Abstract
Tumor cells release NKG2D ligands to evade NKG2D-mediated immune surveillance. The purpose of our investigation was to explore the cellular mechanisms of release used by various members of the ULBP family. Using biochemical and cellular approaches in both transfectant systems and tumor cell lines, this paper shows that ULBP1, ULBP2, and ULBP3 are released from cells with different kinetics and by distinct mechanisms. Whereas ULBP2 is mainly shed by metalloproteases, ULBP3 is abundantly released as part of membrane vesicles known as exosomes. Interestingly, exosomal ULBP3 protein is much more potent for down-modulation of the NKG2D receptor than soluble ULBP2 protein. This is the first report showing functionally relevant differences in the biochemistry of the three members of the ULBP family and confirms that in depth study of the biochemical features of individual NKG2D ligands will be necessary to understand and manipulate the biology of these proteins for therapy.
Keywords: Cancer, Cell/Exocytosis, Protein/Secretion, Subcellular Organelles/Vesicles, Immunology, Nkg2d, Ulbp, Exosomes, Human, Innate Immunity
Introduction
NKG2D is an activating immune receptor that can be expressed by most cytotoxic lymphocytes, including NK and CD8+ T cells (1). Engagement of NKG2D by its ligands leads to the activation or co-stimulation of lysis and cytokine secretion (for review, see Ref. 2). In humans, NKG2D ligands (NKG2D-L)5 occur in two families of proteins: the polymorphic family of MHC-I-related chain A/B (MICA/B) and the multigene family of UL16-binding proteins (ULBPs, also known as RAET1A–E). In total, 10 members of this gene family have been described, of which six can be expressed as functional proteins (3). Two members of the ULBP family have a transmembrane region (ULBP4 and -5), like MICA/B, whereas the other ULBP molecules are linked to the cell membrane via glycosylphosphatidylinositol (GPI) anchors. The existence of such a large number of ligands for a single receptor is not fully understood but may reflect a differential role for different ligands in immune surveillance or an evolutionary response to selective pressures exerted by pathogens or cancer.
In general, NKG2D-L are not expressed ubiquitously; instead, they are expressed in response to several types of cellular stress, such as pathogen infection (4), DNA damage (5), proteasome inhibition (6), and tumor transformation (7). For example, MICA/B are expressed in epithelial tumors, melanoma, neuroblastoma, various hematopoietic malignancies, and carcinomas; ULBPs are found in leukemia, gliomas and melanomas. An additional complication is that mRNA can be found in many cells that do not express protein suggesting post-transcriptional regulation of NKG2D-L expression (8–10).
Mice deficient in NKG2D expression show an enhanced susceptibility to the development of tumors (11). However, shedding NKG2D-L as soluble molecules allows tumor cells to evade NKG2D surveillance. Apart from reducing NKG2D-L expression on the tumor cell surface, the release of soluble molecules may also impair immune surveillance by promoting down-regulation of NKG2D (12, 13). In fact, the sustained presence in vivo of NKG2D-L down-modulates the receptor (14, 15), and, at least in mouse models, blocking shedding of NKG2D-L can prevent tumor formation (16). In patients with colorectal or prostate cancer, hepatocellular carcinoma, and neuroblastoma, decreased NKG2D expression and impaired activation of NK cells was associated with high levels of soluble MICA in serum (17–20). Importantly, therapy of patients with chronic myeloid leukemia led to a substantial decrease of soluble MICA levels, accompanied by restored NKG2D expression on CD8+ T cells and NK cells (8). Overall, the release of soluble NKG2D-L by tumor cells has a negative impact on NKG2D-dependent immune surveillance of cancer and suggest that a better understanding of this process may lead to the identification of useful targets for therapy.
Recently, members of the ADAM (a disintegrin and metalloproteinase) family have been identified as key proteases involved in the shedding of some alleles of MICA and MICB (21, 22), and a member of the disulfide isomerase family, ERp5, has also been proposed to play a role in the shedding of MICA (23). ADAMs 10 and 17 have been shown to be involved in proteolytic cleavage of ULBP2 (22), but nothing else is known about the shedding of other ULBPs. Indeed, in general, little is known about the biochemistry and cell biology of the ULBPs other than that they have signals for a GPI anchor (24, 25) and that ULBP3 can associate with microdomains of the membrane rich in sphingolipids and cholesterol (detergent-resistant membranes) (26).
We have analyzed the biochemical features of the ULBPs released to the extracellular media of transfectant and tumor cell lines and here report interesting differences in the kinetics and mechanisms of shedding of the ULBPs. Although ULBP1 was shed at low levels, and ULBP2 was abundantly shed as a soluble protein, ULBP3 was shed with slower kinetics, and, surprisingly, much of this released ULBP3 was found in the membrane of small vesicles known as exosomes. Interestingly, the ULBP released in exosomes potently down-modulated the NKG2D receptor. Overall, these data provide the first evidence of functionally relevant biochemical differences between the three members of the ULBP family linked through a GPI anchor and suggest new approaches to understand the diversity of NKG2D-L.
EXPERIMENTAL PROCEDURES
Cells and Reagents
ULBP1, -2, and -3 constructs were obtained from Dr. Richard Apps (27). The kidney monkey cell line CV1 and 293T cells (ATCC) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, l-glutamine, and antibiotics. Chinese hamster ovary (CHO) cells were maintained in Hams F12 medium with the same supplements. CV1 cells were transfected as described (28). CHO cells were transfected using Lipofectamine 2000 and ULBP expression plasmid mixed (9:1 ratio) with a vector conferring resistance to puromycin (29). Stable transfectants were sorted where necessary and grown in selective medium (8 μg/ml puromycin, Calbiochem).
Antibodies directed against ULBPs were purchased from R&D Systems (Abingdon, UK). BB94 (Batimastat) was a kind gift of British Biotech. Leupeptin, pepstatin A, 1,10-phenanthroline were purchased from Sigma; PMA and ionomycin from Calbiochem. Anti-hamster CD63 hybridoma was provided by Dr. M. Marsh (clone eh1C9b, generated by S. Schmid). NK primary cell lines were generated from healthy donor peripheral blood mononuclear cells as described (30).
Flow Cytometry
105 cells were preincubated in PBS containing 1% bovine serum albumin, 0.1% sodium azide (PBA). Cells were then incubated with mouse monoclonal antibodies and bound antibody was visualized using either phycoerythrin- or fluorescein isothiocyanate-labeled F(ab′)2 fragments of goat anti-mouse Ig (Dako). Samples were analyzed using a FACScan II flow cytometer (Becton Dickinson). Dead cells were excluded from all analyses by staining with propidium iodide.
ELISA
For detection of soluble proteins, cells were incubated for 16–24 h in medium in the absence of serum. Detection of sULBP was performed using a sandwich ELISA procedure. Plates were coated for 16 h at 4 °C with the appropriate polyclonal anti-ULBP1, -2, and -3 antibody (R&D Systems) (5 μg/ml) and blocked with 2% bovine serum albumin-PBS for 2 h at 37 °C. Tissue culture supernatant was added for 2 h at 37 °C. Bound ULBP protein was detected using the appropriate biotinylated goat anti-ULBP (R&D Systems) followed by streptavidin-horseradish peroxidase (Amersham Biosciences), and the assay was developed using the peroxidase substrate system (ABTS, Roche Applied Science). Absorbance was measured at 410 nm with a reference wavelength of 490. Samples were analyzed in duplicates. Under these conditions, the cut-off for detection of recombinant soluble ULBP-Fc constructs (R&D Systems) was ∼1 ng/ml, and the ELISA absorbance values were directly proportional to the concentration of soluble ULBP protein over the range 1 to 100 ng/ml.
Exosome Purification
Exosomes were prepared by sequential centrifugation as in Raposo et al. (31). Exosomes pelleted after centrifugation at 100,000 × g for 2 h were resuspended either in 100 μl of PBS for electron microscopy or directly in sample buffer for Western blot. The amount of protein in exosomes was estimated using Bio-Rad protein assay. The resulting supernatant (soluble fraction) was recovered by precipitation with trichloroacetic acid. Where indicated, further purification of exosomes by flotation on a discontinuous sucrose gradient was performed. A step sucrose gradient (4 ml of 60% sucrose, 3 ml of 30%, and 1 ml of 5%) was layered on top of the exosome suspension, mixed with 2.5 volumes of 85% sucrose in TNE buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 5 mm EDTA). Gradients were centrifuged for 20 h at 100,000 × g. 1-ml fractions were collected, and proteins were further solubilized by adding deoxycholate (final concentration 0.2%).
Western Blot
Cell lysates were prepared by incubation in TNE buffer containing 1% Nonidet P-40 and the protease inhibitors leupeptin, pepstatin, and 1,10-phenanthroline for 30 min at 4 °C. Nuclei were eliminated by centrifugation at 13,000 × g. Lysates, supernatants, and exosome preparations were run on 12% SDS-PAGE gels and transferred to Immobilon-P (Millipore) membrane. The membrane was blocked using PBS containing 0.1% Tween 20 (PBS-T) and 5% nonfat dry milk. Detection of ULBP was performed by incubation with biotinylated goat polyclonal anti-ULBP antibody, followed by horseradish peroxidase-conjugated streptavidin. Proteins were visualized using the ECL system (Amersham Biosciences). Quantitative analysis of the Western blot data were done using NIH ImageJ software.
Electron Microscopy
Samples of exosomes for examination by electron microscopy were prepared by floating a carbon-coated 400 mesh Formvar electron microscopy grid on top of one drop of freshly prepared exosomes for 15 s. The grid was then briefly washed with deionized water and floated on a drop of 2% phosphotungstic acid, pH 7.0. Samples were examined using a Philips CM100 operating at 60 or 80 keV.
Confocal Microscopy
Cells were fixed with 4% paraformaldehyde at 4 °C for 15 min, permeabilized by incubation with 0.1% saponin at room temperature for 10 min, stained with polyclonal anti-ULBP antibodies (R&D Systems), and analyzed by confocal microscopy as described previously (32). Fluorescence images were obtained using a confocal microscope (Leica TCS-NT-UV confocal laser scanning microscope). Images of fixed cells were taken using a 63 × 1.32 objective with the confocal pinhole set to one airy unit. Images were obtained by scanning series of single focal planes across the cell using Leica TCS software. To explore the whole intracellular area, series of sections (total interval z = 2–4 μm) were acquired.
NKG2D Down-modulation
Primary human NK cells, 3–5 days after stimulation with feeders and interleukin-2, were cultured for 24 or 48 h in the presence of supernatants of untransfected CHO cells or CHO-ULBP2 or CHO-ULBP3 transfectants. When using exosome fractions, a total protein of 40–100 ng was added to the NK cells. NKG2D surface expression was monitored by staining with mAb specific for NKG2D (clone 1D11, Santa Cruz Biotechnology) and flow cytometry using a FACScan cytometer running Cellquest software (BD Biosciences).
Cytotoxicity Assays
Cytotoxicity assays were carried out using a one-step fluorimetric assay based on the use of AlamarBlue (Invitrogen) (33).
RESULTS
Differential Kinetics and Shedding of ULBP1, -2, and -3
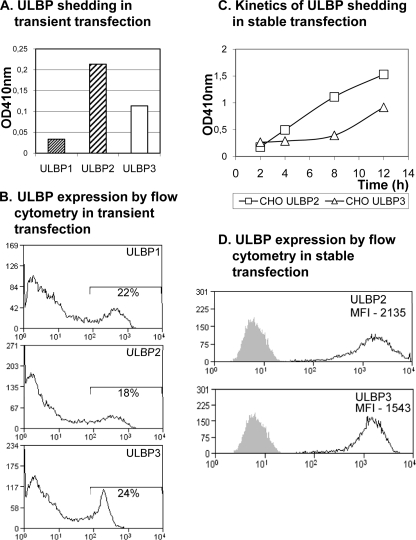
To characterize the release of the three GPI-anchored ULBPs, the amount of ULBP1, -2, and -3 in tissue culture supernatant shed from CV1 and CHO cell transfectants was evaluated using a sandwich ELISA. Initially, to avoid artifacts associated with the selection of particular clones during the generation of stable transfectants, these experiments were done using transient transfection. Interestingly, differential shedding of ULBP1, -2, and -3 expressed in the same cellular background was observed. Although ULBP2 and -3 were detected in large amounts, ULBP1 was only shed at relatively low levels (Fig. 1A). This result was not due to differences in the efficiency of transfection or the levels of expression of the different ULBPs (Fig. 1B). The differential shedding of the various ULBPs was also obvious in a Western blot, implying that the result was not biased by the ELISA assay. These differences in the patterns of release of the three ULBPs from cells suggested that different mechanisms could mediate the release of these molecules. Given the low level of shedding of ULBP1, the next sets of experiments focused on detailed study of the shedding of ULBP2 and ULBP3, and analysis of stably transfected CHO cells revealed that the kinetics of accumulation of ULBP2 and ULBP3 in the supernatant were different; large amounts of ULBP2 were detected after only brief incubations, whereas the accumulation of ULBP3 occurred at a slower rate (Fig. 1, C and D).
FIGURE 1.
Different kinetics of release of ULBP1, -2, and -3. A, CHO cells were transiently transfected with ULBP1, ULBP2, and ULBP3. 24 h later, supernatants were collected and analyzed using a sandwich ELISA. B, cellular ULBP expression was analyzed by flow cytometry. C, kinetics of release of ULBP2 and ULBP3 in stably transfected CHO cells. ULBP release was assessed by ELISA after the cells had been in culture for 2, 4, and 8 h. Data shown here are representative from three experiments. Analysis of ULBP transfected CV1 cells gave similar results (not shown). D, flow cytometry analysis of ULBP2 and ULBP3 expression by stable transfectants.
Metalloproteases and the Cleavage of ULBP2 and ULBP3
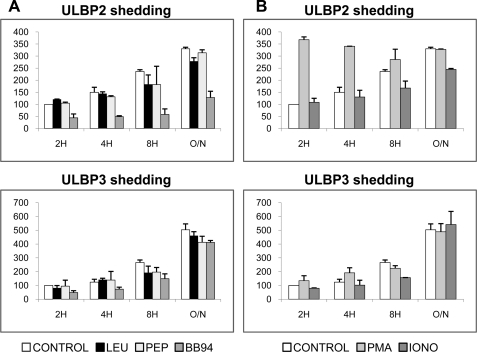
It has been reported that the release of ULBP2 is inhibited by metalloprotease inhibitors (34) but that metalloprotease inhibition had no effect on surface expression of ULBP1 or ULBP3. We extended these studies using a panel of protease inhibitors to explore the ability of inhibitors of different classes of proteases to affect ULBP2 and ULBP3 shedding in time course experiments (Fig. 2). Leupeptin is an inhibitor of serine and cysteine proteinases; pepstatin is a potent inhibitor of various aspartic proteinases; BB94 inhibits both matrix metalloproteases and some members of the ADAM family of proteases, which have been shown to affect ULBP2 release. Although leupeptin and pepstatin did not affect the shedding of ULBP2 or -3, BB94 dramatically decreased the shedding of ULBP2 from both CHO cells (Fig. 2A) and CV1 cells (data not shown). At short times of incubation, BB94 weakly inhibited ULBP3 release, but no clear effect was apparent after 16 h of incubation. These results confirmed that metalloproteases mediate shedding of ULBP2 and suggest that the release of ULBP3 depends only partially on metalloproteases.
FIGURE 2.
ULBP2 and -3 are released by the action of metalloproteases, but other mechanisms are also involved. A, CHO transfectants were treated with 1 μm leupeptin (LEU), 1 μm pepstatin (PEP) and a broad metalloproteinase inhibitor, 10 μm BB94. Data are expressed as percentage shedding of the untreated control at 2 h. The inhibition of the release of ULBP2 by the metalloproteinase inhibitor BB94 was the only statistically significant change (t test, p < 0.001), whereas ULBP3 release was only weakly, and not significantly, inhibited by BB94. B, CHO cells were treated with 100 ng/ml of PMA and 5 μm of ionomycin (IONO). The increase of ULBP2 shedding mediated by PMA is statistically significant (p < 0.005). Cells were incubated in the presence of the indicated compounds for 2, 4, 8, and 12 h. Detection of soluble ULBPs in tissue culture supernatant was performed by ELISA.
Analysis of the effect of pharmacological stimulation on the release of ULBP2 and -3 revealed more differences between the release of these molecules. Phorbol esters (PMA) activate ADAM17-mediated shedding of proteins (known to be involved in ULBP2 shedding) (22, 35). Ca2+ ionophores (ionomycin) activate ADAM10 (36) and, at long incubation times, exosome release (37). PMA treatment increased ULBP2 shedding, although the effect disappeared at longer time points but had little effect on ULBP3 release. Ionomycin weakly stimulated the release of the ULBPs only at longer times (Fig. 2B).
These results are consistent with published data, suggesting the involvement of ADAM proteases in release of ULBP2 (22). Overall, these data also imply that the release of ULBP2 and -3 depend on distinct cellular pathways.
ULBP3 Is Released in Exosomes
To understand the differences in the shedding of the various ULBPs, a biochemical characterization of the molecules released to the supernatant was performed using both transfectants and tumor cell lines.
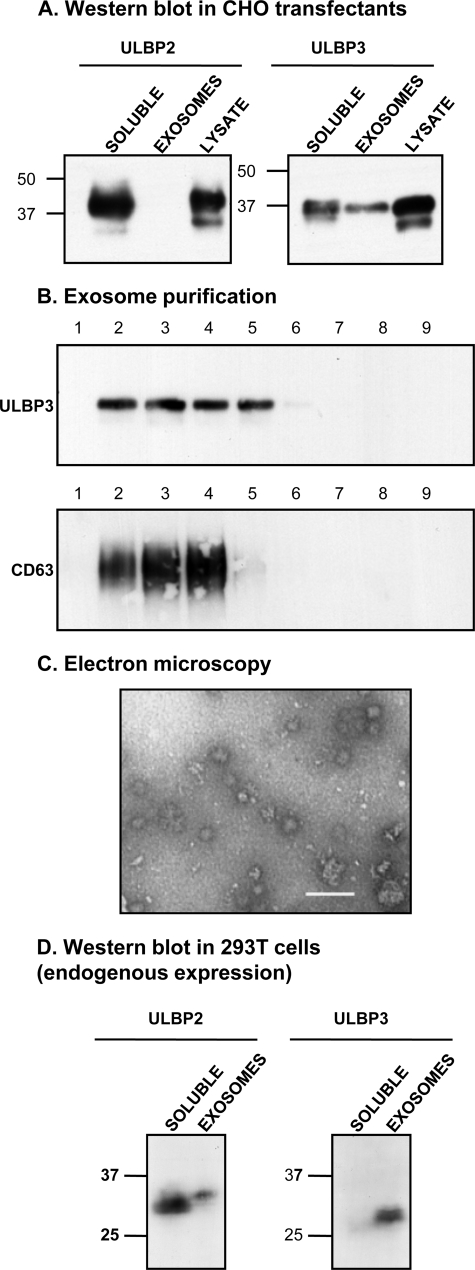
Supernatants were fractionated to separate proteins present as soluble monomeric forms from those present in microvesicles called exosomes. Exosomes are small vesicles (30–100 nm) secreted to the extracellular medium after fusion of the endocytic structures known as multivesicular bodies with the plasma membrane (38). Purification of exosomes involves several steps of sequential centrifugation at different speeds to separate them from other membrane fragments and organelles. Exosomes pellet after centrifugation at 100,000 × g and, therefore, can be distinguished from soluble proteins (31). Exosomes and soluble proteins were isolated from supernatants of CHO-ULBP2 and -ULBP3 transfectants and analyzed by Western blot. ULBP3 but not ULBP2 was detected in the 100,000 × g pellet (Fig. 3A), and these fractions included CD63, a cellular marker of late endosomes and multivesicular bodies known to localize in exosomes (39). ULBP3 was also observed in exosomes purified from CV1 transfectants (supplemental Fig. 1). The presence of ULBP3 in exosomes was confirmed by further purification in a sucrose gradient. As shown in Fig. 3B, ULBP3 was found in the low density fractions where purified exosomes typically migrate, co-fractionating with CD63. Exosome preparations, visualized by electron microscopy, contained vesicles of ∼50 nm (Fig. 3C). To eliminate the possibility that the results above were an artifact of transfected cells, the same analysis was performed in the tumor cell line 293T, endogenously expressing ULBP2 and ULBP3. ULBP3 was released in exosomes in this tumor cell line, whereas ULBP2 was mainly observed in the supernatant after centrifugation at 100,000 × g (Fig. 3D).
FIGURE 3.
ULBP3 is released in exosomes both in transfectants and the tumor cell line 293T. A, the exosome fraction and soluble proteins from CHO-ULBP2 and -3 transfectants were purified after 24 h in culture as described under “Experimental Procedures” and compared with total lysate from the same cells in Western blot analysis. Similar results were obtained on analysis of CV1-ULBP3 cells (supplemental Fig. 1). ULBP2 was shed mainly as a soluble protein, whereas ULBP3 was released both as a soluble protein and in exosomes. B, fractionation of exosomes in a sucrose gradient shows co-migration with CD63. C, exosomes from CHO-ULBP3 cells were negatively stained with 2% phosphotungstic acid and analyzed by electron microscopy. As expected, nano-sized vesicles from 30 to 120 nm were observed. Bar, 100 nm. D, analysis of tissue culture supernatant from the 293T cell line, which expresses ULBP2 and ULBP3 endogenously, confirmed the results obtained in the transfectant system. Soluble and exosome fractions were analyzed by Western blot. ULBP2 is mainly shed as a soluble protein, whereas ULBP3 is mainly released in exosomes.
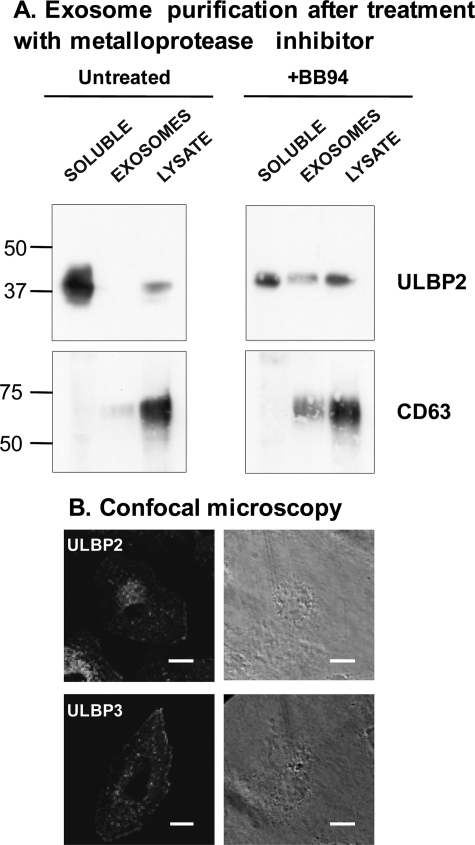
These data demonstrate that ULBP2 is mainly shed as a truncated soluble protein, whereas ULBP3 is released in exosomes. Both ULBP2 and ULBP3 are GPI-anchored proteins and reside in regions of membranes rich in sphingolipids and cholesterol, known as detergent-resistant membranes.6 Recruitment to detergent-resistant membranes is a mechanism described for incorporation of proteins into exosomes (40); thus, why would ULBP3 but not ULBP2 be preferentially included into exosomes? We hypothesized that ULBP2 is not excluded from exosomes but rather is more susceptible to metalloprotease attack and is therefore lost by proteolysis before inclusion into exosomes. To test this idea, the soluble and exosome fractions of supernatants of ULBP2 transfectants, untreated or treated with metalloprotease inhibitors, were analyzed for the presence of ULBP2 by Western blot. Fig. 4A shows that treatment with BB94 augments the amount of ULBP2 present in exosomes, confirming this hypothesis: whereas in untreated cells, only 2.77% of the released ULBP2 protein was present in exosomes, in BB94-treated cells, this percentage augmented to 31.89%, and a 30% reduction of soluble protein could be observed (from 97.23% in untreated to 68.11% in BB94). Treatment of ULBP3 transfectants with BB94 produced a decrease in the amount of shed soluble ULBP3 but not of the exosomal protein (data not shown). In confocal microscopy, the pattern of distribution within the cell was different for ULBP2 and ULBP3, suggesting that these molecules may also differ in intracellular trafficking (Fig. 4B and supplemental Fig. 2).
FIGURE 4.
ULBP2 and ULBP3 can be recruited to exosomes and have different susceptibility to metalloproteases. A, the exosome fraction and soluble proteins from CHO-ULBP2 and -3 transfectants, either untreated or treated with the metalloprotease inhibitor BB94, were purified as in Fig. 3A. Western blot analysis of ULBP molecules and CD63 is shown. B, CV1-ULBP2 and CV1-ULBP3 transfectants were analyzed by confocal microscopy. Images show a single focal plane across the cell (depth of the plane 0.2035 μm) using the 63× objective. The series of images corresponding to the same cell are shown in supplemental Fig. 2. Scale bar, 25 μm.
Both Soluble ULBP2 and Exosomal ULBP3 Can Induce Down-regulation of NKG2D
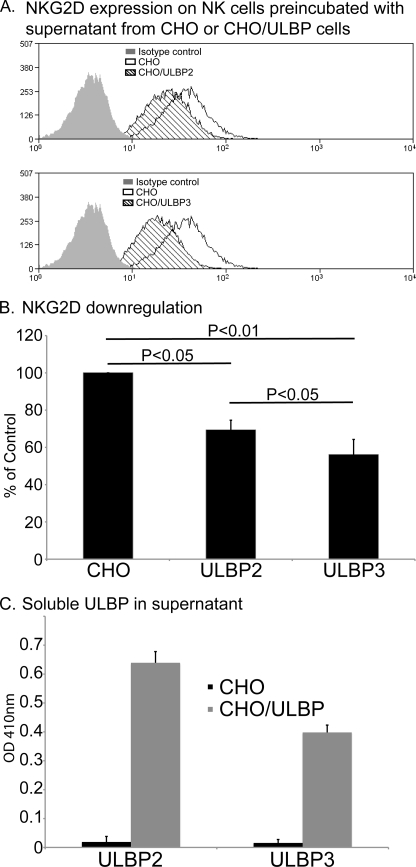
The release of soluble MICA, as well as promoting immune evasion by reducing the cell surface expression of NKG2D-L on tumors, has also been reported to trigger a systemic down-regulation of the NKG2D receptor on NK cells and CD8+ T cells (12, 17, 41). Thus, it was of interest to compare the effect of the addition of supernatants containing ULBP2 (mainly soluble) and ULBP3 (released in exosomes) on cell surface expression of NKG2D on primary human NK cells. To avoid the confounding effects of factors such as transforming growth factor-β on lymphocyte proliferation and NKG2D expression (42–44), supernatants from untransfected CHO cells and CHO transfectants expressing either ULBP2 or ULBP3 were used for these experiments. A representative example of this type of experiment is shown in Fig. 5. Both ULBP2- and ULBP3-containing supernatants could induce decreased cell surface NKG2D expression. A t test of the data from multiple experiments shows that these effects are statistically significant. It is interesting to note that the effect of ULBP3 is consistently stronger than that of ULBP2, especially because the amount of ULBP2 in the supernatants was always higher than that of ULBP3 (Fig. 5C). The simplest interpretation of these data is that this reflects differences in how the two ULBP species exist in the supernatant: soluble, probably monomeric ULBP2 versus multivalent ULBP3 molecules in exosome membranes.
FIGURE 5.
Both ULBP2 and ULBP3 containing supernatants provoke down-regulation of NK cell NKG2D. A, supernatants collected from CHO cells (control) and ULBP-transfected CHO cells were incubated with primary NK cells, prepared from healthy donors, for 48 h. Cell surface NKG2D was then quantitated by flow cytometry. B, t test of five experiments performed with two different donors. Data are expressed as percentage of NKG2D expression observed on NK cells incubated in medium alone. Down-modulation of NKG2D is significant after treatment with both ULBP2 and ULBP3 proteins (p < 0.05 and p < 0.01, respectively). C, ELISA assay to quantitate the amounts of ULBP2 and ULBP3 supernatants used in the down-regulation experiments.
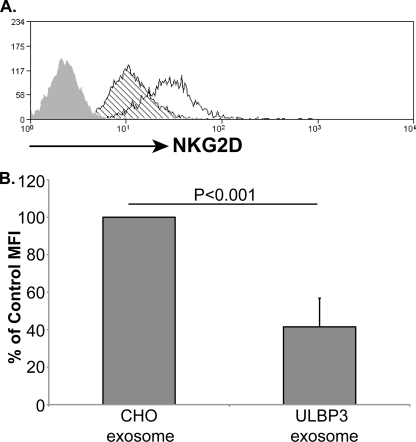
To confirm that the down-modulation of NKG2D receptor, observed in the experiments above, was due to the effect of exosomal protein and not to any soluble protein present in the supernatant, it was important to test whether purified ULBP3-containing exosomes could affect cell surface expression of NKG2D on NK cells. Fig. 6 shows that exposure to exosomes purified from ULBP3 transfectants triggered a marked reduction in cell surface NKG2D expression. Because purified exosomes from untransfected CHO cells had no effect on receptor expression, down-modulation of NKG2D depended specifically on the presence of exosomal ULBP3.
FIGURE 6.
Purified exosomes containing ULBP3 down-modulate NKG2D. A, exosomes were incubated with primary NK cells for 24 h. Cell surface NKG2D was then quantitated by flow cytometry. B, t test of four experiments performed with two different donors. MFI, mean fluorescence intensity.
Exosomal ULBP3 Can Compromise NKG2D-mediated Cell Cytotoxicity
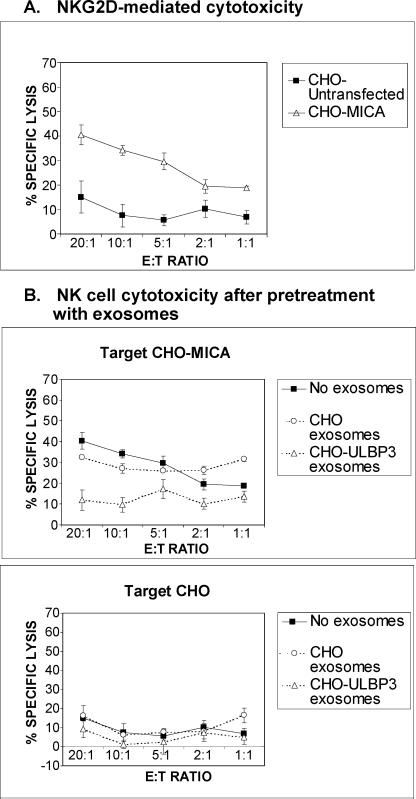
The fact that exosomal protein potently down-modulated NKG2D suggested that incubation with ULBP3-containing exosomes might impair cytotoxic responses. To test this hypothesis, exosomes purified from ULBP3 transfectants and untransfected control cells were incubated with primary NK cells before including these effectors in cytotoxicity assays. As expected, human NK cells lysed MICA-transfected CHO cells in an NKG2D-dependent manner, while untransfected CHO cells were not efficiently lysed (Fig. 7A). Preincubation with ULBP3-containing exosomes resulted in a clear cut reduction of the NKG2D-mediated lytic activity of NK cells against cells expressing MICA (Fig. 7B). These results demonstrate that ULBP3-containing exosomes but not other exosomes devoid of NKG2D-L inhibit NKG2D-mediated responses.
FIGURE 7.
Purified exosomes containing ULBP3 are potent modulators of NK cell NKG2D-mediated cytotoxicity. A, primary NK cells recognize CHO-MICA transfectants in an NKG2D-dependent fashion. B, effect of purified exosomes on NK cell cytotoxicity against CHO-MICA and untransfected CHO. Purified exosomes from either untransfected or ULBP3-transfected CHO cells (40–100 ng total protein) were incubated with primary NK cells for 24 h. After preincubation with exosomes, NK cells were used as effectors in a cytotoxicity assay (measured using a fluorimetric assay). E:T, effector:target ratio.
DISCUSSION
A recurrent question in the field of the NKG2D system is why are there so many ligands for one, invariant receptor. The existence of such a large number of ligands for a single receptor is not fully understood, but the dominant model is that the important role of these molecules in immune surveillance has subjected them to strong evolutionary pressure to adapt to the different challenges presented by pathogens or cancer (7, 45). In this context, the up-regulation of different ligands with diverse biochemical properties may constitute a mechanism to target specific cellular pathways in response to the different challenges; however little is known about the biochemistry and cell biology of the ULBP family. In this manuscript, we report a study of the biochemistry of release of ULBP1, ULBP2, and ULBP3 from tumor cells. Striking differences in both the kinetics of release and the species of protein produced were observed: truncated soluble protein versus membrane-bound. These data constitute the first demonstration of differences in the biochemistry of the GPI-linked members of the ULBP family and could be of crucial importance to understand the process of escape from the immune system by stressed cells up-regulating NKG2D-L, such as pathogen-infected and cancer cells.
ULBP2 is known to be released by the action of metalloproteinases (34) including members of the ADAM family (22). Using pharmacological agents to stimulate or inhibit different classes of protease, we reproduced the published data with ULBP2 and showed that inhibition of multiple classes of proteases including metalloproteases did not significantly affect ULBP3 release. Furthermore, the kinetics of ULBP3 shedding were not comparable to those of ULBP2, and the release of ULBP3 was much slower. Moreover, an increase in ULBP3 shedding could be observed after a long exposure to ionomycin. As this increase did not occur at short incubation times, this suggested not an effect on ADAM10 but on the process of exosome formation and release.
Analysis of the biochemistry of released ULBP2 and ULBP3 gave a definitive response; ULBP3 was found to be released in exosomes. This was observed in various transfectants and in cells endogenously expressing the protein; thus, we infer that this is due to the characteristics of the protein and not to the cell line. In the transfected cells, a small proportion of ULBP3 can also be observed in the soluble fraction; thus, a truncated monomeric form is generated in addition to the membrane-bound ULBP3 molecule. More work needs to be done to evaluate the spatio-temporal characteristics of these two processes. The observation that ULBP2 can be included in exosomes, but it is more susceptible to metalloprotease attack is consistent with the sequence differences between ULBPs in the stalk region and further study is needed to define the proteolytic site for ULBP2.
The expression of a particular NKG2D-L does not always imply cytotoxic attack toward the cell. Instead, the fate of the target cell depends on the post-transcriptional and post-translational modifications that direct the protein for surface expression, retention, or shedding. Some NKG2D-L do not even reach the surface after translation (46); in other cases, the protein recycles into endosomal compartments (47); and finally, NKG2D-L are known to be released to the surrounding medium (12, 34). In light of our data, it seems plausible to speculate that expression of a particular ULBP would lead to different amounts of protein shed to the supernatant and as different protein species, more or less potent in NKG2D down-modulation. The functional consequences of this differential release would result in the complex spectrum of situations that are observed in cancer patients, some expressing large amounts of NKG2D-L, some not expressing any. For this reason, it will be of crucial importance to study the particular cell biological properties of the individual NKG2D-L.
Elevated levels of soluble MICA have been detected in the sera of patients suffering from various types of cancer and often correlate with a poor prognosis for the patient (48). This relationship is likely to be related to the known effects of MICA/B shedding: a reduction in cell surface density of NKG2D ligands leading to a reduced susceptibility to NKG2D-mediated cytotoxicity and systemic down-regulation of NKG2D on NK cells and CD8+ T cells in cancer patients. The occurrence and significance of serum ULBP molecules has not been well studied, but soluble ULBPs have been detected in the serum of patients with hematological malignancies (34) and colorectal cancer.7 The data presented here show that incubation with supernatants containing either soluble ULBP2 or exosomal ULBP3 molecules also leads to significant down-regulation of cell surface NKG2D expression. Interestingly, incubation of NK cells with the ULBP3-containing supernatant triggers significantly more NKG2D down-regulation (Fig. 5) than the ULBP2 culture supernatant despite containing much less ULBP protein. A goal of future research will be to investigate whether this might be related to differences in how the two ULBP species exist in the supernatant: soluble, probably monomeric ULBP2 versus multivalent ULBP3 molecules in exosome membranes and whether this is also true in patient sera.
The ability of soluble NKG2D-L to provoke NKG2D down-regulation and compromised NK and cytotoxic T lymphocytes function suggests that blockade of NKG2D-L release might be a useful addition to immunological approaches for cancer therapy, but an understanding of the mechanisms involved in the release of soluble NKG2D-L from tumor cells is crucial for the development of effective strategies to block the shedding of these proteins. Metalloproteases are key enzymes mediating proteolytic cleavage of both MICA/B and ULBP2 molecules (22, 47). However, the data in this manuscript now show that NKG2D-L molecules can be released from a tumor cell in more than one way. ULBP3 is not released from cells by proteolysis but rather as a full-length molecule in exosomes. Thus, effective blockade of the accumulation of soluble NKG2D-L in patient sera will require the use of multiple strategies.
In conclusion, we have demonstrated here that different ULBP proteins are released in diverse amounts as different species and that these released ULBP proteins can trigger NKG2D down-regulation. These findings offer the possibility of starting novel approaches to try to understand the regulation of the fate of a cell expressing NKG2D-L. For example, the study could be extended to analyze the effect of different stimuli that up-regulate particular NKG2D-L (6) on the release of that molecule.
Supplementary Material
Acknowledgments
We thank R. Apps for the ULBP constructs; N. Miller for assistance with cell sorting and F. Colucci and his group, M. Field, J. Kaufman, A. Kelly, S. Powis, P. Roda-Navarro and J. Trowsdale for helpful discussions.
This work was supported in part by the Medical Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
L. Fernandez-Messina, unpublished data.
I. Jafferji, L. Fernandez-Messina, S. Aguera-Gonzalez, M. Vales-Gomez, R. McGilvray, L. G. Durrant, J. Trowsdale, and R. A. Eagle, manuscript in preparation.
- NKG2D-L
- NKG2D ligands
- GPI
- glycosylphosphatidylinositol
- CHO
- Chinese hamster ovary
- MHC
- major histocompatibility complex
- MICA/B
- MHC-I-related chain A/B
- ULBP
- UL16-binding proteins
- PBS
- phosphate-buffered saline
- ELISA
- enzyme-linked immunosorbent assay
- PMA
- phorbol esters.
REFERENCES
- 1.Raulet D. H. (2003) Nat. Rev. Immunol. 3, 781–790 [DOI] [PubMed] [Google Scholar]
- 2.Moretta A., Bottino C., Vitale M., Pende D., Cantoni C., Mingari M. C., Biassoni R., Moretta L. (2001) Annu. Rev. Immunol. 19, 197–223 [DOI] [PubMed] [Google Scholar]
- 3.Radosavljevic M., Cuillerier B., Wilson M. J., Clément O., Wicker S., Gilfillan S., Beck S., Trowsdale J., Bahram S. (2002) Genomics 79, 114–123 [DOI] [PubMed] [Google Scholar]
- 4.López-Botet M., Angulo A., Gumá M. (2004) Tissue Antigens 63, 195–203 [DOI] [PubMed] [Google Scholar]
- 5.Gasser S., Orsulic S., Brown E. J., Raulet D. H. (2005) Nature 436, 1186–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valés-Gómez M., Chisholm S. E., Cassady-Cain R. L., Roda-Navarro P., Reyburn H. T. (2008) Cancer Res. 68, 1546–1554 [DOI] [PubMed] [Google Scholar]
- 7.Nausch N., Cerwenka A. (2008) Oncogene 27, 5944–5958 [DOI] [PubMed] [Google Scholar]
- 8.Boissel N., Rea D., Tieng V., Dulphy N., Brun M., Cayuela J. M., Rousselot P., Tamouza R., Le Bouteiller P., Mahon F. X., Steinle A., Charron D., Dombret H., Toubert A. (2006) J. Immunol. 176, 5108–5116 [DOI] [PubMed] [Google Scholar]
- 9.Zwirner N. W., Dole K., Stastny P. (1999) Hum. Immunol. 60, 323–330 [DOI] [PubMed] [Google Scholar]
- 10.Nice T. J., Coscoy L., Raulet D. H. (2009) J. Exp. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra N., Tan Y. X., Joncker N. T., Choy A., Gallardo F., Xiong N., Knoblaugh S., Cado D., Greenberg N. M., Raulet D. H. (2008) Immunity 28, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groh V., Wu J., Yee C., Spies T. (2002) Nature 419, 734–738 [DOI] [PubMed] [Google Scholar]
- 13.Salih H. R., Antropius H., Gieseke F., Lutz S. Z., Kanz L., Rammensee H. G., Steinle A. (2003) Blood 102, 1389–1396 [DOI] [PubMed] [Google Scholar]
- 14.Wiemann K., Mittrücker H. W., Feger U., Welte S. A., Yokoyama W. M., Spies T., Rammensee H. G., Steinle A. (2005) J. Immunol. 175, 720–729 [DOI] [PubMed] [Google Scholar]
- 15.Oppenheim D. E., Roberts S. J., Clarke S. L., Filler R., Lewis J. M., Tigelaar R. E., Girardi M., Hayday A. C. (2005) Nat. Immunol. 6, 928–937 [DOI] [PubMed] [Google Scholar]
- 16.Wu J. D., Atteridge C. L., Wang X., Seya T., Plymate S. R. (2009) Clin. Cancer Res. 15, 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doubrovina E. S., Doubrovin M. M., Vider E., Sisson R. B., O'Reilly R. J., Dupont B., Vyas Y. M. (2003) J. Immunol. 171, 6891–6899 [DOI] [PubMed] [Google Scholar]
- 18.Wu J. D., Higgins L. M., Steinle A., Cosman D., Haugk K., Plymate S. R. (2004) J. Clin. Invest. 114, 560–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinushi M., Takehara T., Tatsumi T., Kanto T., Groh V., Spies T., Kimura R., Miyagi T., Mochizuki K., Sasaki Y., Hayashi N. (2003) Int. J. Cancer 104, 354–361 [DOI] [PubMed] [Google Scholar]
- 20.Raffaghello L., Prigione I., Airoldi I., Camoriano M., Levreri I., Gambini C., Pende D., Steinle A., Ferrone S., Pistoia V. (2004) Neoplasia 6, 558–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutet P., Agüera-González S., Atkinson S., Pennington C. J., Edwards D. R., Murphy G., Reyburn H. T., Valés-Gómez M. (2009) J. Immunol. 182, 49–53 [DOI] [PubMed] [Google Scholar]
- 22.Waldhauer I., Goehlsdorf D., Gieseke F., Weinschenk T., Wittenbrink M., Ludwig A., Stevanovic S., Rammensee H. G., Steinle A. (2008) Cancer Res. 68, 6368–6376 [DOI] [PubMed] [Google Scholar]
- 23.Kaiser B. K., Yim D., Chow I. T., Gonzalez S., Dai Z., Mann H. H., Strong R. K., Groh V., Spies T. (2007) Nature 447, 482–486 [DOI] [PubMed] [Google Scholar]
- 24.Cosman D., Müllberg J., Sutherland C. L., Chin W., Armitage R., Fanslow W., Kubin M., Chalupny N. J. (2001) Immunity 14, 123–133 [DOI] [PubMed] [Google Scholar]
- 25.Onda H., Ohkubo S., Shintani Y., Ogi K., Kikuchi K., Tanaka H., Yamamoto K., Tsuji I., Ishibashi Y., Yamada T., Kitada C., Suzuki N., Sawada H., Nishimura O., Fujino M. (2001) Biochem. Biophys. Res. Commun. 285, 235–243 [DOI] [PubMed] [Google Scholar]
- 26.Eleme K., Taner S. B., Onfelt B., Collinson L. M., McCann F. E., Chalupny N. J., Cosman D., Hopkins C., Magee A. I., Davis D. M. (2004) J. Exp. Med. 199, 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apps R., Gardner L., Traherne J., Male V., Moffett A. (2008) Hum. Reprod. 23, 2535–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valés-Gómez M., Browne H., Reyburn H. T. (2003) BMC Immunol. 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Luna S., Ortín J. (1992) Methods Enzymol. 216, 376–385 [DOI] [PubMed] [Google Scholar]
- 30.Wills M. R., Ashiru O., Reeves M. B., Okecha G., Trowsdale J., Tomasec P., Wilkinson G. W., Sinclair J., Sissons J. G. (2005) J. Immunol. 175, 7457–7465 [DOI] [PubMed] [Google Scholar]
- 31.Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valés-Gómez M., Reyburn H. T. (2006) J. Mol. Biol. 363, 908–917 [DOI] [PubMed] [Google Scholar]
- 33.Nociari M. M., Shalev A., Benias P., Russo C. (1998) J. Immunol. Methods 213, 157–167 [DOI] [PubMed] [Google Scholar]
- 34.Waldhauer I., Steinle A. (2006) Cancer Res. 66, 2520–2526 [DOI] [PubMed] [Google Scholar]
- 35.Hooper N. M., Karran E. H., Turner A. J. (1997) Biochem. J. 321, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horiuchi K., Le Gall S., Schulte M., Yamaguchi T., Reiss K., Murphy G., Toyama Y., Hartmann D., Saftig P., Blobel C. P. (2007) Mol. Biol. Cell 18, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savina A., Furlán M., Vidal M., Colombo M. I. (2003) J. Biol. Chem. 278, 20083–20090 [DOI] [PubMed] [Google Scholar]
- 38.Stoorvogel W., Kleijmeer M. J., Geuze H. J., Raposo G. (2002) Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 39.Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J. (1998) J. Biol. Chem. 273, 20121–20127 [DOI] [PubMed] [Google Scholar]
- 40.de Gassart A., Geminard C., Fevrier B., Raposo G., Vidal M. (2003) Blood 102, 4336–4344 [DOI] [PubMed] [Google Scholar]
- 41.Vetter C. S., Lieb W., Bröcker E. B., Becker J. C. (2004) Br. J. Cancer 91, 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castriconi R., Cantoni C., Della Chiesa M., Vitale M., Marcenaro E., Conte R., Biassoni R., Bottino C., Moretta L., Moretta A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4120–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clayton A., Mitchell J. P., Court J., Mason M. D., Tabi Z. (2007) Cancer Res. 67, 7458–7466 [DOI] [PubMed] [Google Scholar]
- 44.Clayton A., Mitchell J. P., Court J., Linnane S., Mason M. D., Tabi Z. (2008) J. Immunol. 180, 7249–7258 [DOI] [PubMed] [Google Scholar]
- 45.Eagle R. A., Trowsdale J. (2007) Nat. Rev. Immunol. 7, 737–744 [DOI] [PubMed] [Google Scholar]
- 46.Borchers M. T., Harris N. L., Wesselkamper S. C., Vitucci M., Cosman D. (2006) Am. J. Physiol. Lung Cell Mol. Physiol. 291, L222–231 [DOI] [PubMed] [Google Scholar]
- 47.Agüera-González S., Boutet P., Reyburn H. T., Valés-Gómez M. (2009) J. Immunol. 182, 4800–4808 [DOI] [PubMed] [Google Scholar]
- 48.Holdenrieder S., Stieber P., Peterfi A., Nagel D., Steinle A., Salih H. R. (2006) Cancer Immunol. Immunother. 55, 1584–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.