Abstract
In response to hypertension, the heart manifests robust hypertrophic growth, which offsets load-induced elevations in wall stress. If sustained, this hypertrophic response is a major risk factor for systolic dysfunction and heart failure. Extensive research efforts have focused on the progression from hypertrophy to failure; however, precise understanding of underlying mechanisms remains elusive. Recently, autophagy, a process of cellular cannibalization, has been implicated. Autophagy is activated during ventricular hypertrophy, serving to maintain cellular homeostasis. Excessive autophagy eliminates, however, essential cellular elements and possibly provokes cell death, which together contribute to hypertension-related heart disease.
Keywords: Autophagy, Cardiac Muscle, Cardiac Hypertrophy, Lysosomes, Protein Turnover, Heart Failure, Hypertension, Remodeling
Introduction
Hypertension is a leading risk factor for mortality worldwide (1). Astonishingly, in 2000, >25% of the world's adult population was hypertensive (2). The hypertensive population is projected to reach 1.56 billion in 2025, a 60% increase over 25 years. Based on data from the latest National Health and Nutrition Examination Survey, 27% of American adults have hypertension as defined by a systolic blood pressure of 140 mm Hg or higher and/or a diastolic blood pressure of 90 mm Hg or higher (3). Moreover, recent studies highlight an important association between cardiovascular event rates and pre-hypertension, a state affecting >30% of the American population and defined as a systolic blood pressure between 120 and 139 mm Hg and/or a diastolic blood pressure between 80 and 89 mm Hg (3, 4). Because of the fact that hypertension provokes no symptoms, it is often neglected; it is estimated that one-third of hypertensive patients are not aware of their condition. Incomplete awareness plus barriers to treatment on both the patient and the care provider sides of the healthcare equation together add up to a disease with extraordinarily high prevalence and rising incidence. Indeed, it is difficult to conceive of an illness (hypertension plus pre-hypertension) that afflicts >60% of adults in the United States.
Mounting epidemiological evidence demonstrates a linear and independent relationship between hypertension and CVD,2 the leading cause of death worldwide (5). Ischemic heart disease and cerebrovascular disease account for more than one-fifth of mortality in both developing and developed countries (5). In the United States, one in three adults has one or more types of CVD, and a death results every 37 s (6). In 2008, the direct and indirect costs of CVD were estimated at $450 billion, an enormous burden to our economic and public health systems.
Mechanisms of Remodeling in Hypertensive Heart Disease
Increased afterload precipitates increases in systolic wall stress within the pumping left ventricle. According to Laplace's law, wall stress is directly proportional to pressure and chamber size and inversely proportional to ventricular wall thickness (7). In response to high blood pressure, left ventricular wall thickness increases, normalizing wall stress. As post-mitotic cells, cardiomyocytes show negligible, if any, potential for proliferation (8), and hypertrophic growth is the only means to increase cardiac mass.
The phenotype of the hypertrophied heart has been divided into two broad categories. In the setting of pressure overload, such as in hypertension and aortic stenosis, new sarcomeres are added in parallel, and this lateral myocyte growth leads to wall thickening and preserved chamber volume, a process termed concentric hypertrophy (9). Under conditions of volume overload, such as in aortic insufficiency or arteriovenous shunting, sarcomeres are added in series, resulting in longitudinal cell growth. The result is ventricular wall thickening accompanied by chamber dilation, a process termed eccentric hypertrophy.
LVH is thought to be an adaptive short-term response that offsets elevations in ventricular wall stress and consequent excessive oxygen demand. Like many things in biology, however, the long-term consequences of a short-term fix are maladaptive; chronic LVH is a major risk factor for systolic dysfunction and heart failure (10). Despite extensive studies to decipher mechanisms governing the transition from LVH to heart failure, they remain elusive. On a bright note, numerous preclinical studies have demonstrated that abrogation of the hypertrophic response is well tolerated and even beneficial (11).
Remodeling of the adult heart entails alterations in the equilibrium between protein synthesis and protein degradation. In post-mitotic cardiomyocytes, which are largely irreplaceable, the fidelity of protein quality control is critical; accumulation of misfolded molecules and protein aggregates is toxic, triggering adverse responses and cell death. The major cellular mechanism for clearing toxic protein aggregates, along with long-lived proteins and dysfunctional organelles, is autophagy. In the context of cardiology, autophagy and lysosomal pathways have been recognized for many years both in human heart failure and in a variety of models of heart disease (12–16).
Autophagy, Cellular Self-eating
Autophagy is an evolutionarily conserved catabolic mechanism that maintains cellular homeostasis in a variety of contexts (17, 18). Under basal conditions, autophagy is required to degrade long-lived proteins and dysfunctional organelles. Under conditions of stress, such as starvation and hypoxia, autophagy is activated, promoting cell survival by releasing energy substrates via degradation of cellular constituents and by eliminating defective or damaged organelles (19). However, taken too far, excessive and uncontrolled autophagic activation can lead to depletion of essential molecules and organelles, triggering autophagic cell death (20, 21).
Three distinct types of autophagy have been described (17). Microautophagy refers to lysosome-mediated direct engulfment and degradation of cytosolic materials. In chaperone-mediated autophagy, misfolded proteins are translocated to the lysosome via the cytosolic chaperone HSP70. Among all types of autophagy, macroautophagy (hereafter termed autophagy) is the most common and important pathway to degrade long-lived proteins and the only means to clear dysfunctional organelles.
In the presence of ample extracellular nutrients, growth factors, such as insulin and insulin-like growth factor 1, stimulate glucose uptake and promote anabolic reactions. In this context, autophagy is suppressed. In response to starvation, induced either by the shortage of nutrients or by defects in growth factor pathways, autophagy is rapidly activated to replenish ATP. In addition to conditions of nutrient deprivation, enhanced levels of autophagy are seen in microbial invasion, misfolded protein accumulation, cancer, neurodegenerative disorders, and cardiac diseases (19).
Autophagy is a complex process that occurs in a stepwise manner divided into four stages: induction, nucleation, expansion, and maturation/retrieval (Fig. 1) (22, 23). To date, genetic screenings in yeast have identified 32 autophagy-related (ATG) genes that together regulate autophagosome formation (23). First, an isolated membrane (phagophore) is localized to the phagophore assembly site, where induction and nucleation take place. Expansion of the phagophore involves addition of several regulatory elements and engulfment of cytoplasmic constituents. The autophagosome, the hallmark of autophagy, is formed as a distinctive double-membrane vesicle 0.5–1.5 μm in diameter.
FIGURE 1.
Autophagic machinery. In the absence of the suppressive influence of TOR kinase, dephosphorylated ATG13 interacts with ATG1 and ATG17, forming a kinase complex that initiates autophagy. The class III PI3K complex comprises the lipid kinase Vps34, the regulatory subunits Beclin 1/ATG6 and ATG14, and Vps15, which together generate phosphatidylinositol 3-phosphate, leading to nucleation of the autophagosome. Following this, two ubiquitin-like conjugation systems generate ATG16-ATG5-ATG12 and LC3-II-PE complexes, expanding the phagophore and forming the autophagosome. Before fusion with a lysosome to form an autolysosome, most ATG proteins are recycled under the regulation of ATG9 and ATG18. Beclin 1/ATG6 also interacts with UVRAG, Vps34, and Vps15 to form another complex, which is involved in the maturation and trafficking of the autophagosome. Hydrolases within the lysosome degrade the inner membrane of autophagosome and the engulfed cargo. The resulting constituents, including sugars, amino acids, and lipids, are then released to the cytosol. IGF1, insulin-like growth factor 1; 3-MA, 3-methyladenine.
One of the most important upstream regulators of autophagy is the protein TOR (24). In the presence of abundant nutrients and intact insulin signaling, class I PI3K is activated to phosphorylate its downstream target AKT. AKT then activates TOR. Active TOR phosphorylates Atg13, inhibiting its interaction with Atg1, a critical step during autophagy induction (25). During starvation, Atg13 binds Atg1 and Atg17 to promote induction of autophagy.
The Atg1-Atg13-Atg17 complex is best characterized in yeast; the mammalian counterpart manifests slightly different properties (26). Under nutrient-rich conditions, mTOR interacts with the ULK1 (mammalian ATG1), mammalian ATG13, and FIP200 complex and phosphorylates ULK1 and mammalian ATG13 to inhibit autophagy. In starvation, dissociation of mTOR causes activation of ULK1, which phosphorylates mammalian ATG13 and FIP200 to promote autophagy. A class III PI3K complex is then recruited to the assembly site to stimulate nucleation, and the lipid kinase Vps34 is attached to the phagophore membrane through Vps15. Additionally, Beclin 1/ATG6 and ATG14 in this complex regulate Vps34 kinase activity. The activity of this lipid kinase complex is crucial for recruitment of additional ATG proteins to complete autophagosome formation.
Two ubiquitin-like systems contribute to the formation of the autophagosome. ATG12 is activated by a ubiquitin E1-like enzyme, ATG7, and transferred to a ubiquitin E2-like enzyme, ATG10. ATG12 is then covalently conjugated to ATG5, and the resulting ATG12-ATG5 complex interacts with ATG16. In the other ubiquitin-like system, LC3 (mammalian homolog of ATG8) is first cleaved by ATG4 to expose a C-terminal glycine. This processed LC3-I is then activated by ATG7, the E1-like enzyme. After being transferred by the E2-like enzyme ATG3, LC3-I is attached to a PE molecule and localized to the phagophore membrane (LC3-II). Lipidated LC3-II migrates faster than LC3-I upon SDS-PAGE, and its abundance is commonly used to assess autophagic activity. Most of the ATG proteins are ultimately recycled after the maturation of the autophagosome through a pathway involving ATG2, ATG9, and ATG18 (22).
Finally, the autophagosome fuses with a lysosome to form an autolysosome. There, the engulfed cargo and the inner membrane of the former autophagosome are degraded by acid hydrolases. The resulting small molecules, including sugars, amino acids, and lipids, are released to the cytosol through permeases. Thus, working in concert, two kinase systems (ATG1-ATG13 and class III PI3K), two ubiquitin-like systems (ATG5-ATG12 and LC3-II-PE), and a retrieval/maturation system complete the autophagy flux pathway.
Autophagy under Basal Conditions and in Setting of Starvation
The critical importance of autophagy under basal conditions is highlighted by cell death in its absence. Using an small interfering RNA approach, Nakai et al. (27) depleted ATG7 from NRVMs. With reduced autophagic activity, NRVMs manifested a classical hypertrophic response, including increased cell size and enhanced expression of atrial natriuretic factor. The investigators then found that loss of autophagy in the heart in vivo triggered LVH, cardiac dysfunction, and eventually heart failure (27). The importance of basal autophagy is highlighted further by three reports addressing Danon disease (28–30). In this disorder, cardioskeletal myopathy arises due to deficiency of LAMP2, a lysosomal membrane protein. In the absence of LAMP2, fusion of autophagosomes with lysosomes is blocked, leading to accumulation of long-lived proteins and consequent myopathy. Together, these results point to the important housekeeping role in the heart of constitutive autophagy.
During fasting for 3 days, progressive declines in heart weight are observed, consistent with activation of catabolic pathways (31). Using autophagy reporter mice, Kanamori et al. (31) reported that autophagy is induced progressively during this period. Although partial inhibition of autophagy does not affect cardiac function under fed conditions, autophagy suppression during starvation results in reduced cardiac ATP content and impaired heart performance. Similar findings have been reported for NRVMs in culture (32). Collectively, these results indicate that autophagy plays a critical protective role during nutrient deprivation in cardiomyocytes.
Autophagy in Progression from Hypertrophy to Failure
Growth of myocytes is accomplished by increases in protein synthesis, formation of new sarcomeres, and remodeling of existing cellular elements. In initial phases, anabolic processes predominate; indeed, some evidence suggests that catabolic processes may be suppressed early on. For example, in a short-term study by Pfeifer et al. (16), a 10-min infusion of the β-adrenergic agonist isoproterenol in rat hearts reduced the cellular content of autophagic vacuoles by 50%. Similarly, in another model of pressure overload, autophagy was reported to be suppressed 1 week after TAC (27).
Eventually, a new anabolic/catabolic steady-state equilibrium is established, albeit at a higher level, with balance achieved between synthetic and degradative processes. In this context, increases in autophagy have been reported. Our group, for example, reported that pressure overload induces autophagy as early as 24 h post-TAC (33). We found that partial suppression of autophagic activity using Beclin 1+/− animals blunted load-induced pathological remodeling. Conversely, overexpression of Beclin 1, with associated increases in autophagy, amplified adverse remodeling (33). Along these same lines, Porrello et al. (34) reported that autophagy is up-regulated, along with hypertrophic growth of NRVMs, in the setting of long-term treatment with angiotensin II. Nakai et al. (27) reported that autophagy is increased 4 weeks after TAC in mice.
At one level, it seems paradoxical that a mechanism of protein degradation would be activated during cell growth. However, hypertrophic remodeling involves more than simply the addition of proteins; there are substantial alterations in the content of numerous sarcomeric components, for example. Thus, although the overall result is an increase in cell size, activation of degradative pathways may be required in the processing of existing cellular elements.
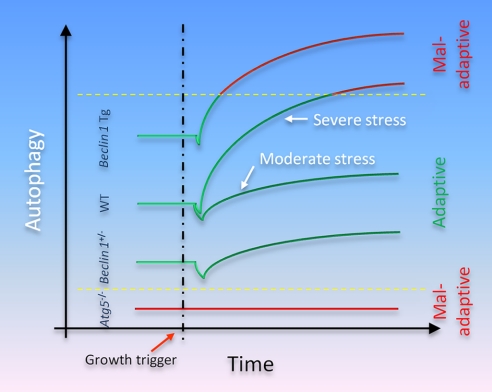
The kinetics of autophagic activation in load-induced hypertrophy is poorly defined. As noted above, different groups have reported different time courses of change, although up-regulated autophagy is a consistent finding. Differences in surgical model, animal strain, and genetic manipulation all likely contribute. For example, Nakai et al. (27) performed banding using a 26-gauge needle and observed cardiac dysfunction and mortality at 4 weeks. In our studies, a smaller 28-gauge needle was used to induce severe TAC, and cardiac dysfunction and increased mortality were observed within days (33). It is possible that severe pressure stress leads to emergence of more robust autophagy and quicker transition into a zone where autophagy is maladaptive (Fig. 2).
FIGURE 2.
Model of the role(s) of autophagy in hypertensive heart disease. In the basal state, constitutive levels of autophagy are required for cell survival, especially in post-mitotic cells that must survive decades without replication. Near-total inactivation of autophagic activity is maladaptive, promoting cell death. Conversely, less drastic decreases (or conversely, increases) in autophagic activity are not associated with untoward events. In the setting of growth stimulation, both anabolic and catabolic processes are activated. During the initial phase, the former predominates, and cell growth ensues. In fact, some evidence suggests that the catabolic autophagic pathway may be transiently suppressed in early phases of growth. Ultimately, however, a new steady state emerges where levels of autophagic flux are increased. Depending on the strength of the growth stimulus (and the genetic context where autophagy is suppressed either completely or partially or amplified), the resulting autophagic activity is either adaptive or maladaptive. In some contexts, hypoxia may contribute to the induction of maladaptive autophagy, as well. WT, wild type; Tg, transgenic.
A number of theories have been espoused to explain the transition from stable compensated hypertrophy to systolic dysfunction and decompensated heart failure. A common element among these theories is myocyte dropout by cell death. Kostin et al. (35) examined explanted hearts from 19 patients with end-stage heart failure and found evidence for each of the three major types of cell death, with autophagic death as the prominent one. Similar findings have been made by Knaapen et al. (36), who reported that as many as 0.3% of cardiomyocytes in end-stage failing human hearts display features of autophagic cell death.
However, despite accumulating evidence linking autophagic cell death to the pathogenesis of heart failure, some uncertainty remains regarding whether increased autophagy is an epiphenomenon or a causative factor. Recent studies using animal models have been performed to address this important question. In a hamster model of spontaneous cardiomyopathy, Miyata et al. (37) found significant accumulations of autophagic vacuoles within cardiomyocytes. Administration of granulocyte colony-stimulating factor suppressed autophagy and partially rescued the cardiac phenotype. Using a Beclin 1 haploinsufficiency model, we showed that decreased autophagic activity protects cardiomyocytes from death and preserves cardiac performance post-TAC (33). Conversely, Beclin 1 transgenic overexpression in the heart accelerated the transition from hypertrophy to failure. Our results suggest that autophagy is a maladaptive process in response to hemodynamic stress. Conversely, Nakai et al. (27) reported deteriorated cardiac function post-TAC in the setting of ATG5 inactivation, and they proposed that autophagy plays a protective role.
Whereas each of these studies reported load-induced activation of cardiomyocyte autophagy, the experimental findings led to opposing conclusions regarding whether autophagy is adaptive or maladaptive. At one level, this is not surprising, as the dual nature of autophagy is a recurring theme in other organ systems and disease states (38). However, important insights can be gleaned from careful comparison of these two studies (39).
First, we have postulated that the physiological impact of autophagy exists as a continuum, and a window of optimal autophagic activation is critical to the maintenance of cellular homeostasis and function (Fig. 2) (39). Indeed, several lines of evidence suggest that basal levels of autophagy are adaptive (40–42), whereas stress-related increases in autophagy can be maladaptive. Nakai et al. (27) employed a model in which Atg5 inactivation presumably led to near-complete elimination of constitutive autophagy. In contrast, Beclin 1+/− mice have only a 50% reduction in autophagic flux. Thus, important functions carried out by basal levels of constitutive autophagy were lost in Atg5-deficient hearts but not in Beclin 1+/− hearts. We have proposed that in the setting of pressure overload, wild-type mice mount an autophagic response that is sufficiently robust to elicit a maladaptive response (39). Atg5-deficient mice cannot increase autophagy and remain in the autophagy-deficient maladaptive range. In Beclin 1+/− mice, the increase in autophagic activity is blunted, maintaining activity closer to the adaptive zone. In Beclin 1 transgenic mice, load-induced autophagic activity shifts yet further into the maladaptive range. Consistent with this notion, some evidence suggests that increased Beclin 1 expression, as occurs in hearts subjected to pressure overload (33), is indicative of maladaptive autophagy (32).
Multiple mechanisms have been proposed to explain the activation of cardiac autophagy (39, 43). Okada et al. (44) reported that ER stress is triggered in response to TAC-induced increases in hemodynamic load. Klionsky and co-workers (45) showed that ER stress per se is sufficient to stimulate Atg1 kinase activity and induce autophagy in yeast. Kouroku et al. (46) found that the PERK/eukaryotic initiation factor 2α branch of the ER stress pathway mediates autophagy induction in a model of neurodegenerative disease.
On the other hand, aggregated protein complexes alone may be a proximal trigger of cardiac autophagy. We have observed robust accumulation of polyubiquitinated proteins co-localized to autophagic active sites in the left ventricle (47). We showed that polyubiquitinated proteins are sufficient to induce autophagy in NRVMs. Consistent with our findings, Depre et al. (48) found increased proteasome expression and activity in load-stressed heart. Collectively, accumulation of protein aggregates may induce ubiquitination pathways and ER stress, which, in turn, trigger autophagy.
Autophagy in Myocardial Ischemia
Ischemic heart disease is a major contributor to the aggregate burden of heart disease around the world (2, 6), and hypertension is a major risk factor in its development (2). As part of this, evidence suggests that myocyte ischemia contributes to the pathogenesis of hypertensive heart disease in the absence of coronary atherosclerosis. During cardiac growth, coronary angiogenesis is induced robustly to meet increased metabolic and oxygen demands. Indeed, intricate coordination between cell growth and angiogenesis is critical to the entire remodeling process. Izumiya et al. (49) attenuated angiogenesis using a decoy vascular endothelial growth factor receptor and found that hearts were predisposed to decompensation after TAC. Similarly, Shiojima et al. (50) reported defective angiogenesis in response to prolonged hypertrophic stimulation. Using a model of inducible cardiomyocyte-specific expression of a constitutively active AKT1 mutant, these investigators found that expression of AKT1 for 2 weeks induced cardiac hypertrophy with preserved contractility. However, the transgenic mice developed dilated cardiomyopathy at 6 weeks, coincident with decreases in capillary density (50). In this setting, autophagy may be suppressed initially by AKT1-mediated increases in mTOR activity. However, continued growth may ultimately exhaust angiogenic potential, resulting in limitations in nutrient and oxygen supply; these latter events may, in turn, induce autophagy.
Autophagy is activated in response to myocardial ischemia. Studying an ex vivo rabbit heart model 30 years ago, Decker and Wildenthal (51) showed that 40 min of hypoxia triggers significant autophagosome and autolysosome formation. Ultrastructural analysis revealed autophagosomes in close proximity to swollen and fragmented mitochondria (51). In rodents in vivo, Matsui et al. (32) reported dramatic up-regulation of autophagosome formation 30 min after ischemia-inducing surgery. Together, these results strongly suggest that autophagy is activated by myocardial ischemia. Going further, these investigators reported that suppression of autophagy using 3-methyladenine enhanced myocyte death triggered by glucose deprivation, an in vitro mimic of tissue ischemia (32). These observations suggest that ischemia-induced autophagy is protective by replenishing depleted energy stores and ridding the cell of dysfunctional potentially toxic organelles.
Recent work has revealed that inactivation of hypoxia-inducible factor 1α in fibroblasts blunts hypoxia-induced autophagy (52). A role for AMPK (AMP-activated protein kinase) in ischemia-induced autophagy has been posited, as well (32). Thus, these molecules may participate in autophagy that promotes cell survival by eliminating dysfunctional mitochondria, which would otherwise release reactive oxygen species and pro-apoptotic mediators.
Autophagy in I/R
Mounting evidence suggests that reoxygenation following ischemia triggers a second wave of autophagic activation. Decker and Wildenthal (51) detected numerous autophagosomes after 20 min of hypoxia and 30 min of reperfusion compared with 20 min of hypoxia alone. In a chronic I/R model in swine, Yan et al. (53) reported that six episodes of reduced coronary flow followed by 12 h of reperfusion elicited increased expression of elements of the autophagy pathway. In addition, they found that apoptosis was suppressed when autophagy was up-regulated. Similar findings have been reported in the cardiac myoblast cell line H9c2, HL-1 cells, NRVMs, and isolated rat hearts (54–56).
Divergent results have emerged regarding whether autophagy is adaptive or maladaptive in the context of I/R injury. Matsui et al. (32) found dramatic up-regulation of Beclin 1 following I/R in mice. In Beclin 1+/− animals, autophagy was significantly attenuated compared with wild-type controls. Interestingly, Beclin 1+/− mice manifested less infarction and suppressed apoptosis (32). Consistent findings were reported by Valentim et al. (55) in both NRVMs and adult cardiomyocytes; I/R activated cell death and autophagy, and suppression of autophagy with 3-methyladenine improved cell viability. Collectively, these data lend support to the notion that autophagy induced by I/R is detrimental.
In contrast, Hamacher-Brady et al. (56) reported that simulated I/R induced cell death in HL-1 cells in concert with increases in autophagy. Augmentation of autophagy with rapamycin or by Beclin 1 overexpression was protective. Gurusamy et al. (54) reported that adaptation by repeated brief episodes of ischemia activated autophagy and that wortmannin blocked autophagy induction and abolished the cell-protective effects. Yitzhaki et al. (57) used an adenosine receptor agonist to mimic simulated I/R and found that autophagy is required for cardioprotection in both HL-1 cells and NRVMs. In aggregate, these data suggest that autophagy during simulated I/R is a prosurvival adaptive response. Clearly, more work is warranted to elucidate the role of autophagy as protective or detrimental during I/R.
The functional role of autophagy in I/R injury is likely to be complex (58), and several mechanisms have been proposed (59). Structural studies identified Beclin 1 as a BH3-only protein, which raises an important issue, as BH3-only proteins are generally pro-apoptotic (60). This is consistent with the observation that Beclin 1 interacts with the prosurvival molecule Bcl-2 to inhibit its action. Based on this, it is tempting to speculate that Beclin 1 induction, as occurs in I/R, stimulates autophagy and possibly cell death through its BH3 domain.
Another molecule involved in I/R-induced cell death is Bnip3, a downstream target of hypoxia-inducible factor 1α. In the heart under basal conditions, Bnip3 is expressed at negligible levels, but its abundance is massively up-regulated following I/R (61). Ablation of Bnip3 does not elicit an obvious cardiac phenotype (61). However, Diwan et al. (61) reported preserved left ventricular systolic performance and diminished I/R-induced cardiac dilatation in Bnip3-deficient mice. In contrast, overexpression of Bnip3 induced progressive ventricular dilation and impaired systolic performance possibly due to increased apoptosis. These results suggest that Bnip3 promotes I/R-induced cell death. Consistent with this, Hamacher-Brady et al. (62) provided evidence that Bnip3 induces mitochondrial fragmentation and autophagy. Suppression of Bnip3 using a dominant-negative mutant protected against I/R injury. Collectively, these findings suggest that Bnip3 contributes to cell death during I/R injury, positioning Bnip3 as a potential therapeutic target.
Conclusions and Perspective
The association between hypertension and cardiovascular disease is long-established. Aspects of the underlying pathogenesis are starting to be unveiled, and autophagy has emerged as a significant contributor. Whereas basal constitutive autophagy is indispensable to maintain cellular homeostasis, autophagy is activated in response to the cellular stresses occurring in hypertensive heart disease. In some contexts, autophagy is protective and adaptive, providing energy resources and molecular building blocks to promote cell function and survival. Under other circumstances, however, autophagic activity is maladaptive, promoting disease pathogenesis and cell death (Fig. 2).
Precedent exists (for example, in oncology) for a requirement of finely tuned autophagic activation; too little autophagy and too much autophagy can each be detrimental. Despite considerable progress in our understanding of the molecular machinery of autophagy, our understanding of its adaptive versus maladaptive effects lags. Looking to the future, detailed elucidation of the mechanisms and effects of autophagy in hypertensive heart disease will be essential to our long-term goal of targeting this mechanism for therapeutic gain.
Supplementary Material
Acknowledgments
We are grateful to members of the Hill laboratory for valuable comments.
This work was supported, in whole or in part, by National Institutes of Health Grants HL-075173, HL-080144, HL-090842, HL-072016, and HL-097768. This work was also supported by American Heart Association Grant 0640084N and the American Heart Association-Jon Holden DeHaan Foundation. This is the first of five articles in the “Biochemistry in Medicine: Hypertension Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- CVD
- cardiovascular disease
- LVH
- left ventricular hypertrophy
- TOR
- target of rapamycin
- mTOR
- mammalian TOR
- PI3K
- phosphatidylinositol 3-kinase
- PE
- phosphatidylethanolamine
- NRVM
- neonatal rat ventricular myocyte
- TAC
- transverse aortic constriction
- ER
- endoplasmic reticulum
- I/R
- ischemia/reperfusion.
REFERENCES
- 1.Ezzati M., Lopez A. D., Rodgers A., Vander Hoorn S., Murray C. J. (2002) Lancet 360, 1347–1360 [DOI] [PubMed] [Google Scholar]
- 2.Kearney P. M., Whelton M., Reynolds K., Muntner P., Whelton P. K., He J. (2005) Lancet 365, 217–223 [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Wang Q. J. (2004) Arch. Intern. Med. 164, 2126–2134 [DOI] [PubMed] [Google Scholar]
- 4.Vasan R. S., Larson M. G., Leip E. P., Evans J. C., O'Donnell C. J., Kannel W. B., Levy D. (2001) N. Engl. J. Med. 345, 1291–1297 [DOI] [PubMed] [Google Scholar]
- 5.Lopez A. D., Mathers C. D., Ezzati M., Jamison D. T., Murray C. J. (2006) Lancet 367, 1747–1757 [DOI] [PubMed] [Google Scholar]
- 6.Lloyd-Jones D., Adams R., Carnethon M., De Simone G., Ferguson T. B., Flegal K., Ford E., Furie K., Go A., Greenlund K., Haase N., Hailpern S., Ho M., Howard V., Kissela B., Kittner S., Lackland D., Lisabeth L., Marelli A., McDermott M., Meigs J., Mozaffarian D., Nichol G., O'Donnell C., Roger V., Rosamond W., Sacco R., Sorlie P., Stafford R., Steinberger J., Thom T., Wasserthiel-Smoller S., Wong N., Wylie-Rosett J., Hong Y. (2009) Circulation 119, 480–486 [DOI] [PubMed] [Google Scholar]
- 7.Burch G. E., Ray C. T., Cronvich J. A. (1952) Circulation 5, 504–513 [DOI] [PubMed] [Google Scholar]
- 8.Bergmann O., Bhardwaj R. D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B. A., Druid H., Jovinge S., Frisén J. (2009) Science 324, 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill J. A., Olson E. N. (2008) N. Engl. J. Med. 358, 1370–1380 [DOI] [PubMed] [Google Scholar]
- 10.Levy D., Garrison R. J., Savage D. D., Kannel W. B., Castelli W. P. (1990) N. Engl. J. Med. 322, 1561–1566 [DOI] [PubMed] [Google Scholar]
- 11.Frey N., Katus H. A., Olson E. N., Hill J. A. (2004) Circulation 109, 1580–1589 [DOI] [PubMed] [Google Scholar]
- 12.Sybers H. D., Ingwall J., DeLuca M. (1976) Recent Adv. Stud. Cardiac Struct. Metab. 12, 453–463 [PubMed] [Google Scholar]
- 13.Decker R. S., Wildenthal K. (1980) Am. J. Pathol. 98, 425–444 [PMC free article] [PubMed] [Google Scholar]
- 14.Decker R. S., Decker M. L., Herring G. H., Morton P. C., Wildenthal K. (1980) J. Mol. Cell. Cardiol. 12, 1175–1189 [DOI] [PubMed] [Google Scholar]
- 15.Dämmrich J., Pfeifer U. (1983) Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 43, 287–307 [DOI] [PubMed] [Google Scholar]
- 16.Pfeifer U., Föhr J., Wilhelm W., Dämmrich J. (1987) J. Mol. Cell. Cardiol. 19, 1179–1184 [DOI] [PubMed] [Google Scholar]
- 17.Klionsky D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 18.Cecconi F., Levine B. (2008) Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroemer G., Levine B. (2008) Nat. Rev. Mol. Cell Biol. 9, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Nat. Rev. Mol. Cell Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 22.Xie Z., Klionsky D. J. (2007) Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 23.Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009) Dev. Cell 17, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamada Y., Sekito T., Ohsumi Y. (2004) Curr. Top. Microbiol. Immunol. 279, 73–84 [DOI] [PubMed] [Google Scholar]
- 25.Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan E. Y. (2009) Sci. Signal. 2, pe51. [DOI] [PubMed] [Google Scholar]
- 27.Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., Nishida K., Hori M., Mizushima N., Otsu K. (2007) Nat. Med. 13, 619–624 [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lüllmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. (2000) Nature 406, 902–906 [DOI] [PubMed] [Google Scholar]
- 29.Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J. E., Oh S. J., Koga Y., Sue C. M., Yamamoto A., Murakami N., Shanske S., Byrne E., Bonilla E., Nonaka I., DiMauro S., Hirano M. (2000) Nature 406, 906–910 [DOI] [PubMed] [Google Scholar]
- 30.Maron B. J., Roberts W. C., Arad M., Haas T. S., Spirito P., Wright G. B., Almquist A. K., Baffa J. M., Saul J. P., Ho C. Y., Seidman J., Seidman C. E. (2009) J. Am. Med. Assoc. 301, 1253–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanamori H., Takemura G., Maruyama R., Goto K., Tsujimoto A., Ogino A., Li L., Kawamura I., Takeyama T., Kawaguchi T., Nagashima K., Fujiwara T., Fujiwara H., Seishima M., Minatoguchi S. (2009) Am. J. Pathol. 174, 1705–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui Y., Takagi H., Qu X., Abdellatif M., Sakoda H., Asano T., Levine B., Sadoshima J. (2007) Circ. Res. 100, 914–922 [DOI] [PubMed] [Google Scholar]
- 33.Zhu H., Tannous P., Johnstone J. L., Kong Y., Shelton J. M., Richardson J. A., Le V., Levine B., Rothermel B. A., Hill J. A. (2007) J. Clin. Invest. 117, 1782–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porrello E. R., D'Amore A., Curl C. L., Allen A. M., Harrap S. B., Thomas W. G., Delbridge L. M. (2009) Hypertension 53, 1032–1040 [DOI] [PubMed] [Google Scholar]
- 35.Kostin S., Pool L., Elsässer A., Hein S., Drexler H. C., Arnon E., Hayakawa Y., Zimmermann R., Bauer E., Klövekorn W. P., Schaper J. (2003) Circ. Res. 92, 715–724 [DOI] [PubMed] [Google Scholar]
- 36.Knaapen M. W., Davies M. J., De Bie M., Haven A. J., Martinet W., Kockx M. M. (2001) Cardiovasc. Res. 51, 304–312 [DOI] [PubMed] [Google Scholar]
- 37.Miyata S., Takemura G., Kawase Y., Li Y., Okada H., Maruyama R., Ushikoshi H., Esaki M., Kanamori H., Li L., Misao Y., Tezuka A., Toyo-Oka T., Minatoguchi S., Fujiwara T., Fujiwara H. (2006) Am. J. Pathol. 168, 386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothermel B. A., Hill J. A. (2008) Circ. Res. 103, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 42.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 43.Nishida K., Kyoi S., Yamaguchi O., Sadoshima J., Otsu K. (2009) Cell Death Differ. 16, 31–38 [DOI] [PubMed] [Google Scholar]
- 44.Okada K., Minamino T., Tsukamoto Y., Liao Y., Tsukamoto O., Takashima S., Hirata A., Fujita M., Nagamachi Y., Nakatani T., Yutani C., Ozawa K., Ogawa S., Tomoike H., Hori M., Kitakaze M. (2004) Circulation 110, 705–712 [DOI] [PubMed] [Google Scholar]
- 45.Yorimitsu T., Nair U., Yang Z., Klionsky D. J. (2006) J. Biol. Chem. 281, 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R. J., Kominami E., Momoi T. (2007) Cell Death Differ. 14, 230–239 [DOI] [PubMed] [Google Scholar]
- 47.Tannous P., Zhu H., Nemchenko A., Berry J. M., Johnstone J. L., Shelton J. M., Miller F. J., Jr., Rothermel B. A., Hill J. A. (2008) Circulation 117, 3070–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Depre C., Wang Q., Yan L., Hedhli N., Peter P., Chen L., Hong C., Hittinger L., Ghaleh B., Sadoshima J., Vatner D. E., Vatner S. F., Madura K. (2006) Circulation 114, 1821–1828 [DOI] [PubMed] [Google Scholar]
- 49.Izumiya Y., Shiojima I., Sato K., Sawyer D. B., Colucci W. S., Walsh K. (2006) Hypertension 47, 887–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiojima I., Sato K., Izumiya Y., Schiekofer S., Ito M., Liao R., Colucci W. S., Walsh K. (2005) J. Clin. Invest. 115, 2108–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Decker R. S., Wildenthal K. (1980) Am. J. Pathol. 98, 425–444 [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H., Bosch-Marce M., Shimoda L. A., Tan Y. S., Baek J. H., Wesley J. B., Gonzalez F. J., Semenza G. L. (2008) J. Biol. Chem. 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Yan L., Vatner D. E., Kim S. J., Ge H., Masurekar M., Massover W. H., Yang G., Matsui Y., Sadoshima J., Vatner S. F. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13807–13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gurusamy N., Lekli I., Gorbunov N. V., Gherghiceanu M., Popescu L. M., Das D. K. (2009) J. Cell. Mol. Med. 13, 373–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valentim L., Laurence K. M., Townsend P. A., Carroll C. J., Soond S., Scarabelli T. M., Knight R. A., Latchman D. S., Stephanou A. (2006) J. Mol. Cell. Cardiol. 40, 846–852 [DOI] [PubMed] [Google Scholar]
- 56.Hamacher-Brady A., Brady N. R., Gottlieb R. A. (2006) J. Biol. Chem. 281, 29776–29787 [DOI] [PubMed] [Google Scholar]
- 57.Yitzhaki S., Huang C., Liu W., Lee Y., Gustafsson A. B., Mentzer R. M., Jr., Gottlieb R. A. (2009) Basic Res. Cardiol. 104, 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gustafsson A. B., Gottlieb R. A. (2009) Circ. Res. 104, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishida K., Yamaguchi O., Otsu K. (2008) Circ. Res. 103, 343–351 [DOI] [PubMed] [Google Scholar]
- 60.Oberstein A., Jeffrey P. D., Shi Y. (2007) J. Biol. Chem. 282, 13123–13132 [DOI] [PubMed] [Google Scholar]
- 61.Diwan A., Krenz M., Syed F. M., Wansapura J., Ren X., Koesters A. G., Li H., Kirshenbaum L. A., Hahn H. S., Robbins J., Jones W. K., Dorn G. W. (2007) J. Clin. Invest. 117, 2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamacher-Brady A., Brady N. R., Logue S. E., Sayen M. R., Jinno M., Kirshenbaum L. A., Gottlieb R. A., Gustafsson A. B. (2007) Cell Death Differ. 14, 146–157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.