 |
Table 2.
Generations in training process for scenario (b) in Figure 4.
| G | A | A′ | μ12 | μ1 | μ2 | B | a1 | a2 | b1 | b2 | D |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 133 | 11 | 0.5685 | 0.5825 | 0.5868 | 0.4409 | 0.7617 | 0.1982 | 0.0471 | 0.4197 | −751.6648 |
| 2 | 140 | 31 | 0.4899 | 0.5999 | 0.6256 | 0.2563 | 0.3893 | 0.5641 | 0.2501 | −0.0771 | −318.9425 |
| 3 | 140 | 31 | 0.4899 | 0.5999 | 0.6256 | 0.2563 | 0.3893 | 0.5641 | 0.2501 | −0.0771 | −318.9425 |
| 4 | 123 | 58 | 0.3846 | 0.6051 | 0.4342 | 0.2388 | 0.3228 | 0.6245 | 0.4720 | −0.1708 | −154.2835 |
| 5 | 135 | 51 | 0.3740 | 0.8851 | 0.7594 | 0.2460 | 0.5222 | 0.6800 | 0.1758 | −0.1429 | −112.1395 |
| 6 | 133 | 60 | 0.3010 | 0.7808 | 0.4535 | 0.2287 | 0.6118 | 0.7550 | 0.1978 | −0.1140 | −10.4408 |
| 7 | 139 | 56 | 0.3533 | 0.6666 | 0.5943 | 0.2361 | 0.3991 | 0.6825 | 0.3944 | −0.1774 | −8.8121 |
| 8 | 139 | 60 | 0.3094 | 0.7779 | 0.4586 | 0.2329 | 0.6067 | 0.7540 | 0.2011 | −0.1141 | −0.6097 |
| 9 | 139 | 60 | 0.3094 | 0.7779 | 0.4586 | 0.2329 | 0.6067 | 0.7540 | 0.2011 | −0.1141 | −0.6097 |
| 10 | 139 | 60 | 0.3094 | 0.7779 | 0.4586 | 0.2329 | 0.6067 | 0.7540 | 0.2011 | −0.1141 | −0.6097 |
| 11 | 139 | 58 | 0.3241 | 0.7522 | 0.4628 | 0.2351 | 0.5684 | 0.7342 | 0.2307 | −0.1182 | 1.9987 |
| 12 | 139 | 58 | 0.3241 | 0.7522 | 0.4628 | 0.2351 | 0.5684 | 0.7342 | 0.2307 | −0.1182 | 1.9987 |
| 13 | 139 | 59 | 0.3564 | 0.6881 | 0.4891 | 0.2703 | 0.4871 | 0.7786 | 0.4030 | −0.1999 | 4.9859 |
| 14 | 139 | 59 | 0.3564 | 0.6881 | 0.4891 | 0.2703 | 0.4871 | 0.7786 | 0.4030 | −0.1999 | 4.9859 |
| 15 | 139 | 59 | 0.3567 | 0.6883 | 0.4893 | 0.2702 | 0.4862 | 0.7778 | 0.4030 | −0.2001 | 5.0826 |
| 16 | 139 | 59 | 0.3567 | 0.6883 | 0.4893 | 0.2702 | 0.4862 | 0.7778 | 0.4030 | −0.2001 | 5.0826 |
| … | … | … | … | … | … | … | … | … | … | … | … |
| 22 | 140 | 60 | 0.3845 | 0.6764 | 0.5648 | 0.2676 | 0.4536 | 0.7120 | 0.3569 | −0.1580 | 5.9200 |
| 23 | 140 | 60 | 0.3845 | 0.6764 | 0.5648 | 0.2676 | 0.4536 | 0.7120 | 0.3569 | −0.1580 | 5.9200 |
| 24 | 140 | 60 | 0.3845 | 0.6764 | 0.5648 | 0.2676 | 0.4536 | 0.7120 | 0.3569 | −0.1580 | 5.9200 |
| 25 | 140 | 60 | 0.3830 | 0.6683 | 0.5713 | 0.2633 | 0.4420 | 0.7021 | 0.3614 | −0.1544 | 5.9296 |
| 26 | 140 | 60 | 0.3830 | 0.6683 | 0.5713 | 0.2633 | 0.4420 | 0.7021 | 0.3614 | −0.1544 | 5.9296 |
| 27 | 140 | 60 | 0.3830 | 0.6683 | 0.5713 | 0.2633 | 0.4420 | 0.7021 | 0.3614 | −0.1544 | 5.9296 |
| 28 | 140 | 60 | 0.3830 | 0.6683 | 0.5713 | 0.2633 | 0.4420 | 0.7021 | 0.3614 | −0.1544 | 5.9296 |
| 29 | 140 | 60 | 0.3830 | 0.6683 | 0.5713 | 0.2633 | 0.4420 | 0.7021 | 0.3614 | −0.1544 | 5.9296 |
| 30 | 140 | 60 | 0.3830 | 0.6683 | 0.5713 | 0.2633 | 0.4420 | 0.7021 | 0.3614 | −0.1544 | 5.9296 |
5.1. Scenario (a)
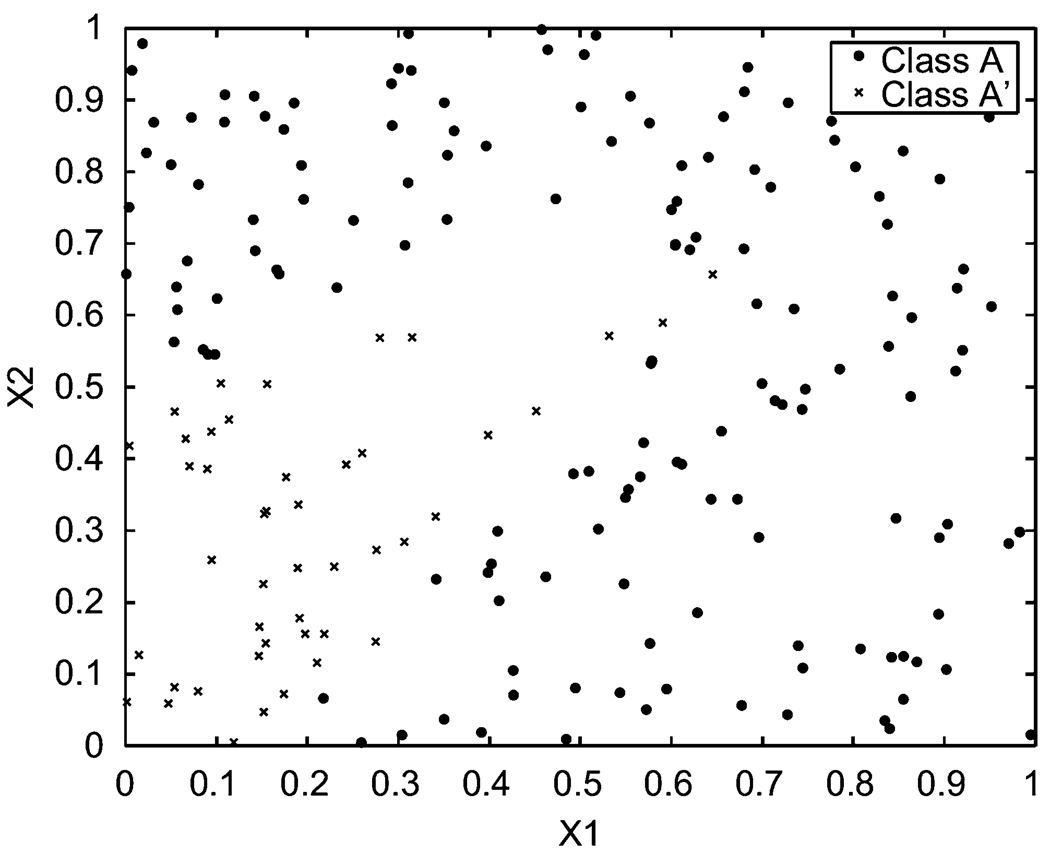
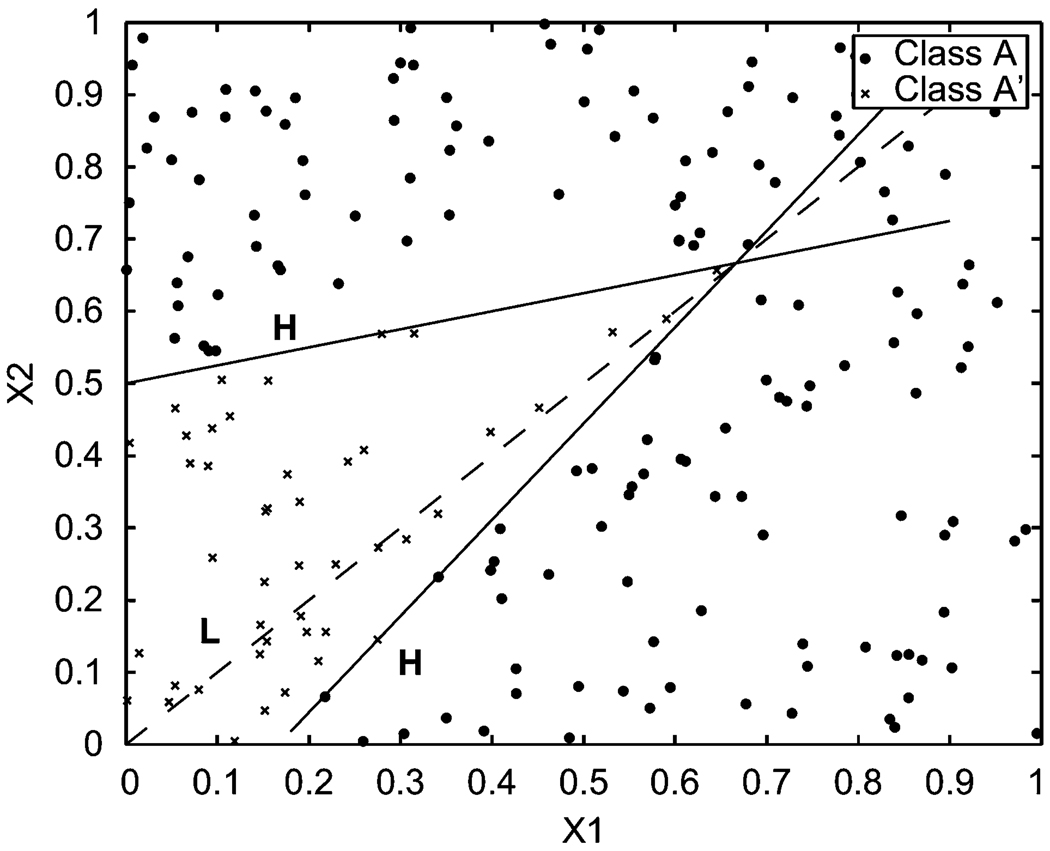
In scenario (a) the preset parameters are μ12 = 0.15, μ1 = 0.20, μ2 = 0.60, a = (0, 0), b = (1, 1), and B = 0.1. The distribution of the data is shown in Figure 2. Class A has 155 points, while class A′ has 45 points. The program for scenario (a) in Figure 2 stops at the 50th generation. The output of the classifier provides the standardized parameter estimates u12 = 0.1389, u1 = 0.1802, u2 = 0.5460, â = (0, 0), b̂ = (1, 1), and B̂ = 0.0917. The classifying boundary found in the last generation is shown in Figure 3.
Figure 2.
Training data set (a), μ12 = 0.15, μ1 = 0.20, μ2 = 0.60, B = 0.1, a1 = 0, a2 = 0, b1 = 1, b2 = 1.
Figure 3.
Classified training data set (a), u12 = 0.1389, u1 = 0.1802, u2 = 0.5460, B = 0.0917, a1 = 0, a2 =0, b1 = 1, b2 = 1.
In Table 1, the second column is the number of sample points that have been correctly classified in class A by the temporary best boundary obtained in that generation, while the third column is the number of sample points that have been correctly classified in class A′. The fourth through seventh columns are the current estimated values of parameters μ12, μ1, μ2, and B corresponding to one of the best chromosomes in each generation. The eighth column contains the penalized total Choquet signed distances from the sample points in the data set to the hyperplane corresponding to one of the best chromosomes in each generation, as described in Section 3.
In Table 1, at generation 33 the classifier has found a good chromosome whose corresponding classifying boundary can separate the training data without any misclassification, is presented. However, according to the stopping condition, the program does not stop until the counter w of the stopping controller reaches 10.
5.2. Scenario (b)
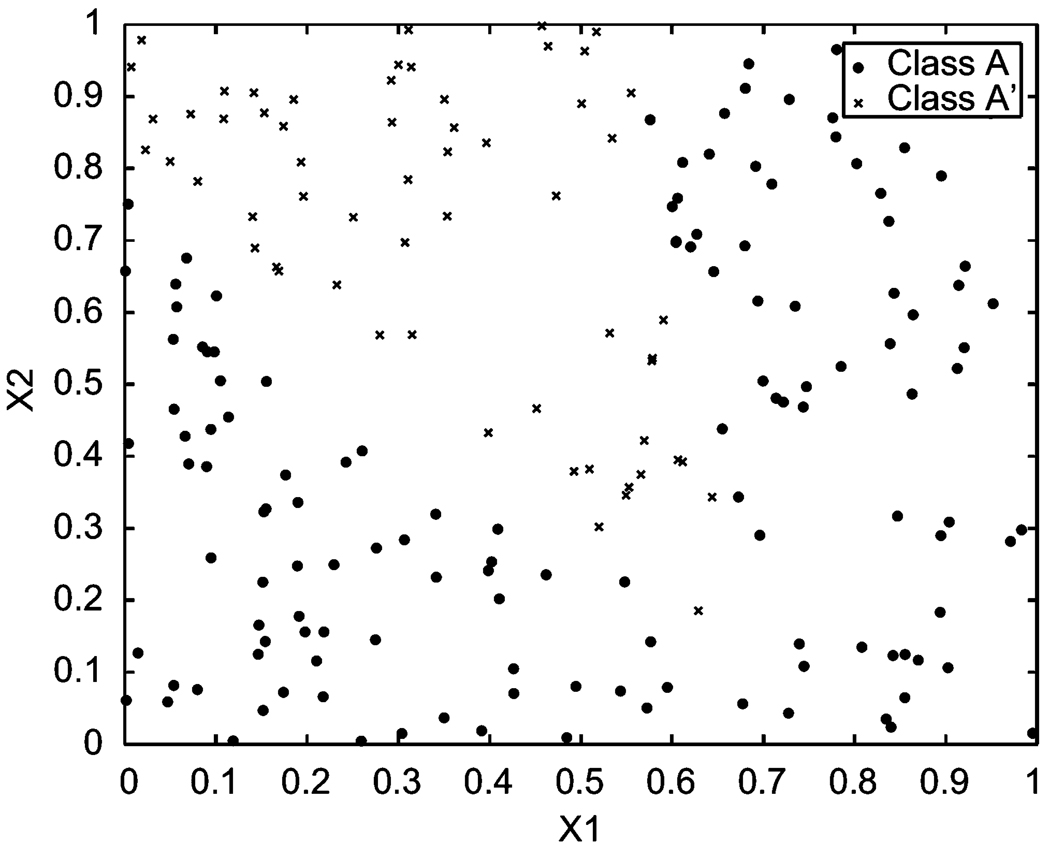
The preset parameters for scenario (b) are μ12 = 0.15, μ1 = 0.60, μ2 = 0.20, a = (0.2, 0.85), b = (0.85, −0.60), and B= 0.12. As shown in Figure 4, class A has 140 points, while class A′ has 60 points. The output of the classifier provides the standardized parameter estimates u12 = 0.3830, u1 = 0.6683, u2 = 0.5713, â = (0.4420, 0.7021), b̂ = (0.3614, −0.154), and B̂ = 0.2633 when the program stops after 30 generations.
Figure 4.
Training data set (b), μ12 = 0.15, μ1 =0.60, μ2 = 0.20, B = 0.12, a1 = 0.2, a2 = 0.85, b1 = 0.85, b2= −0.60.
In Table 2, the fourth through eleventh columns present the current estimates of parameters μ12, μ1, μ2, B, a1, a2, b1, and b2. The twelfth column lists the penalized total Choquet signed distances from the sample points in the data set to the hyperplane corresponding to one of the best chromosomes in each generation, as described in Section 3. The program stops at generation 30 for scenario (b) data in Figure 4. The classifying boundary found in the last generation is shown in Figure 5.
Figure 5.
Classified training data set (b), u12 = 0.3830, u1 = 0.6683, u2 = 0.5713, B = 0.2633, a1 = 0.4420, a2 = 0.7021, b1 = 0.3614, b2= −0.154.
Table 3 summarizes the final results for both scenarios that shows no misclassified sample points.
Table 3.
Classified sample data for scenarios (a) and (b).
| Scenario | (a) | (b) | ||
|---|---|---|---|---|
| Class | A | A′ | A | A′ |
| Classified in A | 155 | 0 | 140 | 0 |
| Classified in A′ | 0 | 45 | 0 | 60 |
6. Case studies
Our previous study [12] applied a special case of the generalized Choquet-integral approach, and demonstrated that the Choquet-integral classification approach is better than other available methods, such as Bayes, Neural Networks, HLM, and Nearest Neighbor, in terms of classification accuracy. Here our case studies compare implementation of this generalized approach with our previous approach using one of our artificial and one of the UCI data sets.
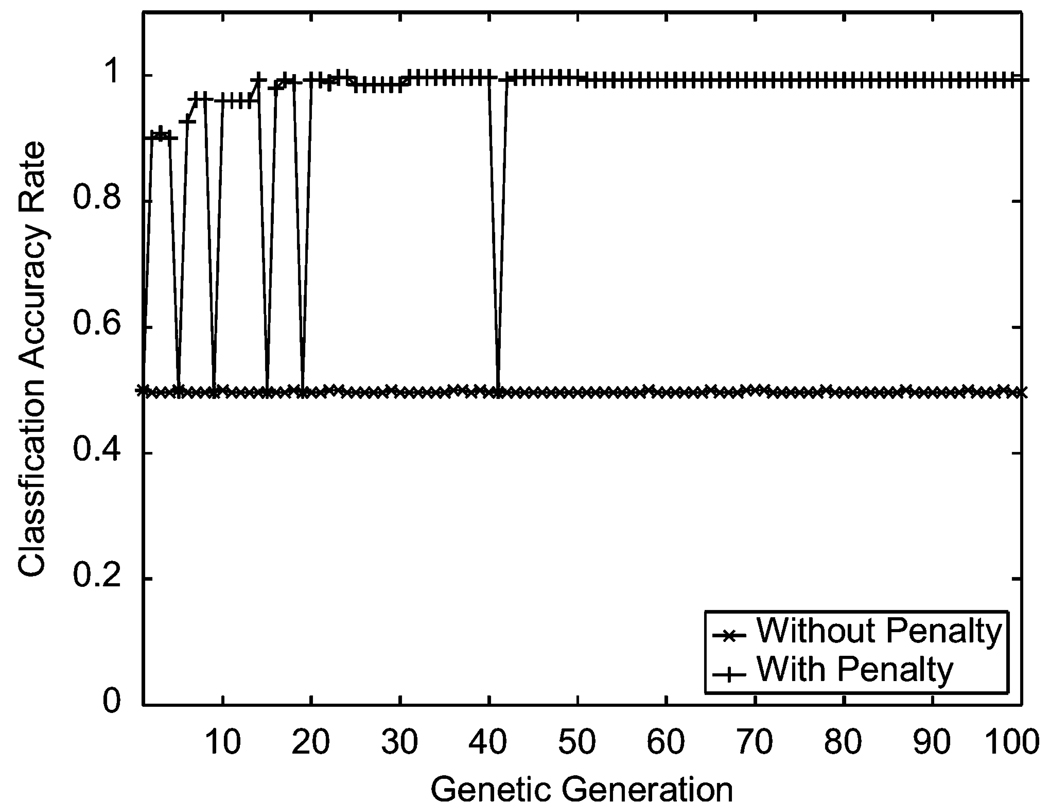
As discussed earlier, our previous approach only tolerates the projection line through the origin, lacks an automatic selection of the least misclassification rate, and does not penalize the misclassified points. In contrast, our current approach dramatically improved the classification accuracy rate by solving the three identified issues. For simplicity we call our previous approach “without penalty” and the current one “with penalty.” For classification performed on the same data used in the simulation scenario (b) where the projection line L is not through the origin, the current approach dramatically increases the classification accuracy rate to 100% by almost 50%, especially as the genetic evolution stabilized after 40 generations (see Figure 6).
Figure 6.
Classification accuracy rate comparison on the simulated data in scenario (b) where the projection line is not through origin.
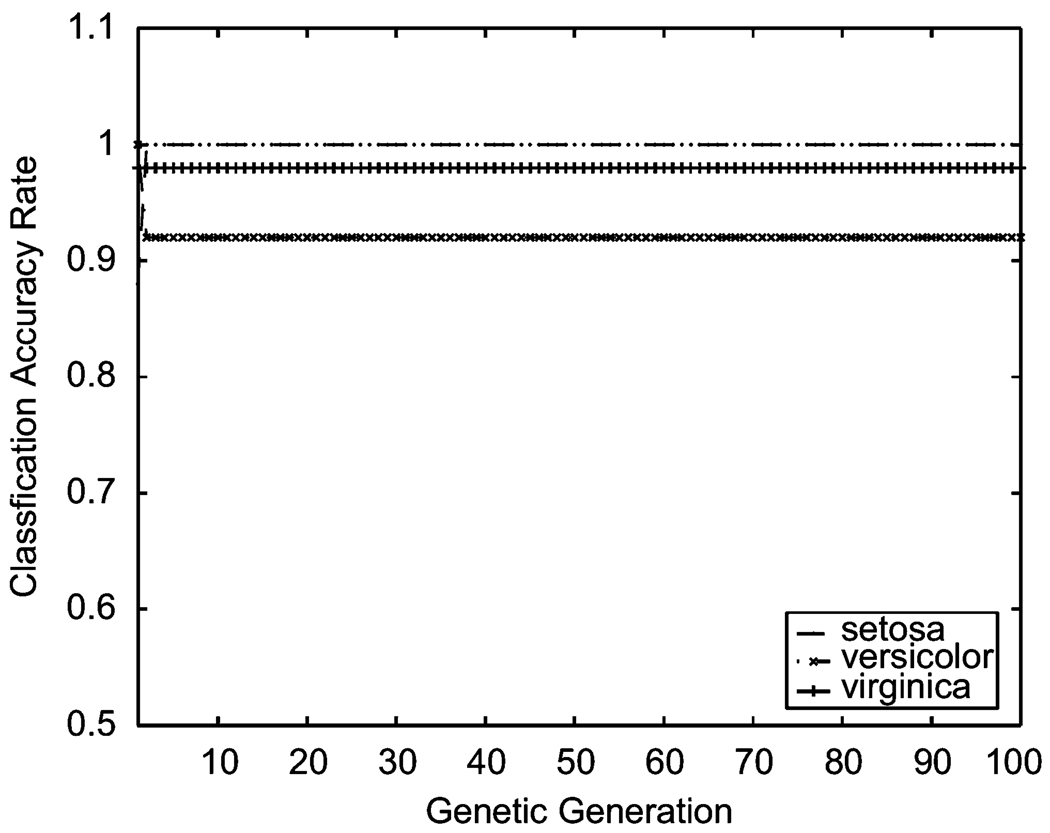
Considering real multi-class situations, we utilized the IRIS data from UCI [32]. These data include three classes (three IRIS species: Setosa, Versicolor, and Virginica) with 50 samples each and four-dimensional features (the length and the width of sepal and petal). The empirical results indicated that the classification accuracy rates of our current with-penalty approach reached 100%, 98%, and 93% for Setosa, Virginica, and Versicolor, respectively, after just a few genetic generations (see Figure 7).
Figure 7.
Classification accuracy rate comparison on a multi-classifier example, IRIS data.
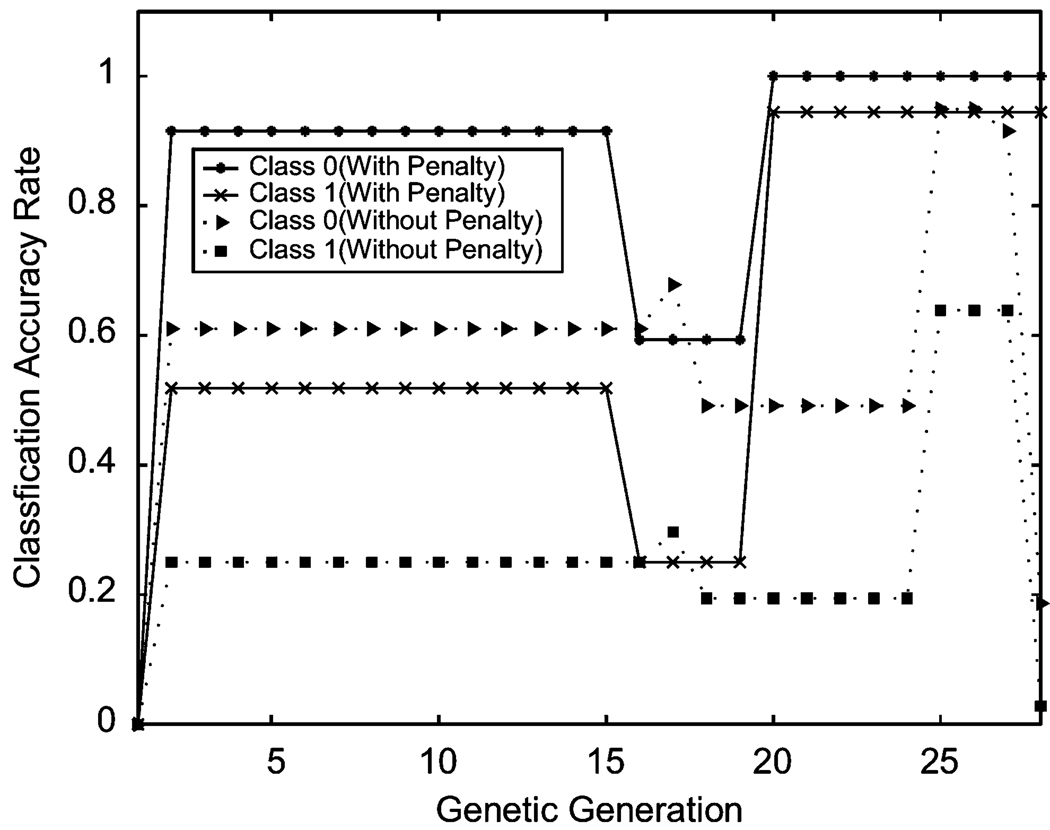
High dimensionality is another common feature in real-world pattern recognition. To address this issue using Choquet classification, we used the Pima Indians Diabetes data set from the UCI repository [32], which consists of 2 classes and eight-dimensional features with 768 samples. The outcome from our current with-penalty approach shows that over 20 genetic generations the classification accuracy rates reached 100% and 98% for each class. The results from the without-penalty approach were unsatisfactory with accuracy rates below 50% and quite unstable (see Figure 8). This comparison demonstrates the superiority of the generalized Choquet approach over the previous without-penalty technique.
Figure 8.
Classification accuracy rate comparison on eight-dimensional Pima Indians diabetes data set.
In addition, we have compared our current approach with nine typical classification methods on the previous two data sets (IRIS and Pima Indians Diabetes) and also on the Wisconsin Breast Cancer, Haberman’s Survival, and Blood Transfusion Service Center data from the same repository. The Breast Cancer data set includes 2 classes and 9 features, comprising 699 records. The Survival data has 3 attributes and 306 patients, with 2 survival status (the patient survived 5 years or longer, or died within 5 years), while the Blood Transfusion data consist of 5 attributes and 748 donors with two categories, donating and nondonating blood. For this comparison, 100% training data for each data set was used to evaluate these nine classification methods.
We have summarized the comparison results for each method in Table 4. Among these methods, the first two are Bayes-based methods: NaiveBayes [33] is a simple probabilistic classifier based on applying Bayes’ theorem with strong (naive) independence; BayesNet classifier is based on the Bayes networks that are composed of the prior probability distribution of the class node and a set of local networks. NB-tree [34] is the tree-based classification method, which is the decision tree with NaiveBayes classifiers at the leaves. Classification Via Regression [35] is the meta-based method, using regression techniques, where class is binarized and one regression model is built for each class value. Radial basis function (RBF) network and sequential minimal optimization (SMO) are the function-based classification methods [36, 40]. RBF networks is a radial basis function network, which uses K-means clustering algorithm to learn either a logistic regression (discrete class problems) or linear regression (numeric class problems). SMO is the one that utilizes sequential minimal optimization algorithm for training a support vector classifier using polynomial or RBF kernels. Fuzzy Lattice Reasoning (FLR) and Fuzzy Decision Tree (FDT) are fuzzy-based classification methods [37–39]. FLR is the classifier that uses the notion of fuzzy lattices for creating a reasoning environment. We also compared our results with those obtained from FDT which is a popular and powerful technique of learning from fuzzy examples, and can be a benchmark for fuzzy classifiers. The best accuracy achieved on each data set, measured by the misclassification rates, is presented in bold in Table 4.
Table 4.
Misclassification rates (E) of selected classifiers on five empirical data sets.
| Method | IRISc (%) | Breast Cancerc (%) |
Pima Indians Diabetesc (%) |
Haberman’s Survivalc (%) |
Blood Transfusionc (%) |
|---|---|---|---|---|---|
| NaiveBayesa | 4.0 | 3.9 | 23.7 | 24.2 | 25.0 |
| BayesNeta | 5.3 | 2.7 | 21.7 | 25.8 | 24.6 |
| NBtreea | 2.7 | 2.7 | 25.7 | 22.9 | 20.5 |
| Classification Via Regressiona | 2.0 | 2.3 | 22.7 | 25.5 | 19.8 |
| SMOa | 3.3 | 3.0 | 22.5 | 25.2 | 23.8 |
| RBF networka | 2.7 | 3.6 | 25.6 | 24.8 | 21.8 |
| Decision tablea | 4.0 | 3.6 | 22.4 | 25.8 | 23.8 |
| Fuzzy Lattice Reasoning (FLR)a Classifier | 3.3d | 0.7d | 19.1d | 38.2d | 32.4d |
| Fuzzy Decision Treeb | 4.0 | 3.0 | 20.1 | 22.8 | 19.1 |
| Choquet Distance based Classifier with Penalty | 2.7 | 0.0e | 0.0f | 26.5 | 4.95g |
Tested with default parameter settings in Weka3.6.0 [41].
Tested with default parameter settings in FID3.4 [42].
Classification of all empirical data sets used 100% training set.
Results when ρ = 0.75, ρ ∈ [0.5, 1] (when ρ = 1, EIris = 0%, EBreast = 0%, EPima = 0%, EHaberman = 2%, EBlood = 11%).
Missing data were preprocessed using multiple imputation procedure in SAS9.2 [43]. Simple logistic regression test showed that no significant pair-wise interaction exists in Breast Cancer and no interaction was added in.
Four significant pair-wise interactions (p < 0.05) were found using logistic regression test and added in Choquet classification.
All pair-wise interactions were included, as the number of attributes is relatively small and three interactions are significant (p < 0.05) using logistic regression tests.
The overall results indicate that our current approach is competitive and can be regarded as one of the best classifiers. Especially for the Wisconsin Breast Cancer, Pima Indians diabetes, and Blood Transfusion data, our approach dramatically outperformed all other alternative methods compared, in terms of the least misclassification rate. For the Haberman’s Survival data sets, our approach is below but close to the least classification rate achieved by FLR when its vigilance value is 0.75, in contrast to the poor performance of the nine alternate methods in the Pima Indians and Blood Transfusion data classification. For the IRIS data, our approach ranked at the second with NBTree and RBF network, less than the average misclassification rate (3.5%) among the nine alternative methods compared. Although our approach costs relatively longer time than other methods in the Wisconsin Breast Cancer, Pima Indians, and Blood Transfusion data set classification, it has even equivalent or better performance than FLR with its extreme vigilance value of 1 (see notes under Table 4). This may indicate a trade-off between the accuracy and the time efficiency, and there may indeed exist the interactions among the features of these data sets, which our approach may best fit. Therefore we believe that the time cost of our approach is tolerable in terms of the highest accuracy achieved and its overall performance (Table 5).
Table 5.
Parameter settings and parameter estimates of Choquet classification for UCI data sets.
| IRIS | |||||||||||
| Result | Iris-setosa 50 | ||||||||||
| Iris-versicolor 46 | |||||||||||
| Iris-virginica 54 | |||||||||||
| Maximum generation | 50 | ||||||||||
| Population size | 800 | ||||||||||
| Estimated parameters | |||||||||||
| μ1 | μ2 | μ3 | μ4 | μ12 | μ13 | μ14 | μ23 | μ24 | μ34 | ||
| 0.5619 | 0.9547 | 0.5916 | 0.5982 | 0.7096 | 0.4955 | 0.0144 | 0.0998 | 0.4394 | 0.5824 | ||
| a1 | a2 | a3 | a4 | ||||||||
| 0.4801 | 0.8875 | 0.0157 | 0.0216 | ||||||||
| b1 | b2 | b3 | b4 | ||||||||
| 0.6452 | 0.8214 | 0.9827 | 0.9485 | ||||||||
| Bound 1 | Bound 2 | ||||||||||
| 0.5521 | 0.0061 | ||||||||||
| Breast Cancer | |||||||||||
| Result | Class 2: 458 | ||||||||||
| Class 4: 241 | |||||||||||
| Maximum generation | 50 | ||||||||||
| Population size | 500 | ||||||||||
| Estimated parameters | |||||||||||
| μ1 | μ2 | μ3 | μ4 | μ5 | μ6 | μ7 | μ8 | μ9 | |||
| 0.8451 | 0.8227 | 0.5142 | 0.0909 | 0.334 | 0.3888 | 0.0653 | 0.2295 | 0.4545 | |||
| a1 | a2 | a3 | a4 | a5 | a6 | a7 | a8 | a9 | |||
| 0.4239 | 0.2228 | 0.7837 | 0.3085 | 0.7508 | 0.8591 | 0.7499 | 0.5009 | 0.5088 | |||
| b1 | b2 | b3 | b4 | b5 | b6 | b7 | b8 | b9 | |||
| 0.1023 | 0.653 | 0 | 0.1168 | 0.7906 | 0.1127 | 0.6651 | 0.1907 | 0.4844 | |||
| Bound | |||||||||||
| 0 | |||||||||||
| PIMA Indian Diabetes | |||||||||||
| Result | Class 0: 500 | ||||||||||
| Class 1: 268 | |||||||||||
| Maximum generation | 30 | ||||||||||
| Population size | 500 | ||||||||||
| Estimated parameters | |||||||||||
| μ1 | μ2 | μ3 | μ4 | μ5 | μ6 | μ7 | μ8 | μ18 | μ38 | ||
| 0.7926 | 0.3335 | 0.2957 | 0.1944 | 0.7915 | 0.5507 | 0.3857 | 0.2681 | 0.2116 | 0.4178 | ||
| μ47 | μ57 | ||||||||||
| 0.6373 | 0.3459 | ||||||||||
| a1 | a2 | a3 | a4 | a5 | a6 | a7 | a8 | ||||
| 0.6552 | 0.848 | 0.0261 | 0 | 0.5307 | 0.3932 | 0.594 | 1 | ||||
| b1 | b2 | b3 | b4 | b5 | b6 | b7 | b8 | ||||
| 0.2585 | 0.0478 | 0.6574 | 0.5944 | 0.6699 | 0.6137 | 0.4581 | 0.2821 | ||||
| Bound | |||||||||||
| 0 | |||||||||||
| Haberman | |||||||||||
| Result | Class 1: 306 | ||||||||||
| Class 2: 0 | |||||||||||
| Maximum generation | 100 | ||||||||||
| Population size | 800 | ||||||||||
| Estimated parameters | |||||||||||
| μ1 | μ2 | μ3 | μ12 | μ13 | μ23 | ||||||
| 0.5552 | 0.5801 | 0.5771 | 0.4166 | 0.5097 | 0.5705 | ||||||
| a1 | a2 | a3 | |||||||||
| 0.4785 | 0 | 0.4025 | |||||||||
| b1 | b2 | b3 | |||||||||
| 0 | 0 | 0.4261 | |||||||||
| Bound | |||||||||||
| 0.2361 | |||||||||||
| Blood Transfusion | |||||||||||
| Result | Class 0: 607 | ||||||||||
| Class 1: 141 | |||||||||||
| Maximum generation | 50 | ||||||||||
| Population size | 800 | ||||||||||
| Estimated parameters | |||||||||||
| μ1 | μ2 | μ3 | μ4 | μ12 | μ13 | μ14 | μ23 | μ24 | μ34 | ||
| 0.2225 | 0.555 | 0.7359 | 0 | 0.6594 | 0 | 0.3674 | 0.6044 | 0.4796 | 0.5643 | ||
| a1 | a2 | a3 | a4 | ||||||||
| 0.0412 | 0.3753 | 0.837 | 0.1273 | ||||||||
| b1 | b2 | b3 | b4 | ||||||||
| 0.139 | 0.653 | 0.5246 | 0.2834 | ||||||||
| Bound | |||||||||||
| 0 | |||||||||||
7. Summary
Based on our previous research on Choquet classification, this paper addressed three unsolved issues through theoretical discussion, simulation experiments, and empirical case studies. This research used 2-class classification as an example for the simplicity of theoretical illustration, and also extended to multi-class multidimensional situations. The current generalized Choquet-integral classification can allow for the projection line at any location, automatic search for the least misclassification rate based on Choquet distance, and penalty on misclassified points. This improvement expands the functionality of Choquet-classification in solving more flexible real-world classification problems and also practically enhances the classification accuracy and power.
Choquet integral has recently been applied to acoustic event classification [44], image analysis [45], image processing [46, 47], voice recognition [48], traffic surveillance [49], and temperature prediction [50]. Our case studies extended the generalized Choquet classification to the biological and medical areas. Our future studies will continue in this line of research by emphasizing the practical value of the Choquet-integral classification.
Acknowledgments
This research was supported in part by National Institutes of Health grants R01 DA023653, DA014661, MH065668, and Layman Awards Foundation.
Contributor Information
Hua Fang, Email: jfang2@unl.edu.
Honggang Wang, Email: hwang1@umassd.edu.
Kimberly Andrews Espy, Email: kespy2@unl.edu.
Zhenyuan Wang, Email: zhenyuanwang@mail.unomaha.edu.
References
- 1.Quinlan JR. C4.5: Programs for Machine Learning. San Mateo, CA: Morgan Kaufmann; 1993. [Google Scholar]
- 2.Breiman L, Friedman J, Olshen R, Stone C. Classification and Regression Trees. New York: Chapman & Hall; 1984. [Google Scholar]
- 3.Friedman J. Multivariate adaptive regression splines (with discussion) Annals of Statistics. 1982;19:1–141. [Google Scholar]
- 4.Yuan Y, Shaw MJ. Induction of fuzzy decision trees. Fuzzy Sets and Systems. 1995;69:125–139. [Google Scholar]
- 5.Kohonen T. Self-Organizing Maps. Heidelberg, Germany: Springer; 1995. [Google Scholar]
- 6.Sarle WS. Neural networks and statistical models; Presented at 19th Annual SAS Users Groups International Conference; Cary, NC. 1994. [Google Scholar]
- 7.Spooner TJ, Ordonez R, Maggiore M, Passino KM. Stable Adaptive Control and Estimation for Nonlinear Systems: Neural and Fuzzy Approximator Techniques. Wiley, NY: 2002. [Google Scholar]
- 8.Grabisch M. The representation of importance and interaction of features by fuzzy measures. Pattern Recognition Letters. 1996;17:567–575. [Google Scholar]
- 9.Grabisch M, Nicolas JM. Classification by fuzzy integral: performance and tests. Fuzzy Sets and Systems. 1994;65:255–271. [Google Scholar]
- 10.Keller JM, Yan B. Possibility expression and its decision making algorithm; Presented at 1st IEEE International Conference on Fuzzy Systems; San Diego, CA. 1992. [Google Scholar]
- 11.Mikenina L, Zimmermann HJ. Improved feature selection and classification by the 2-additive fuzzy measure. Fuzzy Sets and Systems. 1999;107:197–218. [Google Scholar]
- 12.Xu K, Wang Z, Heng PA, Leung KS. Classification by nonlinear integral projections. IEEE Transactions on Fuzzy Systems. 2003;11:187–201. [Google Scholar]
- 13.Liu M, Wang Z. Using generalized Choquet integral projections; Presented at IFSA 2005.2005. [Google Scholar]
- 14.Wang Z. A new model of nonlinear multiregression by projection pursuit based on generalized Choquet integrals; Presented at FUZZ-IEEE2002; Hawaii. 2002. [Google Scholar]
- 15.Wang H, Sharif H, Wang Z. A new classifier based on genetic algorithm; Presented at IPMU 2006.2006. [Google Scholar]
- 16.Leung K, Wong M, Lam W, Wang Z, Xu K. Learning nonlinear multiregression networks based on evolutionary computation. IEEE Transactions on Systems, Man, and Cybernetics. 2002;32:630–643. doi: 10.1109/TSMCB.2002.1033182. [DOI] [PubMed] [Google Scholar]
- 17.Xu K, Wang Z, Heng P, Leung K. Using a new type of nonlinear integral for multi-regression: an application of evolutionary algorithms in data mining; Presented at IEEE SMC’98.1999. [Google Scholar]
- 18.Wang Z, Wong M, Fang J, Xu K. Nonlinear nonnegative multiregressions based on Choquet integrals. International Journal of Approximate Reasoning. 2000;25:71–87. [Google Scholar]
- 19.Choquet G. Theory of capacities. Annales de l’Institut Fourier. 1953;5 [Google Scholar]
- 20.Wang Z, Klir GJ. Fuzzy Measure Theory. New York: Plenum Press; 1992. [Google Scholar]
- 21.Wang Z, Klir GJ, Wang W. Monotone set functions defined by Choquet integral. Fuzzy Sets and Systems. 1996;81:241–250. [Google Scholar]
- 22.Wang Z, Klir GJ. Choquet integrals and natural extensions of lower probabilities. International Journal of Approximate Reasoning. 1997;16:137–147. [Google Scholar]
- 23.Wang Z. Convergence theorems for sequences of Choquet integrals. International Journal of General Systems. 1997;26:133–143. [Google Scholar]
- 24.Wang Z, Leung K, Klir GJ. Integration on finite sets. International Journal of Intelligence Systems. 2006;21:1073–1092. [Google Scholar]
- 25.Wang Z, Yang R, Heng P, Leung K. Real-valued Choquet integrals with fuzzy-valued integrand. Fuzzy Sets and Systems. 2006;157:256–269. [Google Scholar]
- 26.Wang Z, Xu K, Heng P, Leung K. Indeterminate integrals with respect to nonadditive measures. Fuzzy Sets and Systems. 2003;138:485–495. [Google Scholar]
- 27.Wang Z, Xu K, Wang J, Klir G. Using genetic algorithms to determine nonnegative monotone set functions for information fusion in environments with random perturbation. International Journal of Intelligent Systems. 1999;14:949–962. [Google Scholar]
- 28.Wang W, Wang Z, Klir GJ. Genetic algorithms for determining fuzzy measures from data. Journal of Intelligent & Fuzzy Systems. 1998;6:171–183. [Google Scholar]
- 29.Goldberg E. Genetic Algorithms in Search, Optimization, and Machine Learning. Reading, MA: Addison-Wesley; 1989. [Google Scholar]
- 30.Mitchell M, Melanie X. Introduction to Genetic Algorithms. Cambridge, MA: MIT Press; 1996. [Google Scholar]
- 31.Denneberg D. Non-Additive Measure and Integral. Dordrecht: Kluwer Academic Publishers; 1994. [Google Scholar]
- 32.Asuncion A, Newman DJ. UCI Machine Learning Repository. Irvine, CA: University of California, School of Information and Computer Science; 2007. http://www.ics.uci.edu/~mlearn/MLRepository.html. [Google Scholar]
- 33.John GH, Langley P. Estimating continuous distributions in Bayesian classifiers; Presented at 11th Conference on Uncertainty in Artificial Intelligence; Morgan Kaufmann, San Mateo. 1995. [Google Scholar]
- 34.Kohavi R. Scaling up the accuracy of naive-Bayes classifiers: a decision tree hybrid; Presented at Second International Conference on Knowledge Discovery and Data Mining. [Google Scholar]
- 35.Frank E, Wang Y, Inglis S, Holmes G, Witten IH. Using model trees for classification. Machine Learning. 1998;32:63–76. [Google Scholar]
- 36.Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK. Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Computation. 2001;13:637–649. [Google Scholar]
- 37.Kaburlasos VG, Athanasiadis IN, Mitkas PA, Petridis V. Fuzzy lattice reasoning (FLR) classifier and its application on improved estimation of ambient ozone concentration, International. Journal of Approximate Reasoning. 2007;45:152–188. [Google Scholar]
- 38.Yuan Y, Shaw MJ. Induction of fuzzy decision trees. Fuzzy Sets and Systems. 1995;69(2):125–139. [Google Scholar]
- 39.Fuzzy EBY. software. http://www.eecs.berkeley.edu/~zhiheng/
- 40.Oyang Y, Hwang SC, Qu Y, Chen C, Chen ZW. Data classification with radial basis function networks based on a novel kernel density estimation algorithm. IEEE Transactions of Neural Networks. 16(1):225–236. doi: 10.1109/TNN.2004.836229. [DOI] [PubMed] [Google Scholar]
- 41. http://www.cs.waikato.ac.nz/ml/weka/
- 42. http://www.cs.umsl.edu/~janikow/fid/fid34/download.htm.
- 43.SAS Institute Inc. SAS/STAT User’s Guide, version 9.1. Cary, NC: SAS Institute Inc.; 2003. [Google Scholar]
- 44.Temko A, Macho D, Nadeu C. Fuzzy integral based information fusion for classification of highly confusable non-speech sounds. Pattern Recognition. 2008;41:1814–1823. [Google Scholar]
- 45.Strauss O, Comby F. Variable structuring element based fuzzy morphological operations for single viewpoint omnidirectional images. Pattern Recognition. 2007;40:3578–3596. [Google Scholar]
- 46.Bloch I, Ralescu A. Directional relative position between objects in image processing: a comparison between fuzzy approaches. Pattern Recognition. 2003;36:1563–1582. [Google Scholar]
- 47.Pham TD. An image restoration by fusion. Pattern Recognition. 2003;34:2403–2411. [Google Scholar]
- 48.Ramachandran RP, Farrell KR, Ramachandran R, Mammone RJ. Speaker recognition — general classier approaches and data fusion methods. Pattern Recognition. 2002;35:2801–2821. [Google Scholar]
- 49.Li X, Liu Z, Leung K. Detection of vehicles from traffic scenes using fuzzy integrals. Pattern Recognition. 2002;35:967–980. [Google Scholar]
- 50.Yang R, Wang Z, Heng P, Leung K. Fuzzified Choquet integral with fuzzyvalued integrand and its application on temperature prediction. IEEE Transactions on Systems, Man, and Cybernetics. 2008;38:367–380. doi: 10.1109/TSMCB.2007.911377. [DOI] [PubMed] [Google Scholar]